ABSTRACT
Dengue virus (DENV) has been expanding its range to temperate areas that are not usually affected, where the spread of vectors has been facilitated by global trade and climate change. In Europe, there have been many cases of DENV imported from other regions in the past few years, leading to local outbreaks of DENV among people without travel history. Here we describe the epidemiological and molecular investigations of three transmission events locally acquired DENV infections caused by serotypes 1, 2 and 3, respectively, in the Latium Region from August to November 2023. Next-generation or Sanger sequencing was used to obtain the whole genomes, or the complete E-gene of the viruses, respectively. The structure of the DENV-1 and DENV-3 sequences was analysed to identify amino acid changes that were not found in the closest related sequences. The major cluster was supported by DENV-1 (originated in South America), with 42 autochthonous infections almost occurring in the eastern area of Rome, probably due to a single introduction followed by local sustained transmission. Seven DENV-1 subclusters have been identified by mutational and phylogenetic analysis. Structural analysis indicated changes whose meaning can be explained by the adaptation of the virus to human hosts and vectors and their interactions with antibodies and cell receptors.
KEYWORDS: Dengue, arbovirus, genomic surveillance, whole-genome sequencing, molecular epidemiology
Introduction
Dengue is an infection caused by the dengue virus (DENV), which is carried and spread by Aedes spp mosquitoes. It affects more than 100 countries in tropical and subtropical regions (Southeast Asia, Africa, the West Pacific and the Americas) [1]. The DENV burden is increasing globally in parallel with the expansion of – Aedes spp mosquitoes in new areas [1,2]. These dynamics, combined with the increased mobility of people and goods, increased the risk of imported cases in non-endemic areas and the following local spread and autochthonous outbreaks [3–7]. DENV is a single-stranded RNA virus that belongs to the Orthoflavivirus genus. It has four different types (DENV-1, DENV-2, DENV-3 and DENV-4) that are related but not identical: the types share about two-thirds of their genome (65-70% similarity in amino acid sequence) [1,8,9]. The infection has different clinical presentations: most people could be asymptomatic or pauci-symptomatic; symptoms include mild fever, severe headache, retro-orbital pain, myalgia and arthralgia, nausea, and rash. Severe presentation, occurring in a minority of cases, may include abdominal pain or tenderness, vomiting, fluid accumulation, bleeding, tiredness or restlessness and liver enlargement [1]. The main risk factor for severe clinical evolution is secondary infection with a serotype different from the one that caused the first infection through antibody-dependent enhancement, which can lead to severe dengue [1,9,10]. DENV spreads mainly through the bite of a female mosquito from the Aedes spp. genus; the most common one is Aedes aegypti, but Aedes albopictus can also carry the virus [11]. The latter is found in some European countries, including Italy [2,12]. Since the presence and movement of DENV depend on the presence and movement of the mosquitoes and, therefore, on the weather conditions (e.g. temperature, rainfall and humidity [13]), climate change may have helped the virus spread more widely and cause more infections. Specifically, temperature and precipitation are vital in the dynamics of mosquito populations and significantly impact virus transmission by providing water habitats for larvae and pupae. Consequently, the rise in global average temperatures and increased humidity have resulted in longer lifespans for adult mosquitoes, a reduced viral incubation period within vectors and more frequent blood-feeding intervals, leading to quicker replication and enhanced fitness of DENV. The relationship between DENV infections and weather conditions varies by location and socio-economic context: infection is endemic in countries where a positive correlation between DENV prevalence and rainfall has been consistently observed. Conversely, dry conditions in urban areas might pose an epidemic risk due to water stored in unprotected reservoirs attracting Ae. Egypti vectors [14]. DENV poses significant economic and social challenges in affected regions. Dengue poses a significant economic burden, with global costs estimated at around 306 billion international dollars between 2020 and 2050 [15]. This burden is especially harsh in lower-middle-income countries with struggling healthcare systems. Socially, dengue outbreaks can overwhelm medical facilities, disrupt daily life and cause substantial morbidity and mortality, particularly among vulnerable groups. These points highlight the urgent need for effective prevention and control measures to reduce the impact of dengue.
In fact, from 2000 to 2019, the number of reported cases increased by 10 times (from 500,000 to 5.2 million), according to the World Health Organization. A record high was seen in 2019, with cases reported from 129 countries around the world. After a decrease in dengue cases during the COVID-19 pandemic, new peaks were seen in 2023 and in the first months of 2024 with numerous outbreaks, even in areas that did not have DENV before [16]. In this situation, rapid and correct diagnosis, and monitoring of possible cases (both imported and autochthonous) are becoming increasingly important. In Italy, the management of DENV infection is guided by the five-year National Plan for Prevention, Surveillance and Response to Arboviruses (PNA 2020-2025) [17], with the main goal of finding cases fast and accurately to get more information about the epidemic situation in terms of contact tracing and mosquito control. Genomic monitoring combined with epidemiological analysis is a useful way to understand how DENV spreads and moves in a specific area, to get more details about the epidemic, and to determine the extent of local transmission to inform public health actions. Here we report, from an epidemiological and genetic perspective, outbreaks of local DENV that happened in the Lazio region (central Italy) in the period from August to November 2023. We obtained whole-genome sequences (WGS) and complete Envelope (E) gene sequences for mutation, phylogenetic and structural analysis to find out the transmission links and define the adaptive changes in the studied clusters. E proteins are essential for multiple steps of infection and are the outermost proteins in flaviviruses virion [18]. They contain motifs that are essential for cellular binding and are directly related to the infection process of the flavivirus. Identification of E variants in autochthonous cases can be functional to further studies on viral adaptations to local conditions, such as vectors and climate, and to correlate structural envelope modifications with vaccine efficacy.
Materials and methods
Data collection
The DENV case definition was based on the guidelines of the National Health System [17] and the Regional Surveillance Plan for Arboviruses. Based on the clinical and epidemiological criteria, any patient with suspicion of dengue infection should be reported to the Regional Service for the Epidemiology, Surveillance, and Control of Infectious Diseases (SeRESMI) – Lazio region within 24 h, and blood samples should be sent to the Regional Reference Laboratory (RRL) at the Virology Laboratory of the Lazzaro Spallanzani National Institute for Infectious Diseases (INMI) in Rome. Each case had demographic, clinical and epidemiological data collected.
Virological investigations
DENV detection and sequencing
At the RRL, DENV infection was diagnosed by initially screening with a rapid immunochromatographic test (DENGUE Duo Standard Q™, SD biosensor), which detects NS1 antigen and IgM and IgG antibodies against NS1 at the same time. Even in the case of a negative screening test, the diagnostic protocol includes the execution of two molecular tests and a serological immunofluorescence test. Nucleic acid extraction for molecular analysis was done by the QIAsymphony™ automatic extractor using the DSP Virus/Pathogen Kit (QIAGEN, Hilden, Germany), and a specific real-time PCR was used to identify the four DENV serotypes using the method previously described by Santiago et al. [19]. In addition, all samples were tested with a home-made Pan-Flavivirus nested PCR targeting the NS5 gene. Positive PCR samples were sequenced by WGS or complete E gene sequencing, depending on the viral load of the available samples. Samples exhibiting a high viral load (Ct < 27) were processed using NGS to acquire Whole Genome sequences, whereas samples with a lower viral load (Ct range: 27-32) underwent the Sanger protocol to obtain the full E gene sequence. Further details about primers/probes used in specific real-time applications by Santiago et al., as well as the E gene sequencing amplification protocol, are available in supplementary file 1. WGS was done by the amplicon-based method using two serotype-specific primer pools, according to Su et al. [20]. Libraries were then made from 10 to 100 ng of DNA using the Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instructions. Sequencing was done on the Gene Studio S5 Prime Sequencer (Thermo Fisher Scientific) to get about 5 × 105 reads per sample. A custom-made PCR was developed to obtain the full-length E gene of the DENV genome for Sanger sequencing. The method was based on a nested RT–PCR with overlapping amplicons of the second round: the SuperScript III Platinum one-step qRT-PCR System (Thermo Fisher Scientific) and DreamTaq Hot Start PCR Master Mix (Thermo Fisher Scientific) were used for the amplification of the first and second rounds, respectively.
Sequence reconstruction and phylogenetic analysis
A custom Bash script was used to reconstruct DENV genomes. Raw reads were compared to four reference sequences for each DENV serotype from NCBI RefSeq (accession numbers: NC_001477, NC_001474, NC_001475 and NC_002640). The reference with the highest coverage was selected, and the reads were mapped to it using BWAsoftware (version 0.7.17). Mapped reads were cut at primer positions, and consensus calling was done using samtools (version 1.15.1). Then, variant analysis was performed to find genetic variations in the reconstructed genomes. Consensus sequences were aligned with the chosen reference using MAFFT algorithm (version 7.427) and consensus variants were identified using snp-sites (version 2.5.1), which enabled the detection of nucleotide mutations and other genetic changes. The snpEff tool (version 5.2) was used to annotate the variants and distinguish between synonymous and non-synonymous mutations, providing information on the functional impact of genetic variations in the DENV genomes. Consensus sequences from autochthonous and imported cases of DENV-1 and DENV-3, as well as representative complete genomic sequences from NCBI and GISAID nucleotide collections, were selected for further analysis. This dataset includes all DENV-1 and DENV-3 genomic sequences collected worldwide so far, including in Italy. GISAID and NCBI databases were searched to find the most similar DENV genomes to INMI patients’ sequences, and their mutational profiles. Multiple sequence alignment was done with MAFFT (version 7.271). A preliminar phylogenetic analysis was conducted to find most similar sequences and those sequences representative worldwide for country and collection date. Final phylogenetic tree was then constructed using INMI whole genomes and E complete gene sequences with IQ-TREE software (version 1.6.12): transition model with empirical base frequencies and invariable sites was chosen with ModelFinder approach, and the best tree was obtained after performing 5000 bootstrap ultrafast replicates. Bootstrap nodes with support values above 80 were selected and marked. Nucleotide and amino acid mutations of the E gene in INMI patients sequences compared to the most similar DENV genomes were detected and recorded with a homemade Python script.
Quality control
Quality control checks ensured accurate and reliable results. For molecular diagnostics, nucleic acid extraction was done with internal controls, and positive and negative controls validated real-time PCR assay performance to avoid contamination. NGS sequence quality was evaluated using metrics such as read depth (minimum 50X) and genome coverage, with further sequencing or sample exclusion for those not meeting the standards. Variant calling accuracy was confirmed by inspecting BAM files. In Sanger sequencing, overlapping amplicons were generated and checked for alignment to correctly assemble full-length E gene sequences, resolving ambiguous base calls by re-sequencing relevant regions. Sequences from NGS and Sanger methods were compared for reliability, and phylogenetic analyses used well-established models with high bootstrap support to ensure tree reliability.
Structural models
DENV-1 (from INMI-A19 and INMI-A11 patients), DENV-3 (from INMI-A15/16 patients), and reference sequences (for proteins M and E) were aligned using the Clustal Omega web server [21]. The 3D protein structures embedded in the membrane were constructed by homology modelling using the web tool Swiss-model [22]. The templates were selected based on the following criteria: highest identity, highest coverage, and highest crystal resolution. Based on these criteria, the template used for modeling the proteins was the cryoEM tetramer with the pdb entry 3J2P [23]. The protein models were protonated at pH = 7 and inserted in a lipid bilayer to model the membrane structure. The membrane orientation was done as described in the OPM database [24]. The lipid bilayer membrane was modelled with the CHARMM-GUI web server [25] using the following membrane composition reported in the literature [26]: palmitoyl-oleoyl phosphatidylcholine (POPC) 60%, palmitoyl-oleoyl phosphatidylethanolamine (POPE) 30% and palmitoyl-oleoyl phosphatidylserine (POPS) 10%. The system was solvated with water, using the TIP3P model [27], and neutralized properly with the “gmx genion” GROMACS tool [28]. A full protein viral envelope model was also built, based on the 3J27 cryoEM structure [23]. This model was generated by the replication of symmetry-related copies of the hexamer, for a total of 360 E and M proteins. It was performed by Chimera [29] sym command, applying the BIOMT matrices from the original pdb file.
Statistical analysis
For the most frequently found serotype, the likely infection sites for each sequenced patient were determined. All cases that were spatially related within a common area of one square kilometre were identified and represented on a geographical map using PowerBI software.
Ethics statement
Because of Regulation (EU) 2016/679, we did not get the participants’ informed consent as there was a public health emergency during an infectious disease epidemic (art. 54: processing special categories of personal data may be needed for public health reasons without consent of the data subject). The manuscript contains data obtained during the epidemiological investigation as part of the institutional duties of the SeRESMI and Lazio Regional Health Authorities (LRHA), with the aim of defining the outbreak and appling appropriate control measures. For the same reasons, we did not need approval from the INMI Institutional Review Board. The interventions on the patients were carried out only on the basis of their conditions, and not for study purposes.
Results
Epidemiological description of DENV outbreaks
On August 18, 2023, SeRESMI received a report of a possible DENV case with a fever and maculopapular rash. The case tested positive for anti-DENV IgM and IgG, and a serum sample was sent to the RRL, where RT–PCR confirmed DENV-1. The case had no history of travel, contact with travellers, or other potential exposures, and was considered autochthonous. On August 22, the LRHA issued an alert note to all hospital emergency departments, healthcare facilities, and general practitioners in the Lazio region to advise them to include dengue fever in the differential diagnosis of patients who presented with acute fever syndrome and skin rashes, arthralgias, or myalgias, even if they had not travelled to endemic countries [3]. After the detection of the first confirmed autochthonous case, biological samples of all suspected cases were sent to the RRL for immediate diagnosis using rapid tests and additional serological and molecular analyses.
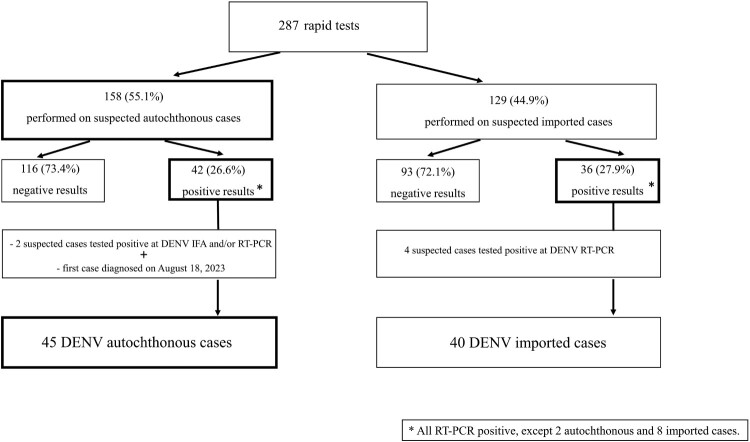
As shown in Figure 1, a total of 287 rapid tests for DENV infection were performed, with 78 (27.1%) tests resulting in positive. Six additional cases were diagnosed when directly tested by immunofluorescence and/or RT–PCR. Of the total of 85 positive cases (including the first diagnosed case), 40 were diagnosed as imported and 45 as autochthonous from August 18, 2023, to November 30, 2023. Of the 45 autochthonous cases, 41 (91.1%) were eventually classified as confirmed cases, while 4 (8.9%) met the criteria for probable DENV cases.
Figure 1.
Algorithm for suspected DENV cases diagnosed in the Lazio region, Italy, from 23 August 2023–30 November 2023, following the first diagnosed autochthonous case.
The investigation among the 45 autochthonous cases showed two outbreaks from local spread and one episode of family spread from an imported case. The largest outbreak, sustained by DENV-1, affected 42 people in or near the central urban area of Rome and its suburbs. Four were probable cases. The first case had symptoms on August 2, and the last one had symptoms on November 7, 2023.
The second outbreak, sustained by DENV-3, affected two people who visited the province of Latina (60 km south of Rome) in the 15 days before having symptoms on August 31, 2023. They did not travel to Rome or abroad recently and tested positive for DENV NS1 antigen rapid test; RT–PCR found DENV-3.
The third episode, sustained by DENV-2, affected a person who had symptoms on September 12, 2023, and tested positive for DENV NS1 antigen, IgM and IgG on September 20, 2023; RT–PCR found DENV-2. The investigation found that the case lived with a symptomatic case, who came back from an endemic country, and tested positive for IgG on August 31, 2023; by retesting the sample from this possible index case, NS1 antigen was found, and RT–PCR identified DENV-2 (Epidemic curve of autochthonous DENV cases by date of onset of symptoms in Lazio region is displayed in Supplementary Figure 2).
Table 1 shows the epidemiological and clinical features of the 45 autochthonous probable/confirmed cases and the 57 imported cases reported in Latium from January 1, 2023 to December 31, 2023. The median age of autochthonous and imported cases differed significantly (50 vs 32 years: t = −5.247, P < 0.0001). Regarding clinical presentation and evolution, autochthonous and imported cases differed significantly in the presence of skin rashes (71% vs. 35%; OR: 4.55, 95% C.I. 1.96–10.56, P = 0.0004) and in the hospitalization rate (51% vs. 72%; OR: 0.40, 95% C.I. 0.18-0.93; P = 0.00325). All cases were fully recovered. The median delay in diagnosis in days from symptoms onset to DENV lab testing was 7 days for autochthonous cases and 5 for imported ones (t = −2.774; P = 0.0066). Only one of the autochthonous cases was epidemiologically linked to a confirmed DENV case, imported from Indonesia. There were no differences observed in demographic characteristics or clinical presentation among different DENV serotypes.
Table 1.
Characteristics of the 102 DENV cases reported in Lazio, January–December 2023.
| Characteristics | Autochthonous | Imported |
|---|---|---|
| Female, n (%) | 27 (60) | 31 (54%) |
| Age, years; median (IQR) | 50 (37-68) | 32 (26-42) |
| Onset symptoms, n (%) | ||
| Fever | 44 (98) | 57 (100) |
| Arthralgia | 31 (69) | 35 (61) |
| Rash | 32 (71) | 20 (35) |
| Asthenia/headache | 35 (78) | 41 (72) |
| Nausea/vomiting | 15 (33) | 21 (37) |
| Myalgia | 20 (44) | 27 (47) |
| Retro-orbital pain | 11 (24) | 19 (33) |
| Conjunctivitis | 3 (7) | 1 (2) |
| Days from symptoms’ onset to lab testing; median (IQR) | 7 (6-9) | 5 (3-6) |
| Hospitalization, n (%) | 23 (51) | 41 (72) |
| Laboratory results | ||
| N. real-time PCR positive (%) | 39 (87) | 53 (93) |
| N. NS-1 Antigen positive (%) | 34 (76) | 49 (86) |
| N. IgM positive (%) | 40 (89) | 28 (49) |
Among the 57 cases of DENV classified as imported, 28 (48%) cases resulted to be imported by a South-East Asian country, 16 (28%) cases reported a recent travel to a Caribbean country, 7 (12%) to a South American and 6 (11%) to a Sub-Saharan African country. Most cases were sustained by DENV-3 (25; 43,9%), followed by DENV-2 (19; 33,3%) and DENV-1 (9; 15,8%). For 4 cases (7%) the viral serotype was not available.
Genetic characterization and phylogenesis analyses
We obtained genomic sequences from a total of 41 DENV cases (28 cases of DENV-1 and 13 cases of DENV-3). For DENV-1 cases, we sequenced 24 samples (14 full E gene sequences and 12 WGS) from the local outbreak cases and 4 samples (one full E gene sequence and 3 WGS) from imported cases. For DENV-3 cases, we sequenced 2 samples (WGS) from autochthonous cases and 11 samples (WGS) from imported cases.
The only local case of DENV-2 was not sequenced, because the blood sample had a very low viral load (Ct > 32).
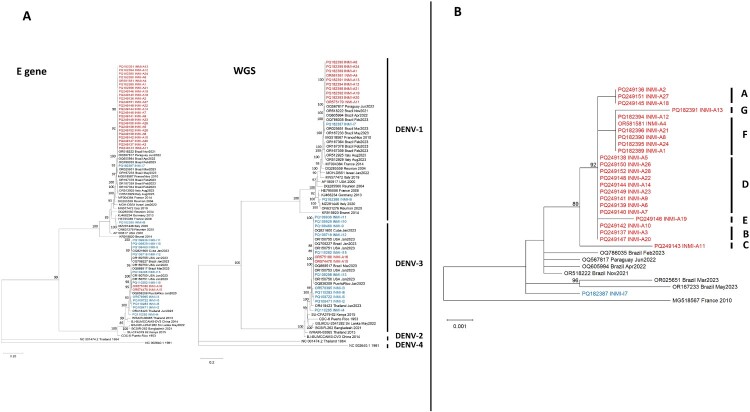
We performed phylogenetic analysis with complete E gene sequences and WGS, as shown in Figure 2 (panel A). All sequences from local DENV-1 cases clustered together in a distinct branch, with no phylogenetic connection to imported sequenced DENV-1 cases. The local DENV-1 cluster seemed to be related to a Brazilian sequence published in GenBank and obtained from a sample collected in November 2021 (accession ID: OR518222). Similar to the local DENV-1 cases, local DENV-3 clustered together, and no phylogenetic connection was found with the imported cases. All imported cases were mixed among different regions of the world.
Figure 2.
Panel A: Phylogenetic analysis based on the complete E gene and WGS of autochthonous and imported cases of DENV-1 and DENV-3. Sequences from autochthonous cases are highlighted in red, and those from imported cases are highlighted in blue. Bootstrap values > 80 are reported aside the corresponding nodes. Panel B: Enlarged detail of the phylogenetic tree of complete E gene sequences from autochthonous DENV-1 cases (red). The identified subclusters are indicated by capital letters. Bootstrap values > 80 are reported aside the corresponding nodes.
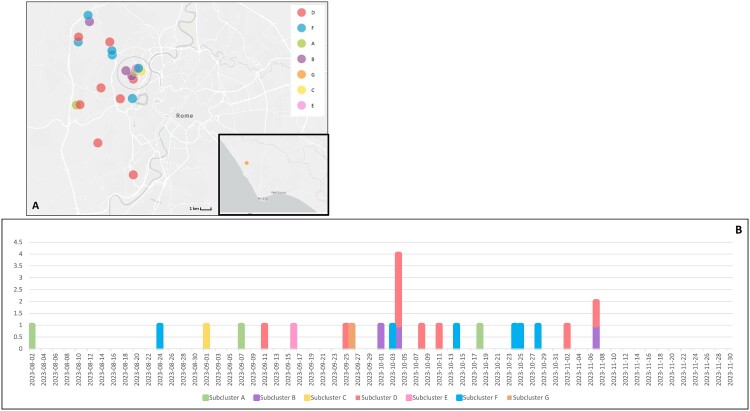
We built a specific DENV-1 phylogenetic tree using the complete E gene sequences of the 26 local cases (Figure 2, panel B). As shown, all DENV-1 sequences branched from an initial node with significant bootstrapping, and we identified seven sub-clusters (named clusters A, B, C, D, E, F and G). Subclusters C and G were separated from the other sequences with significant bootstrap values. Moreover, we analysed the geographical distribution of cases belonging to the DENV-1 cluster. Figure 3 shows the geographical distribution of the clusters identified with the phylogenetic analysis. We identified 10 separate areas, nine of which were in the east of the urban area of Rome, while only one was near the city of Anzio, south of Rome.
Figure 3.
Geographical distribution of DENV-1 sequenced cases. Each dot represents cases that occurred within an area of 1 square kilometre. Colours represent clusters identified by phylogenetic analysis. The epidemic curve shows the appearance of individual cases in chronological order based on the first appearance of symptoms. The cases are represented with the same colour as the cluster they belong to.
Mutational analysis of the E gene
The nucleotide sequences of 24 autochthonous sequenced cases of DENV-1 were compared to the NCBI reference sequence (Accession number: NC_001477). Twelve nucleotide mutations were observed in the E gene region, distributed in different patterns across the study sequences, defining seven different subclusters (clusters A to G). Table 2 shows all mutations that define clusters or ambiguous positions between sequences. By stratifying the 26 E gene sequences based on such nucleotide mutations, the nucleotide mutational patterns among the studied sequences corresponded to the subclusters identified by the phylogenetic analysis.
Table 2.
Mutational analysis of E gene sequences from autochthonous DENV-1 cases.
| Nucleotide position | 42 | 77 | 162 | 360 | 679 | 729 | 918 | 930 | 938 | 1218 | 1239 | 1356 | Sub-cluster |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NC_001477_1974 | A | A | C | G | T | T | C | G | C | C | C | T | ref |
| OR518222_Brazil_Nov2021 | T | T | nearest neighbour | ||||||||||
| INMI-A2 | C | T | C | A | |||||||||
| INMI-A27 | C | T | C | ||||||||||
| INMI-A18 | C | T | Y | C | |||||||||
| INMI-A3 | C | T | Y | B | |||||||||
| INMI-A10 | C | T | Y | ||||||||||
| INMI-A20 | C | T | Y | ||||||||||
| INMI-A11 | A | C | T | A | A | A | T | C | |||||
| INMI-A5 | C | T | D | ||||||||||
| INMI-A6 | C | T | |||||||||||
| INMI-A7 | N | N | C | T | |||||||||
| INMI-A9 | C | T | |||||||||||
| INMI-A14 | C | T | |||||||||||
| INMI-A22 | C | T | |||||||||||
| INMI-A23 | C | T | |||||||||||
| INMI-A26 | C | T | |||||||||||
| INMI-A28 | C | T | |||||||||||
| INMI-A19 | G | C | C | T | E | ||||||||
| INMI-A1 | C | F | |||||||||||
| INMI-A4 | C | ||||||||||||
| INMI-A8 | C | ||||||||||||
| INMI-A12 | N | N | C | ||||||||||
| INMI-A21 | C | ||||||||||||
| INMI-A24 | C | ||||||||||||
| INMI-A13 | G | T | C | C | G |
More in detail, while all the sequences showed the T729C mutation (except the INMI-A17 sequence which had a degenerate N base), 17 out of 24 sequences (71%) of subclusters A, B, C and D shared the synonymous C918T substitution, while 9 sequences of subclusters F and G did not have this substitution. The subcluster A sequences revealed, in addition to the two mutations above, also the synonymous T1356C substitution; only the INMI-A18 sequence (phylogenetically belonging to subcluster A) had a degenerate Y base at nucleotide position 1239, as well as the sequences of subcluster B.
The sequences of subclusters C and E (one sequence only for each subcluster) and the two sequences of subcluster G were those with the greatest number of nucleotide mutations. Most of the nucleotide mutations found in the study sequences were synonymous, except for two non-synonymous substitutions found in the INMI-A19 and INMI-A11 sequences: T679C and C938A, leading to the amino acid substitutions S227P and A313D in the E protein, respectively.
Regarding the autochthonous cases of DENV-3, the two sequences obtained for INMI-A15 and INMI-A16 were identical. Comparing them with the NCBI reference sequence (Accession number: NC_001475), two unique non-synonymous mutations were found: nucleotide substitutions G1541T and C2346T, leading to A203S and T471I aminoacidic substitutions in the E protein, respectively.
Structural analysis
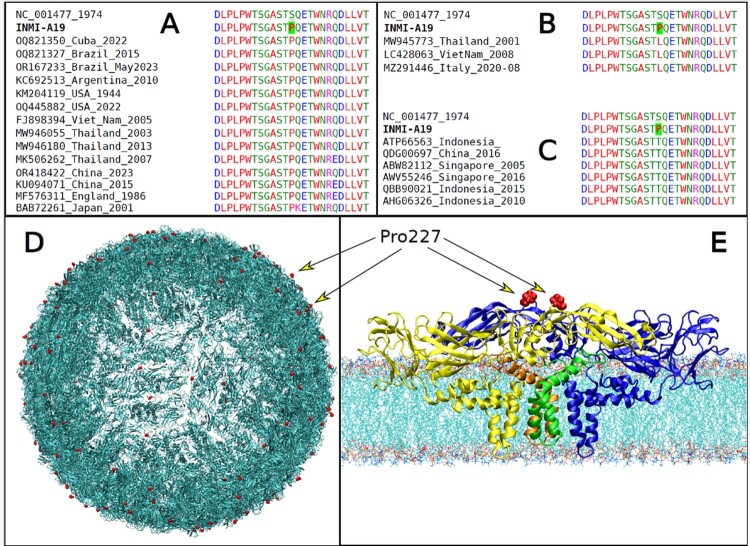
We first looked at the S227P change in the DENV-1 E protein of the patient INMI-A19 from a structural perspective, because this change was not found in the most similar sequences worldwide. Interestingly, the same P227 residue was found in several unrelated cases at different times and places (Figure 4A), indicating its possible biological importance. Also, the S227L and S227T variations were seen in sequenced DENV-1 genomes (Figure 4B,C show alignments for S227L and S227T, respectively), suggesting this residue plays a key role in how the virus adapts. Consistent with these genetic findings, residue 227 was situated on the surface of the viral envelope, near the area where DENV antibodies were shown to bind [30], as illustrated in Figure 5D,E, where Pro227 has been marked with a red colour in both the envelope model and the E-M tetramer model on the membrane, respectively. Furthermore, residue 227 is next to the premembrane (prM) structural protein in the immature DENV virion and could affect the interaction with prM-reactive antibodies [31].
Figure 4.
Colour-coded multiple sequence alignment with Clustal Omega [18] in the region of the pS227P variant for the INMI-A19 case. Comparison of INMI sequence, reference and representative genome sequences with S227P, S227L and S227T variants are shown in panels A, B and C, respectively. The pS227P variant in the INMI-A19 case is highlighted with a green background. (D) Model of the DENV envelope. The surface is composed of a network of 360 E and M proteins. The variant pS227P variant in the E protein of DENV-1, found in INMI patient INMI-A19, is highlighted in red colour; (E) Model of the E-M tetramer complex in membrane. The two E proteins are coloured yellow and blue, while the two M ones are green and orange. The two Pro227 are shown in red, as in panel A.
Figure 5.
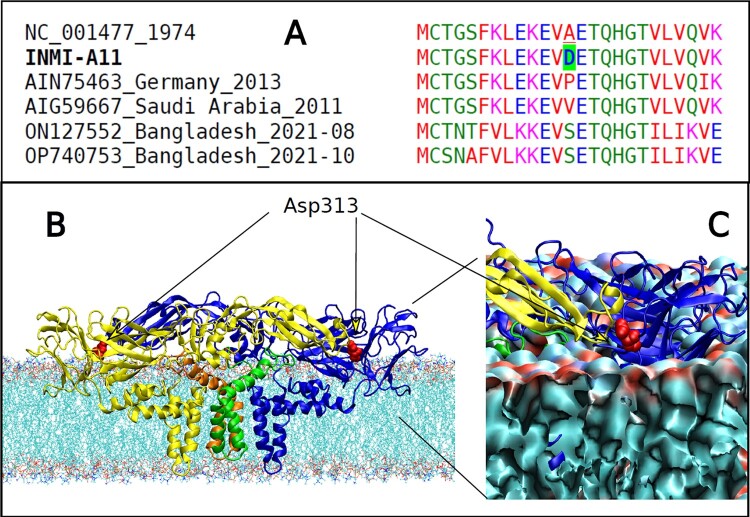
(A) Colour-coded multiple sequence alignment with Clustal Omega [18] in the region of the A313D variant for INMI-A11 case. Comparison of INMI sequence, reference and representative genome sequences with A313P, A313V and A313S variants are shown. (B) Model of the DENV E-M tetramer complex in membrane. The A313D variant in the E protein of DENV-1, found in case INMI-A11, is highlighted in red colour; (C) Enlargement of the D313 region showing the interface proximity to the residue with the polar lipid heads in the membrane.
Another unusual change in the E protein of the Lazio DENV-1 cluster was A313D, found only in the case INMI-A11. It was not in the closest sequences worldwide or in any other DENV-1 genome. But A313P, A313V and A313S variants were found in Europe, the Middle East and Asia, respectively (Figure 5A). Residue 313 of the E protein was at the membrane interface of the envelope (Figure 5). The negative charge of Asp313, replacing the non-polar alanine, was near the polar lipid heads of the membrane (Figure 5C), likely altering the virus-protein-membrane interaction. Residue 313 is also close to residues 310 and 311, which have been reported to affect antibody interactions [32].
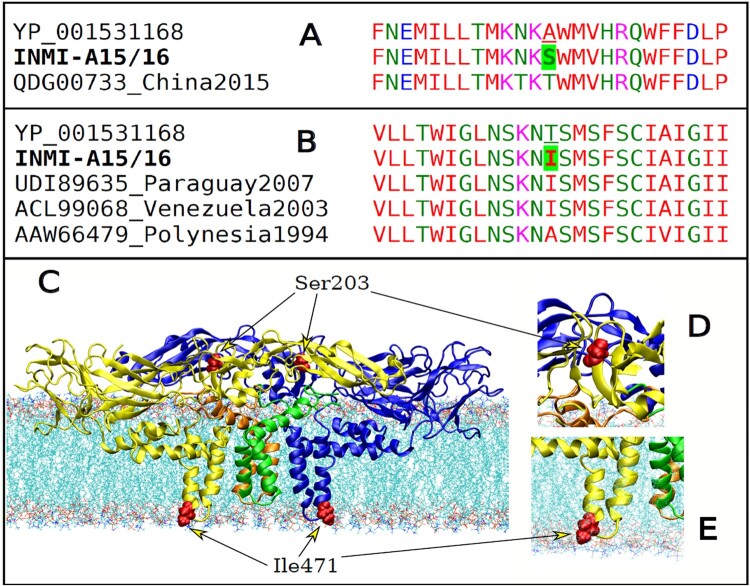
The E gene of the local DENV-3 cases had two non-synonymous mutations: A203S and T471I. These were not in the global sequences most closely related to them. We only saw a different mutation at 203, A203T, in a Chinese case in 2015 (Figure 6A). The same Ile471 was in cases from Venezuela and Paraguay in 2003 and 2007 (Figure 6B), and the T471A mutant was in Polynesia in 1994 (Figure 6B).
Figure 6.
(A, B) Colour-coded multiple sequence alignment with Clustal Omega [18], for INMI-A15 and – A16 cases, in the region of the A203S and T471I variants are shown in panels A and B, respectively. (C) Model of the DENV E-M tetramer complex in the membrane. The A203S and T471I variants in the E protein of DENV-3 are highlighted in red colour. (D) Enlargement of the S203 region. (E) Enlargement of the I471 region, showing the proximity of the residue with the polar lipid heads in the inner viral membrane.
From a structural point of view, residue 203 was found in an accessible position on the envelope surface (Figure 6C,D). On the other hand, the nonpolar Ile471, located in the inner envelope interface, substituted the polar Thr residue present in the DENV-3 reference. In this case, the rearrangement of the E protein could also be monitored by Molecular Dynamics simulations.
Discussion
In this paper, we describe distinct episodes of autochthonous DENV transmission in the Lazio region in central Italy. Blood samples were obtained from patients who were diagnosed as DENV-infected by the Laboratory of Virology of INMI Spallanzani during the summer and autumn of 2023, and genetic analysis was performed on the DENV strains isolated from these samples. The strains, isolated from distinct autochthonous outbreaks, were classified into three different serotypes, namely DENV-1, DENV-2, and DENV-3. A complete phylogenetic reconstruction analysis was carried out for DENV-1 and DENV-3 strains since the viral load of the only autochthonous DENV-2 case was not compatible with sequencing. The analysis revealed that the DENV-1 sublineage originated in South America (probably from Brazil) and the DENV-3 sublineage in the USA. These molecular findings demonstrate that original DENV infections in Italy were detected soon after importation. Italy, and the city of Rome in particular, is exposed to a relatively high risk of DENV infections, as the virus may be imported from areas where DENV infection is endemic, along with the ever-increasing ease of travel for tourism, and a competent vector is present.
Interestingly, the results from the phylogenetic analysis show that the DENV-1 sequences cluster into the same genetic branch (bootstrap > 80) without any other interspersed imported sequences.
Again, when the DENV-1 sequences were characterized for the presence of mutational events in the E gene of the viral genome, they could be classified into at least seven genetically distinct subclusters. The spatial distribution of genetic subclusters has been investigated, observing that almost all cases occurred in the eastern area of Rome and that most mutational events occurred within the same one-square-kilometre area in the center of Rome. These latter molecular findings are of interest for different reasons. They suggest, from a genetic point of view, that these sequences are all related to a single introduction. The observations also lead to the hypothesis that local transmission can have contributed to the genetic micro-evolution of the virus compared to the ancestral imported strain, thus suggesting the presence of a possible undetected local circulation in the metropolitan area of Rome. This latter consideration could also explain what was found about the genetic peculiarities of the autochthonous DENV-3 cases. It cannot be excluded that these cases may not have been isolated, but may instead represent the peak of a larger, undiagnosed local microcirculation of DENV-3 strains. However, following the diagnosis of these autochthonous cases, the Local Health Unit of Latina identified approximately 300 people who had stayed in the area of potential exposure during the same period. All these contacts underwent active surveillance, and none developed symptoms; additionally, 10% underwent voluntary screening, and none tested positive for either DENV antibodies or antigens (unpublished data). Moreover, vector control has been intensified following indication of the Italian National Plan against arboviral infections [17], mostly directed at Aedes albopictus (competent vector for DENV present in Central Italy). Control measures were based on the use of larvicides, adulticides and the removal of breeding sites [3].
Again, it is of particular interest to genetically investigate the DENV-1 subclusters more in-depth. The outbreak strains are often homogeneous. This is not unexpected because viral propagation at the outbreak level may occur very rapidly and in a very short time, thus leading to rapid clonal expansion with a low probability of genetic diversification. In the Rome outbreak, the DENV-1 sequences belonged to genetic subclusters defined by at least one sequence diverging from the ancestral imported strain by at least one mutation in the E gene of the viral genome. Although this subcluster definition cannot be aligned with the criteria being applied to designate sublineages as proposed by official taxonomy, it is evident that the DENV-1 sequences in Rome form, at least phylogenetically, multiple branches and that they are not interspersed within international clusters.
Furthermore, focusing on non-synonymous substitutions in the E gene region, we found that two sequences of DENV-1 showed aminoacidic substitutions in positions having possible biological relevance, i.e. a possible influence in the further evolutionary process of changing the virus. Additional studies are needed to better explain such possible implications affecting the interaction between antibodies, vectors and viral antigens. In particular, the S227P and A313D mutations were not present in worldwide DENV-1 sequences phylogenetically closest to the sequences of the 2023 Rome outbreak, thus leaving the possibility that the acquisition of these mutations could have occurred during the local transmission chain.
From an epidemiologic point of view, multiple reasons could be the basis of the 2023 DENV-1 outbreak in Rome. First, some urbanistic and social peculiarities of the city and the affected neighbourhood. Historical buildings, which are mainly represented in this area, are often built around an internal courtyard, which may host shared gardens and fountains, providing favourable habitats for vectors and significantly increasing vector density. Second, Rome's parks and gardens are among the largest of the European cities, thanks to the presence of rich aristocratic villas spread all over the city centre, which also may favour the life cycle of the vector. In Italy, the activity of the vector mosquitoes is mainly restricted to the summer season (usually between June and October), when outbreaks could be triggered by the arrival of imported cases from endemic areas and may result in secondary cases. Nevertheless, it must be considered that the important climate change that temperate regions of the world are experiencing is also enlarging the period of vector viability, and therefore the probability of sustaining an autochthonous circulation may progressively increase.
This perspective may raise some concern about some nature-based solutions (such as urban greening achieved by growing public gardens, urban forests, parks and street trees), which aim to protect or restore an ecosystem and are emerging as a promising tool for improving the health and well-being of a constantly increasing urban population. In fact, while urban greening efforts have undeniable benefits for human health and the biological communities inhabiting these green zones, disease vector populations may also be affected, possibly promoting greater pathogen transmission and the emergence of infectious diseases such as DENV, West Nile fever, malaria, leishmaniosis and tick-borne diseases [33].
Other considerations may also be suggested. In fact, central Rome inhabitants usually report a higher average income compared to other areas; touristic travels to tropical endemic areas may be more frequent with consequently higher imported DENV cases, which may constitute the driver for autochthonous sustained circulation. Median age was higher (50 – IQR: 37-68; 32-IQR: 26-42) for autochthonous/non-travel associated DENV compared to imported cases. This finding is possibly related to the lower median age of individuals who travel to endemic tropical or subtropical destinations for tourism or professional reasons [34].
Up to the beginning of December 2023, over 5 million cases and over 5000 DENV-related deaths have been reported from 86 countries/territories globally. In Europe, autochthonous/non-travel associated DENV cases have been reported during 2023 from France (45 cases) and Spain (3 cases) [35]. In Italy, 82 cases were reported during the same period, in the North (Lombardy) and Central (Lazio) areas, with no epidemiologic or phylogenetic correlation among them [3,4,36].
In this scenario, given the increasing impact of DENV infection on public health, our findings strengthen the importance of an integrated surveillance system that should be able to promptly identify autochthonous arbovirus transmission and implement control measures. The integration of passive surveillance with different surveillance tools (such as laboratory-based surveillance, syndromic surveillance, or novel data streams) combined with entomological surveillance may facilitate the detection, response, and control of arboviruses spreading, including DENV. Viral variant identification and characterization could play a role in a prompt evaluation of vaccine and antibody efficacy. Finally, information and training for health care professionals might be useful to achieve early diagnosis and reporting of some emerging infectious diseases that are designated to become endemic in European countries.
Supplementary Material
Funding Statement
This study was supported by the Italian Ministry of Health “Ricerca Corrente – Linea 1 – Progetto on emerging and re-emerging infections – INMI L. Spallanzani I.R.C.C.S” funding and by Horizon Europe Framework Programme (grant agreement number 101137192, AVITHRAPID).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
DENV-1 and DENV-3 sequences of complete genome and E gene region are submitted to NCBI Nucleotide database with the following accession numbers: OR575040, OR575123, OR574470, OR575180, OR578395, OR575179, PQ108471, PQ110285, PQ109722, PQ110283, PQ108460, PQ109929, PQ109936, PQ109718, PQ109298, PQ110282, PQ249136-PQ249152; PQ182387-PQ182396.
References
- 1.Paz-Bailey G, Adams LE, Deen J, et al. Dengue. Lancet. 2024 Jan 24;403(10427):667–682. doi: 10.1016/S0140-6736(23)02576-X. Epub ahead of print. PMID: 38280388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z, Bambrick H, Frentiu FD, et al. Projecting the future of dengue under climate change scenarios: progress, uncertainties and research needs. PLoS Negl Trop Dis. 2020 Mar 2;14(3):e0008118. doi: 10.1371/journal.pntd.0008118. PMID: 32119666; PMCID: PMC7067491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Carli G, Carletti F, Spaziante M, et al. Outbreaks of autochthonous Dengue in Lazio region, Italy, August to September 2023: preliminary investigation. Euro Surveill. 2023 Nov;28(44):2300552. doi: 10.2807/1560-7917.ES.2023.28.44.2300552. PMID: 37917030; PMCID: PMC10623645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassaniti I, Ferrari G, Senatore S, et al. Preliminary results on an autochthonous dengue outbreak in Lombardy Region, Italy, August 2023. Euro Surveill. 2023 Sep;28(37):2300471. doi: 10.2807/1560-7917.ES.2023.28.37.2300471. PMID: 37707980; PMCID: PMC10687988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barzon L, Gobbi F, Capelli G, et al. Autochthonous dengue outbreak in Italy 2020: clinical, virological and entomological findings. J Travel Med. 2021 Dec 29;28(8):taab130. doi: 10.1093/jtm/taab130. PMID: 34409443; PMCID: PMC8499737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermeulen TD, Reimerink J, Reusken C, et al. Autochthonous dengue in two Dutch tourists visiting Département Var, southern France, July 2020. Euro Surveill. 2020 Oct;25(39):2001670. doi: 10.2807/1560-7917.ES.2020.25.39.2001670. Erratum in: Euro Surveill. 2020 Oct;25(40): PMID: 33006305; PMCID: PMC7531074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vita S, Bordi L, Sberna G, et al. Autochthonous dengue fever in 2 patients, Rome, Italy. Emerg Infect Dis. 2024 Jan;30(1):183–184. doi: 10.3201/eid3001.231508. Epub 2023 Nov 15. PMID: 37967518; PMCID: PMC10756386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merakou C, Amendola A, Fortuna C, et al. Diagnosis of imported dengue and Zika virus infections in Italy from November 2015 to November 2022: laboratory surveillance data from a national reference laboratory. Viruses. 2023 Dec 28;16(1):50. doi: 10.3390/v16010050. PMID: 38257751; PMCID: PMC10818496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riccò M, Peruzzi S, Balzarini F, et al. Dengue fever in Italy: The “eternal return” of an emerging arboviral disease. Trop Med Infect Dis. 2022 Jan 13;7(1):10. doi: 10.3390/tropicalmed7010010. PMID: 35051126; PMCID: PMC8782038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silveira LTC, Tura B, Santos M.. Systematic review of dengue vaccine efficacy. BMC Infect Dis. 2019 Aug 28;19(1):750. doi: 10.1186/s12879-019-4369-5. PMID: 31455279; PMCID: PMC6712597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaffner F, Mathis A.. Dengue and dengue vectors in the WHO European region: past, present, and scenarios for the future. Lancet Infect Dis. 2014 Dec;14(12):1271–1280. doi: 10.1016/S1473-3099(14)70834-5. Epub 2014 Aug 26. PMID: 25172160. [DOI] [PubMed] [Google Scholar]
- 12.Benelli G, Wilke ABB, Beier JC.. Aedes albopictus (Asian Tiger Mosquito). Trends Parasitol. 2020 Nov;36(11):942–943. doi: 10.1016/j.pt.2020.01.001. Epub 2020 Feb 6. PMID: 32037135. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, Clements AC, Williams G, et al. A threshold analysis of dengue transmission in terms of weather variables and imported dengue cases in Australia. Emerg Microbes Infect. 2013 Dec;2(12):e87. doi: 10.1038/emi.2013.85. Epub 2013 Dec 18. PMID: 26038449; PMCID: PMC3880872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutheneni SR, Morse AP, Caminade C, Upadhyayula SM.. Dengue burden in India: recent trends and importance of climatic parameters. Emerg Microbes Infect. 2017 Aug 9;6(8):e70. doi: 10.1038/emi.2017.57. PMID: 28790459; PMCID: PMC5583666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S., Cao Z., Jiao L., et al. The global economic burden of dengue in 2020–2050: estimates and projections for 141 countries and territories. Lancet. 2024 Jan 16;16(8):935–941. Preprint. [Google Scholar]
- 16.World Health Organization . Disease outbreak news; dengue – global situation 2023 Dec 21 [cited 2024 Sep 12] Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON498.
- 17.Italian Ministry of Health [Internet] . Piano Nazionale di prevenzione, sorveglianza e risposta alle Arbovirosi (PNA) 2020-2025 [Internet]. November 2019. [cited 2024 Sep 12] Available from: https://www.salute.gov.it/imgs/C_17_pubblicazioni_2947_allegato.pdf.
- 18.Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ.. The 3.8Å resolution cryo-EM structure of Zika virus. Science. 2016 Apr 22;352(6284):467–470. doi: 10.1126/science.aaf5316. Epub 2016 Mar 31. PMID: 27033547; PMCID: PMC4845755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santiago GA, Vergne E, Quiles Y, et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis. 2013 Jul 11;7(7):e2311. doi: 10.1371/journal.pntd.0002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su W, Jiang L, Lu W, et al. A serotype-specific and multiplex PCR method for whole-genome sequencing of dengue virus directly from clinical samples. Microbiol Spectr. 2022 Oct 26;10(5):e0121022. doi: 10.1128/spectrum.01210-22. Epub 2022 Sep 12. PMID: 36094197; PMCID: PMC9602986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madeira F, Pearce M, Tivey ARN, et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022 July 5;50(W1):W276–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2 July 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Ge P, Yu X, et al. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomize AL, Todd SC, Pogozheva ID.. Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. 2022 Jan;31(1):209–220. doi: 10.1002/pro.4219. Epub 2021 Nov 8. PMID: 34716622; PMCID: PMC8740824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo S, Cheng X, Lee J, et al. CHARMM-GUI 10 years for biomolecular modeling and simulation. J Comput Chem. 2017 Jun 5;38(15):1114-1124. doi: 10.1002/jcc.24660. Epub 2016 Nov 14. PMID: 27862047; PMCID: PMC5403596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber RG, Marzinek JK, Boon PLS, et al. Bond, computational modelling of flavivirus dynamics: The ins and outs. Methods. 2021;185:28–38. ISSN 1046-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekka M, Lennart N.. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A. 2001;105(43):9954–9960. doi: 10.1021/jp003020w [DOI] [Google Scholar]
- 28.Van Der Spoel D, Lindahl E, Hess B, et al. GROMACS: fast, flexible, and free. J Comput Chem. 2005 Dec;26(16):1701–1718. doi: 10.1002/jcc.20291. PMID: 16211538. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Meng W, Horton M, et al. Potent neutralizing antibodies elicited by dengue vaccine in rhesus macaque target diverse epitopes. PLoS Pathog. 2019 Jun 6;15(6):e1007716. doi: 10.1371/journal.ppat.1007716. PMID: 31170257; PMCID: PMC6553876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowd KA, Sirohi D, Speer SD, et al. prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis. Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2218899120. doi: 10.1073/pnas.2218899120. Epub 2023 Jan 13. PMID: 36638211; PMCID: PMC9933121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarker A, Dhama N, Gupta RD.. Dengue virus neutralizing antibody: a review of targets, cross-reactivity, and antibody-dependent enhancement. Front Immunol. 2023 Jun 2;14:1200195. doi: 10.3389/fimmu.2023.1200195. PMID: 37334355; PMCID: PMC10272415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fournet F, Simard F, Fontenille D.. Green cities and vector-borne diseases: emerging concerns and opportunities. Euro Surveill. 2024 Mar;29(10):2300548. doi: 10.2807/1560-7917.ES.2024.29.10.2300548. PMID: 38456216; PMCID: PMC10986671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travel trends by age demographic: age as the key factor in tourism. [cited 2024 Sep 12] Available from: https://mize.tech/blog/travel-trends-by-age-demographic-age-as-the-key-factor-in-tourism/.
- 35.ECDC . Communicable disease threats report, 10–16 December 2023, week 50 [cited 2024 Sep 12] Available from: https://www.ecdc.europa.eu/en/publications-data/communicable-disease-threats-report-10-16-december-2023-week-50.
- 36.ISS . National surveillance system of arboviral diseases: regular bulletins [cited 2024 Sep 12] Available from: https://www.epicentro.iss.it/en/arboviral-diseases/surveillance.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DENV-1 and DENV-3 sequences of complete genome and E gene region are submitted to NCBI Nucleotide database with the following accession numbers: OR575040, OR575123, OR574470, OR575180, OR578395, OR575179, PQ108471, PQ110285, PQ109722, PQ110283, PQ108460, PQ109929, PQ109936, PQ109718, PQ109298, PQ110282, PQ249136-PQ249152; PQ182387-PQ182396.