Abstract
Background
Aerobic glycolysis is a tumor cell phenotype and a hallmark in cancer research. The alternative splicing of the pyruvate kinase M (PKM) gene regulates the expressions of PKM1/2 isoforms and the aerobic glycolysis of tumors. Polypyrimidine tract binding protein (PTBP1) is critical in this process; however, its impact and underlying mechanisms in colorectal cancer (CRC) remain unclear. This study aimed to investigate the role of PTBP1 crotonylation in CRC progression.
Methods
The crotonylation levels of PTBP1 in human CRC tissues and cell lines were analyzed using crotonylation proteomics and immunoprecipitation. The main crotonylation sites were identified by immunoprecipitation and immunofluorescent staining. The glycolytic capacities of CRC cells were evaluated by measuring the glucose uptake, lactate production, extracellular acidification rate, and glycolytic proton efflux rate. The role and mechanism of PTBP1 crotonylation in PKM alternative splicing were determined by Western blot, quantitative real-time PCR (RT-qPCR), RNA immunoprecipitation, and immunoprecipitation. The effects of PTBP1 crotonylation on the behaviors of CRC cells and CRC progression were assessed using CCK-8, colony formation, cell invasion, wound healing assays, xenograft model construction, and immunohistochemistry.
Results
The crotonylation level of PTBP1 was elevated in human CRC tissues compared to peritumor tissues. In CRC tissues and cells, PTBP1 was mainly crotonylated at K266 (PTBP1 K266-Cr), and lysine acetyltransferase 2B (KAT2B) acted as the crotonyltranferase. PTBP1 K266-Cr promoted glycolysis and lactic acid production, increasing the PKM2/PKM1 ratio in CRC tissues and cells. Mechanistically, PTBP1 K266-Cr enhanced the interaction of PTBP1 with heterogeneous nuclear ribonucleoprotein A1 and A2 (hnRNPA1/2), thus affecting the PKM alternative splicing. PTBP1 K266-Cr facilitated CRC cell proliferation, migration, and metastasis in vitro and in vivo. Pathologically, a high level of PTBP1 K266-Cr was associated with poor prognosis in CRC patients.
Conclusions
Crotonylation of PTBP1 coordinates tumor cell glycolysis and promotes CRC progression by regulating PKM alternative splicing and increasing PKM2 expression.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05793-5.
Keywords: Colorectal cancer, PTBP1, Crotonylation, PKM, Alternative splicing
Introduction
Tumor cells prefer aerobic glycolysis beyond oxidative phosphorylation, even under sufficient oxygen supply conditions, consuming more glucose and producing more lactic acid than normal cells, which eventually results in rapid tumor growth, also known as the “Warburg effect” [1, 2]. Many studies have shown that aerobic glycolysis is related to the progression of CRC, becoming a potential target for CRC diagnosis and treatment [3, 4].
Pyruvate kinase (PK), one of the rate-limiting step enzymes of glycolysis, has four different isoforms, i.e., PKL, PKR, PKM1, and PKM2. These isoforms are distributed in different tissues and cells and have respective functions [5]. The PKLR gene encodes PKL and PKR, while PKM1 and PKM2 are encoded by the pyruvate kinase M (PKM) gene. The PKM gene contains two mutually exclusive exons, exon 9 and exon 10, which encode two distinct isoforms of PKM, i.e., PKM1 and PKM2, respectively [6]. One of the mechanisms controlling the glycolytic phenotype is the tight regulation of alternative splicing of PKM transcripts [7]. PKM2, but not PKM1, is almost universally re-expressed in cancer and promotes aerobic glycolysis [8].
Regulation of PKM alternative splicing impacts the metabolic reprogramming in cancer cells. Human polypyrimidine tract binding protein (PTBP1) has four RNA recognition motifs (RRM), including RRM1, RRM2, RRM3, and RRM4 [9]. RRM binds to the pyrimidine-rich regions of RNA sequences and participates in the regulation of alternative splicing [10]. In cancer cells, upregulated PTBP1 binds to heterogeneous nuclear ribonucleoprotein (hnRNP) family members hnRNPA1 and hnRNPA2, forming a triple complex. It can exclude exon 9 by binding to sequences flanking exon 9, typically on intron 8 of PKM precursor mRNA (pre-mRNA) [11], leading to increased expression of PKM2 [12–14]. In recent years, the effect of post-translational modifications (PTMs) on the function of PTBP1 protein has attracted increasing attention. Deacetylation of PTBP1 can regulate PKM alternative splicing, resulting in reduced enrichment of PTBP1 on PKM pre-mRNA in breast cancer [15]. Nevertheless, there are limited reports on the role of PTMs in PTBP1 regulation of PKM alternative splicing in CRC.
Previous studies have shown that lysine crotonylation (Kcr) of metabolism- and glycolysis-related enzymes in CRC can affect enzyme activities and glycolysis through altered interactions with proteins [16, 17]. Crotonylation is closely related to the regulation of tumor gene transcription. Histone crotonylation can specifically mark active promoters and potential enhancers and activate gene regulatory elements to regulate gene expression [18–20]. Crotonylation of non-histone proteins can lead to the development of tumors by regulating chromatin remodeling, cell cycle, and DNA damage repair [21–23]. Nonetheless, the role of crotonylation in alternative splicing needs further study. Herein, we characterized the functional crotonylation site of PTBP1 in CRC, finding that increased PTBP1 K266-Kcr level modulated glucose metabolism in CRC cells. Moreover, hypercrotonylation of PTBP1 regulated PKM alternative splicing and increased the ratio of PKM2/PKM1 by increasing the binding of PTBP1 to PKM pre-mRNA, ultimately promoting CRC growth. Our findings on crotonylation of PTBP1 and its regulation of pre-mRNA splicing process provide new insights into the role of PTM in CRC progression.
Materials and methods
Patients and tissue sampling
A total of 80 patients diagnosed with CRC in Shanxi Provincial Academy of Traditional Chinese Medicine (Taiyuan, China) were included in this study. The tumoral and adjacent non-tumor colorectal tissues were harvested from all the patients during radical tumor excision surgeries. A small piece (approximately 0.5 cm3) of tumor tissue and the matching adjacent tissue were taken from the surgically resected colon specimen, immediately frozen in liquid nitrogen, and stored in freezers at − 80 ℃.
The research protocol was approved by the Ethics Committee of Shanxi Provincial Academy of Traditional Chinese Medicine (approval No. 2019-06KY005). The entire experimental protocol complied with the institutional guidelines.
Cell lines
HEK293T and human CRC cell lines, including HCT116, SW620, and SW480, were purchased from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). HEK293T cells were maintained at 37 ℃ and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen) supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma-Aldrich). HCT116 cells were cultured in McCoy’s 5A medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin and maintained at 37 ℃ in 5% CO2. SW620 and SW480 cells were cultured in Leibovitz’s L-15 medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 ℃ with 100% air.
Crotonylation proteomics
Four pairs of fresh CRC tissues and paired paracancerous tissues were processed for proteome profiling with the technical support of Jingjie PTM BioLab (Hangzhou, China). After protein extraction, trypsin digestion, and pan antibody-based PTM enrichment that were performed as previously described [17], the tryptic peptides were analyzed using liquid chromatography-mass spectrometry (LC–MS) on a nano-elution UHPLC system (Bruker Daltonics). MaxQuant search engine (V.1.6.6.0, http://www.maxquant.org/) software was applied to analyze the raw data. Tandem mass spectra were searched against the Human SwissProt concatenated with the reverse decoy database. The false discovery rate (FDR) was adjusted to < 1%.
Bioinformatics analysis
The criterion of fold-change selection was 1.5. For enrichment of protein domain analysis, the InterPro database (a tool providing functional analysis of protein sequences by classifying them into families and predicting the presence of domains and important sites) was searched, and a two-tailed Fisher’s exact test was employed to test the enrichment of differentially expressed proteins against all identified proteins. The database of Clusters of Orthologous Groups of proteins (COG/KOG, http://www.ncbi.nlm.nih.gov/COG) was used to classify the function of differentially modified proteins. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was utilized to annotate protein pathways. KEGG online service tool KAAS (V.2.0, http://www.genome.jp/kaas-bin/kaas_main) was used to annotate the submitted proteins, after which the annotated proteins were matched into the corresponding pathway in the database through KEGG mapper V2.5 (http://www.kegg.jp/kegg/mapper.html). Protein–protein interaction (PPI) network analysis of PTBP1 and hnRNP proteins was accomplished by the STRING database (https://cn.string-db.org/).
Cell transfection
Cell transfection for KAT2B siRNA was carried out by Lipofectamine RNAiMAX. Briefly, cells were harvested after 48 h of transfection. The KAT2B siRNA sequence was as follows: TAGCCATGCCCTAGCTGCTCATGTT.
PTBP1-knockdown HCT116 cell line was generated using a lentivirus-mediated delivery system. The shPTBP1 sequence was ligated into the HBLV-U6-ZsGreen-Puro (Hanbio) plasmid, after which it was co-transfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. Lentiviral supernatant was harvested 42 h after initial plasmid transfection. HCT116 cells were infected by viral supernatant, and cell pools were selected by 1 mg/mL puromycin for 1 week. The shPTBP1 sequence was as follows: 5′-ACTTGTGTCACTAACGGACCGTTTA-3.
To generate PTBP1-rescued cell lines, the cDNAs of Flag-tagged PTBP1 WT, PTBP1 K266Q, or PTBP1 K266R mutant were subcloned into pQCXIH retrovirus vector. All transfections were carried out using LipoFiter 3.0 (Hanbio, Shanghai, China) based on a 1:1 ratio of LipoFiter 3.0 (μg): total DNA (μg).
Immunofluorescent staining (IF)
Cells were seeded onto glass coverslips in 6-well plates and were treated with anacardic acid (Selleck, S7582) or siKAT2B. Next, they were fixed with 4% formaldehyde for 15 min and permeabilized with 0.3% TritonX‐100 in PBS for 10 min. After blocking for 1 h with blocking buffer (5% bovine serum albumin in 0.1% Triton/PBS), cells were incubated with anti-PTBP1 K266cr (1:50) or KAT2B (1:100) overnight at 4 °C. Following washing with PBS, cells were incubated with Alexa Fluor 594-conjugated AffiniPure Goat Anti-Rabbit IgG (Abclonal, 1:200). The nuclei were counterstained with DAPI (1:100).
Measurement of glucose uptake
Glucose uptake by cells was measured using a glucose uptake colorimetric assay kit (ab136955, Abcam) according to the manufacturer’s protocol. Briefly, PTBP1 WT, PTBP1 K266Q, and PTBP1 K266R cells were seeded into a 96-well plate at a density of 2000 cells per well and starved in a serum-free culture medium overnight. Cells were then incubated with the glucose analog 2-deoxyglucose (2-DG), and NADPH was generated by oxidation of the accumulated 2-DG6P, resulting in the oxidation of a substrate. The OD values at 412 nm were measured on a microplate reader in a kinetic mode.
Measurement of lactate production
The PTBP1 WT, K266Q, and K266R cells were seeded into a 6-well plate for 48 h. The cells and culture media were collected for intracellular and extracellular lactate content analysis following the manufacture’s instruction of lactate assay kit (A019-2–1, Jiancheng Biotech, Nanjing, China).
Extracellular acidification rate (ECAR) and glycolytic proton efflux rate (glycoPER) assays
ECAR and glycoPER were determined using XF glycolysis stress test kit and XF glycolysis rate assay kit on a Seahorse instrument (XF24, Agilent). As the same pre-treatment of ECAR and glycoPER, the sensor cartridge was hydrated a day before the test. PTBP1 WT, PTBP1 K266Q, and PTBP1 K266R in HCT116 cells were plated at a density of 1.0 × 104/well in an XF24 plate for overnight culture and then changed into the Seahorse detection liquid at 37 °C for 60 min without CO2 supply.
To measure ECAR, glucose, oligomycin, and 2-DG were diluted into XF media and loaded into the cartridge to achieve final concentrations of 10 mM, 1 µM and 50 mM, respectively. To measure glycoPER, Rot/AA and 2-DG were loaded in sensor cartridge at the final concentrations of 0.5 μM and 50 mM, respectively. Finally, the ECAR and glycoPER were obtained by data normalization processing. The glycolysis, glycolytic capacity, and glycolytic reserve levels were calculated based on ECAR data, while the basal glycolysis and compensatory glycolysis were calculated based on glycoPER data.
RT-qPCR
For gene expression studies, total RNAs from PTBP1 WT, K266Q, and K266R-rescued HCT116 cells were isolated using Trizol (Invitrogen, Carlsbad, CA, USA), and each RNA sample was reverse transcribed using Servicebio® RT First Strand cDNA Synthesis Kit (G3330, Servicebio, Wuhan, China), followed by PCR assays. All RT-qPCR reactions were run in three independent experiments. Along with GAPDH serving as an internal reference, constitutive exon (exon 11) normalization was performed for PKM1 and PKM2 expression analysis. The primer sequences of PKM isoforms were selected as previously reported by Choksi et al. [15], and the primer sequence of the PTBP1-binding site was from the report by Chen et al. [11] (Table S2).
Western blot
After extracting total proteins with RIPA buffer (Thermo Scientific, USA) and 1 mM PMSF, all proteins were separated on 10% SDS-PAGE (Invitrogen, USA) gels, transferred onto PVDF membranes (Millipore, USA) and then blocked using 5% skim milk. The proteins were detected by incubating with primary antibodies overnight, incubated with horseradish peroxidase-labeled secondary anti-mouse or anti-rabbit (1:2000) (ZSGB-Bio, Beijing, China) antibodies at room temperature, and then finally visualized by the ECL detection kit (Solarbio, China). The following commercial antibodies were used: anti-FLAG (Sigma, F1804, USA), anti-PanKcr (PTM, 501, China), anti-β-Actin (Biogot, AP0060, China), hnRNPA2 (ab283592, Abcam), anti-PTBP1 (57,246, CST); KAT2B Rabbit pAb (A0066), hnRNPA1 Rabbit mAb (A11564), PKM1-specific Rabbit pAb (A18800) and PKM2-specific Rabbit pAb (A18799) were from Abclonal, Wuhan, China. The anti-PTBP1 K266cr antibody was synthesized by ABclonal (Wuhan, China).
Immunoprecipitation (IP) and co-immunoprecipitation (Co-IP) assays
The Pierce Classic IP Kit (Thermo Scientific, 26,146, USA) was used to perform IP and Co-IP assays following the manufacturer’s protocols. Briefly, coupling resin was cross-linked with 10 μg antibody for 2 h, and antibody-coupled resin was incubated with protein lysates overnight. After washing the resin, antigen complexes were eluted using an elution buffer. Subsequently, elution proteins were evaluated by western blotting.
RNA immunoprecipitation (RIP)
Cells were harvested and crosslinked with 1% formaldehyde at room temperature for 10 min and then quenched with 1 M glycine. Next, they were rinsed with PBS two times and lysed with lysis buffer (0.1 M HEPES, 0.1 M KCl, 5 mM MgCl2, 0.5% NP40, 1 mM DTT, 1 × protease inhibitors). The supernatants were used for immunoprecipitation with anti-PTBP1 or non-specific rabbit IgG at 4 °C overnight. Protein-RNA complexes were added with protein-RNA elution buffer (0.1 M Tris–HCl pH 8, 0.01 M EDTA, 1% SDS) and RNA was eluted from the beads in elution buffer (100 mM NaHCO3, 1% SDS), after which the crosslinks were reversed in de-crosslinking buffer (500 mM NaCl, 2 mM EDTA, 20 mM Tris–HCl pH 6.8, 0.5 mg/mL Proteinase K) at 65 °C. RNA was then extracted by Trizol (Invitrogen, Carlsbad, CA, USA), and the PTBP1-binding site on intron 8 of PKM was quantified by RT-qPCR. Equal amounts of input RNA from each sample were used to generate standard curves, and % input values were calculated based on the standard curves.
Cell counter kit-8 (CCK8) and colony formation assays
In order to perform the CCK8 assay, the PTBP1 WT, K266Q, and K266R-rescued HCT116 cells were suspended and seeded into 96-well plates at 5000 cells per well. After the treatment with 10 μL of CCK8 solution (HY-K0301, MCE, USA) for 1 h, the absorbance of each well was measured at 450 nm using a microplate reader (Bio-Rad, USA).
In order to perform a colony formation assay, 1000 rescued HCT116 cells were seeded into 6-well plates for 14 days. Cells were fixed with 4% paraformaldehyde for 20 min, stained with 0.1% crystal violet, and photographed.
Cell invasion assay and wound healing assay
To perform cell invasion assay, the PTBP1 WT, K266Q, and K266R-rescued HCT116 cells (2 × 104 cells/well) were seeded into the upper chamber of transwell covered with Matrigel glue (8 µm pore size, 3422, Corning, Shanghai, China). A complete medium containing 20% FBS was added into the lower chamber. After further culturing for another 48 h, the invaded cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet, and photographed.
The rescued HCT116 cells were seeded in six-well plates and cultured to perform wound healing assay. Scratch wounds were made using plastic pipette tips (100 μL pipette tips, Eppendorf) when the cells reached 80% confluency in each well. The progression of cell migration was photographed at 24 h and 48 h, respectively.
Xenograft assay
All the animals were housed in an environment with a temperature of 22 ± 1 ºC, a relative humidity of 50 ± 1%, and a light/dark cycle of 12/12 h. The PTBP1 rescued WT, K29Q, and K29R HCT116 cells were trypsinized and counted. Next, 4 × 106 cells were resuspended in McCoy’s 5A medium and injected into the right anterior armpit of 4-week-old male BALB/c nude mice (Gempharmatech Co., Ltd., Nanjing, China). One week after injection, the length and width of the tumor were measured using a digital caliper every two days, after which the mice were sacrificed, and the tumors were dissected and photographed. The weight and volume of tumors were measured. The volume calculated formula was V = Length Width2/6.
The animal use protocol was approved by the Animal Experimentation Ethics Committee of Shanxi Provincial Academy of Traditional Chinese Medicine (approval No. 2023KY-07013) and abided by the standards of the National Institute of Health Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry
Parts of the harvested CRC tissues and peritumor tissues were fixed with 10% formalin, routinely processed, embedded in paraffin wax, and cut into 5 μm sections. The tissue sections were then deparaffinized, rehydrated, treated with antigen retrieval, and incubated with primary PTBP1 K266-Cr antibody (1:200) antibody at 4 ℃ for overnight. Tissue sections were washed and incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Zhong Shan Jin Qiao, Beijing, China) for 20 min at room temperature. Subsequently, after incubating with 3,3′-diaminobenzidine (DAB), sections were counterstained with hematoxylin and sealed with coverslips. Grade criteria for tumor tissues and adjacent normal tissues were classified based on PTBP1 K266-Cr antibody staining. Staining intensity was classified as 0 (lack of staining), 1 (mild staining), 2 (moderate staining), and 3 (strong staining). The percentage of staining was designated 1 (< 25%), 2 (25–50%), 3 (51–75%), or 4 (> 75%). The semi-quantitative score was calculated by multiplying these two values on a scale of 0–12. A score of 0−4 was classified as low expression, and a score of > 4 was classified as high expression.
Statistical analysis
All photographed cells were quantified using the ImageJ software. SPSS 19.0 software and GraphPad Prism were used for statistical analyses. Data were expressed as mean ± standard deviation (SD) from at least three independent experiments. Comparison between groups was performed using unpaired or paired Student t-test, one-way ANOVA followed by Dunnett’s multiple comparisons test, or two-way ANOVA as appropriate. Categorical data were calculated using Chi-square or Fisher's exact test. A P-value of < 0.05 represented statistical significance.
Results
The crotonylation modification of PTBP1 is elevated in CRC tissues
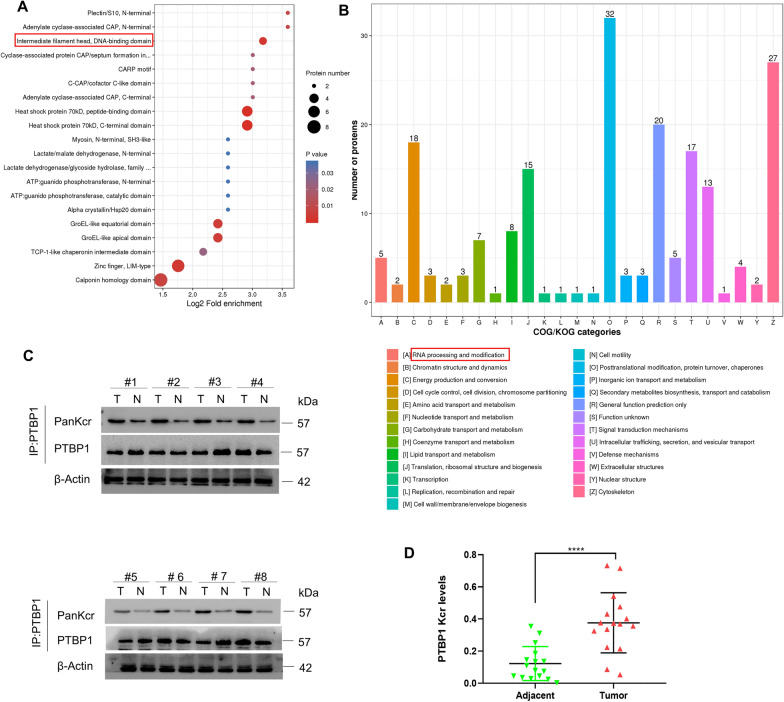
A quantitative Kcr proteomic study was conducted in human CRC tissues and paired peritumoral tissues to elucidate the role of protein Kcr in CRC. Through domain enrichment analysis of differentially modified Kcr proteins, we found that the Kcr in certain protein domains showed a trend of significant enrichment, such as the DNA-binding domain (Fig. 1A). Results of COG/KOG functional classification statistics indicated that these enriched proteins, including PTBP1, are involved in various biological processes, particularly that of RNA processing and modification (Fig. 1B). As shown in Table S1, PTBP1 protein had a higher T/N ratio compared with the other crotonylated proteins.
Fig. 1.
Crotonylation modification of PTBP1 was elevated in CRC. A Bubble diagram of protein domain enrichment at differentially modified Kcr protein. B COG/KOG categories of proteins corresponding to differential crotonylation modification sites. C The Kcr levels of PTBP1 were determined by immunoprecipitation. Eight representative pairs of samples. D Quantification of relative PTBP1 Kcr levels in the 16 pairs of samples tested. The intensity of WB was quantified by ImageJ, followed by statistical analysis. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
To further verify the results of Kcr proteomics, the expression levels of PTBP1 Kcr were analyzed by immunoprecipitation method in 16 pairs of CRC tumor tissues and the matched paracancer tissues (Fig. 1C). Relative quantitative results for CRC tissues showed a notable elevation in the PTBP1 Kcr level compared to adjacent normal tissues (Fig. 1D). Meanwhile, Western blot was used to analyze the PTBP1 Kcr expression in a panel of CRC cell lines (HCTI16, SW480, and Sw620). Results showed that the Kcr of PTBP1 could be detected in all three CRC cell lines (Fig. S1). These findings collectively suggest that PTBP1 Kcr is upregulated in CRC.
PTBP1 is crotonylated at K266 and KAT2B is the major crotonyltranferase for PTBP1
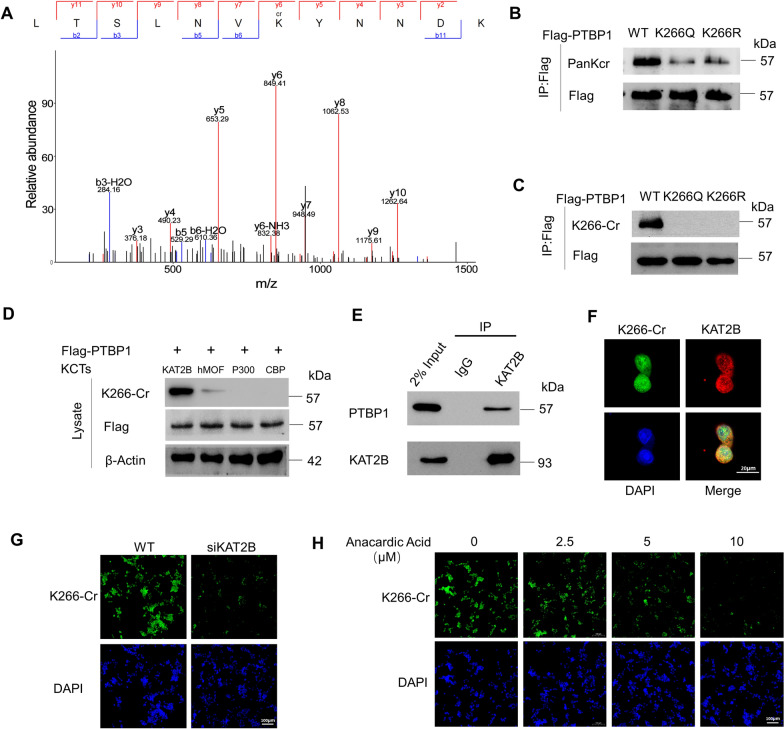
LC–MS/MS analysis of CRC tissue lysates was conducted to identify the crotonylation sites of PTBP1, and results showed that lysine (K) residue 266 was the major crotonylation site of PTBP1 in CRC (Fig. 2A). The physiological function of acylation is related to the charge of lysine residues. Positively charged lysine residues often involve protein–protein interaction and catalytic activity. Crotonylation can robustly neutralize the positive charge of lysine, thus affecting many aspects of protein function [24]. To clarify the role of PTBP1 K266-Cr, we further produced an electrically neutral K266 glutamine mutant (K266Q) as the protein hypercrotonylation mimic. Meanwhile, K266 lysine was mutated to an electrically positive arginine (K266R) to mimic the decrotonylation of PTBP1 K266. Compared with the PTBP1 wild-type (WT) cells, both K266R and K266Q mutations greatly affected the recognition of lysine acylation groups by specific antibodies, thus decreasing PTBP1 Kcr in HCT116 cells (Fig. 2B; Fig. S2A). To further investigate the K266 crotonylation site, an antibody labeled “K266-Cr” was generated to precisely detect the crotonylated PTBP1 at the K266 site. Dot blot determined the specificity of the K266-Cr antibody (Fig. S2B). IF and Western blot analyses demonstrated the applicability of the K266-Cr antibody in CRC cell lines and CRC tissues, respectively (Fig. S2C, D). The IP analysis using the K266-Cr antibody revealed a significant decrease of PTBP1 Kcr in HCT116 cells expressing K266 Q/R compared to the WT cells (Fig. 2C; Fig. S2E), suggesting that K266 is the primary crotonylation site of PTBP1 in CRC.
Fig. 2.
PTBP1 was crotonylated at K266 and KAT2B is the major crotonyltranferase for PTBP1. A Crotonylated PTBP1 K266 was identified by a tandem mass spectrum. The identified peptide is shown. B PTBP1 WT, K266Q, or K266R tagged by Flag were transfected into HCT116 cells. PTBP1 Kcr level was detected by a pan-Kcr antibody. C Crotonylation of PTBP1 WT, K266Q, or K266R was detected by PTBP1 K266-Cr antibodies. D HEK293T cells were co-transfected with indicated plasmids, and cell lysates were harvested for western blot with indicated antibodies. E Endogenous interaction between PTBP1 and KAT2B was detected by co-IP in HEK293T cells. F HCT116 cells were transfected with KAT2B and stained with PTBP1 K266-Cr antibody (green) and anti- KAT2B antibody (red). DAPI (blue), nucleus. Scale bar, 20 µm. G Endogenous KAT2B was knocked down with siRNA in HCT116 cells and then PTBP1 K266-Cr was detected by IF. DAPI (blue), nucleus. Scale bar, 100 µm. H HCT116 cells were treated with the indicated dose of Anacardic Acid. Then the PTBP1 K266-Cr levels were detected by IF
To further identify the crotonyltranferases (KCTs) for PTBP1 K266, PTBP1 was co-transfected with CBP, p300, KAT2B, or human males absent on the first (hMOF) in HEK293T cells, revealing that KAT2B mainly facilitated the crotonylation of PTBP1 at K266 (Fig. 2D). As shown in Fig. 2E, endogenous PTBP1 directly interacted with KAT2B. Subsequent IF showed the interaction of PTBP1with KAT2B in the nucleus of HCT116 cells (Fig. 2F). By knocking down KAT2B using siRNA or treating HCT116 with a KAT2B inhibitor, anacardic acid, we observed a significant reduction of PTBP1 K266-Cr, as confirmed by IF assays (Fig. 2G, H; Fig. S2F). Overall, the above results support that KAT2B acts as a crotonyltransferase to promote PTBP1 K266-Cr.
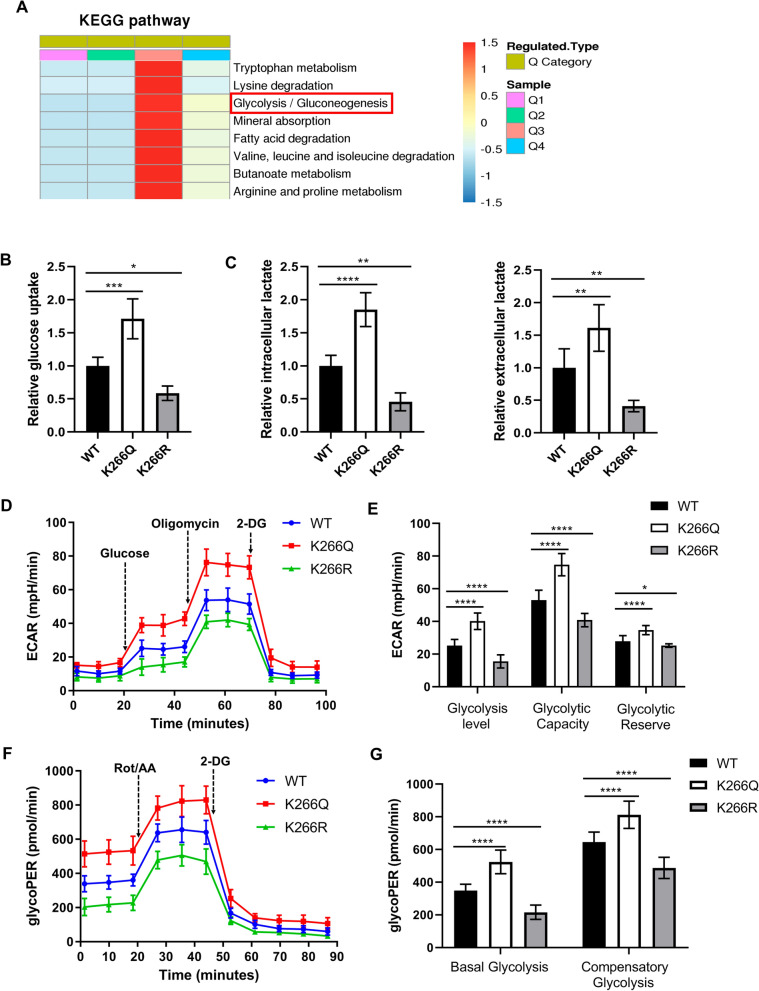
PTBP1 K266-Cr promotes glycolysis in CRC cells
In order to further identify the hierarchical clustering of crotonylation proteomics based on protein functional classification, we divided the differentially modified Kcr sites into Q1 (< 0.667), Q2 (0.667–0.769), Q3 (1.3–1.5), Q4 (> 1.5), according to their differential multiples. The cluster heat map of each Q group based on KEGG enrichment revealed that crotonylation may have an essential role in glycolytic pathways in CRC (Fig. 3A). To investigate the effect of PTBP1 K266-Cr on glycolysis, we knocked down endogenous PTBP1 followed by re-expressions of PTBP1 WT, PTBP1 K266Q and K266R in HCT116 cells (Fig. S3A). As shown in Fig. 3B, C, the glucose uptake and lactate production in PTBP1 K266Q cells significantly increased, while they decreased in PTBP1 K266R rescued cells, compared to WT cells. These results indicate that PTBP1 K266 Cr increases glycolytic flux in CRC cells.
Fig. 3.
PTBP1 K266-Cr promotes glycolysis in CRC cells. A Cluster analysis heat map based on KEGG classification enrichment. B Relative glucose uptake in PTBP1 WT, K266Q and K266R-rescued HCT116 cells. C Relative intracellular and extracellular lactate in PTBP1 WT, K266Q and K266R-rescued HCT116 cells. D Kinetic profiles of ECAR in PTBP1 WT, K266Q and K266R-rescued cells. E Quantification of glycolysis level, glycolytic capacity and glycolytic reserve in cells. F Kinetic profiles of glycoPER in HCT116 cells expressing PTBP1 WT, K266Q and K266R. G Quantification of basal glycolysis and compensatory glycolysis from the kinetic profiles of glycoPER. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Bars, mean ± SD (n = 5)
Next, the ECAR and glycoPER of HCT116 cells were tested using a Seahorse XFe24 energy analysis system to examine the glycolytic capacities of the cells. Compared to PTBP1 WT cells, cells with K266Q displayed higher ECAR and glycoPER values, while cells possessing K266R showed the lowest ECAR and glycoPER values (Fig. 3D, F). Quantitative statistics revealed that the glycolysis level, glycolytic capacity, and glycolytic reserve were increased in K266Q cells and decreased in K266R cells (Fig. 3E). Consistently, both the basal glycolysis and compensatory glycolysis of K266Q cells were significantly increased, significantly differing from WT and K266R cells (Fig. 3G). Thus, these results reveal that hypercrotonylation of PTBP1 facilitates glycolysis in CRC cells.
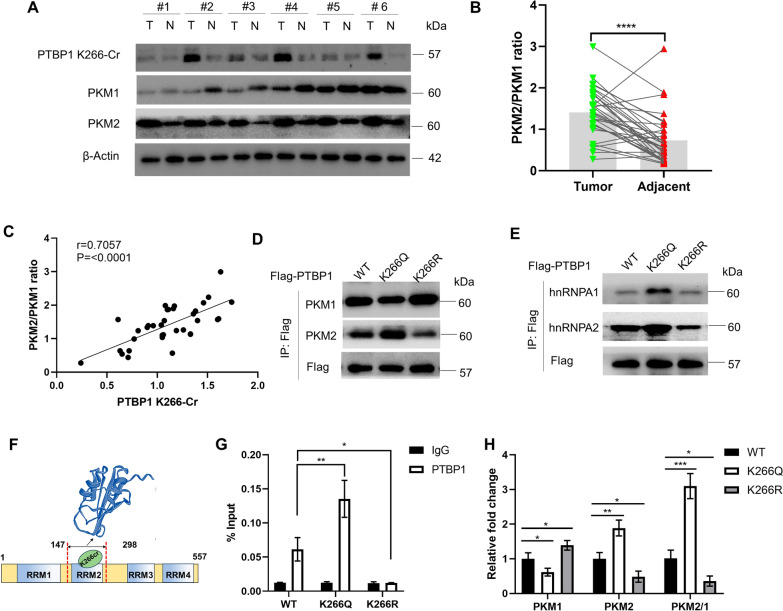
PTBP1 K266-Cr regulates PKM alternative splicing by affecting the affinity for pre-mRNA
High expression of PKM2 relative to PKM1 is one of the critical factors for cancer cells to gain a metabolic advantage over normal cells [7]. Therefore, PKM alternative splicing may have an important role in the metabolic preference of cancer cells. In order to investigate the role of PTBP1 K266-Cr in the regulation of PKM alternative splicing, the expression levels of PTBP1 K266-Cr and PKM isoforms in CRC tissues were first examined. We used 35 pairs of human CRC samples and adjacent normal tissues to perform western blotting analysis. Compared with the matched peritumoral tissues, the expression levels of PTBP1 and K266-Cr in the tumor tissues were increased, and the PKM2 level was also relatively elevated, while the PKM1 level was significantly decreased (Fig. 4A; Fig. S4A). In addition, the PKM2/PKM1 ratio was significantly higher in the tumor tissues than in the matched peritumoral tissues (Fig. 4B). Remarkably, in tumor tissues, the PTBP1 K266-Cr level was positively correlated with the PKM2/PKM1 ratio (r = 0.7057, P < 0.0001) (Fig. 4C). Consistently, in HCT116 cells, compared with the PTBP1 WT cells, K266Q-rescued cells had significantly enhanced expression of PKM2 and elevated PKM2/PKM1 ratio, while K266R cells did not show such changes (Fig. 4D; Fig. S4B). These findings suggest that PTBP1 K266-Cr regulates the switching of PKM isoforms from PKM2 to PKM1 at the protein level in CRC.
Fig. 4.
PTBP1 K266-Cr regulates PKM alternative splicing by affecting the affinity for pre-mRNA. A Levels of PTBP1 K266-Cr, PKM1 and PKM2 were determined by direct western blotting after CRC tissues were lysed. Relative protein levels were normalized to β-actin. Shown are six pairs of samples. B Quantification of relative PKM2/PKM1 ratio in the 35 pairs of samples tested. C Correlation between PTBP1 K266-Cr level and PKM2/PKM1 ratio in the tested 35 CRC samples. D Expression of PKM isoforms in PTBP1 WT, K266Q and K266R-rescued HCT116 cells by western blot. E PTBP1 WT, K266Q and K266R-rescued HCT116 cells were lysed and immunoprecipitated with anti-Flag antibody. hnRNPA1and hnRNPA2 proteins were detected by western blot. F Schematic representation of RRMs for PTBP1 containing K266. Three-Dimensional Structure of PTBP1 RRM2 is from PDB (ID:1sjr). G Binding of PTBP1 on intron 8 of PKM pre-mRNA in the IP relative to IgG was assessed via RT-qPCR (n = 3). H RT-qPCR of PKM isoforms in PTBP1 WT, K266Q and K266R-rescued HCT116 cells (n = 3). The PKM1/PKM2 ratio was calculated based on their relative mRNA levels. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Bars, mean ± SD
As a component of hnRNPs complex, PTBP1 acted as a hub protein interacting with hnRNPA1, hnRNPA2, and other hnRNP proteins, as was identified using PPI network analysis based on STRING database (Fig. S4C). To characterize the effect of PTBP1 K266-Cr on the stability of the triple complex (PTBP1/hnRNPA1/hnRNPA2), we examined the interaction of PTBP1 with hnRNPA1/2 by co-IP, finding that K266Q had stronger interactions with hnRNPA1 and hnRNPA2 (Fig. 4E). We further investigated the relation between K266 and PTBP1 RRMs based on (PDB:1sjr) [25]. As shown in Fig. 4F, the RRM2 of PTBP1, where the K266 is located, contributed substantially to RNA binding. To inspect the effect of PTBP1 K266-Cr on its affinity for PKM pre-mRNA, the RIP assay of PTBP1 was performed in PTBP1 WT, K266Q, and K266R-rescued HCT116 cells. As a result, enrichment of PTBP1 on intron 8 of PKM pre-mRNA was enhanced in K266Q-rescued cells. On the contrary, the binding of K266R at this site was much weaker than in the WT cells (Fig. 4G). Subsequently, PKM isoform RNA expression was measured in HCT116 cells by RT-qPCR. K266Q-rescued HCT116 cells exhibited PKM1 downregulation and PKM2 upregulation at the transcript level, while the K266R cells showed the opposite results (Fig. 4H). Meanwhile, the PKM1/PKM2 ratio was remarkably elevated in K266Q cells, whereas this ratio was reduced in K266R cells, compared with the WT cells (Fig. 4H). Overall, these results confirm that PTBP1 K266-Cr leads to an increase in its affinity for PKM pre-mRNA and ultimately modulates the PKM1/PKM2 ratio by regulating PKM alternative splicing.
PTBP1 K266-Cr promotes CRC cell proliferation, migration, invasion, and tumor growth
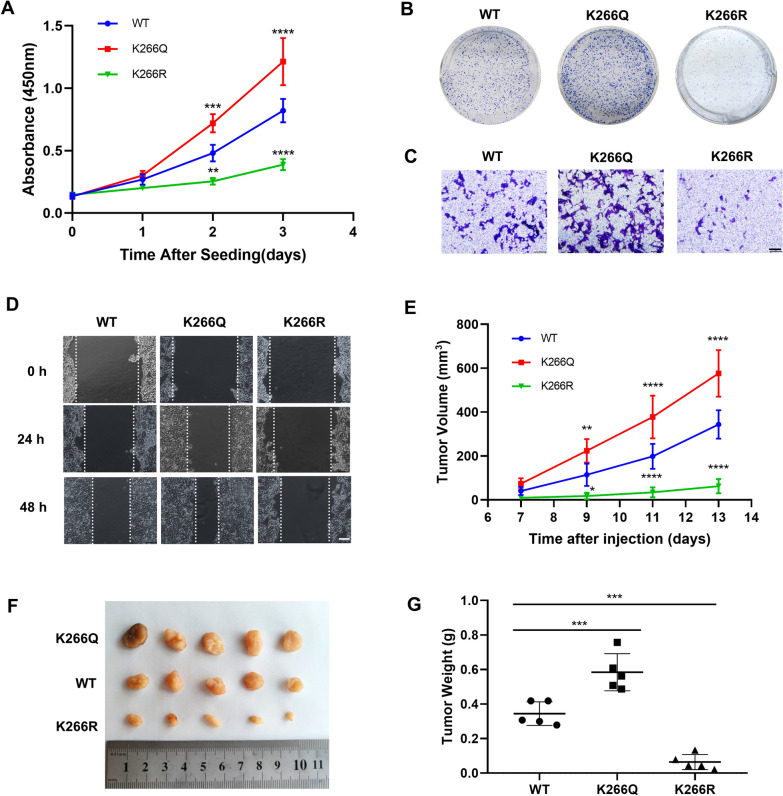
Next, we further studied the effect of PTBP1 K266-Cr on CRC progression, finding that the proliferation rate of K266Q-rescued cells was significantly higher than that of WT cells, while it was relatively decreased in K266R-rescued cells (Fig. 5A). Colony formation assay showed that the colony growth of K266Q-rescued cells was enhanced compared with that of WT cells, while the growth of K266R-rescued cells was on the contrary. (Fig. 5B; Fig. S5A). In the transwell assay and wound healing experiment, K266Q-rescued cells showed enhanced migration and invasion abilities, while K266R-rescued cells showed decreased migration and invasion (Fig. 5C, D; Fig. S5B). These results confirm that PTBP1 K266-Cr is vital in CRC tumor growth in vitro.
Fig. 5.
PTBP1 K266-Cr promoted CRC cell proliferation, invasion, migration and tumorigenesis. A PTBP1 WT, K266Q and K266R-rescued HCT116 cells were seeded at the same number. Cell proliferation was determined by CCK8 assay. B Colony formation assay of PTBP1 WT, K266Q and K266R-rescued HCT116 cells. Representative images of colony are shown. C Representative images of invaded cells in the transwell assay. Scale bar: 100 μm. D Cell migration ability was assessed by wound healing assay in HCT116 cells. Scale bar: 100 μm. E Xenograft experiment was performed in BALB/c nude mice injected with PTBP1 WT, K266Q and K266R-rescued HCT116 cells. Measurement of diameters of tumors was carried out every 2 days one week after injection, and tumor volumes were calculated. F Thirteen days later, mice were sacrificed, and tumors were dissected and photographed. G Measurement of weight of tumors. Dots represent weight of WT, K266Q and K266R tumors (n = 5). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Bars, mean ± SD
Xenografts assay was performed to determine the physiological influence of PTBP1 K266-Cr on tumor growth in vivo. Rescued cells were subcutaneously injected into 4-week-old BALB/c nude mice. The tumor size (length and width) was measured every 2 days after one week of injection. Strikingly, injection of K266Q cells promoted tumor growth, whereas injection of K266R cells caused smaller tumors in the xenograft model than the tumors caused by WT cell injection (Fig. 5E, F). Compared with the WT group, the tumor weight increased in the K266Q group but decreased in the K266R group (Fig. 5G). These results further confirm that PTBP1 K266-Cr substantially enhances the tumor growth of CRC.
PTBP1 K266-Cr correlates with poor prognosis in CRC
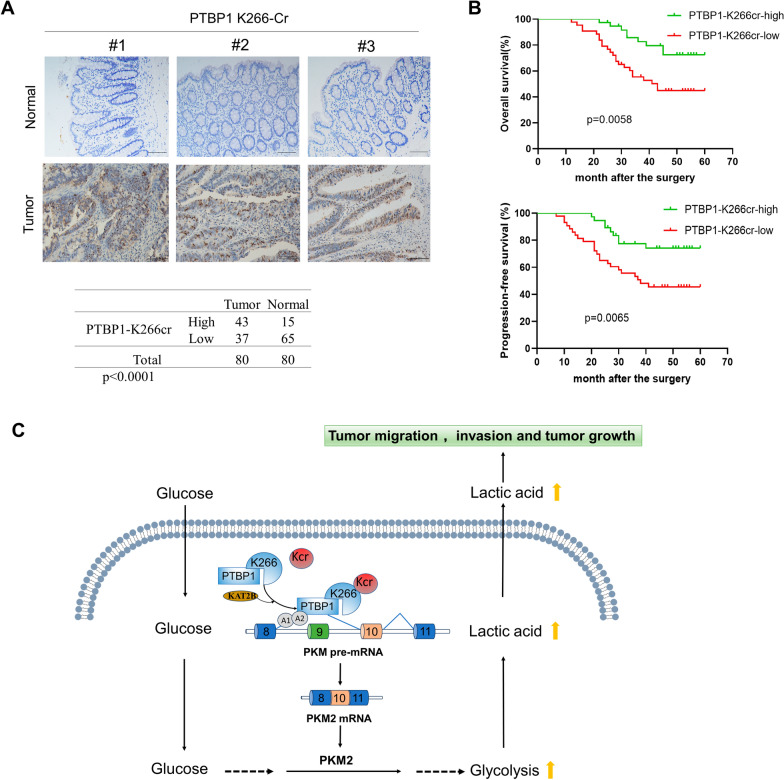
To examine whether PTBP1 K266-Cr participates in the clinicopathological characteristics and prognosis of CRC, we performed immunohistochemical staining on the 80 pairs of CRC tissues and the adjacent peritumor tissues. Based on the levels of PTBP1 K266-Cr positive signals, the staining results were classified into 0 – 12 scales. Statistical analysis proved a significant increase of PTBP1 K266-Cr levels in the tumor tissues compared with the adjacent tissues (p < 0.0001), as shown in a representative of three pairs of samples (Fig. 6A). In addition, higher PTBP1 K266-Cr levels in the CRC tissues were closely related to late TNM stage and distant metastasis, whereas there were no significant differences with regard to age, gender, tumor size, tumor differentiation, and lymph node metastasis (Table 1). Meanwhile, CRC patients with higher PTBP1 K266-Cr levels had a poorer prognosis in CRC patients who displayed a shorter median survival for OS and for PFS (Fig. 6B). The results above indicated that enhanced PTBP1 K266-Cr predicted poor prognosis in patients with CRC.
Fig. 6.
PTBP1 K266-Cr correlates with poor prognosis in CRC. A Three representative samples were shown and the statistical analysis of all samples. Scale bar: 100 μm. A total of 80 paired CRC tumor tissues and adjacent normal tissues were analyzed. B The overall survival and progression-free survival were analyzed by the log-rank test using the Kaplan–Meier analysis. C Working model illustrates that PTBP1 crotonylation promotes colorectal cancer progression through alternative splicing-mediated upregulation of PKM2
Table 1.
The relationship between PTBP1 K266-Cr expression and clinicopathologic characteristics in CRC patients
| Parameters | PTBP1 K266-Cr | P-value | |
|---|---|---|---|
| High (n = 43) | Low (n = 37) | ||
| Gender | |||
| Female | 21 | 16 | 0.6581 |
| Male | 22 | 21 | |
| Age (year) | |||
| ≤ 60 | 25 | 17 | 0.3696 |
| > 60 | 18 | 20 | |
| Tumor size (cm) | |||
| ≤ 5 | 22 | 15 | 0.3758 |
| > 5 | 21 | 22 | |
| Tumor differentiation | |||
| Well | 7 | 5 | 0.155 |
| Moderate | 19 | 24 | |
| Poor | 17 | 8 | |
| TNM stage | |||
| I + II | 13 | 20 | 0.0411* |
| III + IV | 30 | 17 | |
| Lymph node metastasis | |||
| N0 | 16 | 22 | 0.072 |
| N1–N2 | 27 | 15 | |
| Distant metastasis | |||
| M0 | 17 | 25 | 0.0147* |
| M1 | 26 | 12 | |
*Statistically significant (P < 0.05)
Discussion
Alternative splicing is one of the crucial posttranscriptional regulatory mechanisms that regulate the translation of mRNA subtypes and produce protein diversity [26]. There is growing evidence that aberrant splicing may result from the PTMs of proteins, such as acetylation, methylation, and ubiquitination of proteins [27–29]. As a novel modification, the role of crotonylation in alternative splicing needs to be further elucidated. Yu et al. [30] revealed that decrotonylation of the alternative splicing factor PHF5A could regulate aging progress through alternative splicing-mediated downregulation of CDK2. However, crotonylation regulation on pre-mRNA splicing in cancer is poorly understood. Herein, we discovered that Kcr of PTBP1 mediated by KAT2B regulates PKM2/PKM1 expression through alternative splicing to promote CRC progression. Our findings revealed that crotonylation of PTBP1 enhances the glycolysis of CRC cells, subsequently promoting CRC progression (Fig. 6C).
PTBP1 is a posttranscriptional gene expression regulator that controls mRNA splicing, protein translation, stability, and localization. PTBP1 has many molecular functions related to RNA metabolism and is a major repressive regulator of alternative splicing, leading to exon skipping in many alternative splicing pre-mRNAs [31]. PTBP1 has a role in various diseases, particularly in some cancers, including colorectal cancer [32]. High expression of PTBP1 has been found to promote CRC invasion through alternative splicing [33]. To date, the role of PTBP1 in alternative splicing has mainly been studied at the protein expression level. Herein, we focused on the role of PTBP1 crotonylation in PKM splicing and CRC progression. The protein quantitation was normalized to remove the effect of protein expression on the modification, revealing that the Kcr level of PTBP1 was upregulated in CRC compared to that in paracancerous tissues (shown in Fig. l C and Table S1). This conclusion was verified by both crotonylation proteomic and immunoprecipitation assays.
K266 was identified as the main Kcr site of PTBP1, and KAT2B was the crotonyltranferase of PTBP1 K266-Cr (Fig. 2A, D). KAT2B, also known as p300/CBP-associated factor (PCAF), is a transcriptional adaptor protein and a histone acetyl-transferase (HAT) that functions as the catalytic subunit of the PCAF transcriptional co-activator complex [34–37]. However, the role of KAT2B in other PTMs, including crotonylation, has not been fully elucidated. Herein, we found that PTBP1 was crotonylated by KAT2B in CRC. Other researchers reported that non-histone NPM1 was also crotonylated by KAT2B in human lung adenocarcinoma cell line H1299 [21], implying that KAT2B possesses evolutionarily conserved KCT activity, at least in cancer cells. Since small-molecule inhibitors targeting KAT2B have become a research hotspot [38], our study provides further insights into the molecular prerequisites for targeting crotonylation-functional tumors.
Enhanced glycolysis in tumor cells leads to the accumulation of lactate. Numerous studies have reported the crucial role of lactate in cancer progression and cell fate determination [39, 40]. Our results revealed that enhanced PTBP1 K266-Cr increased the intracellular and extracellular lactate levels in CRC, which was directly caused by hyperglycolysis (shown in Fig. 3). However, the role of crotonylation in glycolysis remains to be elucidated. According to recent reports, dynamic crotonylation of metabolic enzymes is involved in metabolic regulation and is related to pancreatic tumor and oral squamous cell carcinoma progression [41, 42]. Previously, we found that crotonylation of α enolase (a key enzyme for glycolysis) promotes CRC progression [16]. Our present study revealed that alternative splicing-related PTBP1 crotonylation affects tumor cell glycolysis. We provided a new perspective on the regulatory role and mechanism of crotonylation in tumor cell glycolysis and Warburg effect.
Previous studies on the PTBP1/PKM alternative splicing pathway mainly focused on the effect of the PTBP1 protein level on PTBP1/hnRNPA1/hnRNPA2 trimers. Upregulated oncogene c-Myc in tumor cells can bind to PTBP1/hnRNPA1/hnRNPA2 promoter and facilitate the transcription and high expression of these three genes, leading to elevation of the PKM2/PKM1 ratio [43]. Consistently, high expression of PTBP1 increases glucose consumption, lactate production, and PKM2 expression but decreases PKM1 levels in CRC cells [44, 45]. Our present study showed that, as a critical component of the hnRNPs complex, PTBP1 K266-Cr enhanced the interaction with other components (Fig. 4E). Meanwhile, the affinity of PTBP1 for splicing sites on PKM pre-mRNA was increased (shown in Fig. 4G). The transfer of crotonyl to histone lysine residues effectively neutralizes its positive charge, making chromatin recruitment and transcription factors unable to bind to DNA to achieve the purpose of gene silencing [24, 46]. Thus, it can be speculated that crotonylation may affect the role of PTBP1 in PKM alternative splicing by altering its spatial structure.
Our data showed that high PTBP1 K266-Cr levels in CRC were closely related to poor prognosis in CRC (shown in Fig. 6B), suggesting that PTBP1 K266-Cr has a broader application value as a prognostic marker or therapeutic target in clinical practice. Further studies are needed to determine whether splicing modulators or crotonylation inhibitors can affect CRC specific therapy by targeting PTBP1 K266-Cr. In addition, while it is known that inflammation is associated with tumors, it still needs to be investigated whether some inflammation- and tumor-related pathways, such as the nuclear factor kappa-B (NF-κB) pathway, can benefit from PTBP1 crotonylation.
The present study revealed that PTBP1 K266-Cr increases the PKM2/PKM1 ratio through pre-mRNA alternative splicing, contributing to CRC progression. This mechanism provides a potential strategy for CRC therapy by targeting the PTBP1/PKM alternative splicing axis.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- 2-DG
2-Deoxyglucose
- CCK8
Cell counter kit-8
- Co-IP
Co-immunoprecipitation
- CRC
Colorectal cancer
- ECAR
Extracellular acidification rate
- FDR
False discovery rate
- glycoPER
Glycolytic proton efflux rate
- HAT
Histone acetyl-transferase
- hMOF
Human males absent on the first
- hnRNP
Heterogeneous nuclear ribonucleoprotein
- IF
Immunofluorescent staining
- IP
Immunoprecipitation
- KAT2B
Lysine acetyltransferase 2B
- KCTs
Crotonyltranferases
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC–MS
Liquid chromatography-mass spectrometry
- PCAF
P300/CBP-associated factor
- PPI
Protein–protein interaction
- PTBP1
Polypyrimidine tract binding protein
- PTMs
Post-translational modifications
- RIP
RNA immunoprecipitation
- RRM
RNA recognition motifs
Author contributions
JH and XW designed the experiments. HC, XW, SH, YG, GL and LG performed the experiment. LG, FZ, ZW, JS and NL interpreted and analyzed the data. JH drafted the manuscript. JC, NL and JS supervised the study and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (82202622, 82002063), Shanxi Medical Key Science and Technology Project Plan of China (2020XM01), Basic Research Program of Shanxi Province (202203021212070), and partially by grant of Health Commission of Shanxi Province Scientific research subject (2022123) and Shanxi Traditional Chinese Medicine Administration Scientific Research Project (2023ZYYC004).
Availability of data and materials
The datasets downloaded, generated, and analyzed during the present study are available from the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025709.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with both the Declarations of Helsinki and Istanbul. The experimental protocols were approved by the Ethic Committee of Shanxi Provincial Academy of Traditional Chinese Medicine (Approval No. 2019-06KY005). All participants were informed and consented. The animal experiments were approved by the Animal Experimentation Ethics Committee of Shanxi Province Academy of Traditional Chinese Medicine (Approval No. 2023KY-07013). Animal care and experimental procedures were performed according to the criteria outlined in NIH guidelines. The maximal tumor size permitted by their ethics committee was 1000 mm3. All the tumor lesions in this study were below 1000 mm3. All the animals were housed in a specific pathogen-free environment, including bedding, caging systems, and diet.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jian-Yun Shi, Email: shijianyun0418@126.com.
Ning Li, Email: lining205310@126.com.
Ji-Min Cao, Email: caojimin@sxmu.edu.cn.
References
- 1.Intlekofer AM, Finley L. Metabolic signatures of cancer cells and stem cells. Nat Metab. 2019;1(2):177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tekade RK, Sun X. The Warburg effect and glucose-derived cancer theranostics. Drug Discov Today. 2017;22(11):1637–53. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Chen Q, Zhou Y, et al. S100A2 promotes glycolysis and proliferation via GLUT1 regulation in colorectal cancer. FASEB J. 2020;34(10):13333–44. [DOI] [PubMed] [Google Scholar]
- 4.Wang NN, Zhang PZ, Zhang J, et al. Penfluridol triggers mitochondrial-mediated apoptosis and suppresses glycolysis in colorectal cancer cells through down-regulating hexokinase-2. Anat Rec (Hoboken). 2021;304(3):520–30. [DOI] [PubMed] [Google Scholar]
- 5.Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23(11):560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takenaka M, Noguchi T, Sadahiro S, et al. Isolation and characterization of the human pyruvate kinase M gene. Eur J Biochem. 1991;198(1):101–6. [DOI] [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–3. [DOI] [PubMed] [Google Scholar]
- 8.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–6. [DOI] [PubMed] [Google Scholar]
- 9.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–31. [DOI] [PubMed] [Google Scholar]
- 10.Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, Leote AC, D’Artista L, Gallage S, Seehawer M, Carroll T, Dharmalingam G, Wee KB, Mellone M, Pombo J, Heide D, Guccione E, Arribas J, Barbosa-Morais NL, Heikenwalder M, Thomas GJ, Zender L, Gil J. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. 2018;34(1):85-102.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19(3):346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valcárcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7(11):R705–8. [DOI] [PubMed] [Google Scholar]
- 13.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107(5):1894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70(22):8977–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choksi A, Parulekar A, Pant R, et al. Tumor suppressor SMAR1 regulates PKM alternative splicing by HDAC6-mediated deacetylation of PTBP1. Cancer Metab. 2021;9(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou JY, Cao J, Gao LJ, et al. Upregulation of α enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem Biophys Res Commun. 2021;578:77–83. [DOI] [PubMed] [Google Scholar]
- 17.Hou JY, Gao LJ, Shen J, et al. Crotonylation of PRKACA enhances PKA activity and promotes colorectal cancer development via the PKA-FAK-AKT pathway. Genes Dis. 2023;10(2):332–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Wei W, Liu Y, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 2017;3:17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabari BR, Tang Z, Huang H, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2018;69(3):533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Wan J, Zhan J, et al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27(7):946–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei W, Mao A, Tang B, et al. Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J Proteome Res. 2017;16(4):1743–52. [DOI] [PubMed] [Google Scholar]
- 23.Hou JY, Zhou L, Li JL, Wang DP, Cao JM. Emerging roles of non-histone protein crotonylation in biomedicine. Cell Biosci. 2021;11(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Sabari BR, Panchenko T, et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol Cell. 2016;62(2):181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson PJ, Monie TP, Szendröi A, et al. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12(9):1631–43. [DOI] [PubMed] [Google Scholar]
- 26.Zong Z, Li H, Yi C, Ying H, Zhu Z, Wang H. Genome-wide profiling of prognostic alternative splicing signature in colorectal cancer. Front Oncol. 2018;8:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reda A, Hategan LA, McLean T, et al. Role of the histone variant H2A.Z.1 in memory, transcription, and alternative splicing is mediated by lysine modification. Neuropsychopharmacology. 2024;49(8):1285–1295. 10.1038/s41386-024-01817-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azhar M, Xu C, Jiang X, et al. The arginine methyltransferase Prmt1 coordinates the germline arginine methylome essential for spermatogonial homeostasis and male fertility. Nucleic Acids Res. 2023;51(19):10428–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan W, Qiu Y, Xu Y, Liu Y, Miao Y. Ubiquitination and ubiquitin-like modifications as mediators of alternative pre-mRNA splicing in Arabidopsisthaliana. Front Plant Sci. 2022;13: 869870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu AQ, Wang J, Jiang ST, et al. SIRT7-induced PHF5A decrotonylation regulates aging progress through alternative splicing-mediated downregulation of CDK2. Front Cell Dev Biol. 2021;9: 710479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi H, Nishimura J, Kagawa Y, et al. Significance of polypyrimidine tract-binding protein 1 expression in colorectal cancer. Mol Cancer Ther. 2015;14(7):1705–16. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZN, Liu D, Yin B, et al. High expression of PTBP1 promote invasion of colorectal cancer by alternative splicing of cortactin. Oncotarget. 2017;8(22):36185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274(3):1189–92. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19(2):1202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sfakianos AP, Raven RM, Willis AE. The pleiotropic roles of eIF5A in cellular life and its therapeutic potential in cancer. Biochem Soc Trans. 2022;50(6):1885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26(37):5341–57. [DOI] [PubMed] [Google Scholar]
- 38.Liu M, Zhang K, Li Q, et al. Recent advances on small-molecule bromodomain-containing histone acetyltransferase inhibitors. J Med Chem. 2023;66(3):1678–99. [DOI] [PubMed] [Google Scholar]
- 39.Liu N, Luo J, Kuang D, et al. Lactate inhibits ATP6V0d2 expression in tumor-associated macrophages to promote HIF-2α-mediated tumor progression. J Clin Invest. 2019;129(2):631–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu CE, Hu Y, Zhou P, et al. Lactate and TGF-β antagonistically regulate inflammasome activation in the tumor microenvironment. J Cell Physiol. 2021;236(6):4528–37. [DOI] [PubMed] [Google Scholar]
- 41.Yin X, Zhang H, Wei Z, et al. Large-scale identification of lysine crotonylation reveals its potential role in oral squamous cell carcinoma. Cancer Manag Res. 2023;15:1165–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Zhu L, Qin ZY, et al. Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression. Cell Rep. 2023;42(7): 112666. [DOI] [PubMed] [Google Scholar]
- 43.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumazaki M, Shinohara H, Taniguchi K, Takai T, Kuranaga Y, Sugito N, Akao Y. Perturbation of the Warburg effect increases the sensitivity of cancer cells to TRAIL-induced cell death. Exp Cell Res. 2016;347(1):133–42. [DOI] [PubMed] [Google Scholar]
- 45.Cheng C, Xie Z, Li Y, Wang J, Qin C, Zhang Y. PTBP1 knockdown overcomes the resistance to vincristine and oxaliplatin in drug-resistant colon cancer cells through regulation of glycolysis. Biomed Pharmacother. 2018;108:194–200. [DOI] [PubMed] [Google Scholar]
- 46.Wang ZA, Kurra Y, Wang X, Zeng Y, Lee YJ, Sharma V, et al. A versatile approach for site-specific lysine acylation in proteins. Angew Chem Int Ed Engl. 2017;56(6):1643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets downloaded, generated, and analyzed during the present study are available from the corresponding author upon reasonable request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD025709.