Abstract
Background:
Helicobacter pylori (H. pylori) is a known gastro-intestinal pathogen but implicated in extra-gastric diseases. The relationship between H. pylori infection and type 2 diabetes (T2DM) remains insufficiently elucidated, particularly in terms of molecular mediators such as microRNAs (miRNAs) and messenger RNAs (mRNAs).
Objective:
We aimed to characterize expression pattern of insulin signalling mRNAs and targeted miRNAs in T2DM patients exposed to H. pylori infection.
Methods:
We conducted a cross-sectional study among patients diagnosed with type 2 diabetes mellitus and were aged 18 to 60 years. Overnight fasting blood samples were collected and processed for plasma and serum. The plasma samples were used for glucose estimation and the serum used for H. pylori IgG screening. Total RNA was extracted from the serum with commercial kit, and mRNAs and miRNAs quantified by RT-qPCR with specific primers and under predetermined amplification conditions. Clinical data were obtained from medical records of patients.
Results:
Among 351 patients enrolled, 267 (76.1%) were females, 224 (63.8%) were married, and 79 (22.5%) had tertiary education. Expression level of insulin receptor mRNA was significantly lower in H. pylori positive T2DM patients compared to H. pylori negative (P < .05). There was no evidence of a difference in insulin receptor substrate 1 mRNA level (P > .05). Although not statistically significant, the expression levels of miRNA-222 and miRNA-155 in the patients exposed to H. pylori were higher than that of the unexposed group (P > .05).
Conclusions:
We found a significantly reduced serum insulin receptor messenger RNA level and higher levels of miRNA-222 and miRNA-155 in H. pylori exposed T2DM patients. The findings suggest a possible role of the infection in insulin signalling alteration in the patients.
Keywords: H. pylori, IgG seropositivity, insulin receptor, mRNA, T2DM
Introduction
Type 2 diabetes mellitus (T2DM) represents a significant global health burden, characterized by insulin resistance and chronically elevated blood glucose with a global incidence of 10.5% in adults between the ages 20 to 79 years.1,2 The chronic nature of T2DM contributes to a myriad of complications, significantly impairing quality of life and imposing substantial healthcare costs. 3 Evidence suggests a potential association between Helicobacter pylori (H. pylori) infection and the pathogenesis of T2DM.4,5 H. pylori, a Gram-negative bacterium, colonizes the stomach lining and is implicated in various gastrointestinal diseases. Recent studies have hinted at its role beyond gastric pathologies, suggesting a systemic influence that may include the modulation of glucose metabolism.6,7 However, the relationship between H. pylori infection and the dysregulation of metabolic pathways in T2DM remains insufficiently elucidated, particularly in terms of molecular mediators such as microRNAs (miRNAs) and messenger RNAs (mRNAs).
MicroRNAs and messenger RNAs play crucial roles in regulating gene expression and have been implicated in the pathogenesis of a wide range of diseases, including T2DM. Several studies have revealed the role of miRNAs in the regulation of insulin signalling, and the regulatory points include the insulin receptor expression, and downstream insulin substrate one-half expression and the PI3K-AKT pathway.8-10 miRNAs are small non-coding RNAs that modulate genes expression post-transcriptionally and are involved in the regulation of glucose and lipid metabolism. 11 The potential interaction between H. pylori infection and the dysregulation of these molecular mediators in T2DM patients has not been extensively studied. Understanding these relationships could illuminate novel pathways through which H. pylori influences the development and progression of T2DM and might identify new therapeutic targets or diagnostic markers.
The aim of this study is to characterize serum levels of mRNAs and miRNAs in T2DM patients with and without H. pylori exposure. Our hypothesis was that, compared to T2DM alone, H. pylori infection is associated with higher levels of miRNAs; miRNA-222 and miRNA-155, and lower levels of insulin receptor and insulin receptor substrate 1 mRNAs. Our study aspires to advance our understanding of the complex interplay between infectious agents and chronic metabolic diseases such as T2DM, which could pave the way for the development of innovative diagnostic tools and therapeutic strategies.
Methods
Study design and participants
This was a cross-sectional design conducted at the National Diabetic Management and Research Centre in Korle Bu Teaching Hospital, Accra. The centre runs only out-patient services. On average, 45 patients visit the clinic daily. The centre’s catchment population include residents in Greater Accra Region and its environs. Research assistants stationed at the centre approached patients attending the clinic and explained the study. Patients were enrolled if they were diagnosed with type 2 diabetes mellitus and were aged 18 to 60 years. Pregnant and breast-feeding mothers as well as type 1 diabetic patients were excluded from the study. Similarly, patients diagnosed with chronic infectious disease such as Hepatitis B and HIV, and other chronic non-communicable diseases including cancers were excluded from the study. A total of 351 patients were enrolled, of which 300 were screened for serum H. pylori IgG.
Data collection and blood sample processing
Well-structured questionnaire was used to obtain socio-demographic and clinical data. Medications were retrieved from hospital folders after approval has been sought from the clinic administrator. Aseptically, using a syringe and needle, 5 ml whole blood sample was collected from each participant by venipuncture and transferred into fluoride and serum separator tubes. The samples were processed for plasma and serum. To obtain serum, the blood in gel separator tubes were allowed to sit undisturbed for at least 30 minutes to clot. The tubes were centrifuged at 1000 to 2000 × g for 10 to 15 minutes. A pipette was used to carefully remove the serum without disturbing the gel or blood cells. Serum was transferred into labelled Eppendorf tubes and stored at −20 °C until ready to be used. Samples in the fluoride tubes were span at 1000 to 2000 × g for 10 to 15 minutes. Plasma obtained was collected into labelled Eppendorf tubes and stored at −20 °C until ready to be used.
Laboratory analysis
Fully automated chemistry analyser, (BC 200 Mindray Chemistry Analyzer, China) was used to analyse the glucose level in plasma following manufacture’s protocol. Briefly, the stored plasma was thawed on ice and automatically analysed on calibrated chemistry autoanalyzer with a control. A commercial rapid diagnostic test kit was used to screen for H. pylori exposure by detecting the presence of IgG antibodies in the serum. The serum was removed from the freezer and allowed to thaw on ice. A sample of the serum taken with disposable pipette was released into sample applicator of the test cassette and buffer applied following the manufacturer’s protocol.
Extraction of serum total RNA
A total of 15 serum H. pylori IgG positive and 15 negative samples were randomly selected for extraction of total RNA using commercial kit (Zymol Research Quick-RNA Miniprep Plus Kit, USA). The serum was thawed and 140 µl pipetted into an Eppendorf tube and homogenate prepared for RNA extraction with QIAamp Mini column, following manufacturer’s protocol. The purity and concentration of eluted total RNA was determined using nanodrop and stored at −80 °C until ready to be used.
Amplification of miRNAs and mRNAs using RT-qPCR
A total of 12 serum H. pylori IgG positive and 12 negative were used for miRNAs and mRNAs quantification. Both the positive samples (cases), negative samples (controls) and internal controls were set in duplicate. The RT-qPCR plate protocol and cycling conditions were designed using the Quant Studio™ Design & Analysis Software. The reagents: Luna Warm Start® RT Enzyme Mix, 2X Luna Universal One-Step Reaction Mix, nuclease-free water and primer set constitute the reaction master mix. Primers used targeted IR, Irs-1 miRNA-222 and miRNA-155, and housekeeping genes were RNU6B and GAPDH. The serum levels of miRNAs and mRNAs were determined by RT-qPCR using specific primers and Luna Warm Start® RT Enzyme Mix, 2X Luna Universal One-Step Reaction Mix. The RNA samples and PCR reagents were placed on ice and allowed to thaw.
The master mix was prepared following the instructions of the manufacturer. A single PCR reaction mixture of 20.0 µl contained 10.0 µl of Luna Universal One-Step Reaction Mix (2X), 1.0 µl Luna WarmStart RT Enzyme Mix (20X), 1.0 µl of 10 µM of each forward and reverse primers of a target gene or housekeeping gene, and 4.0 µl of deionized water. The PCR 96-well micro-plate was pre-chilled on plate block and 8.0 µl of reaction master mix was pipetted into each well, and 3.0 µl of total RNA from the samples was added to the designated wells. Each sample PCR reaction set was duplicated along with duplicated internal control. Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was used as internal control for mRNA and RNU6B for the miRNAs. The plates were briefly span at 1000 × g for 10 seconds and amplified with QuantStudio 5 Real-Time PCR System using predetermined amplifications conditions.
RT-qPCR cycling conditions
The cycle conditions for the targeted genes are presented in Table 1. Conditions for all the steps are the same with exception of the annealing temperature and duration for the respective genes (targets). Each target gene was run for 45 cycles.
Table 1.
RT-qPCR cycle conditions for miRNAs and miRNAs amplification.
| Step | Temperature | Time |
|---|---|---|
| Reverse transcription | 55 °C | 10 min |
| Initial denaturation | 95 °C | 1 min |
| Denaturation | 95 °C | 10 s |
| Annealing /Extension | IR mRNA (54 °C) Irs-1 mRNA (56 °C), miRNA-155 (52 °C) miRNA 222 (57 °C) |
30 s |
| Melting curve | 60 °C | 30 s |
Primer sets for RT-qPCR
The levels of miRNA-222, miRNA-155, and insulin receptor and insulin substrate 1 mRNAs in serum of patients with positive or negative H. pylori IgG were quantified using reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) under predetermined cycle conditions with specific primers. The primer sets for the amplification are shown Table 2.
Table 2.
Primers for miRNAs and mRNAs quantification using RT-qPCR.
| Target gene | Primer: | Sequences |
|---|---|---|
| miRNA-222 | Forward: | 5′-CCCTCAGTGGCTCAGTAG-3′ |
| Reverse: | 5′-CCACCAGAGACCCAGTAG-3′ | |
| miRNA-155 | Forward: | 5′-CGCGTTAATGCTAATCGTGATA-3′ |
| Reverse: | 5′-AGTGCAGGGTCCGAGGTATT-3′ | |
| Insulin receptor | Forward: | 5′-TTT TCG TCC CCA GGC CAT C-3′ |
| Reverse: | 5′-GTC ACA TTC CCA ACA TCG CC-3′ | |
| Insulin receptor substrate 1 | Forward: | 5′-TTTGAAGACCATAACCCACCAC-3′ |
| Reverse: | 5′-ATTACACCAGTTCGTCCCTTTC-3′ | |
| GADPH | Forward: | 5′-GTCTCCTCTGACTTCAACAGCG-3′ |
| Reverse: | 5′-ACCACCCTGTTGCTGAGCCAA-3′ | |
| RUN6B | Forward: | 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse: | 5′AACGCTTCACGAATTTGCGT-3′ |
GADPH and RUN6B primers were used as endogenous control for mRNAs and miRNAs, respectively.
Data analysis
Background characteristics were summarized using frequency and proportion for categorical variables. Continuous variables were summarized using mean and standard deviation or median and interquartile interval as appropriate. Continuous data was checked for normality and Welch t-test was performed for continuous outcomes which failed the normally test. Multivariable logistic regression model was used to identify factors independently associated with odds of H. pylori IgG sero-positivity. Differential expression of miRNAs and mRNAs was determined by fold change using the formula 2−ΔΔct. P < .05 was considered significant for all statistical analyses. All analyses were performed using Stata 17 SE (StataCorp, College Station, TX, USA).
Results
Socio-demographic characteristics of study participants
Among the 351 patients enrolled, 267 (76.1%) were females, 224 (63.8%) were married, 79 (22.5%) had tertiary education (Table 3). About 243 (69.1%) of the participants were gainfully employed. Almost 95% of the study participants used filtered water as source of drinking water.
Table 3.
Socio-demographic characteristics of study participants.
| Characteristics | No. of T2DM patients (% of total) (N = 351) |
|---|---|
| Sex: | |
| Female | 267 (76.1) |
| Male | 84 (23.9) |
| Marital status: | |
| Single | 127 (36.2) |
| Married | 224 (63.8) |
| Education: | |
| None | 40 (11.4) |
| Primary/JHS | 104 (29.6) |
| Secondary/SSS/SHS | 128 (36.5) |
| Tertiary | 79 (22.5) |
| Occupation: | |
| Employed | 243 (69.1) |
| Unemployed | 108 (30.9) |
| Household size: | |
| ⩽ 4 | 176 (50.1) |
| > 4 | 175 (49.9) |
| Sources of drinking water: | |
| Only filtered water | 333 (94.9) |
| Other sources | 18 (5.1) |
n = subgroup. Single = widow, widower and divorced; Other sources = drinking from Tap water, well water, protected and unprotected borehole water.
Serum levels of mRNAs and miRNAs in T2DM patients with and without H. pylori exposure
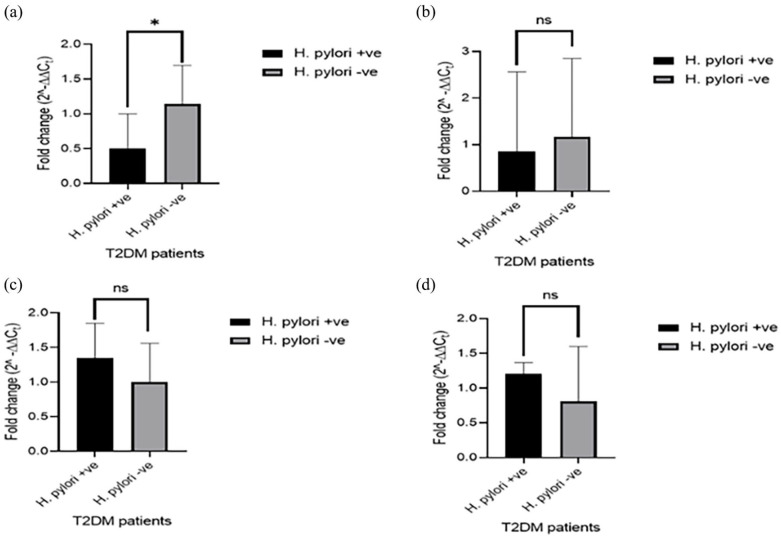
Expression levels of serum mRNAs and miRNAs in T2DM patients is shown in Figure 1. Expression level of insulin receptor mRNA was significantly lower in the exposed T2DM patients than the unexposed (P < .05). However, the difference in insulin receptor substrate 1 mRNA level in the exposed patients compared with the unexposed was not statistically significant (P > .05). Although not statistically significant, the expression levels of miRNA-222 and miRNA-155 in the patients exposed to H. pylori were higher than that of the unexposed group (P > .05).
Figure 1.
Serum mRNAs and miRNAs level in H. pylori-exposed T2DM patients: (a) expression level of insulin receptor mRNA, (b) expression level of insulin receptor substrate 1 mRNA, (c) expression level of miRNA-222 and (d) expression level of miRNA-155.
*P < .05.
Risk factors for H. pylori infection
Table 4 shows the association of the socio-demographic characteristics and source of drinking water with H. pylori infection. The study reports H. pylori seropositive IgG prevalence of 74.3% among Ghanaian type 2 diabetic patients. Among the males, 77.9% were exposed to H. pylori compared to the female counterpart (73.3%), however, the exposure was not associated with sex of the patients (OR = 1.24, 95%CI = 0 0.67 - 2.29, P > .05) and when adjusted for other confounding factors (OR = 1.19, 95%CI = 0.63 - 2.24, P > .05). The study showed no association between educational status (OR = 1.51, 95%CI = 0.82 - 2.79, P > .05) and source of drinking water (OR = 1.80, 95%CI = 0.63 - 5.12, P > .05) with H. pylori infection when adjusted for other confounders.
Table 4.
Risk factors for H. pylori infection.
| Characteristic | HP exposure Frequency, n (%) |
Unadjusted OR (95%CI) |
P-value | Adjusted OR (95%CI) |
p-value |
|---|---|---|---|---|---|
| Total | 223/300 (74.3) | ||||
| Sex | |||||
| Female | 165/225 (73.3) | 1 | 1 | ||
| Male | 58/75 (77.3) | 1.24 (0 0.67 - 2.29) |
.493 | 1.19 0.63 - 2.24 |
.582 |
| Education | |||||
| None/Primary/JHS | 92/130 (70.8) | 1 | 1 | ||
| Secondary | 77/98 (78.6) | 1.51 0.82 - 2.79 |
.184 | 1.46 0.79 - 2.72 |
.230 |
| Tertiary | 54/72 (75.0) | 1.24 0.64 - 2.38 |
1.17 0.61 - 2.28 |
||
| Source of drinking water | |||||
| Only filtered water | 213/284 (75.0) | 1 | 1 | ||
| Other sources | 10/16 (62.5) | 1.80 (0.63 - 5.12) |
.271 | 1.56 0.53 - 4.56 |
.414 |
A total of 300 patients were screened for H. pylori (HP) exposure. n = proportion of infection among the subgroups; other sources = tap water, borehole water, protected and unprotected well water.
Discussion
Current study reports altered expression of mRNAs and miRNAs in H. pylori exposed T2DM patients compared to the unexposed patients. Lower level of insulin receptor mRNA was found in the serum of H. pylori-exposed patients than the unexposed counterparts. Serum levels of both miRNA-222 and miRNA-155 were higher in the exposed patients than the unexposed, although the difference between the levels was not statistically significance.
Studies have reported upregulation of miRNA-155 during H. pylori infection in both in vivo and in vitro, and the activator protein–1 (AP-1) pathway was found to drive miRNA-155 induction.12-14 miRNA-555 level was strongly correlated with insulin resistance in T2DM patients infected with hepatitis C, 15 and levels of miRNAs in serum were reported to have a good diagnostic value for diabetic nephropathy and urinary microalbumin. 16 In an in vitro study, miRNA-222 expression level inversely correlated with insulin receptor substrate-1 (IRS-1) expression in liver cells. 13 Several studies have associated increased expression of miRNAs with obesity, insulin resistance, familial hyperchosterolemia, and new onset of diabetes mellitus in patients,13,17-19 and the expression patterns were found to be a potential biomarker for diagnosing metabolic diseases. Similarly, downregulation of miRNA-222 was reported in patients with weight loss. 14
Nevertheless, a decreased level of miRNA-155 was reported in peripheral blood mononuclear cells of T2DM patients with diabetic neuropathy compared to both healthy controls and T2D patients without diabetic neuropathy. 15 Also, a low level of miRNA-155 was found in T2DM patients with diabetic retinopathy compared with T2D patients without the condition.20,21
Dysregulation of insulin signalling and metabolic syndrome have been linked to the suppressive role of miRNAs, 8 and H. pylori infection-associated altered expression of miRNAs is well documented in several disease conditions.8,22 However, the link between the H. pylori exposure and dysregulation of host insulin signalling and metabolic syndrome is not clear.
A study demonstrated that a direct interaction of miRNA-1222 with 3’ UTR of IRS-1 mRNA underpins insulin signalling impairment, 13 and its inhibitory action on Akt phosphorylation and glucose transporter 4 (Glut 4) expression also reduced glucose uptake by cells. 8 Available research evidence indicated an important role of miRNA-155 in the pathogenesis of T2DM and its complications, and involvement of the miRNA in the disease conditions was attributed to its ability to prevent glycaemic control. 23 Identifying factors that are involved in the control of optimal serum glucose in T2DM patient prevent complication.
The current study identifies altered serum levels of miRNAs/RNA in T2DM patients with H. pylori exposure as a possible crucial molecular factor in glycaemic control. Previous studies only associated the infection with T2DM and provided a limited information on the possible mechanistic role of the infection in the dysregulation of glucose metabolism resulting T2DM. Our study provides evidence of altered molecular markers that can potentially be explored for diagnosis and direct personal medicine. Not considering other infectious agents in the analysis was a limitation of the study. Other pathogens potentially regulate gene expression through epigenetics or miRNAs regulations. Investigations using cell lines in controlled environment will eliminate such possible confounders. The small sample size hindered statistical analysis to establish a direct relationship between the infection, glucose level and expression levels of miRNAs and mRNAs.
Type 2 diabetes mellitus management plan focuses on the control of blood glucose with the aim of maintaining macro and microvascular health. Currently, drugs approved for T2DM management is generalized across patients, nevertheless the ultimate goal is to individualize glycaemic control interventions and treat concurrent comorbidities. 24 Personalized treatment plan for glycaemic control in T2DM patients recognizes factors that needed to be significantly controlled. 25 Although, this study did not associate any environmental factor with the H. pylori infection, identification of environmental factors that directly influence biological response of patients is crucial. Our study has revealed H. pylori exposure (74.3%) as a factor to be considered in glycaemic control. The reported altered expression of miRNAs/mRNAs in the H. pylori exposed patients warrants further investigations into miRNA therapy along with H. pylori eradication to promote personalized medicine in T2DM patients.
Conclusion
The current study reports for the first-time altered levels of miRNAs/mRNAs among H. pylori IgG seropositive Ghanaian patients diagnosed with T2DM. This study suggests a crucial role of H. pylori infection in poor glycaemic control in T2DM. Hence, the need for further investigations with larger sample size and explore the possibilities for miRNA therapy along with H. pylori eradication, promoting individual medicine.
Acknowledgments
We thank members of Quaye Laboratory, Department of Biochemistry, Cell and Molecular Biology, West African Centre for Cell Biology of Infectious Pathogens, University of Ghana for their support in running the assays.
Declarations
Ethics Approval and Consent to Participate: Ethical approval was obtained from the Ethical and Protocol Review Committee of the School of Biomedical and Allied Health Sciences (SBAHS) with an ethics identification number SBAHS/AA/MLAB/10823555/2022-2023. Participants who agreed to participate in the study signed informed consent form.
Consent for Publication: Not applicable.
Authors Contributions: Samuel Bosomprah: Conceptualization; Formal analysis; Writing – review and editing; Supervision. Emmanuel Ayitey Tagoe: Conceptualization; Data curation; Formal analysis; Original draft; Writing – review and editing. Osbourne Quaye: Data curation; Writing – review & editing. Pius Agyenim Boateng: Data curation; Original draft; Writing – review & editing. Jael Acquah Appiah: Data curation; Original draft; Writing – review & editing.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of Data and Material: The authors confirm that the data supporting the findings of this study are available within the article.
References
- 1. Westman EC. Type 2 diabetes mellitus: a pathophysiologic perspective. Front Nutr. 2021;8:707371-707375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Magliano D, Boyko E. IDF Diabetes Atlas 10th edition scientific committee. In: IDF DIABETES ATLAS [Internet]. 10th ed. International Diabetes Federation; 2021:35914061. [PubMed] [Google Scholar]
- 3. Gebremariam GT, Biratu S, Alemayehu M, et al. Health-related quality of life of patients with type 2 diabetes mellitus at a tertiary care hospital in Ethiopia. PLoS One. 2022;17:e0264199-NaN15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kouitcheu Mabeku LB, Noundjeu Ngamga ML, Leundji H. Helicobacter pylori infection, a risk factor for type 2 diabetes mellitus: a hospital-based cross-sectional study among dyspeptic patients in Douala-Cameroon. Sci Rep. 2020;10:12141-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bener A, Ağan AF, Al-Hamaq AOAA, et al. Prevalence of Helicobacter pylori infection among type 2 diabetes mellitus. Adv Biomed Res. 2020;9:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malamug LR, Karnchanasorn R, Samoa R, Chiu KC. The role of Helicobacter pylori seropositivity in insulin sensitivity, beta cell function, and abnormal glucose tolerance. Scientifica. 2014;2014:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wawro N, Amann U, Butt J, et al. Helicobacter pylori seropositivity: prevalence, associations, and the impact on incident metabolic diseases/risk factors in the population-based KORA study. Front Public Health. 2019;7:96-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palihaderu P, Mendis B, Premarathne J, et al. Therapeutic potential of miRNAs for type 2 diabetes mellitus: an overview. Epigenetics Insights. 2022;15:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riolo G, Cantara S, Marzocchi C, Ricci C. miRNA targets: from prediction tools to experimental validation. Methods and protocols. 2020;4:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61-69. [DOI] [PubMed] [Google Scholar]
- 11. Zhang J, Li S, Li L, et al. Exosome and exosomal MicroRNA: trafficking, sorting, and function. Genom Proteom Bioinform. 2015;13:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140-4147. [DOI] [PubMed] [Google Scholar]
- 13. Ono K, Igata M, Kondo T, et al. Identification of microRNA that represses IRS-1 expression in liver. PLoS One. 2018;13:e0191553-NaN13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortega FJ, Mercader JM, Catalán V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59:781-792. [DOI] [PubMed] [Google Scholar]
- 15. El Samaloty NM, Hassan ZA, Hefny ZM, Abdelaziz DHA. Circulating microRNA-155 is associated with insulin resistance in chronic hepatitis C patients. Arab J Gastroenterol. 2019;20:1-7. [DOI] [PubMed] [Google Scholar]
- 16. Bai X, Luo Q, Tan K, Guo L. Diagnostic value of VDBP and miR-155-5p in diabetic nephropathy and the correlation with urinary microalbumin. Exp Ther Med. 2020;20:86-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mir FA, Mall R, Iskandarani A, et al. Characteristic MicroRNAs linked to dysregulated metabolic pathways in Qatari adult subjects with obesity and metabolic syndrome. Front Endocrinol. 2022;13:937089-937113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeinali F, Aghaei Zarch SM, Jahan-Mihan A, et al. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and type 2 diabetes mellitus: a case control study. PLoS One. 2021;16:e0251697-NaN13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baker MA, Davis SJ, Liu P, et al. Tissue-specific MicroRNA expression patterns in four types of kidney disease. J Am Soc Nephrol. 2017;28:2985-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazzeo A, Beltramo E, Lopatina T, et al. Molecular and functional char-acterization of circulating extracellular vesicles from diabetic patients with and without retinopathy and healthy subjects. Exp Eye Res. 2018;176:69-77. [DOI] [PubMed] [Google Scholar]
- 22. Jiang Q, Wang Y, Hao Y, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jankauskas SS, Gambardella J, Sardu C, Lombardi A, Santulli G. Functional role of miR-155 in the pathogenesis of diabetes mellitus and its complications. Noncoding RNA. 2021;7:39-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perreault L, Skyler JS, Rosenstock J. Novel therapies with precision mechanisms for type 2 diabetes mellitus. Nat Rev Endocrinol. 2021;17:364-377. [DOI] [PubMed] [Google Scholar]
- 25. Azzam MM, Ibrahim AA, Abd El-Ghany MI. Factors affecting glycemic control among Egyptian people with diabetes attending primary health care facilities in Mansoura District. Egypt J Intern Med. 2021;33:33-43. [Google Scholar]