Abstract
Breast cancer is the most prevalent and deadliest cancer among women globally, representing a major threat to public health. In response, the World Health Organization has established the Global Breast Cancer Initiative framework to reduce breast cancer mortality through global collaboration. The integration of big data analytics (BDA) and precision medicine has transformed our understanding of breast cancer's biological traits and treatment responses. By harnessing large-scale datasets – encompassing genetic, clinical, and environmental data – BDA has enhanced strategies for breast cancer prevention, diagnosis, and treatment, driving the advancement of precision oncology and personalised care. Despite the increasing importance of big data in breast cancer research, comprehensive studies remain sparse, underscoring the need for more systematic investigation. This review evaluates the contributions of big data to breast cancer precision medicine while addressing the associated opportunities and challenges. Through the application of big data, we aim to deepen insights into breast cancer pathogenesis, optimise therapeutic approaches, improve patient outcomes, and ultimately contribute to better survival rates and quality of life. This review seeks to provide a foundation for future research in breast cancer prevention, treatment, and management.
Keywords: Breast cancer, big data, precision medicine, personalised treatment, disease management
Introduction
The burden of breast cancer is increasing globally. In 2020, among the newly diagnosed 19.3 million cancer patients, 2.26 million cases were diagnosed as breast cancer, surpassing lung cancer as the most common type of cancer worldwide.1–3 Breast cancer not only ranks as the fourth leading cause of cancer deaths worldwide but also represents the primary cause of female mortality. 3 Due to its high incidence and mortality rate, breast cancer has become a global challenge and poses a significant threat to women's health. In recent years, the mortality rate among Western populations, especially in young age groups, has declined due to advancements in breast cancer diagnosis and treatment.4,5 However, breast cancer remains the leading cause of cancer-related deaths in women globally. 6 In Europe, 66 women are diagnosed with breast cancer every hour, with 18 unfortunate cases resulting in death. 7 Without adequate control measures, it is estimated that the incidence of new breast cancer cases will rise by 26.7%, and the number of deaths will increase by 25.2% over the next 20 years. 7 In the United States, breast cancer is the second leading cause of cancer-related deaths after lung cancer. 8 According to the American Cancer Society, the average risk of developing breast cancer in U.S. women is approximately 13%, meaning that 1 in every 8 women will be diagnosed with breast cancer, and approximately 1 in 39 women (approximately 3%) will die from breast cancer.8,9 By 2022, there would have been approximately 287,850 new cases of invasive breast cancer, with 43,250 women dying from the disease. 10 In China, statistics show that in 2022, there were approximately 357,200 cases of female breast cancer and 75,000 deaths, making it one of the leading causes of cancer-related deaths among women. 11 In Malaysia, breast cancer remains the most common type of cancer in both women and men, with 8371 new cases, accounting for approximately 31.3% of the overall incidence rate.1,12 The 5-year incidence rate is approximately 36,754 cases, with a mortality rate of approximately 11.1%, which is second only to lung cancer (15.1%) and colorectal cancer (11.7%). 13 Therefore, the prevention, management, and treatment of breast cancer are urgent and require adequate attention.
The World Health Organization has released the Global Breast Cancer Initiative (GBCI), a key measure to address the global breast cancer issue. 14 Its primary objective is to assist governments and international organisations in formulating practical and feasible policies to improve breast cancer incidence and mortality rates among women. The core goal of this proposal is to reduce the global breast cancer mortality rate by 2.5% annually, with a vision to save 2.5 million lives by 2040. 14 The initiative aims to achieve a reduction in cancer incidence through a series of measures that enhance awareness of breast cancer risk factors and related hazards, as well as screening levels. The GBCI, jointly established by the U.S. National Center for Chronic Disease Prevention and Health Promotion and the Centers for Disease Control and Prevention, consists of three core components: early health promotion screening, timely breast cancer diagnosis, and comprehensive breast cancer management. 15 These pillars collectively form the strategic foundation for the initiative to achieve its ambitious goals. The GBCI is a new concept driven by data and prevention, which will bring revolutionary changes to breast cancer screening and treatment in the future. 16 With the active participation of stakeholders and the achievement of the grand objectives of the GBCI, it becomes crucial to research how to utilise big data analytics (BDA) to improve the effectiveness and accuracy of breast cancer interventions.
In recent years, with the rise of BDA and precision medicine, there has been a profound shift in the understanding of the biological characteristics and treatment response of breast cancer. By integrating the BDA technology into the GBCI, researchers and clinical doctors can strengthen existing breast cancer prevention, diagnosis, and management strategies, gaining a deeper understanding of the molecular mechanisms driving the development and progression of breast cancer, thereby advancing the goal of precision oncology and personalised medicine. BDA has become a revolutionary tool in healthcare, providing unprecedented opportunities to leverage diverse data types to understand disease mechanisms, treatment responses, and patient outcomes.17,18 However, despite the increasing research on precision therapy for breast cancer, studies involving breast cancer big data remain limited. There has yet to be a systematic and comprehensive discussion on breast cancer big data and precision medicine. Therefore, this review aims to systematically summarise and analyse breast cancer big data and its role in precision medicine to provide insights and guidance for relevant research, thus promoting continued advancements in precision oncology and personalised patient care.
Foundations of big data
The rapid development of information and communication technology has given rise to significant changes in the digital domain, forcing most industries to adapt to this digital trend and consider digitalisation an indispensable requirement. 19 The core of digital transformation lies in digitisation, which begins with the digitalisation process, i.e., the conversion of analogue signals (physical behaviours and observations) into digital form using digital technology. 20 Subsequently, digitisation involves utilising and manipulating this digitalised information, generating impacts from organisational and societal perspectives. At the core of digitisation lies BDA, which processes large datasets and transforms them into actionable knowledge for decision-making. Big data originates from massive datasets resulting from information exchange between multiple systems 21 and possesses the following seven ‘V’ characteristics22–24:
Volume (size): Big data involves managing and analysing massive amounts of information, typically ranging from several terabytes (TB) to several exabytes (EB), requiring substantial processing and storage capabilities.
Variety (complexity): Big data is presented in various forms, including structured data (e.g., databases), semi-structured data (e.g., logs and XML), and unstructured data (e.g., social media posts and video content).
Velocity (speed): Big data is generally generated in real-time or near-real-time, necessitating instant analysis and processing to derive valuable insights.
Veracity (quality): The veracity of big data refers to the accuracy and reliability of data, ensuring the trustworthiness and quality of data sources.
Variability (flexibility): The content, scale, and speed of big data are constantly changing, requiring adaptability for effective analysis and processing.
Value (knowledge): The ultimate goal of big data is to extract useful information and insights from datasets to provide robust support and directional guidance for decision-making and innovation activities.
Valence (connectedness): Another characteristic of big data is the connectivity between datasets.
Due to the significant characteristics of large data volume and complexity, traditional processing methods have become extremely difficult. Recently, significant achievements have been made in analysing and mining massive amounts of data by humans. The entire process of in-depth interpretation, mining, and analysis of these large-scale, high-dimensional, diverse, and rapidly generated data sets is collectively called BDA. 25
The digital advancement in the medical field is referred to as Medicine 4.0, which has promoted the integration of robotics, 3D printing, the internet, and artificial intelligence (AI), leading to the development of modern medical innovative applications. 26 These applications cover numerous domains, from on-demand medical services provided by online markets to remote medical solutions enabling remote interactions. Moreover, BDA plays a crucial role in virtual reality applications during diagnosis and treatment practices, surgical procedures, and wearable medical devices for continuous health monitoring.27,28 The core of the effectiveness of these technologies lies in generating accurate, reproducible, and reliable data. Therefore, data collection has become the cornerstone of medical institutions. As clinical communities increasingly recognise the importance of data, institutions intensify their efforts to generate and collect data that aligns with technological advancements. Consequently, healthcare systems are inundated with vast amounts of data related to patients, marking a significant shift in medical management and decision-making processes. 29
The exponential growth of big data brings both opportunities and challenges to healthcare. By 2025, the digital universe will reach 175ZB, 30 while the healthcare sector already generated 2314EB of data in 2020. 31 These data are collected from various sources, covering clinical records, medical images, genomics, environmental data, and personal or societal behavioural information from various organisations and departments. However, various challenges must be addressed before they can be effectively applied to healthcare decision-making processes (Table 1). The inconsistency in data formats, naming conventions, and collection methods hinders direct comparisons between different datasets. Traditional databases face challenges in integrating multifaceted medical information. Although electronic health records digitise the storage of patient medical information, they still need help with a series of problems such as interoperability, long data collection times, data redundancy, and complex document processing flows.32,33 Therefore, applying modern digital technologies becomes crucial in addressing these challenges.
Table 1.
General challenges in data collection, centralisation and management in healthcare settings.
| Type | Challenge | Description | References |
|---|---|---|---|
| Data collection | Quality of data collected | The quality of healthcare data collected poses a significant limitation that should be taken seriously into consideration. The data collected may be unstructured, improper, or non-standardised in some cases. Therefore, the research organisation and industry must exert additional effort to convert this information into usable and meaningful data. Due to the serious constraints on healthcare data quality, errors and variations in the results should be excluded. | Asri et al. 34 |
| Precision health data trustworthiness | The trustworthiness of health data is challenging to maintain because it is complex and diverse. Checking the credibility of health data becomes more challenging due to the increasing size of health data, distributed storage of data, and a massive number of data sources, including Internet of Medical Things devices. Trustworthy data sources, such as government agencies and reputed organisations, follow health data governance policies, allowing for the inspection of data through metadata and associated information. However, with the advent of the Internet of Medical Things (IoMT), it is difficult to manage and maintain the reliability of health data due to the possibility of extracting data from faulty or improperly configured IoMT devices. | Thapa and Camtepe 35 | |
| Data collection method | The issues that arise in the collection and interpretation of data are focused on ensuring scientific rigour, validity, and reliability. These concerns include addressing the challenges that arise when combining data from multiple sources, such as differences in data capture methods or sample characteristics. Additionally, the unique features of each study, including variations in methods of recruiting patients, can result in differing challenges during the data collection process. These challenges can pose significant threats to the scientific quality of the data and may impact the interpretation of the findings. Therefore, it is crucial to develop strategies to manage and avoid these challenges to ensure that the collected data is of high quality and accurately reflects the intended outcomes. | Holden et al. 36 | |
| Data processing and centralisation | Data heterogeneity | Organisations such as hospitals, pharmacies, and medical centres have data in different systems and settings, making it difficult to use this large amount of data effectively. These organisations require a common data warehouse that can provide them with homogeneous information to manage the data. The main challenge in utilising healthcare data is the heterogeneity of healthcare data which makes it difficult to search, integrate, and extract information. To implement personalised and precise treatment, it is essential to achieve interoperability. However, the inability to exchange information between cancer diagnosis and treatment systems is currently limiting data-driven clinical practice. To address this challenge, it is necessary to develop a global system that can formalise and harmonise the different data models, classifications, thesauri, vocabularies, terminologies, and ontologies used in different systems. | Asri et al., 34 Hong et al. 37 |
| Data management | Data breach | For 12 consecutive years, the healthcare industry had the highest data breach cost of any industry. In 2022, the healthcare industry is paying an average of US$10.10 million for data breaches. In the future, the security policies in healthcare organisations should be reviewed and provide security solutions for cloud computing. | IBM 38 |
| Information security | Cloud computing has the potential to benefit e-health services, but there are still concerns about information security. The complexity of security problems in the cloud models requires additional investments to implement data management policies. Basic data must be registered and approved by relevant people to take necessary measures for predictable or informative events. Healthcare cloud computing has several issues, including data transmission and access control, the integrity of data protection, and loss of physical control over personal information. Data mobility, data breaches, and the inability to locate or process data are also major challenges. Identity and access management, authentication, and Internet-based access are other significant concerns in healthcare cloud computing. The use of cybernetic management solutions is required to securely transmit data and protect devices from breaches and unauthorised access. To ensure data security and trust in cloud computing for healthcare organisations, it is recommended to use encryption and data protection mechanisms to authenticate authorised users and licensing. | Mehraeen et al. 39 | |
| Data analytics | Lack of good governance and annotated data | Many current healthcare applications are limited by inadequate quality control, data standardisation, and sample size. Radiomics studies typically utilise images captured using various scanning devices from different manufacturers. The lack of standardised protocols for data acquisition and reconstruction parameters leads to significant variations. Consequently, to distinguish signal from noise in medical images, approved methodologies are required, necessitating the standardisation of image preprocessing, tissue segmentation, feature calculation, and statistical methodologies. To address these issues, a range of technical challenges must be overcome. | Hong et al. 37 |
| Big data analytics | Although healthcare systems typically store accurate information, it may not always be up to date. Therefore, retrieving, analysing, and comparing big data is necessary to make timely and precise decisions based on real-time data processing, which can be crucial for the life or death of patients. To prevent and detect infections as early as possible and ultimately save lives, companies must invest in big data analytics, which entails acquiring staff, such as data scientists, and resources, as well as purchasing data analytics technologies. Additionally, medical organisations must be convinced to adopt big data analytics. However, utilising data mining and big data analytics requires specialised expertise and knowledge, making it a costly undertaking for companies to hire such individuals. | Asri et al. 34 | |
| Data sharing | Ransomware | The danger in information sharing is the lack of privacy in patients’ data. Sensitive medical data is now accessible through computers and mobile devices that often lack extensive security and cause ransomware attacks. The main attacking purpose of ransomware is financial gain, where hospitals have big healthcare data storage and security flaws in IT processes. Attackers use ransomware to block access or encrypt victims’ files and demand ransom in exchange for the decryption key or to restore access. | Olivier et al. 40 |
| Consent management | There are challenges in managing the consent of health data especially during data sharing and data linkage. Consent is mandatory for handling health data, governed by ethical guidelines and legislation, to protect privacy, confidentiality, and autonomy. There are three types of consent: explicit, implicit, and opt-out consent. Consent management solutions have been proposed, including tools for modelling consent, a repository for storing it, and a data access management component. Data sharing and linkage pose challenges addressed through static and dynamic consent approaches, with dynamic consent allowing for two-way communication and subject control over consent. Trust is a way to increase consent approvals as trust and privacy concerns are inversely proportional. | Thapa and Camtepe 35 |
Big data-driven precision medicine: integration of multi-omics and methodologies
Big data-driven precision medicine has emerged in the current medical science field. The key in this field is to achieve personalised medical treatment and smarter healthcare services by analysing large amounts of data and adopting innovative technologies. The combination of multi-omics plays a decisive role by integrating rich omics data such as genomics, transcriptomics, proteomics, and metabolomics to reveal the molecular mechanisms of disease development, providing critical support for precision medicine. 41 Over the past decade, due to the rapid advancements in short-read sequencing technologies (such as the Illumina platform) and long-read sequencing technologies (such as PacBio and Oxford Nanopore platforms), there has been a deeper understanding of the complex interactions within biological systems and the molecular and physical phenomena they entail. 42 Next-generation sequencing (NGS) technologies are being used to generate large datasets for interpreting human genetics and the regulatory mechanisms at the molecular level. This includes identifying single nucleotide polymorphisms through genome-wide association studies, exploring disease progression through transcriptome analysis and full transcriptome-wide associations, and discovering the interactions and impacts between food and diseases through microbiome-wide association studies. Since 2013, at least 14 countries have launched government-funded genomics medicine initiatives, with total investments reaching up to $4 billion. However, this amount is relatively small compared to the Precision Medicine Initiative in China. The initiative aims to sequence the genomes of 100 million individuals by 2030, with a funding of $9.2 billion. 43
The cost of NGS has dropped from millions to a few thousand dollars, 44 which will help comprehensively generate genetic data for all diseases, including cancer and rare diseases. It is gratifying that the human genetic variation profiling 45 from various countries and The Cancer Genome Atlas 46 (TCGA; https://www.cancer.gov/ccg/research/genome-sequencing/tcga), these digital catalogues involving cancer genome changes have been established and opened. Furthermore, significant breakthroughs have been made in collecting proteomic, metabolomic, and other data from millions of patients through techniques such as mass spectrometry, nuclear magnetic resonance, and imaging. 47 Combining this data with mathematical algorithms can lead to more effective classification based on specific phenotypes. Using big data has also offered unprecedented opportunities for comprehending pharmacology and toxicology. For instance, through integrated multi-omics analysis, Lv et al. 48 demonstrated how cadmium exposure influences the survival and function of granulosa cells via multiple signalling pathways. This data was extensively stored and analysed, laying a data foundation for future toxicity evaluations. Xu et al. 49 discovered that bisphenol A and its structural analogues possess varying potentials to induce mitochondrial dysfunction and apoptosis. This research approach, based on big data and integrated multi-omics, enables researchers to understand complex pathological mechanisms and the effects of drugs on cells at a broader molecular level, thereby providing a foundation for personalised treatment strategies in precision medicine. Therefore, the critical task for the future is to integrate this omics data and other relevant data and further perform genetic association and AI training for predictive analysis based on a multi-omics approach, thus revealing the molecular principles behind disease progression and treatment effects. In the future, these multi-omics technologies can be developed and transformed into user-friendly and reliable tools for routine clinical operations, thus achieving support for clinical decision-making and personalised treatment as the goal for all patients with chronic diseases, especially those diagnosed with cancer.
Apart from the extensive use of NGS technology, numerous other research techniques are progressing swiftly in precision medicine research. These approaches have demonstrated significant research potential and practical application value in various disease settings, particularly oncology research: (1) Functional diagnostic technologies such as live tumour cell detection, molecular precise analysis of tumour responses, and device-based in situ methods have made significant progress. 50 The crux of this approach lies in its independence from genomic information, instead directly observing the cell behaviour in specific environments. 50 Consequently, functional diagnostic tests can evaluate the patient sensitivity to chemotherapy drugs or targeted therapies, offering the foundation for personalised treatment strategies. (2) Liquid biopsy is a non-invasive technique for detecting tumours by analysing circulating tumour DNA or other biomarkers in bodily fluids such as blood. 51 It enables real-time monitoring of dynamic changes in tumours. 52 Compared to traditional tissue biopsies, liquid biopsy can detect the risks of recurrence or metastasis at earlier stages, providing more timely interventions. 52 Liquid biopsy can assist in identifying resistance to cancer treatment, allowing for timely adjustments in treatment strategies, which is critical for achieving precision medicine. (3) Although NGS dominates genomic research, gene chips remain essential in precision medicine. One significant application of gene chip technology is the screening for potential therapeutic targets and biomarkers, which is crucial for the design of personalised therapies. 53 With advances in technology, the accuracy and reproducibility of gene chips have significantly improved, resulting in a broader application in clinical research. 53 (4) The application of epigenetics in precision medicine is becoming increasingly widespread. Researchers can identify epigenetic changes associated with specific diseases by detecting epigenetic markers, thus providing new insights for precision treatment. 54 These approaches offer significant support for personalised therapies through various avenues, driving the advancement of individualised healthcare. In precision medicine research, the diversification of research methods not only enriches data sources but also dramatically enhances the reliability and effectiveness of the research. Looking ahead, with the continuous development and integration of these advanced technologies, they provide multiple perspectives for furthering precision medicine, contributing to the realisation of personalised treatments.
In addition, big data-driven precision medicine also relies on advanced data science methods. Anomaly detection is a critical challenge when processing large-scale medical data. The method proposed by Li et al. 55 demonstrates how to overcome these challenges through BDA, ensuring the reliability and accuracy of detection results. By processing, cleaning, transforming, exploring, dimensionality reduction, pattern searching, visualisation, and reporting meaningful information and insights can be extracted from massive multi-omics data, thus optimising the healthcare system. 56 However, despite significant advancements in multi-omics integration technologies, some challenges still need to be addressed, such as poor data collection quality, high data heterogeneity, low data reliability, and lack of adequate data management and annotation (Table 1). These issues will impact clinical decision-making. Despite the challenges, many successful cases have shown that using advanced analytical technologies, such as machine learning and AI, is very effective in medical applications for cancer (especially breast cancer) (Table 2). Additionally, programming languages such as R and Python are used to write scripts, algorithms, and software; big data tools such as Hadoop and Apache Spark, as well as high-performance computing clusters accessed through grid computing infrastructure, are significant contributors to the digitisation of multidimensional medical data. 56 Therefore, future research directions should include optimising data integration and analysis algorithms, expanding the scale of samples, and establishing more rigorous quality control standards.
Table 2.
Application of digitisation in healthcare sectors by focusing on cancers.
| Type | Application | Type of Cancer | Description | References |
|---|---|---|---|---|
| Early detection | Detection of breast cancer using full-field digital mammography (FFDM) | Breast cancer | FFDM consists of a dedicated electronic detector system that captures and displays the X-ray signals on a computer rather than film. This technology separates the process of image acquisition from that of image display, and because the signal for each pixel is digitally stored, the same image can be manipulated to show different brightness and contrast combinations, allowing radiologists to more easily see through denser tissues. Softcopy display of digital mammography allows adjustment of magnification, brightness, and contrast after the mammography examination, thus enabling a more detailed examination of questionable areas without necessarily requiring additional imaging. | Newman 57 |
| Screening high-risk women using magnetic resonance imaging (MRI) | Breast cancer | Magnetic Resonance Imaging (MRI) is a non-invasive and non-ionising radiation technology for early detection of breast cancer. Some studies have shown greater than 90% sensitivity in detecting lesions using contrast-enhanced MR imaging. Further improvements in specificity and optimal acquisition protocols, examination and interpretation standards are ongoing areas of investigation. Digitisation has enabled the use of MRI for early detection of breast cancer without using ionising radiation. | Newman 57 | |
| Diagnosis | Classification of benign-malignant patterns in digital mammograms | Breast cancer | Computer-aided diagnosis (CAD) based on support vector machine recursive feature elimination, correlation bias reduction algorithm, and Q similarity-based learning algorithm is applied to real clinical mammograms and resulted in an accuracy of 98.16%, a sensitivity of 98.63%, specificity of 97.80%, and computational time of 2.2 s for breast cancer diagnosis. | Eltrass and Salama 58 |
| Classification of melanoma (fatal skin cancer) and nevus (non-cancer, benign) | Skin cancer | An intelligent data-driven system with image processing technique utilises Gaussian filter, improved K-mean clustering and Support vector machine (SVM) to classify skin cancer into melanoma and nevus with 96% accuracy. | Khan et al. 59 | |
| Prognosis | Machine learning-based data mining approach for the prediction of breast cancer survivability | Breast cancer | With a large dataset of >200,000 cases, a decision tree with 10-fold cross-validation methods showed a prediction accuracy of 93.6% accuracy, providing relative prediction ability for cancer prognosis. | Delen et al. 60 |
| Treatment | Deep-learning assisted auto-digitisation method for interstitial needles | Gynaecological cancer | A deep-learning assisted auto-digitisation method for interstitial needles was used in the treatment planning of 3D CT image-based interstitial high dose-rate brachytherapy (HDRBT) of gynaecological cancer. The digitisation process took ∼5 min to complete, and the achieved accuracy and efficiency made the method clinically attractive. | Jung et al. 61 |
Precision medicine and AI: the hope for innovation in healthcare
Precision medicine, a healthcare approach tailored to individual differences, has gained significant attention. Its fundamental principle is to deliver personalised diagnostic and treatment plans by analysing a patient's genomic data, environmental influences, and lifestyle factors. The rapid advancement of AI technology has further propelled the field, offering powerful tools for large-scale data analysis, personalised treatment recommendations, and the early prediction and prevention of diseases, showcasing immense potential in healthcare.
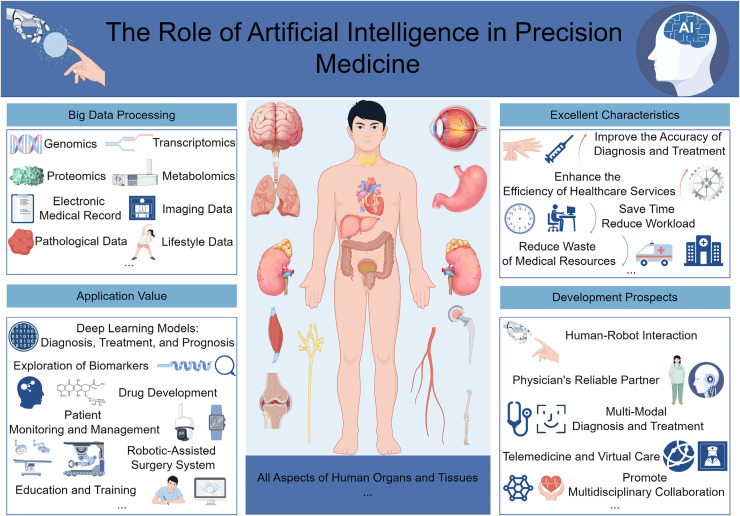
Firstly, AI has demonstrated exceptional application value in the precision medicine approach for cardiovascular diseases. A comprehensive review study indicates that in applying AI models in the cardiovascular field, predictive studies constitute 50%, diagnostic studies 21%, and phenotypic analysis, and risk stratification each 14%. 62 Additionally, the most widely used machine learning algorithms are logistic regression (36%), random forests (32%), and support vector machines (25%). 62 This suggests that AI technology has significantly advanced in the diagnosis, prognostic prediction, risk stratification, and treatment planning of cardiovascular diseases. Secondly, the application of AI in genomics and oncology has also garnered significant attention from researchers. A study reported that the role of AI in drug discovery and personalised treatment is becoming increasingly prominent, particularly in the analysis of gene expression and variant data, AI models have demonstrated an accuracy of up to 99%, a sensitivity of 100%, and a specificity of 96% in evaluating tumours. 63 AI technology can analyse multi-dimensional clinical and biological data to identify new therapeutic targets and optimise treatment outcomes. By integrating big data and AI, personalised treatment plans can be provided for patients. This approach reduces the trial-and-error process, enhances treatment efficiency and effectiveness, and minimises the risk of side effects. Additionally, AI has demonstrated outstanding performance in disease prevention and early warning. AI algorithms can rapidly identify early signs of disease by analysing large-scale medical data, including patients’ medical histories, genomic data, and lifestyle information. This enables early prediction of disease occurrence, providing clinicians with solid evidence for early intervention and prevention strategies. 64 Finally, the widespread application of AI in precision medicine not only enhances the accuracy of diagnosis and treatment but also drives improvements in the efficiency of healthcare services. A study noted that AI-assisted systems can reduce diagnostic time costs, lessen the workload of physicians, and significantly improve the utilisation efficiency of medical resources. 65 These cases illustrate that AI, by optimising data processing workflows, enhancing diagnostic precision, providing personalised treatment recommendations, and conducting predictive analyses, has significantly bolstered the practicality of precision medicine. This advancement dramatically contributes to progress in the medical field and highlights AI's central role and vast potential for development in future medical practice (Figure 1).
Figure 1.
The application and prospects of artificial intelligence technology in precision medicine (drawn using the figdraw platform; https://www.figdraw.com/).
The application of big data in breast cancer
1. Approaches for breast cancer
Breast cancer, as a common malignant tumour disease, faces many challenges in its treatment and management, including personalised therapy, clinical trial design, and accurate assessment of the condition. The application of big data technology provides a new path for solving such problems. Cancer datasets exhibit unique attributes compared to other fields, such as smaller size and high heterogeneity, revealing multiple dimensions of cellular systems and biological activities. 66 Cancer research involves the analysis of five main data types: molecular omics data, perturbation phenotypic data, molecular interaction data, imaging data, and textual data. 66 Due to the limited amount of each type of data and their high heterogeneity, new computational methods are needed to integrate data from different dimensions and queues, which will help better understand the complexity of breast cancer from multiple biological perspectives.
To better leverage big data and enhance its accuracy, defining the screening criteria for breast cancer patients becomes particularly critical. Hence, it is essential to contemplate establishing stringent inclusion and exclusion criteria for subjects to ensure the precision, reproducibility, and scientific validity of breast cancer big data research findings. Inclusion criteria: (1) Confirmed cases: All patients diagnosed with breast cancer through pathology should be included, encompassing both primary and recurrent breast cancer. (2) Age and gender: These two factors deserve consideration, and stratified analysis should be performed to ensure comprehensive coverage. While breast cancer primarily affects adult females, the biological characteristics of males and underage patients should not be overlooked. (3) Treatment types: Include breast cancer patients who have undergone surgery, radiotherapy, chemotherapy, targeted therapy, or endocrine therapy. Detailed records of specific treatment regimens should be maintained, including treatment methods, drug names, dosages, and duration. Data on different treatment regimens should be categorised for subsequent continuous monitoring and in-depth analysis of treatment response characteristics. (4) Follow-up information: Patients with follow-up records should be included to ensure the data's long-term reliability and dynamic assessment. Follow-up data should encompass survival status, survival time, recurrence status, and distant metastasis, facilitating the analysis of prognostic factors and evaluation of treatment effectiveness. (5) Data completeness: Establish a comprehensive and diversified dataset that includes clinical information, pathological data, radionics, and genetic testing results to ensure the thoroughness of the analysis.
Exclusion criteria: (1) Presence of other malignancies: To avoid data confusion and ensure consistency, patients diagnosed with other types of malignancies should be excluded before the confirmation of breast cancer or during the study period. (2) Severe comorbidities: Exclude patients with severe comorbidities that may affect research outcomes, such as uncontrolled cardiovascular diseases, liver dysfunction, or renal failure. (3) Ethical safety: Exclude data from patients who did not provide informed consent, ensuring that the research complies with ethical standards. In addition, patient data that involves sensitive personal information and cannot guarantee anonymisation should also be excluded.
A solid scientific foundation is established for constructing high-quality breast cancer datasets by strictly adhering to inclusion and exclusion criteria. This approach provides a reliable basis for subsequent statistical analysis and clinical research. These criteria ensure the data's accuracy and representativeness and enhance overall data quality and research rigour. In the future, this dataset can be instrumental in exploring the pathogenesis, treatment responses, and prognostic factors of breast cancer. Additionally, it will facilitate in-depth investigations into the characteristics of various breast cancer subtypes and the development of personalised treatment strategies.
Big data holds significant potential in enhancing breast cancer diagnosis, treatment, and management. By analysing extensive patient information and outcomes datasets, researchers can gain new insights into the disease and develop more effective treatment methods. By integrating multi-omics data, researchers can identify gene mutations and molecular biomarkers related to breast cancer, providing clues for early diagnosis and personalised treatment. For instance, gene expression profiling can help determine tumour subtypes, guiding treatment selection; proteomic and metabolomic analyses can provide detailed information on the tumour microenvironment, revealing potential mechanisms of treatment resistance and disease progression. Big data also plays a crucial role in predicting the risk of breast cancer incidence. Through technologies like machine learning and AI, researchers can utilise various data, including genetic information, family history, and lifestyle factors, to identify individuals who may benefit from early screening or preventive interventions, thus developing predictive models to identify individuals at higher risk of breast cancer. 67 For example, Rabiei and their team conducted an in-depth analysis using machine learning methods on demographic, laboratory, and mammographic data, finding that integrating multiple risk factors into predictive models of breast cancer can more effectively diagnose the disease promptly and help formulate more targeted care plans. 67 Big data technology enables researchers to analyse vast amounts of gene expression data, revealing key pathways and genes influencing cancer development. The study by Xue et al. 68 revealed the oncogenic role circRNA_0000326 in breast cancer through BDA, promoting tumour development by regulating the miR-9-3p/YAP1 axis. Big data also plays an indispensable role in predicting breast cancer prognosis. By employing bioinformatics approaches, researchers can integrate multi-omics and clinical data to identify various factors influencing survival and construct an accurate prognosis prediction model. These models guide clinical practitioners, enabling them to tailor treatment plans based on the actual conditions of patients, thereby enhancing treatment effectiveness. For instance, the study by Jiang et al. 69 found that a prognosis model based on cuproptosis-related lncRNA could effectively predict the survival time and immune microenvironment characteristics of breast cancer patients, demonstrating the application of big data in constructing precision medicine tools. This data-driven approach not only aids in the in-depth study of breast cancer biology but also lays the groundwork for personalised treatment strategies in precision medicine. Through the integration of clinical, genetic, and molecular-based predictions, oncology is poised to enter a new stage in the field of precision oncology, thus achieving true precision oncology. 70
Furthermore, big data also plays a crucial role in drug development and clinical trials for breast cancer. 71 Through in-depth analysis of large-scale data on patient treatment outcomes and drug responses, research teams have the opportunity to identify the best treatment regimens and design more successful experimental projects. This lays a solid foundation for further exploration and discovery of new drug targets and treatment strategies, accelerating the drug development process. Additionally, clinical trials based on real-world data help evaluate the efficacy and safety of drugs, providing patients with a broader range of treatment options. In breast cancer treatment management, big data has a profound impact on personalised medicine. Tailoring treatment plans based on the analysis of patients’ genes and other essential information not only improves treatment effectiveness but also reduces related adverse reactions.
2. Breast cancer databases
The breast cancer database is constructed by numerous contributing factors, including the researchers’ contributions, laboratory research, charitable organisations/institutions, projects from various countries, and participation in international collaborative alliances (Table 3). These databases gather many datasets, including patient demographics, molecular-level data, tumour characteristics, treatment plans, and outcomes. Their diverse objectives include clinical research, personalised medicine, and quality improvement. From locally relevant localised datasets to comprehensive, integrated repositories spanning multiple technological platforms and covering thousands of patients, these provide valuable resources for breast cancer research.
Table 3.
Breast cancer databases.
| Database Name | Organisation | Location/Region | Purposes | Type of Data | Data Source | Sample size | Follow-up | Accessibility | References |
|---|---|---|---|---|---|---|---|---|---|
| ONCOPOOL | European Commission Framework 5 Project | Europe | To retrospectively compile the database of primary operable invasive breast cancers treated in the 1990s in 10 European breast cancer Units | Patient characteristics, tumour characteristics, pathology, therapies and outcomes | 12 European breast cancer Units in 10 European states | 16,944 cases (with follow-up data) | 10 years period | No. Data stored at Central Database at Nottingham City Hospital | Blamey et al. 72 |
| The Breast Cancer Gene Expression Miner (bc-GenExMiner) database | Swiss Institute of Bioinformatics (SIB) and the University of Lausanne | Switzerland | To explore and analyse gene expression data from breast cancer patients, and to improve gene prognostic analysis performance by using the same bioinformatics process. |
|
21 public data sets encompassing 8 different microarray platforms and 12 DNA chips. The sources of publicly available breast cancer gene expression data sets are such as Gene Expression Omnibus (GEO), ArrayExpress, and Stanford microarray database. | N/A | N/A | Publicly available breast cancer gene expression data sets | Jézéquel et al. 73 |
| Breast Cancer Data Base, Sweden 2.0 (BCBaSe 2.0) | Swedish Breast Cancer Group | Sweden | To provide a resource for researchers studying breast cancer, with the ultimate goal of improving patient care and outcomes. The database is used to investigate risk factors, treatment options, and survival rates, among other topics. | Demographic information, tumour characteristics, treatment regimens, and outcomes | Swedish Cancer Registry and National Board of Health and Welfare | Over 400,000 patients | 10 years | Accessible to researchers who meet certain criteria, such as obtaining ethical approval and signing a data transfer agreement. The database is managed by the Swedish Breast Cancer Group, which reviews all requests for data access. | Wadsten et al. 74 |
| The Cancer Genome Atlas (TCGA) Database | National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) | The United States | To improve diagnostic methods, treatment standards, and prevent cancer. | Genomic, transcriptomic, epigenomic, and proteomic data, as well as clinical and demographic information for cancer patients. | Tumour and normal tissue samples from patients with different types of cancer | Over 11,000 patients | N/A | Publicly available through the TCGA Data Portal (https://portal.gdc.cancer.gov/) operated by the Genomic Data Commons (GDC). | Tomczak et al., 46 Wang et al. 75 |
| METABRIC | Molecular Taxonomy of Breast Cancer International Consortium | Canada and the United Kingdom | To determine the molecular characteristics of breast tumours for the best treatment process. | Gene expression, copy number, gene mutation, and clinical data | Tumour and normal samples, 10 types of breast cancer subtypes | 2509 primary breast tumours and 548 matched normal sample | N/A | Publicly available through the cBioPortal FOR CANCER GENOMICS Datasets (https://www.cbioportal.org/study/summary?id=brca_metabric). | Curtis et al., 76 Pereira et al. 77 |
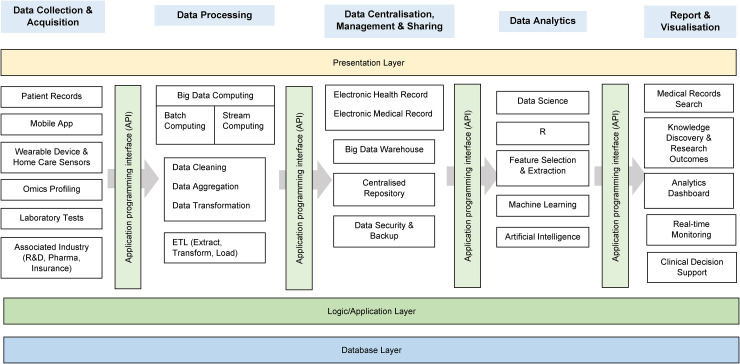
Several vital components constitute a comprehensive breast cancer database in the modern digital infrastructure (Figure 2). These components include traditional electronic health records of patients, mobile applications integrating Internet of Things technology, data obtained from laboratories and patient homes, data analysis tools capable of processing large-scale datasets and identifying patterns, and web-based interfaces providing convenient access and visualisation of information for end-users (including doctors and researchers) (Table 3). The structure must be carefully designed to create an efficient database that facilitates collaboration among researchers (Table 4). This design scheme should integrate cutting-edge technological methods and focus on interoperability and compatibility with established standards. Additionally, it should encompass robust data protection and privacy measures to ensure the security of patient information.
Figure 2.
Big data flow in medical research with a modern digital approach and three-level database architecture.78,79
Table 4.
General components and requirements of breast cancer databases.
| Type | Items | Descriptions | References |
|---|---|---|---|
| Components | Electronic health records | Provide a comprehensive view of patent history and treatment and facilitate data sharing between healthcare providers and researchers. | Thompson et al. 80 |
| Mobile app | Enable the bi-directional collection of data such as patient-reported data such as symptoms, treatment side effects, and educational materials and support resources to the patient. | Alford et al. 81 | |
| Data analysis tools | Machine learning algorithms and visualisation software can used to analyse and interpret breast cancer data by identify trends, patterns and insights difficult to discern from raw data. | Anklesaria et al. 82 | |
| Web-based interfaces | A user-friendly and up-to-date way to access and interact with the database to search and filter data, generate report and share information with other stakeholders is important. | Tsangaris et al. 83 | |
| Requirements | Database structure design | A good database should have a good balance between the ‘defined purpose’, ‘functions’ and ‘software technology’, starts with the design process of determining the purpose of database, organising the information into tables, turning information items into columns, set up table relationship (one-to-one, one-to-many, many-to-many), define data dictionary and standard the data input. | Yi and Hunt 84 |
| Interoperability with well-defined, comprehensive standards | Ability of a database to share, interdigitate and make use of data between healthcare software systems developed by different engineers, in the perspectives of both hardware deployment and software structure. | Han et al. 85 | |
| Data privacy | While ensuring data integrity and sharing remain uncompromised, measures are taken to prevent data leaks and unauthorised access, ensuring the security and reliability of the database. | Kababji et al., 86 Rehman et al., 87 Khalifeh et al. 88 | |
| Dataset and pattern classification | The ability to evaluate heterogeneous datasets with variability in both genetic and morphological appearance of suspicious breast cancer lessions, and accurately identify and logically classify the clusters separating clinical subtypes such as tumour category, architectural distortions, masses, and calcifications | Markey et al., 89 Tamayo et al. 90 | |
| Computer-aided diagnosis (CAD) | A cost-effective, accurate and non-invasive technique is required to assist in the diagnostic process by having suitable automated algorithms to process and analyse the data captured in the database. Key steps in the algorithm such as pre-processing, feature extraction, dimensionality reduction, feature selection and classification would help in the CAD. | Raghavendra et al. 91 |
However, the application value of big data in breast cancer can only be realised when the data is accessible. Some of the most well-known breast cancer databases include the Surveillance, Epidemiology, and End Results Program in the United States, the European Network for the Study of Adrenal Tumors database in Europe, and the Japan Breast Cancer Society (JBCS) database in Japan. These databases have played crucial roles in advancing our understanding of breast cancer and have improved patient care. It is worth noting that the database of JBCS is considered one of the oldest breast cancer record centres, with its history dating back to data collection in 1975, followed by digitalisation based on the internet in 2004, and operation on the National Clinical Database starting in 2012. 92
3. Online tools for breast cancer
With the development and advancement of internet technology, many online tools have been developed to assist doctors and patients in making breast cancer treatment decisions. PREDICT (https://predict.nhs.uk) is an online tool developed in the United Kingdom that evaluates the effectiveness of adjuvant therapy and patient prognosis after early breast cancer surgery using clinically relevant indicators such as clinical information, pathology and immunohistochemistry, 93 which are widely used in practice. The model can be freely accessed online, and it has been released in multiple languages and validated across various populations. 94 Adjutorium (https://vanderschaar-lab.com/adjutorium/) is another publicly accessible web decision support tool for breast cancer developed by a UK team, which assists early-stage breast cancer women in making adjuvant treatment decisions through machine learning algorithms. 95 The Breast Cancer Index (https://www.breastcancerindex.com/) is a tool developed in the United States primarily to predict the effectiveness of endocrine therapy. It can help predict the risk of recurrence within 5–10 years after diagnosis and whether there may be benefits from long-term (10-year) hormone therapy for early-stage hormone receptor-positive breast cancer. 96 ClinOmicsTrail (https://clinomicstrail.bioinf.uni-sb.de/) is an interactive visualisation analysis tool for stratified breast cancer treatment, which comprehensively evaluates various treatment options through multi-omics data, providing support for oncologists in the process of selecting breast cancer treatment decisions. 97 A Chinese research team has constructed a cell death index model (https://tnbc.shinyapps.io/CDI_Model/) containing 12 gene features using machine learning algorithms, which can predict the clinical prognosis and drug sensitivity after surgery for triple-negative breast cancer, aiding in selecting appropriate treatment plans for patients. 98 The myBeST tool (http://mybestpredict.com/) is a breast cancer model developed based on 13 feature variables capable of predicting the 5-year survival rate of Malaysian female breast cancer patients. 99 Additionally, YouTube video medium is also a potentially important tool for disseminating breast cancer health education knowledge and treatment information; its role cannot be ignored. 100
In summary, an increasing number of online tools play an essential role in breast cancer treatment decision-making. The application of these tools can provide powerful references and support for doctors and researchers, alleviate patients’ panic and anxiety, and assist in personalised care. However, it should be noted that due to significant individual differences among patients, there may be uncertainties or errors in the prediction results, and their implications must be interpreted cautiously. Therefore, in the future, it is necessary further to strengthen the accumulation and research of online tool data, optimise big data algorithms, update model variables promptly to improve prediction accuracy and precision and increase the dissemination of correct breast cancer prevention and treatment information to highlight its better practical value.
Towards precision treatment
1. Breast cancer: from data to insights and decision
Precision treatment for breast cancer, also known as personalised medicine, is a novel approach aimed at customising disease prevention and treatment strategies based on individual patient characteristics (including genetic, environmental, and lifestyle factors). 101 This transformation from the traditional ‘one-size-fits-all’ to a selective approach controlled by individual differences 102 enables healthcare models to provide more efficient and accurate care services for breast cancer patients. 103 Through large patient information and outcomes datasets, researchers can gain new insights into breast cancer and develop more efficient treatment strategies tailored to specific patient groups. 104
The main strategies of precision medicine include predicting disease risks based on subjects’ genetic information, identifying genes related to specific diseases and drug responses, and addressing technical issues in treatment based on patient's genetic and phenotypic information.105,106 However, these requirements rely on the intervention of big data technology, which opens up new possibilities for the successful implementation of precision medicine with its vast, diverse, and fast data processing capabilities. For cancer patients, the health history of cancer families and genetic variations have a significant impact on the prediction, diagnosis, and treatment of their risks. Therefore, the use of big data for intervention analysis becomes particularly critical. Precision treatment involves using advanced genomic technology to analyse the genomic composition of patients’ tumours. This can identify gene mutations that drive tumour growth and provide targeted treatment recommendations. For example, mutations in breast cancer susceptibility genes (BRCA1 and 2) can result in a lifetime risk of breast cancer as high as 45–87%. 106 In addition, other factors such as the patient's age, health condition, and medical history should also be considered. The importance of precision treatment for breast cancer lies in its potential to improve patient prognosis and reduce healthcare costs, bringing better results for patients and healthcare systems. 107 By analysing large datasets related to breast cancer, patients are stratified to determine which subgroups are more likely to respond to specific treatment methods to identify the most promising drugs and develop experimental protocols with a higher chance of success. Based on these results, doctors can promptly adjust treatment plans, select drugs with a strong targeted effect to reduce the risk of adverse reactions, 108 and also help improve the patient's recovery expectations.
A Phase II randomised clinical trial showed that for metastatic breast cancer, utilising targeted therapy through genomic analysis when its gene mutations are associated with anti-tumour activity in clinical trials can improve patient survival rates. 109 Another study indicated that precise diagnosis can be achieved by analysing a large number of breast cancer pathology reports and utilising machine learning models for deep learning, annotation, and image processing. 110 This method reduces the misdiagnosis rate and unnecessary biopsy rate for breast cancer, thus reducing unnecessary repeat examinations and medical costs for patients. Furthermore, a radiology diagnostic model has been developed based on multicentre datasets and clinical pathological risk factors, which successfully predicts the complete response of pathological features after neoadjuvant chemotherapy for breast cancer, providing essential guidance for personalised treatment. 111 These examples demonstrate that by collecting and analysing big data, we can gain a deeper understanding of the macroscopic and microscopic characteristics of breast cancer, thus providing more precise treatment choices for patients. This approach not only improves patient survival rates, reduces healthcare costs, and avoids ineffective treatments but also minimises hospitalisation time to the greatest extent, 109 enhances understanding of the underlying mechanisms of the disease, and develops more effective treatment strategies. Therefore, with the emergence of big data, the detection, treatment, and care of breast cancer have significantly improved, creating conditions for the realisation of precision medicine.
2. Precision treatment for breast cancer: diversity of data types
The precision treatment of breast cancer patients using big data technology requires gathering various types of data, including genetic information, clinical manifestations, lifestyle, treatment methods, and outcomes. 67 Obtaining comprehensive patient data, such as demographics, medical history, and treatment responses, is crucial for understanding individual differences and adjusting corresponding treatments. 112 Through in-depth research on genetic information, we can better understand the genetic susceptibility and variation that influence tumour growth. This will help identify specific mutations or biomarkers to guide targeted therapy better and predict treatment outcomes. 113 Clinical data is used for diagnosing and staging breast cancer, tracking patient responses to treatment, and further building predictive models to identify high-risk patients. Pathological data provides insights into tumour characteristics, aiding in the classification of breast cancer subtypes for personalised interventions. 114 With the progress of genome sequencing work, many new genes have been discovered and validated in diseases, laying the foundation for developing precision treatment plans. Since cancer diagnosis involves handling unbalanced data, considering data-level analysis techniques is crucial for accurate and reliable insights.
Integrating lifestyle data from breast cancer patients plays a crucial role in advancing precision medicine in the widespread use of big data. Precision medicine seeks to customise treatment plans based on each person's unique characteristics, and incorporating lifestyle data provides a critical dimension towards achieving this goal. Inherent lifestyle, such as dietary habits, physical activity, and environmental exposures, are crucial elements that influence breast cancer risk and progression.115,116 Personal inclinations, daily routines, and feedback on lifestyle interventions are all critical components in optimising treatment outcomes. Adjustments to lifestyle, such as family planning, have been shown to affect the cardiometabolic health of breast cancer survivors, further validating the potential for personalised modifications.117,118 Researchers have even proposed a personalised breast cancer risk prediction model that relies on lifestyle habits and health record features. 119 In precision medicine, focusing on patients as the core perspective and integrating lifestyle data can lead to a deeper understanding of the dynamic interactions among genetics, environment, and habits, providing a holistic view of patient health. Current research into cancer prevention lifestyle recommendations aims to understand the specific impact of these recommendations on breast cancer prevention, spanning the entire diagnostic process and emphasising a comprehensive strategy for personalised interventions. 120 In summary, lifestyle data is of significant importance in the precision treatment of breast cancer. Therefore, in breast cancer treatment, lifestyle data collection and application should be given due attention to lay a foundation for improving overall treatment efficacy and patient quality of life.
Collecting and analysing these diverse types of data not only deepens our understanding of breast cancer but also creates conditions for more efficient treatment methods tailored to specific patients. Collaborative efforts to integrate multi-omics, clinical, and lifestyle data are crucial for comprehensively understanding and formulating treatment strategies. Given the dynamic molecular patterns of breast cancer, optimising treatment techniques using multiple types of patient-specific data will help improve the effectiveness of precision medicine.
3. Precision treatment for breast cancer: impact and contributions of models
Owing to the continuous development of big data technology and the rapid advancements in AI technology, particularly the application of large language models (LLMs) and multimodal foundation models (such as deep learning networks), unprecedented prospects have emerged for the precision treatment of breast cancer. These modelling approaches can integrate and analyse diverse data comprehensively, enabling a more accurate assessment of patients’ treatment responses and formulating personalised treatment plans accordingly.
The application of LLMs and multimodal foundation models in precision medicine for breast cancer has received widespread attention. LLMs can process vast medical literature and clinical records to extract valuable information, thereby assisting clinical decision-making. For example, Deng et al. 121 compared ChatGPT-3.5, ChatGPT-4.0, and Claude2 in five critical areas of breast cancer clinical scenarios, finding that GPT-4.0 exhibited excellent performance, highlighting its potential in clinical applications. At the same time, Claude2 showed professional advantages in evaluation and diagnosis. This has positively impacted the demand for optimisation of LLMs tailored to specific domains, promoting the development of AI in breast cancer clinical settings. Changes in body composition have significant implications for the treatment and prognosis of breast cancer. Zhao et al. 122 developed a machine learning model to predict the risk of osteoporosis in breast cancer patients compared to healthy women, thereby contributing to the improvement of disease prevention and treatment strategies. Cancer-related cognitive impairment presents a severe clinical challenge for breast cancer patients. A study showed that by investigating the factors associated with cancer-related cognitive impairment in breast cancer patients, the first risk prediction model for this population was developed – the Scientific Symptom Model 2.0. This model can accurately identify breast cancer patients with cognitive impairment, providing strong evidence and strategies for the early detection, diagnosis, and intervention of such individuals. 123 These are significant manifestations of models in the precision medicine of breast cancer. Meanwhile, multimodal foundation models can integrate various data types, such as imaging, genomics, transcriptomics, and pathology, thereby enhancing the accuracy of breast cancer diagnosis and prognosis prediction. For example, Zhao et al. 124 developed a convolutional neural network model based on a digital pathology and deep learning algorithm framework using a large sample multi-omics cohort, which can predict molecular subtyping, molecular targets, and patient prognosis of triple-negative breast cancer from whole-slide digital pathology images. Wu et al. 125 proposed a multimodal deep-learning radiomics nomogram model designed to estimate the malignancy of breast cancer. The study showed that this model demonstrated exceptional predictive performance when validating multicentre data. Compared to traditional unimodal models, its diagnostic capabilities were significantly enhanced, showcasing great potential for clinical application. 125 Oh et al. 126 developed an LLM-driven multimodal AI model that integrates text and image data, showing significant advantages over traditional single-modal AI models in delineating target areas for breast cancer radiotherapy. Especially in scenarios with insufficient data and the need for external validation, its generalisation performance and data processing efficiency are well demonstrated. This implies that LLMs have broad application potential in integrating multimodal data and may provide strong support for personalised therapies for breast cancer patients.
One significant advantage of multimodal data fusion technology is its ability to capture the complex characteristics of breast cancer from multiple perspectives, thereby enhancing the accuracy of disease classification and prognosis prediction. For example, the multimodal deep neural network model proposed by Sun et al. 127 effectively improved the accuracy of breast cancer prognosis prediction by integrating multidimensional data, highlighting the tremendous potential of deep learning techniques in handling high-dimensional complex data. The patient-derived xenograft model and organoid platform for breast cancer developed by Guillen et al. 128 further demonstrate the application potential of multimodal data in precision medicine for breast cancer. This platform facilitates drug screening and provides suggestions for clinical care through in vivo validation results, which will help advance the clinical translation of precision medicine.
In summary, integrating big data in breast cancer with LLMs and multimodal foundation models provides powerful tools for realising precision medicine. These continuous technological innovations enhance the accuracy of breast cancer diagnosis and prognosis predictions and offer novel perspectives and methods for developing targeted treatment strategies. As technology continues to advance, how to better optimise and apply these models in clinical practice will be an essential research direction in the future.
Challenges and opportunities
With the rapid progress of big data technology and the continuous decrease in costs, an increasing amount of medical data is being accumulated and stored, contributing a massive amount of information and resources to breast cancer research. However, despite this, utilising this data for precision medicine still faces numerous challenges. Firstly, the completeness and quality of medical information are crucial factors determining the credibility of research outcomes. However, the current inconsistency and non-standardisation in data sources and formats directly hinder the efficient use of data. Furthermore, the application of big data technology is constrained by privacy, security, and ethical issues. Therefore, ensuring patients’ privacy and avoiding ethical violations while facilitating data sharing and communication becomes an urgent issue that needs to be addressed. 129 Additionally, the accuracy and consistency of big data are often determined by the quality of the data. In certain situations, erroneous decisions may be made, leading to issues with no clear regulations or guidelines to determine responsibility in cases of service failure or harm caused. 130 Finally, the exploration of breast cancer involves interdisciplinary research. Integrating professional skills and knowledge across various disciplines and comprehensively integrating multiple types of data to collectively advance the continuous development of precision medicine in breast cancer poses a significant challenge. With the development of LLMs, medical research has made significant progress regarding data integration and information processing. However, effectively utilising big data from breast cancer in precision medicine and integrating it with multimodal foundation models for in-depth analysis presents numerous challenges. First, the performance of the models relies on high-quality training datasets; however, medical literature and case reports related to breast cancer often lack uniform evaluation standards, which may weaken the generalisation performance of the models. Secondly, the ‘black box’ nature of LLMs makes it difficult for clinicians to understand and trust their predictions, which poses particular challenges in clinical applications. 131 Furthermore, the integration process of multimodal models faces issues such as computational complexity and resource consumption. Finally, ensuring that the various models can effectively collaborate during the integration process to avoid information loss and conflicts between models is also a pressing challenge that needs to be addressed. 132
Additionally, integrating germline and tumour genetics within breast cancer big data presents a crucial ethical challenge. The integration of germline and tumour genomic data requires in-depth analysis and interpretation on a technical level, as well as careful consideration from both legal and ethical perspectives. In the study of breast cancer, the big data involved often contains susceptible genomic information, which not only pertains to the patients themselves but may also impact the privacy rights of their family members. This issue becomes particularly critical when researching germline cells, as the transmission of genetic information can have potential implications for offspring. Therefore, before collecting and applying such data, it is essential to ensure that patients and their family members are fully informed and consent voluntarily. Researchers also need to balance data sharing and privacy protection to ensure the security and confidentiality of the research data, preventing data breaches and unauthorised use. 133 In integrating germline and tumour genetics research, it is crucial to ensure that patients fully understand the complex genetic information. However, patients often need help to fully grasp the implications of this information, which may lead to unequal information levels during the informed consent process. 134 In current ethical research, simplifying the information transfer process and ensuring patients make informed decisions based on a thorough understanding have become vital issues. 135
However, the opportunities brought by big data in precision medicine for breast cancer research cannot be ignored. With the advancement of wearable devices and remote monitoring technology, the effective collection and interpretation of real-time data provide valuable opportunities for doctors to promptly monitor the health status and treatment feedback of breast cancer patients, thus enabling timely changes in treatment strategies. By combining machine learning and AI technology and using advanced algorithms and computational models, future extensive data analysis will be more intelligent and automated. This will accelerate the identification of biomarkers and the development of new drugs, as well as optimise and design personalised treatment strategies. 136 Emphasising patient participation, interaction, and feedback is a goal of future big data platforms, allowing patients to contribute their data and participate in data quality improvement directly 137 and treatment decision-making processes. This practice not only enhances the transparency and satisfaction of the treatment process but also has the potential to optimise the final treatment outcomes.
Research limitations
Although big data plays a crucial role in advancing precision treatment for breast cancer, there are still some limitations in this study that are worth noting. First, the big data concerning breast cancer is sourced from multiple databases and large-scale datasets. The standardisation among these different data sources and the diversity in data collection methods may affect the reliability of the results and the generalisability of the research conclusions. Second, when processing and analysing large-scale medical data, data privacy and security are critical issues that cannot be overlooked. Data breaches may impact the development of big data. Third, although the big data contains many samples, it remains to be further validated whether these data can adequately represent the characteristics of breast cancer across different regions, ethnicities, and populations. Limitations in the samples may restrict the generalisability of the research findings. Fourth, precision treatment for breast cancer involves the interdisciplinary integration of bioinformatics, medicine, and computer information technology. More collaboration between disciplines and information barriers must be needed to ensure the translation and application of research findings. Fifth, breast cancer big data relies on machine learning and artificial intelligence algorithms for analysis. Therefore, the performance and accuracy of the algorithms will impact the accuracy of predictions and the reliability of clinical applications. Sixth, ethics and regulations are also important topics. While obtaining informed consent from patients regarding data use and sharing is possible, thus protecting their privacy and adhering to relevant laws and regulations, ethical and legal requirements may vary across different countries and regions, creating challenges for cross-border data use and international research collaboration. Despite these hindrances and impacts, these limitations will provide directions for improvement and motivation for the rapid development and research progress of big data in the future field of life and health.
Conclusion
The significant application of big data in the field of cancer has dramatically promoted our understanding of breast cancer. Researchers can identify complex patterns and molecular features related to breast cancer subtypes and treatment responses by comprehensively analysing large-scale datasets involving genetic, clinical, and environmental factors. This discovery provides an essential foundation for precise and personalised treatment strategies and opens up new research directions. Utilising multi-omics big data of breast cancer and developing unique analytical algorithms can help identify new biomarkers and potential therapeutic targets, develop targeted drugs, prioritise treatment effectiveness, and mitigate adverse effects on the body. Wearable medical devices and remote monitoring systems provide a continuous data feedback loop, enabling clinical physicians to track disease progression, monitor treatment effectiveness, and intervene immediately when relevant treatment complications arise. This trend further drives the application of precision medicine in practical healthcare, making it possible to tailor treatment plans according to each patient's characteristics.
However, before realising the full potential of these technologies, several challenges must be overcome, including data interoperability, privacy security, data standardisation, and equitable access to resources. Addressing these issues requires collaborative efforts from academia, industry, and regulatory bodies. The next focus should be on developing more powerful algorithms and analytical tools capable of integrating multiple data sources while managing vast and complex datasets. Additionally, the training and education of healthcare professionals must be enhanced and continuously updated to ensure they possess the necessary skills and knowledge to effectively integrate and interpret complex and diverse data.
In conclusion, with the continuous advancement of big data technology and innovation in research methods, the establishment of robust data management and sharing mechanisms, and strengthened communication and collaboration among clinical physicians, nurses, bioinformaticians, computer scientists, and policymakers, breast cancer precision medicine will be rejuvenated with new vitality, ushering in broader prospects for development and application.
Acknowledgment
The authors wish to express their sincere thanks to Universiti Sains Malaysia for the support provided through the Top-down Research University Grant Scheme (1001/CIPPT/8070033). This seed funding has been instrumental in advancing our work as part of the Breast Cancer Translational Research Program (BCTRP@IPPT).
Footnotes
Contributorship: HZ, HH, CCH, SHC, WKL, and BHY were responsible for drafting the manuscript. BHY and WKL reviewed and approved the final version of the manuscript, as well as its submission to the journal.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Hao Zhang https://orcid.org/0000-0002-0068-6132
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 2.Lei S, Zheng R, Zhang S, et al. Global patterns of breast cancer incidence and mortality: a population-based cancer registry data analysis from 2000 to 2020. Cancer Commun (Lond) 2021; 41: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman L. Oncologic anthropology: global variations in breast cancer risk, biology, and outcome. J Surg Oncol 2023; 128: 959–966. [DOI] [PubMed] [Google Scholar]
- 4.Autier P, Boniol M, La Vecchia C, et al. Disparities in breast cancer mortality trends between 30 European countries: retrospective trend analysis of WHO mortality database. Br Med J 2010; 341: c3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015; 385: 977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardoso F, Kyriakides S, Ohno S, et al. Early breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019; 30: 1194–1220. [DOI] [PubMed] [Google Scholar]
- 7.Berdzuli N. Breast cancer: from awareness to access. Br Med J 2023; 380: 290. [DOI] [PubMed] [Google Scholar]
- 8.Giaquinto AN, Sung H, Miller KD, et al. Breast cancer statistics, 2022. CA Cancer J Clin 2022; 72: 524–541. [DOI] [PubMed] [Google Scholar]
- 9.Giaquinto AN, Miller KD, Tossas KY, et al. Cancer statistics for African American/black people 2022. CA Cancer J Clin 2022; 72: 202–229. [DOI] [PubMed] [Google Scholar]
- 10.Sandhu JK, Kaur A, Kaushal C. Analysis of breast cancer in early stage by using machine learning algorithms: a review. In: 2022 IEEE international conference on current development in engineering and technology (CCET), 2022, pp. 1–7. [Google Scholar]
- 11.Han B, Zheng R, Zeng H, et al. Cancer incidence and mortality in China, 2022. J Natl Cancer Center 2024; 4: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedeta ET, Jobre B, Avezbakiyev B. Breast cancer: global patterns of incidence, mortality, and trends. J Clin Oncol 2023; 41: 10528. [Google Scholar]
- 13.Rhinehart D, Lozier J, Silvestri GA. Not just biology: comparing social determinants of health in patients diagnosed with late-stage lung, breast, and colon cancer. J Clin Oncol 2022; 40: e18582–e18582. [Google Scholar]
- 14.Abila DB, Kangoma G, Kisuza RK, et al. Coverage and socio-economic inequalities in breast cancer screening in low- and middle-income countries: analysis of demographic and health surveys between 2010 and 2019. JCO Glob Oncol 2022. May 5; 8: 59. [Google Scholar]
- 15.Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol 2022; 95: 20211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priyadarshini P, Sarath S, Hemavathy V. Breast cancer awareness package on knowledge, attitude and practice towards breast self examination to prevent breast cancer among women in adopted communities – a pilot analysis. Cardiometry 2022; 22: 471–483. [Google Scholar]
- 17.Kaur P, Sharma M, Mittal M. Big data and machine learning based secure healthcare framework. Procedia Comput Sci 2018; 132: 1049–1059. [Google Scholar]
- 18.Galetsi P, Katsaliaki K, Kumar S. Values, challenges and future directions of big data analytics in healthcare: a systematic review. Soc Sci Med 2019; 241: 112533. [DOI] [PubMed] [Google Scholar]
- 19.Queiroz MM, Pereira SCF, Telles R, et al. Industry 4.0 and digital supply chain capabilities. Benchmarking: Int J 2021; 28: 1761–1782. [Google Scholar]
- 20.Legner C, Eymann T, Hess T, et al. Digitalization: opportunity and challenge for the business and information systems engineering community. Bus Inf Syst Eng 2017; 59: 301–308. [Google Scholar]
- 21.Khanra S, Dhir A, Mäntymäki M. Big data analytics and enterprises: a bibliometric synthesis of the literature. Enterp Inf Syst 2020; 14: 737–768. [Google Scholar]
- 22.Gandomi A, Haider M. Beyond the hype: big data concepts, methods, and analytics. Int J Inf Manage 2015; 35: 137–144. [Google Scholar]
- 23.Debattista J, Lange C, Scerri S, et al. Linked ‘Big’ Data: towards a manifold increase in big data value and veracity. In: 2015 IEEE/ACM 2nd international symposium on big data computing (BDC), 2015, pp.92–98. [Google Scholar]
- 24.Kitchin R. The real-time city? Big data and smart urbanism. Geo J 2014; 79: 1–14. [Google Scholar]
- 25.Saggi MK, Jain S. A survey towards an integration of big data analytics to big insights for value-creation. Inf Process Manag 2018; 54: 758–790. [Google Scholar]
- 26.Awad A, Trenfield SJ, Pollard TD, et al. Connected healthcare: improving patient care using digital health technologies. Adv Drug Delivery Rev 2021; 178: 113958. [DOI] [PubMed] [Google Scholar]
- 27.Popov VV, Kudryavtseva EV, Kumar Katiyar N, et al. Industry 4.0 and digitalisation in healthcare. Materials (Basel) 2022; 15: 2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali F, El-Sappagh S, Islam SR, et al. An intelligent healthcare monitoring framework using wearable sensors and social networking data. Future Gener Comput Syst 2021; 114: 23–43. [Google Scholar]
- 29.Vayena E, Dzenowagis J, Brownstein JS, et al. Policy implications of big data in the health sector. Bull W H O 2018; 96: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rydning DR-JG-J, Reinsel J, Gantz J. The digitization of the world from edge to core. Framingham: Int Data Corp 2018; 16: 1–28. [Google Scholar]
- 31.Karthikeyan S, de Herrera AGS, Doctor F, et al. An ocr post-correction approach using deep learning for processing medical reports. IEEE Trans Circuits Syst Video Technol 2021; 32: 2574–2581. [Google Scholar]
- 32.de la Torre I, González S, López-Coronado M. Analysis of the EHR systems in Spanish primary public health system: the lack of interoperability. J Med Syst 2012; 36: 3273–3281. [DOI] [PubMed] [Google Scholar]
- 33.Cusack CM, Hripcsak G, Bloomrosen M, et al. The future state of clinical data capture and documentation: a report from AMIA’s 2011 policy meeting. J Am Med Inform Assoc 2013; 20: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asri H, Mousannif H, Al Moatassime H, et al. Big data in healthcare: challenges and opportunities. In: 2015 international conference on cloud technologies and applications (CloudTech), 2015, pp.1–7. [Google Scholar]
- 35.Thapa C, Camtepe S. Precision health data: requirements, challenges and existing techniques for data security and privacy. Comput Biol Med 2021; 129: 104130. [DOI] [PubMed] [Google Scholar]
- 36.Holden RJ, McDougald Scott AM, Hoonakker PL, et al. Data collection challenges in community settings: insights from two field studies of patients with chronic disease. Qual Life Res 2015; 24: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong N, Sun G, Zuo X, et al. Application of informatics in cancer research and clinical practice: opportunities and challenges. Cancer Innovation 2022; 1: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.IBM. Cost of a Data Breach Report by IBM. 2022, https://www.ibm.com/reports/data-breach.
- 39.Mehraeen E, Ghazisaeedi M, Farzi J, et al. Security challenges in healthcare cloud computing: a systematic. Glob J Health Sci 2017; 9: 157–168. [Google Scholar]
- 40.Thamer N, Alubady R. A survey of ransomware attacks for healthcare systems: risks, challenges, solutions and opportunity of research. In: 2021 1st Babylon international conference on information technology and science (BICITS), 2021, pp.210–216. [Google Scholar]
- 41.Olivier M, Asmis R, Hawkins GA, et al. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci 2019; 20: 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayega A, Wang YC, Oikonomopoulos S, et al. Transcript profiling using long-read sequencing technologies. Gene Expr Anal: Methods Protoc 2018; 1783: 121–147. [DOI] [PubMed] [Google Scholar]
- 43.Stark Z, Dolman L, Manolio TA, et al. Integrating genomics into healthcare: a global responsibility. Am J Hum Genet 2019; 104: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Service RF. Gene sequencing. The race for the $1000 genome. Science 2006; 311: 1544–1546. [DOI] [PubMed] [Google Scholar]
- 45.Consortium GP. A global reference for human genetic variation. Nature 2015; 526: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomczak K, Czerwińska P, Wiznerowicz M. Review the cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol/Współcz Onkol 2015; 2015: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma X, Fernández FM. Advances in mass spectrometry imaging for spatial cancer metabolomics. Mass Spectrom Rev 2024; 43: 235–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv Z, Hu J, Huang M, et al. Molecular mechanisms of cadmium-induced cytotoxicity in human ovarian granulosa cells identified using integrated omics. Ecotoxicol Environ Saf 2024; 272: 116026. [DOI] [PubMed] [Google Scholar]
- 49.Xu G, Huang M, Hu J, et al. Bisphenol A and its structural analogues exhibit differential potential to induce mitochondrial dysfunction and apoptosis in human granulosa cells. Food Chem Toxicol 2024; 188: 114713. [DOI] [PubMed] [Google Scholar]
- 50.Friedman AA, Letai A, Fisher DE, et al. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 2015; 15: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khansari N. AI machine learning improves personalized cancer therapies. Aust Med J (Online) 2024; 17: 1166–1173. [Google Scholar]
- 52.Low SK, Nakamura Y. The road map of cancer precision medicine with the innovation of advanced cancer detection technology and personalized immunotherapy. Jpn J Clin Oncol 2019; 49: 596–603. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Y, Qing T, Song Y, et al. Standardization efforts enabling next-generation sequencing and microarray based biomarkers for precision medicine. Biomark Med 2015; 9: 1265–1272. [DOI] [PubMed] [Google Scholar]
- 54.Werner RJ, Kelly AD, Issa JJ. Epigenetics and precision oncology. Cancer J 2017; 23: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Li J, Wang C, et al. Outlier detection using iterative adaptive mini-minimum spanning tree generation with applications on medical data. Front Physiol 2023; 14: 1233341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dash S, Shakyawar SK, Sharma M, et al. Big data in healthcare: management, analysis and future prospects. J Big Data 2019; 6: 1–25. [Google Scholar]
- 57.Newman L. Developing technologies for early detection of breast cancer: a public workshop summary. 2000.
- 58.Eltrass AS, Salama MS. Fully automated scheme for computer-aided detection and breast cancer diagnosis using digitised mammograms. IET Image Proc 2020; 14: 495–505. [Google Scholar]
- 59.Khan MQ, Hussain A, Rehman SU, et al. Classification of melanoma and nevus in digital images for diagnosis of skin cancer. IEEE Access 2019; 7: 90132–90144. [Google Scholar]
- 60.Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med 2005; 34: 113–127. [DOI] [PubMed] [Google Scholar]
- 61.Jung H, Shen C, Gonzalez Y, et al. Deep-learning assisted automatic digitization of interstitial needles in 3D CT image based high dose-rate brachytherapy of gynecological cancer. Phys Med Biol 2019; 64: 215003. [DOI] [PubMed] [Google Scholar]
- 62.Mohsen F, Al-Saadi B, Abdi N, et al. Artificial intelligence-based methods for precision cardiovascular medicine. J Pers Med 2023; 13: 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rezayi S SRNK, Saeedi S. Effectiveness of artificial intelligence for personalized medicine in neoplasms: a systematic review. Biomed Res Int 2022; 2022: 7842566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westerlund AM, Hawe JS, Heinig M, et al. Risk prediction of cardiovascular events by exploration of molecular data with explainable artificial intelligence. Int J Mol Sci 2021; 22: 10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong WJ, Wu SH, Cheng MQ, et al. Integration of artificial intelligence decision aids to reduce workload and enhance efficiency in thyroid nodule management. JAMA Netw Open 2023; 6: e2313674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang P, Sinha S, Aldape K, et al. Big data in basic and translational cancer research. Nat Rev Cancer 2022; 22: 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabiei R, Ayyoubzadeh SM, Sohrabei S, et al. Prediction of breast cancer using machine learning approaches. J Biomed Phys Eng 2022; 12: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue XL, Zhao S, Xu MC, et al. Circular RNA_0000326 accelerates breast cancer development via modulation of the miR-9-3p/YAP1 axis. Neoplasma 2023; 70: 430–442. [DOI] [PubMed] [Google Scholar]
- 69.Jiang ZR, Yang LH, Jin LZ, et al. Identification of novel cuproptosis-related lncRNA signatures to predict the prognosis and immune microenvironment of breast cancer patients. Front Oncol 2022; 12: 988680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parikh RB, Gdowski A, Patt DA, et al. Using big data and predictive analytics to determine patient risk in oncology. Am Soc Clin Oncol Educ Book 2019; 39: e53–e58. [DOI] [PubMed] [Google Scholar]
- 71.Zhang P, Brusic V. Mathematical modeling for novel cancer drug discovery and development. Expert Opin Drug Discovery 2014; 9: 1133–1150. [DOI] [PubMed] [Google Scholar]
- 72.Blamey R, Hornmark-Stenstam B, Ball G, et al. ONCOPOOL–A European database for 16,944 cases of breast cancer. Eur J Cancer 2010; 46: 56–71. [DOI] [PubMed] [Google Scholar]
- 73.Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 2012; 131: 765–775. [DOI] [PubMed] [Google Scholar]
- 74.Wadsten C, Wennstig A-K, Garmo H, et al. Data resource profile: breast cancer data base Sweden 2.0 (BCBaSe 2.0). Int J Epidemiol 2021; 50: 1770–1771f. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA). Methods Mol Biol 2016; 1418: 111–141. [DOI] [PubMed] [Google Scholar]
- 76.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pereira B, Chin S-F, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016; 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Kung L, Ting C, et al. Beyond a technical perspective: understanding big data capabilities in health care. In: 2015 48th Hawaii international conference on system sciences, 2015, pp.3044–3053. [Google Scholar]
- 79.Ta V-D, Liu C-M, Nkabinde GW. Big data stream computing in healthcare real-time analytics. In: 2016 IEEE international conference on cloud computing and big data analysis (ICCCBDA), 2016, pp.37–42. [Google Scholar]
- 80.Thompson CA, Kurian AW, Luft HS. Linking electronic health records to better understand breast cancer patient pathways within and between two health systems. eGEMs 2015; 3: 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alford SH, Mahatma R, Amode S, et al. Abstract P6-12-03: development of technology for bi-directional, tailored support of the relationship between breast cancer patients and their providers. Cancer Res 2020; 80: P6-12-03. [Google Scholar]
- 82.Anklesaria S, Maheshwari U, Lele R, et al. Breast cancer prediction using optimized machine learning classifiers and data balancing techniques. In: 2022 6th international conference on computing, communication, control and automation (ICCUBEA), 2022, pp.1–7. [Google Scholar]
- 83.Tsangaris E, Edelen M, Means J, et al. User-centered design and agile development of a novel mobile health application and clinician dashboard to support the collection and reporting of patient-reported outcomes for breast cancer care. BMJ Surg Interv Health Technol 2022; 4: e000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yi M, Hunt KK. Organizing a breast cancer database: data management. Chin Clin Oncol 2016; 5: 45. [DOI] [PubMed] [Google Scholar]
- 85.Han Y, Zhang Y, Vermund SH. Blockchain technology for electronic health records. Int J Environ Res Public Health 2022; 19: 15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kababji S E, Mitsakakis N, Fang X, et al. Evaluating the utility and privacy of synthetic breast cancer clinical trial data sets. JCO Clin Cancer Inform 2023; 7: e2300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rehman MU, Shafique A, Ghadi YY, et al. A novel chaos-based privacy-preserving deep learning model for cancer diagnosis. IEEE Trans Netw Sci Eng 2022; 9: 4322–4337. [Google Scholar]
- 88.Khalifeh S, Georgi J, Shakhatreh S. Design and implementation of a steganography-based system that provides protection for breast cancer patient’s data. In: 2022 56th annual conference on information sciences and systems (CISS), 2022, pp.19–24. [Google Scholar]
- 89.Markey MK, Lo JY, Tourassi GD, et al. Self-organizing map for cluster analysis of a breast cancer database. Artif Intell Med 2003; 27: 113–127. [DOI] [PubMed] [Google Scholar]
- 90.Tamayo P, Slonim D, Mesirov J, et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 1999; 96: 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raghavendra U, Gudigar A, Rao TN, et al. Computer-aided diagnosis for the identification of breast cancer using thermogram images: a comprehensive review. Infrared Phys Technol 2019; 102: 103041. [Google Scholar]
- 92.Tokuda Y. The Japanese breast cancer society breast cancer registry in the national clinical database: current status and future perspectives. Nihon Geka Gakkai Zasshi 2014; 115: 17–21. [PubMed] [Google Scholar]
- 93.Wishart GC, Azzato EM, Greenberg DC, et al. PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res 2010; 12: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Candido dos Reis FJ, Wishart GC, Dicks EM, et al. An updated PREDICT breast cancer prognostication and treatment benefit prediction model with independent validation. Breast Cancer Res 2017; 19: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alaa AM, Gurdasani D, Harris AL, et al. Machine learning to guide the use of adjuvant therapies for breast cancer. Nat Mach Intell 2021; 3: 716–726. [Google Scholar]
- 96.Bartlett J, Sgroi D, Treuner K, et al. Breast cancer index and prediction of benefit from extended endocrine therapy in breast cancer patients treated in the adjuvant tamoxifen–to offer more? (aTTom) trial. Ann Oncol 2019; 30: 1776–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schneider L, Kehl T, Thedinga K, et al. Clinomicstrailbc: a visual analytics tool for breast cancer treatment stratification. Bioinformatics 2019; 35: 5171–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zou Y, Xie J, Zheng S, et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int J Surg 2022; 107: 106936. [DOI] [PubMed] [Google Scholar]
- 99.Nik Ab Kadir MN, Mohd Hairon S, Ab Hadi IS, et al. A comparison between the online prognostic tool PREDICT and myBeST for women with breast cancer in Malaysia. Cancers (Basel) 2023; 15: 2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chai BS, Ingledew P-A. Characteristics assessment of online YouTube videos on radiotherapy for breast cancer. Clin Breast Cancer 2023; 23: e230–e238. [DOI] [PubMed] [Google Scholar]
- 101.König IR, Fuchs O, Hansen G, et al. What is precision medicine? Eur Respir J 2017; 50: 1700391. [DOI] [PubMed] [Google Scholar]
- 102.Beckmann JS, Lew D. Reconciling evidence-based medicine and precision medicine in the era of big data: challenges and opportunities. Genome Med 2016; 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duan X-P, Qin B-D, Jiao X-D, et al. New clinical trial design in precision medicine: discovery, development and direction. Signal Transduct Target Ther 2024; 9: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bettaieb A, Paul C, Plenchette S, et al. Precision medicine in breast cancer: reality or utopia? J Transl Med 2017; 15: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenomics: challenges and opportunities. Ann Intern Med 2006; 145: 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pinker K, Chin J, Melsaether AN, et al. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology 2018; 287: 732–747. [DOI] [PubMed] [Google Scholar]
- 107.Sarhangi N, Hajjari S, Heydari SF, et al. Breast cancer in the era of precision medicine. Mol Biol Rep 2022; 49: 10023–10037. [DOI] [PubMed] [Google Scholar]
- 108.Greenwalt I, Zaza N, Das S, et al. Precision medicine and targeted therapies in breast cancer. Surg Oncol Clin 2020; 29: 51–62. [DOI] [PubMed] [Google Scholar]
- 109.Andre F, Filleron T, Kamal M, et al. Genomics to select treatment for patients with metastatic breast cancer. Nature 2022; 610: 343–348. [DOI] [PubMed] [Google Scholar]
- 110.Panahiazar M, Chen N, Lituiev D, et al. Empowering study of breast cancer data with application of artificial intelligence technology: promises, challenges, and use cases. Clin Exp Metastasis 2022; 39: 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res 2019; 25: 3538–3547. [DOI] [PubMed] [Google Scholar]
- 112.Dlamini Z, Francies FZ, Hull R, et al. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput Struct Biotechnol J 2020; 18: 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harris EER. Precision medicine for breast cancer: the paths to truly individualized diagnosis and treatment. Int J Breast Cancer 2018; 2018: 4809183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fotouhi S, Asadi S, Kattan MW. A comprehensive data level analysis for cancer diagnosis on imbalanced data. J Biomed Inform 2019; 90: 103089. [DOI] [PubMed] [Google Scholar]
- 115.Manni A, El-Bayoumy K. Lifestyle modifications and breast cancer risk. Cancers (Basel) 2023; 15: 2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lofterød T, Frydenberg H, Flote V, et al. Exploring the effects of lifestyle on breast cancer risk, age at diagnosis, and survival: the EBBA-life study. Breast Cancer Res Treat 2020; 182: 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Natalucci V, Marini CF, Lucertini F, et al. Effect of a lifestyle intervention program’s on breast cancer survivors’ cardiometabolic health: two-year follow-up. Heliyon 2023; 9: e21761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baldelli G, Natalucci V, Ferri Marini C, et al. A home-based lifestyle intervention program reduces the tumorigenic potential of triple-negative breast cancer cells. Sci Rep 2024; 14: 2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qi S-a, Kumar N, Xu J-Y, et al. Personalized breast cancer onset prediction from lifestyle and health history information. PLoS One 2022; 17: e0279174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cannioto RA, Attwood KM, Davis EW, et al. Adherence to cancer prevention lifestyle recommendations before, during, and 2 years after treatment for high-risk breast cancer. JAMA Network Open 2023; 6: e2311673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deng L, Wang T, Yangzhang, et al. Evaluation of large language models in breast cancer clinical scenarios: a comparative analysis based on ChatGPT-3.5, ChatGPT-4.0, and Claude2. Int J Surg 2024; 110: 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhao F, Li C, Wang W, et al. Machine learning predicts the risk of osteoporosis in patients with breast cancer and healthy women. J Cancer Res Clin Oncol 2024; 150: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou Z, Ren J, Liu Q, et al. A nomogram for predicting the risk of cancer-related cognitive impairment in breast cancer patients based on a scientific symptom model. Sci Rep 2024; 14: 14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao S, Yan CY, Lv H, et al. Deep learning framework for comprehensive molecular and prognostic stratifications of triple-negative breast cancer. Fundam Res 2024; 4: 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu P, Jiang Y, Xing H, et al. Multimodality deep learning radiomics nomogram for preoperative prediction of malignancy of breast cancer: a multicenter study. Phys Med Biol 2023; 68: 175023.. [DOI] [PubMed] [Google Scholar]
- 126.Oh Y, Park S, Byun HK, et al. LLM-driven multimodal target volume contouring in radiation oncology. arXiv preprint arXiv: 2311.01908 2023. [DOI] [PMC free article] [PubMed]
- 127.Sun D, Wang M, Li A. A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans Comput Biol Bioinf 2018; 16: 841–850. [DOI] [PubMed] [Google Scholar]
- 128.Guillen KP, Fujita M, Butterfield AJ, et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer 2022; 3: 232–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jourquin J, Reffey SB, Jernigan C, et al. Susan G. Komen big data for breast cancer initiative: how patient advocacy organizations can facilitate using big data to improve patient outcomes. JCO Precis Oncol 2019; 3: –9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sebastian AM, Peter D. Artificial intelligence in cancer research: trends, challenges and future directions. Life 2022; 12: 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Luo Z, Xu C, Zhao P, et al. Augmented large language models with parametric knowledge guiding. arXiv preprint arXiv: 2305.04757 2023.
- 132.Morales JC, Carrillo-Perez F, Castillo-Secilla D, et al. Enhancing breast cancer classification via information and multi-model integration. In: Bioinformatics and biomedical engineering: 8th international work-conference, IWBBIO 2020, Granada, Spain, May 6–8, 2020, Proceedings 8, 2020, pp.750–760. [Google Scholar]
- 133.Peppercorn J, Shapira I, Deshields T, et al. Ethical aspects of participation in the database of genotypes and phenotypes of the National Center for Biotechnology Information: the Cancer and Leukemia Group B Experience. Cancer 2012; 118: 5060–5068. [DOI] [PubMed] [Google Scholar]
- 134.Vähäkangas K. Ethical aspects of molecular epidemiology of cancer. Carcinogenesis 2004; 25: 465–471. [DOI] [PubMed] [Google Scholar]
- 135.Lolkema MP, Gadellaa-van Hooijdonk CG, Bredenoord AL, et al. Ethical, legal, and counseling challenges surrounding the return of genetic results in oncology. J Clin Oncol 2013; 31: 1842–1848. [DOI] [PubMed] [Google Scholar]
- 136.Ozer ME, Sarica PO, Arga KY. New machine learning applications to accelerate personalized medicine in breast cancer: rise of the support vector machines. Omics: J Integr Biol 2020; 24: 241–246. [DOI] [PubMed] [Google Scholar]
- 137.Petersen C. The future of patient engagement in the governance of shared data. eGEMs 2016; 4: 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]