Abstract
Background
Primary screening for high-risk human papillomavirus (hrHPV) with cytological triage for women with non-16/18 hrHPV-positive status has become popular in China. However, cytology relies on the subjective judgment of pathologists, leading to inconsistent clinical performance.
Methods
A total of 657 hrHPV-positive women aged 25–64 years were enrolled in this cross-sectional study. All participants underwent colposcopic biopsy after cytology triage, with cytology residual specimens undergoing DNA methylation testing. CIN2+ and CIN3+ sensitivity and specificity were compared between the different triage strategies (n=487): PAX1 methylation (PAX1m) , Glycophorin C methylation (GYPCm), cytology, and combinations between them or with HPV16/18.
Results
The area under the receiver operating characteristic curves (AUCs) for PAX1m and GYPCm in detecting CIN2 or worse (CIN2+) were 0.867 (95% confidence interval [CI]: 0.796–0.937) and 0.873 (95% CI: 0.808–0.938), respectively. The sensitivities of PAX1m and GYPCm were consistent with those of cytology for both CIN2+ and CIN3+ detection. The relative specificities of PAX1m and GYPCm for CIN2+ detection compared to cytology were 2.83 (95% CI: 2.33–2.45) and 3.09 (95% CI: 2.40–3.98), respectively. The relative specificities of combining HPV 16/18 with PAX1m and GYPCm for CIN2+ detection compared to cytology were 3.38 (95% CI: 2.96–3.86) and 3.67 (95% CI: 3.15–4.27), respectively. Compared to low levels of DNA methylation, high levels of PAX1m and GYPCm resulted in odd ratios (ORs) of 57.66 (95% CI: 13.57–409.12, p < 0.001) and 23.87 (95% CI: 6.49–115.42, p < 0.001) for CIN3+, adjusted for HPV 16/18 and cytology results.
Conclusions
PAX1m and GYPCm demonstrated superior ability to identify cervical precancerous lesions and cervical cancer, with AUC values exceeding 0.85. For detecting CIN2+/CIN3+ in women with hrHPV-positive status, DNA methylation (combined with HPV 16/18) showed higher specificity than cytology (combined with HPV 16/18) and is a potential molecular biomarker for detecting cervical (pre)cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-13126-4.
Keywords: PAX1 methylation, GYPC methylation, DNA methylation, Cervical cancer, Cervical intraepithelial neoplasia, High-risk human papillomavirus, Relative sensitivity, Relative specificity
Background
Generally, hrHPV-DNA testing is widely recognized as the primary method for screening cervical (pre)cancer, having replaced cytological examination as the primary screening approach [1]. Persistent hrHPV infection is the primary cause of cervical cancer (CC) [2], with hrHPV testing exhibiting high sensitivity and a negative predictive value for high-grade cervical lesions and CC [3, 4]. Thus, multiple national and regional guidelines recommend hrHPV as the primary screening test for CC [5].
Most hrHPV-positive cases represent transient HPV infections and do not lead to related diseases. Without an appropriate triage strategy, direct referral of all hrHPV cases can result in an unacceptably high rate of vaginal colposcopy, causing anxiety and unnecessary treatments [6]. Triage tests for patients with hrHPV-positive status include HPV 16/18 genotyping, cytology, and p16/Ki-67 cytoimmunochemistry [7]. Among these, the combination of HPV 16/18 and cytology is essential, as it helps reduce the colposcopy referral rate [8]. However, the subjective nature of cytology and the low threshold for atypical squamous cells of undetermined significance (ASC-US) referrals result in many patients undergoing unnecessary colposcopy examinations. The limitation of HPV 16/18 genotyping is that other hrHPV types can also cause serious related diseases. Additionally, with the increasing number of individuals vaccinated against HPV 16/18, the incidence of cervical lesions associated with HPV 16/18 has declined, potentially reducing the effectiveness of this genotyping method [9].
The methylation of host DNA has been shown to have high sensitivity and specificity for cervical intraepithelial neoplasia or worse (CIN2+), especially for invasive cancers [10–14]. The PAX1 gene is a tumor suppressor gene that inhibits the malignant phenotype of cells under the carcinogenic pressure of hrHPV. It activates dual specificity phosphatases 1, 5, and 6, inhibiting the EGF/MAPK signaling pathway to suppress cancer [15]. However, the PAX1 gene often becomes abnormally hypermethylated, silenced, and inactivated, losing its tumor-suppressive function [16, 17]. The application of PAX1m in CC screening and prevention is as follows: (1) triaging women with non-16/18 hrHPV-positive status [18]; (2) predicting the progression of cervical lesions after CIN2/CIN3 conization [19]; (3) assessing its relationship with hrHPV load and p16/Ki67 immunohistochemical staining [19, 20]; (4) assessing the necessity of cervical canal curettage (ECC) [21]; (5) monitoring CC treatment [22]; and (6) prognostic evaluation of CC [23]. The GYPC gene for the human erythrocyte membrane glycoprotein C, also known as glycophorin C, is located on human chromosome 2 and contains multiple exons and introns that encode a protein containing 128 amino acid residues. Significant methylation folding changes in GYPC have been observed in the plasma of patients with ovarian cancer compared to that in the plasma patients without ovarian cancer [24]. To our knowledge, limited research exists on the correlation between Glycophorin C methylation (GYPCm) and CC. GYPCm combined ZSCAN12m can be used for diagnosing uterine cancers with a sensitivity of 90.9% [25].
In this study, we explored the clinical performance of DNA methylation and compared triage strategies for detecting cervical (pre)cancer in women with hrHPV-positive status undergoing outpatient opportunistic cervical screening. We assessed the performance of DNA methylation markers, PAX1m and GYPCm, both alone and in combination with cytology and HPV 16/18 testing.
Methods
Study population and sample collection
This cross-sectional study included women who tested positive for hrHPV during CC screening at hospital outpatient clinics from June to December 2023. The inclusion criteria were as follows: (1) age 25–64 years, (2) not pregnant or lactating, (3) no history of surgical resection for cervical lesions or CC, (4) no history of other cancers, (5) HIV negative, and (6) normal immune function. The exclusion criteria were as follows: (1) samples with insufficient liquid-based cells or ineffective DNA methylation detection, and (2) those who had not completed the pathological diagnosis of colposcopy-directed biopsy.
The specimens used in this study were cervical exfoliated cells collected by a colposcopy physician. Liquid-based cytology (LBC) was conducted first, followed by DNA extraction from the remaining specimens. These specimens were stored at −20℃ and subjected to DNA methylation testing after obtaining the pathological results. The study flow chart is shown in Fig. 1. This study was approved by the Research and Clinical Trial Ethics Committee of Changsha Hospital for Maternal & Child Health Care (No. EC-20230726-02), and all participants provided written informed consent before specimens collection.
Fig. 1.
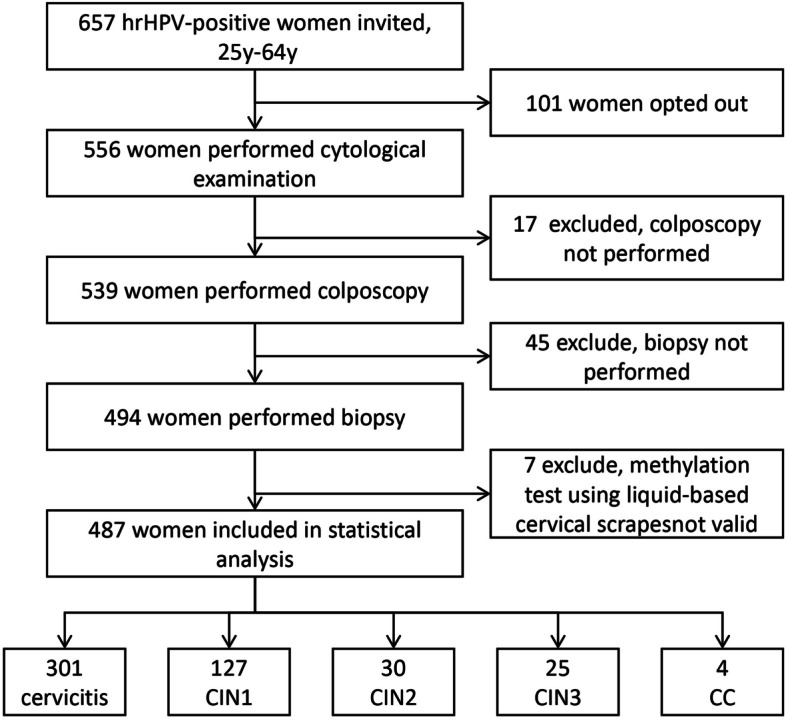
Study flow chart. hrHPV, high-risk Human papillomavirus; CIN1, Cervical intraepithelial neoplasia grade 1; CIN2, Cervical intraepithelial neoplasia grade 2; CIN3, Cervical intraepithelial neoplasia grade 3; CC, Cervical cancer
HrHPV testing
HPV testing was conducted using 21 subtypes (HybriBio Ltd., Guangzhou, China) following the manufacturer’s instructions. The simple procedure involves: (1) amplification of HPV DNA by PCR, (2) hybridization of the DNA amplicon with fixed specificity using HybriBio's proprietary flow hybridization technology, and (3) identification of 21 HPV genotypes by enzyme immunoassay. This study included women who tested positive for 14 hrHPV types and one potentially high-risk type (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, and 68).
Cytology
Those with hrHPV-positive status were advised to undergo cytological examination for triage. Participants were instructed to avoid sexual intercourse for 1 d before sampling and to refrain from vaginal flushing or medication for 3 d before sampling. Cytological sampling was performed at least 5 d after menstruation. Cervical exfoliated cells were preserved in a liquid cell preservation solution (Hologic, MA, USA). Subsequently, the collected cells were processed into thin smears using a ThinPrep cytology analyzer, examined under a microscope, and interpreted according to the 2014 Bethesda system guidelines.
Colposcopy-directed biopsy
For women referred for colposcopy, 1–3 samples of living tissue were taken from the site of the most severe cervical lesion. Four specimens were randomly selected in a counterclockwise direction in cases with a normal colposcopic impression. Biopsy specimens were stained with hematoxylin and eosin and sectioned for initial examination by a pathologist, followed by an independent secondary examination by another pathologist. In cases where the first two assessments were inconsistent, a third pathologist was consulted to make the final determination.
PAX1 and GYPC methylation analysis
The remaining DNA from cervical exfoliated cells were stored at −20℃ and subjected to DNA methylation testing within 3 months. This testing was conducted at a certified laboratory in China (Hoomya Medical Laboratory, Changsha). PAX1m and GYPCm analysis were performed using the cervical cancer gene methylation test kit (Hoomya) according to the manufacturer’s instructions. The COL2A1 gene served as an internal reference, with methylation results calculated as ΔCpgene = Cpgene − CpCOL2A1. The DNA methylation analysis involved the following steps: (1) bisulfite conversion, which transforms unmethylated cytosine (C) into uracil (U) while leaving methylated cytosine unchanged; (2) amplification of 50 cycles using a fluorescence quantitative polymerase chain reaction instrument (LC480; Roche Applied Science, CA, USA); and (3) calculation of ΔCp of the target gene. When the quality control was met, but the target gene was not amplified, the Cp of the target gene was set to 50, and ΔCp calculation was performed.
Statistical analysis
All data were analyzed using R Version 4.3.2. Two-sided p-values < 0.05 were considered statistically significant. Continuous variables, such as age, PAX1m, and GYPCm, were nonnormally distributed and are presented as median (interquartile range, IQR). Categorical variables were recorded as frequencies and percentages. The distribution plots of PAX1m and GYPCm in cervical lesions were generated using the “ggplot2” package, while receiver operating characteristic (ROC) curves were constructed using the “pROC” package. To maximize the Youden index for diagnosing CIN2+, the criteria for determining positive results for PAX1m and GYPCm were set at ΔCp ≤ 10.86 and ΔCp ≤ 5.97, respectively. Conversely, negative results were ΔCp > 10.86 for PAX1m and ΔCp > 5.97 for GYPCm. The sensitivity and specificity of each ΔCp value of PAX1m and GYPCm were obtained from ROC analysis (Supplementary Tables 1and 2). Using the same principle for detecting CIN2+ and CIN3+, PAX1m was classified into three levels using two cut-off values as follows: low (ΔCp > 11), moderate (9 < ΔCp ≤ 11), and high (ΔCp ≤ 9). GYPCm was similarly categorized into low (ΔCp > 6), moderate (4 < ΔCp ≤ 6), and high (ΔCp ≤ 4). The calculation of sensitivity and specificity was performed using the “gmodels” and “DescTools” packages, with the Wilson score method used to estimate 95% confidence intervals (CIs). The 95% CIs for relative sensitivity and specificity were calculated using the formula proposed by Hayen et al. [26]. Odds ratios (ORs) and adjusted ORs for PAX1m and GYPCm levels were estimated using logistic regression.
Results
Patients and histological outcomes
This study included 487 women with hrHPV-positive status, with a median age of 42 years (IQR: 34.5–51.0). Of these, 381 women (78.2%) were infected with one hrHPV subtype, while 7 women (1.4%) were infected with four or more hrHPV subtypes. The positivity rate for HPV 16/18 was 17.9% (87 cases). The rate of cytological abnormalities (≥ ASC-US) was 73.3%. The median ΔCp value for PAX1m was 13.9 (10.4–20.4), and for GYPCm was 14.0 (6.8–20.3). Histological outcomes included 301 cases of cervicitis (61.8%), 127 cases of CIN1 (26.1%), 30 cases of CIN2 (6.2%), 25 cases of CIN3 (5.1%), and 4 cases of CC (0.8%) (Table 1).
Table 1.
Basic characteristic of participants
| level | Percentage | |
|---|---|---|
| n | 487 | |
| Age (median [IQR]) | 42.0 [34.5, 51.0] | |
| HPV infection (%) | Single | 381 (78.2) |
| Double | 79 (16.2) | |
| Triple | 20 (4.1) | |
| Quadruple | 4 (0.8) | |
| Quintuple | 3 (0.6) | |
| HPV16/18 (%) | Negative | 400 (82.1) |
| Positive | 87 (17.9) | |
| Cytology (%) | NILM | 130 (26.7) |
| ASC-US | 218 (44.8) | |
| LSIL | 90 (18.5) | |
| ASC-H/AGC | 36 (7.4) | |
| HSIL | 13 (2.7) | |
| PAX1 methylation (median [IQR]) | 13.9 [10.4, 20.4] | |
| GYPC methylation (median [IQR]) | 14.0 [6.8, 20.3] | |
| Colposcopy (%) | Normal | 6 (1.2) |
| LSIL | 424 (87.1) | |
| HSIL | 56 (11.5) | |
| CC | 1 (0.2) | |
| Pathology (%) | Cervicitis | 301 (61.8) |
| CIN1 | 127 (26.1) | |
| CIN2 | 30 (6.2) | |
| CIN3 | 25 (5.1) | |
| CC | 4 (0.8) |
Abbreviation: IQR Interquartile range, HPV Human papillomavirus, NILM No intraepithelial lesions or malignancy, ASC-US Atypical squamous cells of undetermined significance, LSIL Low-grade squamous intraepithelial lesion, ASC-H Atypical squamous cells cannot exclude HSIL, AGC Atypical glandular cells, HSIL High-grade squamous intraepithelial lesion, CC Cervical cancer, CIN1 Cervical intraepithelial neoplasia grade 1, CIN2 Cervical intraepithelial neoplasia grade 2, CIN3 Cervical intraepithelial neoplasia grade 3
Distribution and AUCs of PAX1m and GYPCm
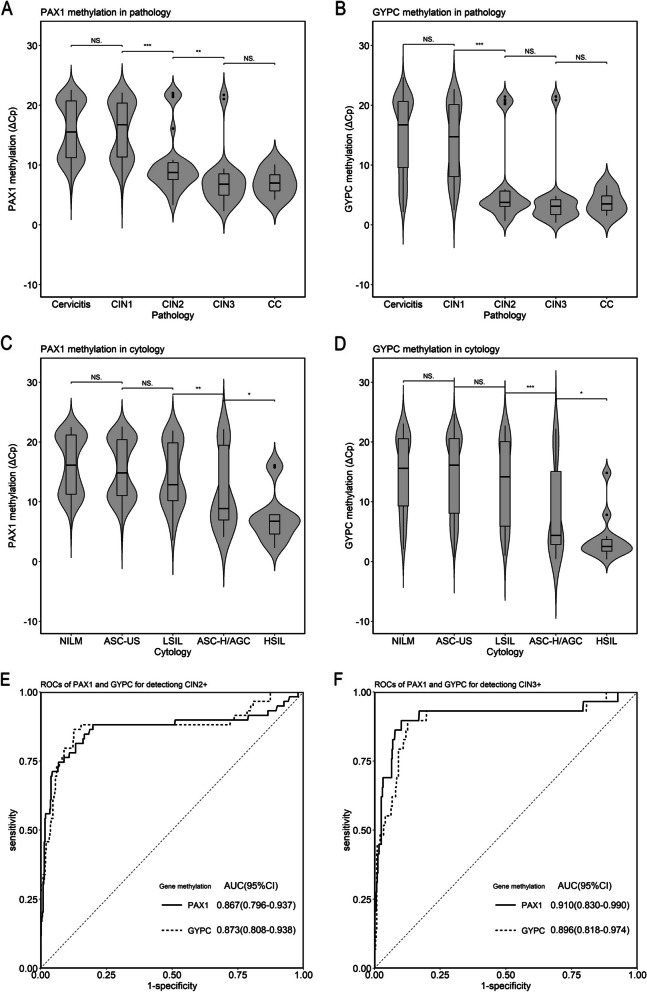
The distribution of ΔCp values for PAX1m and GYPCm in histopathological and cytological results is shown in Fig. 2A–D. Statistically significant differences were observed in the ΔCp values of PAX1m and GYPCm between the LSIL and ASC-H/AGC groups (all p < 0.01). The differences in PAX1m ΔCp values between CIN1 and CIN2, as well as between CIN2 and CIN3, were statistically significant (all p < 0.01). A statistically significant difference in GYPCm ΔCp values was also observed between CIN1 and CIN2 (p < 0.001). The area under the ROC curve (AUC) values for detecting CIN2+ with PAX1m and GYPCm were 0.867 (95% CI: 0.796–0.937) and 0.873 (95% CI: 0.808–0.973), respectively (Fig. 2E). The AUCs of triage strategies for detecting CIN2+ were detailed in Supplementary Table 3. The AUC values for detecting CIN3+ with PAX1m and GYPCm were 0.910 (95% CI: 0.830–0.990) and 0.896 (95% CI: 0.818–0.974), respectively (Fig. 2F), showing good discriminative ability.
Fig. 2.
Violin plots of PAX1 and GYPC methylation distribution in lesions and ROCs plot for detection of CIN2+ and CIN3+. A. Distribution of PAX1 methylation in histopathologic lesions. B. Distribution of GYPC methylation in cytopathologic lesions. C. Distribution of PAX1 methylation in cytological lesions. D. Distribution of GYPC methylation in cytological lesions. E. ROC of GYPC methylation for detecting CIN2+. E. ROCs of PAX1 and GYPC methylation for detecting CIN2+. F. ROCs of PAX1 and GYPC methylation for detecting CIN3+. CIN1, Cervical intraepithelial neoplasia grade 1; CIN2, Cervical intraepithelial neoplasia grade 2; CIN3, Cervical intraepithelial neoplasia grade 3; CC, Cervical cancer; NILM, No intraepithelial lesions or malignancy; ASC-US, Atypical squamous cells of undetermined significance; LSIL, Low-grade squamous intraepithelial lesion; ASC-H, Atypical squamous cells cannot exclude HSIL; AGC, Atypical glandular cells; HSIL, High-grade squamous intraepithelial lesion; ROC, Receiver operating characteristic curve; AUC, Area under the curve; CIN2+, Cervical intraepithelial neoplasia grade 2 or worse; CIN3+, Cervical intraepithelial neoplasia grade 3 or worse. NS., no significance; * p < 0.05; ** p < 0.01; *** p < 0.001
Performance of triage markers for CIN2+ and CIN3+ detection
The sensitivity for detecting CIN2+ and CIN3+ in women with hrHPV-positive status using cytological triage was 84.7% (95% CI: 73.5%–91.8%) and 89.7% (95% CI: 73.6%–96.4%), respectively. The sensitivities of PAX1m and GYPCm were consistent with those of cytology, with relative sensitivities of 1.04 (95% CI: 0.52–2.09) and 1.02 (95% CI: 0.55–1.88) for detecting CIN2+, and 1.04 (95% CI: 0.26–4.15) and 1.00 (95% CI: 0.40–2.52) for detecting CIN3+, respectively. The relative cytological specificities for PAX1m and GYPCm in detecting CIN2+ were 2.83 (95% CI: 2.33–2.45) and 3.09 (95% CI: 2.40–3.98), respectively, while for detecting CIN3+, the relative specificities were 2.74 (95% CI: 2.32–3.24) and 2.98 (95% CI: 2.43–3.66), respectively.
The sensitivity for detecting CIN2+ and CIN3+ in women with hrHPV-positive status using HPV 16/18 combined with cytology triage was 91.5% (95% CI: 81.6%–96.3%) and 96.6% (95% CI: 82.8–99.4), respectively. The sensitivities of PAX1m and GYPCm combined with cytology were consistent with those of HPV 16/18 combined with cytology, with relative sensitivities of 1.01 (95% CI: 0.38–2.71) and 1.00 (95% CI: 0.38–2.61) for detecting CIN2+, and 1.00 (95% CI: 0.37–2.67) and 0.96 (95% CI: 0.24–3.86) for detecting CIN3+, respectively. The relative specificities of PAX1m and GYPCm combined with cytology for detecting CIN2+ were 1.21 (95% CI: 1.17–1.26) and 1.28 (95% CI: 1.24–1.33), respectively. The relative specificities for detecting CIN3+ was 1.19 (95% CI: 1.14–1.23) and 1.26 (95% CI: 1.21–1.30), respectively.
The sensitivity of combining HPV 16/18 with PAX1m and GYPCm for detecting CIN2+ and CIN3+ was consistent, at 93.2% (95% CI: 83.8%–97.3%) and 96.6% (95% CI: 82.8%–99.4%), respectively. The relative specificities of HPV 16/18 combined with PAX1m and GYPCm for detecting CIN2+ were 3.38 (95% CI: 2.96–3.86) and 3.67 (95% CI: 3.15–4.27), respectively. For detecting CIN3+, the relative specificities were 3.26 (95% CI: 2.89–3.67) and 3.54 (95% CI: 3.10–4.05), respectively.
Whether alone or in combination, the ability of DNA methylation to detect CIN2+ and CIN3+ due to cytology is mainly manifested in higher specificity.
ORs of PAX1 and GYPC methylation levels for CIN2+ and CIN3+
The ORs for moderate and high levels of PAX1m compared to low levels for CIN2+ were 7.25 (95% CI: 2.69–20.61) and 97.14 (95% CI: 41.24–261.33), respectively. For CIN3+, the ORs were 4.60 (95% CI: 0.54–38.83) and 113.49 (95% CI: 32.14–722.80), respectively. The ORs for moderate and high levels of GYPCm compared to low levels for CIN2+ were 21.31 (95% CI: 8.75–55.94) and 81.59 (95% CI: 35.11–211.49), respectively. For CIN3+, the ORs were 23.44 (95% CI: 6.51–110.10) and 61.30 (95% CI: 19.64–270.53), respectively.
After adjusting for HPV 16/18 and cytological results, the ORs for high levels of PAX1m compared to low levels for CIN2+ and CIN3+ were 97.14 (95% CI: 41.24–261.33) and 113.49 (95% CI: 32.14–722.80), respectively. The ORs high levels of GYPCm compared to low levels for CIN2+ and CIN3+ were 45.95 (95% CI: 18.52–125.05) and 23.87 (95% CI: 6.49–115.42), respectively.
GYPCm is associated with a higher risk of CIN2+ and CIN3+ at moderate levels, while PAX1m is associated with a higher risk at high levels.
Discussion
This cross-sectional study indicates that molecular DNA methylation analysis is comparable to cytological examination regarding sensitivity for detecting CI2+ and CIN3+ in outpatient women with hrHPV-positive status aged 25–64 years. Specifically, the sensitivity for CIN2+ was approximately 85%for PAX1m, GYPCm and cytology. For CIN3+, the sensitivities were approximately 90%. Notably, the specificity of DNA methylation (> 80%) for CIN2+ was significantly higher than that of cytology (approximately 30%). For CIN3+, the specificity of DNA methylation (> 75%) was also higher than that of cytology (Table 2). Previous studies have shown that with an ASC-US threshold for cytology, the sensitivity is higher and does not significantly differ from that of DNA methylation. However, the specificity of DNA methylation was much higher than that of cytology [13, 27–29]. Among the abnormal cytological results, ASC-US accounted for 44.8%, and 206 cases (94.5%) of 218 ASC-US cases were NILM and CIN1, which may be the reason for the high cytological sensitivity and low specificity (Supplementary Table 4). Currently, the World Health Organization and various countries recommend hrHPV testing as the primary tool for CC screening [5, 30]. Most hrHPV-positive cases are transient infections typically clear within 2 years [31]. Therefore, triage for patients with hrHPV-positive status is essential, primarily cytological triage for patients with non-16/18 hrHPV-positive status [32]. However, there are a number of other methods that are being explored, such as double staining (p16/Ki67) [33], HPV E6/E7 mRNA [34] and DNA ploidy [35]. For the p16/Ki67 triage of hrHPV-positive women with CIN2+ and CIN3+, the sensitivities were recorded at 86.5% and 89.5%, respectively, while the specificities were noted to be 57.5% and 54.0%. Furthermore, the positive predictive values (PPVs) were determined to be 24.4% and 10.9% [33], respectively. Comparing our ‘PAX1m or GYPCm’ data in CIN2+ and CIN3+, the sensitivities were 88.1% and 93.1%, while the specificities were 73.6% and 69.9%, respectively. Additionally, the positive predictive values (PPV) were recorded at 31.5% and 16.4%. Overall, based on the initial literature data regarding p16/Ki67 and our methylation study findings, the outcomes of methylation analysis demonstrated superiority over those of P16/Ki67. However, future comparative studies involving p16/Ki67 or other testing and 'PAX1m or GYPCm' within the same cohort are needed. World Health Organization Recommendations, patients who are HPV 16/18 positive or who have cytology results of ASC-US+ are recommended for referral to colposcopy [5, 36]. We also analyzed and compared the clinical performance of HPV 16/18 combined with cytology and HPV 16/18 combined with DNA methylation. The results showed no significant difference in sensitivity for detecting CIN2+ and CIN3+ between the two approaches. However, the specificity of combining HPV 16/18 with cytology for detecting CIN2+ and CIN3+ was 19.9% (95% CI: 16.3%–23.9%) and 19.4% (95% CI: 16.1%–23.3%), respectively. This indicates that the screening strategy of hrHPV initial screening and HPV 16/18 combined with cytology triage for cervical (pre)cancer still resulted in a large number of women being misdiagnosed with high-grade cervical lesions, resulting in unnecessary referrals to colposcopy and a waste of medical resources. The specificities of combining HPV 16/18 with PAX1m and GYPCm for detecting CIN2+ and CIN3+ exceeded 60% (Table 2). In a real-world population of individuals with hrHPV-positive status, the sensitivity and specificity of HPV 16/18 combined with DNA methylation for detecting CIN2+ were 85.9% and 60.7%, respectively [10], aligning closely with our findings.
Table 2.
Sensitivities and specificities of different markers for CIN2+ and CIN3+ detection in hrHPV-positive women.
| Triage marker |
Sensitivity (%) (n/N) 95%CI |
Specificity (%) (n/N) 95%CI |
PPV (%) (n/N) 95%CI |
NPV (%) (n/N) 95%CI |
Compared with LBC ASC-US | Compared with HPV 16/18 or LBC ASC-US | ||
|---|---|---|---|---|---|---|---|---|
| Relative sensitivity (95%CI) | Relative specificity (95%CI) | Relative sensitivity (95%CI) | Relative specificity (95%CI) | |||||
| For detecting CIN2+ | ||||||||
| LBC ASC-US | 84.7 (50/59) 73.5-91.8 | 28.3 (121/428) 24.2-32.7 | 14.0 (50/357) 10.8-18.0 | 93.1 (121/130) 87.4-96.3 | 1 | 1 | — | — |
| HPV16/18 | 27.1 (16/59) 17.4-38.6 | 83.4 (357/428) 79.6-86.6 | 18.4 (16/87) 11.6-27.8 | 89.2 (357/400) 85.8-91.9 | 0.32 (0.17-0.61) | 2.95 (2.34-3.73) | — | — |
| PAX1m | 88.1 (52/59) 77.5-94.1 | 80.1 (343/428) 76.1-83.7 | 38.0 (52/137) 30.3-46.3 | 98.0 (343/350) 95.9-99.0 | 1.04 (0.52-2.09) | 2.83 (2.33-2.45) | — | — |
| GYPCm | 86.4 (51/59) 75.5-93.0 | 87.4 (374/428) 83.9-90.2 | 48.6 (51/105) 39.2-58.0 | 97.9 (374/382) 95.9-98.9 | 1.02 (0.55-1.88) | 3.09 (2.40-3.98) | — | — |
| HPV 16/18 or LBC ASC-US | 91.5 (54/59) 81.6-96.3 | 19.9 (85/428) 16.3-23.9 | 13.6 (54/397) 10.6-17.3 | 94.4 (85/90) 87.6-97.6 | — | — | 1 | 1 |
| PAX1m or LBC ASC-US | 93.2 (55/59) 83.8-97.3 | 24.1 (103/428) 20.2-28.3 | 14.5 (55/380) 11.3-18.4 | 96.3 (103/107) 9.08-98.6 | — | — | 1.01 (0.38-2.71) | 1.21 (1.17-1.26) |
| GYPCm or LBC ASC-US | 91.5 (54/59) 81.6-96.3 | 25.5 (109/428) 21.6-29.8 | 14.5 (54/373) 11.3-18.4 | 95.6 (109/144) 90.1-98.1 | — | — | 1.00 (0.38-2.61) | 1.28 (1.24-1.33) |
| HPV 16/18 or PAX1m | 93.2 (55/59) 83.8-97.3 | 67.1 (287/428) 62.5-71.3 | 28.1 (55/196) 22.2-34.7 | 98.6 (287/291) 96.5-99.5 | — | — | 1.02 (0.38-2.71) | 3.38 (2.96-3.86) |
| HPV 16/18 or GYPCm | 93.2 (55/59) 83.8-97.3 | 72.9 (312/428) 68.5-76.9 | 32.2 (55/171) 25.6-39.5 | 98.7 (312/316) 96.8-99.5 | — | — | 1.02 (0.38-2.71) | 3.67 (3.15-4.27) |
| PAX1m or GYPCm | 88.1 (52/59) 77.5-94.1 | 73.6 (315/428) 69.2-77.6 | 31.5 (52/165) 24.9-39.0 | 97.8 (315/322) 95.6-98.9 |
0.96 (0.38-2.46) |
3.71 (3.16-4.35) |
||
| For detecting CIN3+ | ||||||||
| LBC ASC-US | 89.7 (26/29) 73.6-96.4 | 27.7 (127/458) 23.8-32.0 | 7.3 (26/357) 5.0-10.5 | 97.7 (127/130) 93.4-99.2 | 1 | 1 | — | — |
| HPV16/18 | 37.9 (11/29) 22.7-56.0 | 83.4 (382/458) 79.7-86.5 |
12.6 (11/87) 7.2-21.2 |
95.5 (382/400) 93.0-97.1 | 0.42 (0.13-1.35) | 3.00 (2.40-3.78) | — | — |
| PAX1m | 93.1 (27/29) 78.0-98.1 | 76.0 (348/458) 71.9-79.7 | 19.7 (27/137) 13.9-27.2 | 99.4 (348/350) 97.9-99.8 | 1.04 (0.26-4.15) | 2.74 (2.32-3.24) | — | — |
| GYPCm | 89.7 (26/29) 73.6-96.4 | 82.7 (379/458) 79.0-85.9 | 24.8 (26/105) 17.5-33.8 | 99.2 (379/382) 97.7-99.7 | 1.00 (0.40-2.52) | 2.98 (2.43-3.66) | — | — |
| HPV 16/18 or LBC ASC-US | 96.6 (28/29) 82.8-99.4 | 19.4 (89/458) 16.1-23.3 | 7.1 (28/397) 4.9-10.0 | 98.9 (89/90) 94.0-99.8 | — | — | 1 | 1 |
| PAX1m or LBC ASC-US | 96.6 (28/29) 82.8-99.4 | 23.1 (106/458) 19.5-27.2 | 7.4 (28/380) 5.1-10.5 | 99.1 (106/107) 94.9-99.8 | — | — | 1.00 (0.37-2.67) | 1.19 (1.14-1.23) |
| GYPCm or LBC ASC-US | 93.1 (27/29) 78.0-98.1 | 24.5 (112/458) 20.7-28.6 | 7.2 (27/373) 5.0-10.3 | 98.2 (112/114) 93.8-99.5 | — | — | 0.96 (0.24-3.86) | 1.26 (1.21-1.30) |
| HPV 16/18 or PAX1m | 96.6 (28/29) 82.8-99.4 | 63.3 (290/458) 58.5-67.6 | 14.3 (28/196) 10.1-19.9 | 99.7 (290/291) 98.1-99.9 | — | — | 1.00 (0.37-2.67) | 3.26 (2.89-3.67) |
| HPV 16/18 or GYPCm | 96.6 (28/29) 82.8-99.4 | 68.8 (315/458) 64.4-72.9 | 16.4 (28/171) 11.6-22.7 | 99.7 (315/316) 98.2-99.9 | — | — | 1.00 (0.37-2.67) | 3.54 (3.10-4.05) |
| PAX1m or GYPCm | 93.1 (27/29) 78.0-98.1 | 69.9 (320/458) 65.5-93.9 | 16.4 (27/165) 11.5-22.8 | 99.4 (320/322) 97.8-99.8 |
0.96 (0.31-2.99) |
3.60 (3.12-4.14) |
||
Abbreviation: CIN2+, Cervical intraepithelial neoplasia grade 2 or worse, CIN3+ Cervical intraepithelial neoplasia grade 3 or worse, hrHPV high-risk Human papillomavirus, PPV Positive predictive value, NPV Negative predictive value, LBC Liquid based cytology, ASC-US Atypical squamous cells of undetermined significance, CI Confidence interval, HPV Human papillomavirus, PAX1m PAX1 methylation, GYPCm GYPC methylation
A meta-analysis of 11 articles found that 819 out of 1,470 patients with CIN2 experienced natural regression within 2 years, with a regression rate of approximately 50% (95% CI: 43%–57%) [37]. We categorized PAX1m and GYPCm into three levels. The high levels of PAX1m and GYPCm had ORs of 113.49 (95% CI: 32.14–722.80) and 61.30 (95% CI: 19.64–270.53) for CIN3+ compared to low levels, respectively (Table 3). This may indicate a positive correlation between the level of DNA methylation and the severity of cervical lesions, as previous studies have reported that the methylation levels of various genes increase with the severity of cervical lesions [19, 20, 38].
Table 3.
ORs of PAX1 and GYPC methylation levels for CIN2+ and CIN3+ in hrHPV-positive women
| Methylation level | OR (95%CI)# | ||
|---|---|---|---|
| Low | Moderate | High | |
| PAX1m n (ΔCp range) | 347 (11.01-22.57) | 77 (9.08-10.99) | 63 (2.8-8.99) |
| CIN2+ vs. <CIN2 | 1 | 7.25 (2.69-20.61) | 97.14 (41.24-261.33) |
| P | — | <0.001 | <0.001 |
| CIN3+ vs. <CIN3 | 1 | 4.60 (0.54-38.83) | 113.49 (32.14-722.80) |
| P | — | 0.130 | <0.001 |
| PAX1m adjusted by HPV16/18 and LBC results | |||
| CIN2+ vs. <CIN2 | 1 | 7.61 (2.78-21.94) | 59.97 (22.91-178.20) |
| P | — | <0.001 | <0.001 |
| CIN3+ vs. <CIN3 | 1 | 4.41 (0.51-37.93) | 57.66 (13.57-409.12) |
| P | — | 0.147 | <0.001 |
| GYPCm n (ΔCp range) | 381 (6.01-24.71) | 51 (4.01-6.00) | 55 (0.37-3.94) |
| CIN2+ vs. <CIN2 | 1 | 21.31 (8.75-55.94) | 81.59 (35.11-211.49) |
| P | — | <0.001 | <0.001 |
| CIN3+ vs. <CIN3 | 1 | 23.44 (6.51-110.10) | 61.30 (19.64-270.53) |
| P | — | <0.001 | <0.001 |
| GYPCm adjusted by HPV16/18 and LBC results | |||
| CIN2+ vs. <CIN2 | 1 | 16.70 (6.44-46.09) | 45.95 (18.52-125.05) |
| P | — | <0.001 | <0.001 |
| CIN3+ vs. <CIN3 | 1 | 13.42 (3.28-6.82) | 23.87 (6.49-115.42) |
| P | — | <0.001 | <0.001 |
Abbreviation: OR Odds ration, CIN2 Cervical intraepithelial neoplasia grade 2, CIN2+ Cervical intraepithelial neoplasia grade 2 or worse, CIN3 Cervical intraepithelial neoplasia grade 3, CIN3+ Cervical intraepithelial neoplasia grade 3 or worse, PAX1m PAX1 methylation, GYPCm GYPC methylation, HPV Human papillomavirus, LBC Liquid based cytology
#OR and 95%CI were calculated using logistic regression
We also evaluated PAX1m and GYPCm combined with cytology as a triage tool for women with hrHPV-positive status. This combination demonstrated the same sensitivity as HPV 16/18 combined with cytology but with higher specificity. The relative specificity for detecting CIN2+ was 1.21 (95% CI: 1.17–1.26) and 1.28 (95% CI: 1.24–1.33), respectively, while the specificity for detecting CIN3+ was 1.19 (95% CI: 1.14–1.23) and 1.26 (95% CI: 1.21–1.30), respectively (Table 2). This approach is also a relatively optimal strategy for triaging non-type hrHPV-positive cases. We had further insight (Supplementary Table 5) and found that, the sensitivity of PAX1m in detecting CIN2+ and CIN3+ combined with GYPCm for "and" was slightly less than that of HPV16/18 combined with cytology ASC-US+, while its specificity was much higher.
This study has several limitations. First, some women with hrHPV-positive status did not complete the entire study, including cytology, colposcopy, or biopsy. This led to the exclusion of their data, which may have introduced bias in the clinical performance indicators. Second, during the analysis, when calculating the relative sensitivity, there were fewer cases of CIN3+, although the sensitivity for detecting CIN3+ was consistent. Finally, as a cross-sectional study, we could not assess the future risk of developing CIN2+ and CIN3+ in women with different DNA methylation and cytology results in the future. Notably, the quality of cytology varies widely in China, especially in the underdeveloped north-west. The advantages offered by DNA methylation analysis are molecularly based, demonstrating robustness and reproducibility. Another advantage of DNA methylation analysis over cytology is sample compatibility, with both self-sampling and HPV residual specimens. Although current methylation technologies may not be suitable for large-scale implementation, technological advances and continued development of methylation analysis are expected to lead to automated and user-friendly analyses suitable for high-throughput testing in laboratories with PCR facilities. Therefore, future real-world studies with larger sample sizes are necessary for tracking and follow-up to provide more reliable evidence-based medical data and to guide the clinical application of DNA methylation.
Conclusion
High levels of PAX1m and GYPCm were effective indicators for identifying CIN3+ (all ORs > 20). The sensitivities of PAX1m, GYPCm, and cytology for detecting CIN2+ and CIN3+ were comparable. Compared to the hrHPV-positive triage strategy (HPV 16/18 combined with cytology), the specificities of HPV 16/18 combined with PAX1m and GYPCm were three times higher. Therefore, DNA methylation may serve as an essential tool for detecting cervical (pre)cancer in women with hrHPV-positive status.
Supplementary Information
Acknowledgments
Not applicable.
Abbreviations
- AGC
Atypical glandular cells
- ASC-H
Atypical squamous cells cannot exclude HSIL
- ASC-US
Atypical squamous cells of undetermined significance
- AUC
Area under the curve
- CC
Cervical cancer
- CIN1
Cervical intraepithelial neoplasia grade 1
- CIN2
Cervical intraepithelial neoplasia grade 2
- CIN3
Cervical intraepithelial neoplasia grade 3
- CIN2+
Cervical intraepithelial neoplasia grade 2 or worse
- CIN3+
Cervical intraepithelial neoplasia grade 3 or worse
- CI
Confidence interval
- DNA
DeoxyriboNucleic Acid
- GYPCm
GYPC methylation
- HPV
Human papillomavirus
- hrHPV
high-risk Human papillomavirus
- HSIL
High-grade squamous intraepithelial lesion
- IQR
Interquartile range
- LBC
Liquid based cytology
- LSIL
Low-grade squamous intraepithelial lesion
- NILM
No intraepithelial lesions or malignancy
- NPV
Negative predictive value
- OR
Odds ration
- PAX1m
PAX1 methylation
- PPV
Positive predictive value
- ROC
Receiver operating characteristic curve
Authors’ contributions
Hong Tao carried out the experiment of the study, read the relevant literatures, performed the statistical analyses, interpreted the results and drafted the manuscript. Fang Yu, Li Yang, Xiaozhu Pei and Saiping Mao were involved in the design of the study, participated to collect specimens, do the experiment and helped to revise the manuscript. Xing Fan were involved in the conception and design of the study and direct the progress of the study. All authors read and approved the final manuscript.
Funding
This work was supported by the Research Project of department of science and technology of Changsha (kh2302012).
Data availability
The datasets used during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This research project was approved by the Research and Clinical Trial Ethics Committee of the Changsha Hospital for Maternal & Child Health Care (No. EC-20230726-02), informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marcus JZ, Cason P, Downs LS Jr, Einstein MH, Flowers L. The ASCCP Cervical Cancer Screening Task Force Endorsement and Opinion on the American Cancer Society Updated Cervical Cancer Screening Guidelines. J Low Genit Tract Dis. 2021;25(3):187–91. [DOI] [PubMed] [Google Scholar]
- 2.Cuschieri KS, Cubie HA, Whitley MW, Gilkison G, Arends MJ, Graham C, McGoogan E. Persistent high risk HPV infection associated with development of cervical neoplasia in a prospective population study. Journal of clinical pathology. 2005;58(9):946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbyn M, Simon M, Peeters E, Xu L, Meijer C, Berkhof J, Cuschieri K, Bonde J, Ostrbenk Vanlencak A, Zhao FH, et al. 2020 list of human papillomavirus assays suitable for primary cervical cancer screening. Clin Microbiol Infect. 2021;27(8):1083–95. [DOI] [PubMed]
- 4.Ejegod DM, Junge J, Franzmann M, Kirschner B, Bottari F, Sideri M, Sandri MT, Bonde J. Clinical and analytical performance of the BD Onclarity™ HPV assay for detection of CIN2+ lesions on SurePath samples. Papillomavirus Res. 2016;2:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO guideline for screening and treatment of cervical pre-cancer lesions for cervical cancer prevention, second edition. www.ncbi.nlm.nih.gov/books/NBK572317/. [PubMed]
- 6.Kyrgiou M, Athanasiou A, Kalliala IEJ, Paraskevaidi M, Mitra A, Martin-Hirsch PP, Arbyn M, Bennett P, Paraskevaidis E. Obstetric outcomes after conservative treatment for cervical intraepithelial lesions and early invasive disease. The Cochrane database of systematic reviews. 2017;11(11):Cd012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanczuk GA, Baxter GJ, Currie H, Forson W, Lawrence JR, Cuschieri K, Wilson A, Patterson L, Govan L, Black J, et al. Defining Optimal Triage Strategies for hrHPV Screen-Positive Women-An Evaluation of HPV 16/18 Genotyping, Cytology, and p16/Ki-67 Cytoimmunochemistry. Cancer Epidemiol Biomarkers Prev. 2017;26(11):1629–35. [DOI] [PubMed] [Google Scholar]
- 8.Cuschieri K, Ronco G, Lorincz A, Smith L, Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders P, et al. Eurogin roadmap 2017: Triage strategies for the management of HPV-positive women in cervical screening programs. Int J Cancer. 2018;143(4):735–45. [DOI] [PubMed] [Google Scholar]
- 9.Arbyn M, Rezhake R, Yuill S, Canfell K. Triage of HPV-positive women in Norway using cytology, HPV16/18 genotyping and HPV persistence. Br J Cancer. 2020;122(11):1577–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiberhuber L, Barrett JE, Wang J, Redl E, Herzog C, Vavourakis CD, Sundström K, Dillner J, Widschwendter M. Cervical cancer screening using DNA methylation triage in a real-world population. Nature medicine. 2024;30(8):2251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molano M, Machalek DA, Tan G, Garland S, Balgovind P, Haqshenas G, Munnull G, Phillips S, Badman SG, Bolnga J, et al. Performance of CADM1, MAL and miR124-2 methylation as triage markers for early detection of cervical cancer in self-collected and clinician-collected samples: an exploratory observational study in Papua New Guinea. BMJ Open. 2024;14(6):e081282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan C, Hu J, Luo T, Dong B, Li H, Wang W, Yan J, Cai H. Analysis of the diagnostic performance of PAX1/SOX1 gene methylation in cervical precancerous lesions and its role in triage diagnosis. Journal of medical virology. 2024;96(5):e29521. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Jin X, Kong L, Liou Y, Liu P, Dong Z, Zhou S, Qi B, Fei J, Chen X, et al. Triage performance of PAX1(m)/JAM3(m) in opportunistic cervical cancer screening of non-16/18 human papillomavirus-positive women: a multicenter prospective study in China. Clin Epigenetics. 2024;16(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao D, Yang Z, Dong S, Li Y, Mao Z, Lu Q, Xu P, Shao M, Pan L, Han X, et al. PCDHGB7 hypermethylation-based Cervical cancer Methylation (CerMe) detection for the triage of high-risk human papillomavirus-positive women: a prospective cohort study. BMC medicine. 2024;22(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI, Chan MWY, Liao CC, Chen CW, Lin WY, et al. Paired Box-1 (PAX1) Activates Multiple Phosphatases and Inhibits Kinase Cascades in Cervical Cancer. Scientific reports. 2019;9(1):9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai HC, Lin YW, Huang TH, Yan P, Huang RL, Wang HC, Liu J, Chan MW, Chu TY, Sun CA, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123(1):161–7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Wang H, Chen S, Fan X, Liu Y, Shi S, Wang R. Reactivation of methylation-silenced PAX1 inhibits cervical cancer proliferation and migration via the WNT/TIMELESS pathway. Molecular carcinogenesis. 2024;63(7):1349–61. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Zhao C, Zhang X, Li J, Zhao Y, Zhang W, Ren L, Wei L. PAX1/JAM3 Methylation and HPV Viral Load in Women with Persistent HPV Infection. Cancers. 2024;16(7):1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Zhao C, Zhao Y, Li J, Wang J, Luo H, Tang Z, Guo Y, Wei L. The role of PAX1 methylation in predicting the pathological upgrade of cervical intraepithelial neoplasia before cold knife conization. Frontiers in oncology. 2022;12:1064722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo H, Lian Y, Tao H, Zhao Y, Wang Z, Zhou J, Zhang Z, Jiang S. Relationship between p16/ki67 immunoscores and PAX1/ZNF582 methylation status in precancerous and cancerous cervical lesions in high-risk HPV-positive women. BMC cancer. 2024;24(1):1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Wu H, Fu K, Shen Y, Li L, Liao Z, Liu Y, Kang Y, Zhang Y. PAX1 methylation as a robust predictor: developing and validating a nomogram for assessing endocervical curettage (ECC) necessity in human papillomavirus16/18-positive women undergoing colposcopy. Clinical Epigenetics. 2024;16(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y, Li X, Zhang M, Yang Y, Ge G, Wang K, Gong Y, Liang Y, Niu H, Ci W. Enhanced Detection of Genitourinary Cancers Using Fragmentation and Copy Number Profiles Obtained from Urinary Cell-Free DNA. Clin Chem. 2021;67(2):394–403. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Liu H, Zhou X, Zhou Y, Zhang Y, Liou YL, Zeng M, Zhu H. PAX1 hypomethylation as a prognostic biomarker for radioresistance of cervical cancer. Clin Epigenetics. 2023;15(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marinelli LM, Kisiel JB, Slettedahl SW, Mahoney DW, Lemens MA, Shridhar V, Taylor WR, Staub JK, Cao X, Foote PH, et al. Methylated DNA markers for plasma detection of ovarian cancer: Discovery, validation, and clinical feasibility. Gynecol Oncol. 2022;165(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans I, Reisel D, Jones A, Bajrami A, Nijjar S, Solangon SA, Arora R, Redl E, Schreiberhuber L, Ishaq-Parveen I, et al. Performance of the WID-qEC test versus sonography to detect uterine cancers in women with abnormal uterine bleeding (EPI-SURE): a prospective, consecutive observational cohort study in the UK. The Lancet Oncology. 2023;24(12):1375–86. [DOI] [PubMed] [Google Scholar]
- 26.Hayen A, Macaskill P, Irwig L, Bossuyt P. Appropriate statistical methods are required to assess diagnostic tests for replacement, add-on, and triage. J Clin Epidemiol. 2010;63(8):883–91. [DOI] [PubMed] [Google Scholar]
- 27.Yang L, Tao H, Lin B, He X, Chen Y, Fan X. Utilization of PAX1 methylation test for cervical cancer screening of non-HPV16/18 high-risk HPV infection in women. Future Oncol. 2023;19(28):1917–27. [DOI] [PubMed]
- 28.Huang M, Wang T, Li M, Qin M, Deng S, Chen D. Evaluating PAX1 methylation for cervical cancer screening triage in non-16/18 hrHPV-positive women. BMC cancer. 2024;24(1):913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Guo R, Lai T, Qiao L, Fu H. The application of PAX1 methylation detection and HPV E6/E7 mRNA detection in cervical cancer screening. J Obstet Gynaecol Res. 2021;47(8):2720–8. [DOI] [PubMed]
- 30.Chinese Cervical Cancer Screening Guidelines (1). Chinese Clinical Journal of Obstetrics and Gynecology. 2023;24(4):437–42.
- 31.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. The Journal of pathology. 2006;208(2):152–64. [DOI] [PubMed] [Google Scholar]
- 32.Clifford GM, Gallus S, Herrero R, Muñoz N, Snijders PJ, Vaccarella S, Anh PT, Ferreccio C, Hieu NT, Matos E, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366(9490):991–8. [DOI] [PubMed] [Google Scholar]
- 33.Wright TC Jr, Stoler MH, Ranger-Moore J, Fang Q, Volkir P, Safaeian M, Ridder R. Clinical validation of p16/Ki-67 dual-stained cytology triage of HPV-positive women: Results from the IMPACT trial. International journal of cancer. 2022;150(3):461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustinucci D, Giorgi Rossi P, Cesarini E, Broccolini M, Bulletti S, Carlani A, D’Angelo V, D’Amico MR, Di Dato E, Galeazzi P, et al. Use of Cytology, E6/E7 mRNA, and p16INK4a-Ki-67 to Define the Management of Human Papillomavirus (HPV)-Positive Women in Cervical Cancer Screening. American journal of clinical pathology. 2016;145(1):35–45. [DOI] [PubMed] [Google Scholar]
- 35.Cang W, Li Q, Gu L, Hong Z, Hu Y, Di W, Qiu L. Clinical Evaluation of DNA Ploidy for the Triage of HPV-Positive Chinese Women During Cervical Cancer Screening. Cancer prevention research (Philadelphia, Pa). 2021;14(3):355–62. [DOI] [PubMed] [Google Scholar]
- 36.Huh WK, Ault KA, Chelmow D, Davey DD, Goulart RA, Garcia FA, Kinney WK, Massad LS, Mayeaux EJ, Saslow D, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol. 2015;136(2):178–82. [DOI] [PubMed] [Google Scholar]
- 37.Tainio K, Athanasiou A, Tikkinen KAO, Aaltonen R, Cardenas Hernandes J, Glazer-Livson S, Jakobsson M, Joronen K, Kiviharju M, et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: systematic review and meta-analysis. BMJ. 2018;360:k499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banila C, Lorincz AT, Scibior-Bentkowska D, Clifford GM, Kumbi B, Beyene D, Wheeler CM, Cuschieri K, Cuzick J, Nedjai B. Clinical performance of methylation as a biomarker for cervical carcinoma in situ and cancer diagnosis: A worldwide study. Int J Cancer. 2022;150(2):290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are available from the corresponding author upon reasonable request.