Abstract
Changes in diet causing ecological stress pose a significant challenge to animal survival. In response, the gut microbiota, a crucial part of the host’s digestive system, exhibits patterns of change reflective of alterations in the host’s food component. The impact of temporal dietary shifts on gut microbiota has been elucidated through multidimensional modeling of both food component and macronutrient intake. However, the broad distribution of wild generalist and the intricate complexity of their food component hinder our capacity to ascertain the degree to which their gut microbiota assist in adapting to spatial dietary variations. We examined variation in patterns of the gut microbial community according to changes in diet and in a colobine monkey with a regional variable diet, the golden snub-nosed monkey (Rhinopithecus roxellana). Specifically, we analyse the interactions between variation in food component, macronutrient intake and the gut microbial community. We compared monkeys from four populations by quantifying food component and macronutrient intake, and by sequencing 16S rRNA and the microbial macro-genomes from the faecal samples of 44 individuals. We found significant differences in the diets and gut microbial compositions, in nutrient space and macronutrient intake among some populations. Variations in gut microbiota composition across distinct populations mirror the disparities in macronutrient intake, with a notable emphasis on carbohydrate. Geographical differences in the diet among of golden snub-nosed monkey populations will result in macronutrient intake variation, with corresponding differences in macronutrient intake driving regional differences in the compositions and abundances of gut microbiota. Importantly, the gut microbiota associated with core digestive functions does not vary, with the non-core gut microbiota fluctuating in response to variation in macronutrient intake. This characteristic may enable species heavily reliant on gut microbiota for digestion to adapt to diet changes. Our results further the understanding of the roles gut microbiota play in the formation of host dietary niches.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-024-00349-w.
Keywords: Macronutrient intake, Dietary niches, Gut microbiota, Weighted gene co-expression network, Primate
Introduction
Dietary habits represent a crucial area of research within the field of animal-environment interactions. Understanding how animals respond to diet changes (changes in the food component) is pivotal to uncovering the mechanisms behind their dispersal and distribution patterns [1]. Thus, species with complex food components and a wide range of distribution have become a hot research group in dietary habit studies [2–4]. Here we use ‘generalist’ to describe widely distributed species with complex food components, displaying variability across temporal and spatial scales. Most research on generalists’ responses to ecological pressures stemming from diet changes has concentrated on host behavioral strategies and physiological adaptations [5–7]. However, recent research has indicated that symbiotic gut microbiota significantly influence the host’s digestive processes [8]. Furthermore, gut microbiota has been shown to help host respond to ecological stress [9, 10]. Therefore, a critical aspect of understanding how gut microbiota support their hosts in the face of ecological stresses due to diet changes lies in examining the relationship between these microbial communities and the temporal or spatial variations in the food components of generalists [11, 12].
Preliminary research has shown that changes in host diet significantly influence the composition of gut microbes [13]. Both temporal and spatial diet changes in generalist have been observed to significantly impact their gut microbiota [14, 15]. For example, the gut microbiota of the Ethiopian geladas (Theropithecus gelada) exhibits seasonal variations that correspond to changes in food components [14]. Research on yaks (Bos grunniens) have shown that the composition of their gut microbiota correlates with the geographical distribution of the host populations, indicating an adaptation to their food intake [16]. However, research focusing solely on the level of food component to explain the impact of diet changes on gut microbiota cannot account for the variability introduced by different food items [14, 17]. The inconsistencies due to changes in food components complicate the quantification of their influence on gut microbiological shifts [14, 17]. This situation limits our exploration of what factors drive gut microbiota to exhibit changes in response to changes in food component.
Fortunately, the development of nutritional ecology has provided new perspectives on the research of dietary habits. Nutritional geometric modeling allows downscaling the complexity of food components to a stable set of three macronutrients, carbohydrate, available protein, and lipid [18]. Thus, the effect of dietary habits on gut microbiota has been further clarified. Studies on temporal diet changes highlight the need for a multidimensional analysis that considers both food components and macronutrient intake [19–21]. Seasonal alterations in food component influence gut microbiota changes through shifts in macronutrient intake [17, 21]. Additionally, spatial variation sculpts a distinctive pattern, with regional food availability disparities causing significant differences in the food components of geographically isolated populations. For example, the mountain gorilla (Gorilla beringei beringei) has significant differences in food components between two geographically separated populations [22]. There were significant differences in the food components of different populations of red colobus monkeys (Procolobus rufomitratus) living from the north to the south of Kibale National Park [23]. But few studies have been able to determine food components and macronutrient intake data for host individuals of gut microbiota in populations from different regions due to the difficulty of quantifying macronutrient intake in wild populations. In particular, how gut microbiota of generalist respond to changes in food components and possibly macronutrient intake as a result of spatial variation will help us to understand what adaptive roles gut microbiota play in changes in host diets.
To address these questions, we selected a widespread species with a generalized diet, serving as an ideal model for examining the interplay between gut microbiota and spatial dietary variation. The golden snub-nosed monkey (Rhinopithecus roxellana), a generalist, inhabits the mixed forests of China’s mountainous central provinces, including Shaanxi, Sichuan, Hubei, and Gansu [24]. Despite being categorized as leaf-eating colobines, golden snub-nosed monkeys consume a diverse range of food items, encompassing leaves, seeds, ripe fruits, buds, bark, twigs, flowers, and lichens [25]. The availability of these diverse foods fluctuates temporally and spatially, with more than a 50% variation in food items across different regions [24]. Our study aimed to determine the impact of spatial dietary differences on the composition, structure, and function of gut microbiota within the same species. We posed two key questions: (1) What differences exist in food component and macronutrient intake among various populations of golden snub-nosed monkeys, and how do these differences affect the gut microbiota’s composition? (2) How does the functionality of the gut microbiota vary in response to changes in food component and macronutrient intake across these populations?
Materials and methods
Data collection
The four study sites were all located in the Qinling Mountains: (i) Shaanxi Foping Guanyin Mountain Nature Reserve (FP) (107°51′~108°01′ E, 33°35′~33°45′ N), (ii) Huangbaiyuan National Nature Reserve (HBY) (107°31′~107°42′ E, 33°42′~33°54′ N), (iii) Changqing National Nature Reserve (HY) (107°17′~107°55′ E, 33°19′~33°44′ N), and (iv) Louguantai National Forest Park (LGT) (108°12′~108°27′ E, 33°47′~34°05′ N). We collected food intake data and took faecal samples from golden snub-nosed monkey individuals in each population during the winter (November to February).
For the FP, LGT and HBY populations, we randomly identified target adult individuals and accurately the food intake data using previously described methods [17]. In brief, we randomly chose one individual per day and conducted continuous observations of the focal animal from dawn to dark to record its feeding data. During the observation session, the distance between the observer and the subject was less than 5 m. We recorded the type, quantity of the food and the amount of time feeding [25]. After the focal individual completed feeding, leftover foods were collected as food item samples. All samples were labelled with the information of the collection time, type, and size. Then, they were immediately weighed within 0.2–3 h and sent to the laboratory for storage before the analysis of their macronutrient components.
We collected faecal samples from individuals with recorded feeding behaviour on the same day for high-throughput sequencing. After the focal individual defecated, we immediately collected the faces with sterile cotton swabs and sterile toothpicks. The sample was then stored in 2 mL centrifuge tubes and frozen in liquid nitrogen before analysis.
Owing to research environment constraints, we were unable to collect individual food intake data for the HY population. However, we estimated the average intake by dividing the total food consumption of the HY group by the number of its members. We randomly obtained faecal samples from 5 adult individuals in the HY population. Samples of golden snub-nosed monkeys collected from different regional populations are shown in the Supplementary Table.
Nutrient analysis
We used previously described techniques to analyse the macronutrients and energy values of each food item [25–28]. The nutrients of each food item were analysed for lipids (L), total non-structural carbohydrates (TNC), starch, neutral detergent fibre (NDF), acid detergent fibre (ADF), acid detergent lignin (ADL), available protein (AP), and ash content. For each food item consumed by the target individuals, we determined the macronutrient intake by multiplying the nutrient content, as analysed in the laboratory, by the quantity of the food item intake. These intakes were then summed to calculate the available protein, fat, and carbohydrate intake for each target individual. Metabolizable energy (ME) was calculated for each macronutrient using conversion factors of AP 17 kJ/g, lipid 37 kJ/g, TNC 16 kJ/g [29] and NDF 9 kJ/g.
DNA extraction and 16S rRNA gene sequencing
The microbial DNA (with a total mass of 1.2–10.0 ng) was isolated from each faecal sample using the MOBIO Pow erSoil DNA Isolation Kit and was quantified with NanoDrop One (Thermo Fisher Scientific, Waltham, MA, USA). The V4 regions of the DNA genes were amplified by using the specific primer 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806R (5’-GGACTACHVGGGTW TCTAAT-3’) [30]. After amplification and detection, the PCR products were mixed in equal density ratios and then were purified with an E.Z.N.A.® Gel Extraction Kit (Omega, USA). The sequencing library was created using NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Beverly, MA, USA) according to the manufacturer’s instruction, and the indexes were added. Next, the library was sequenced on an Illumina Hiseq2500 platform (Magigene Biotechnology Co., Ltd. Guangzhou, China), and the 250 bp paired-end reads were generated. The sequence with ≥ 97% similarity was clustered into the same operational taxonomic units (OTU) by USEARCH (http://www.drive5.com/usearch/). The silva (https://www.arb-silva.de/) database was used to annotate taxonomic information (confidence threshold score ≥ 0.5).
Metagenomic sequencing and gene catalogue construction
The sequencing library was created using NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Beverly, MA, USA). The library was analysed for size distribution by Agilent2100 Bioanalyzer (Agilent, USA), and then were sequenced by Illumia Hiseq2500 platform in MAGIGene Co., Ltd. (Shenzhen, China). Quality control was conducted by Trimmomatic (Version 0.38). The reads aligned to the NCBI nonredundant (NR) database were removed with MEGAHIT (Version 1.2.9). The remaining high-quality reads were used for further analysis. The assembly of reads was conducted using MEGAHIT de novo. For each sample, a series of k-mer values (49 to 87) were used and the optimal one with the longest N50 value were chosen for the remaining scaffolds. The clean data were mapped against scaffolds using MEGAHIT. Unused reads from each sample were assembled using the same parameters. Genes (minimum length of 100 nucleotides) were predicted on scaftigs longer than 500 bp using Prodigal (Version 2.6.3). Then, a non-redundant gene catalogue was constructed with Linclust (Version 2.0) using a sequence identity cut-off of 0.9. To determine the abundance of genes, reads were realigned to the gene catalog with BBMap (Version 37.68, https://sourceforge.net/projects/bbmap). Only genes with 2 mapped reads no less than 2 were considered exist in a sample. The abundance of genes was calculated by counting the number of reads and normalizing by gene length. Genes were then searched in KEGG database (http://www.kegg.jp/kegg/) and CAZy database (http://www.cazy.org/) for annotation.
Statistical analysis
We used Permutational Multivariate Analysis of Variance (PERMANOVA) to compare the differences in the Principal Component Analysis (PCA) results of food intake quality and nutrient space among the four regional populations, utilizing the vegan package in R (Version 4.3.2). We used the Kruskal-Wallis test with Bonferroni correction for multiple post-hoc pairwise comparisons to compare the differences in available protein, lipid, and carbohydrate energy supply among the four regional populations in R (Version 4.3.2).
We used the Kruskal-Wallis test with Bonferroni correction for multiple post-hoc pairwise comparisons to compare the differences in the Shannon index and the Chao1 index of gut microbiota among the four regional populations in R (Version 4.3.2). We used PERMANOVA to compare the differences in the Principal Co-ordinates Analysis (PCoA) results for gut microbiota composition among the four regional populations, utilizing the vegan package in R (Version 4.3.2). We employed the Linear Discriminant Analysis Effect Size (LEfSe) algorithm [31], setting a threshold at LDA > 4, to identify microbial communities that exhibit significant differences across different regions.
We used PCA to compare the differences in the results of the CAZy database annotation for gut microbiota among the four regional populations utilizing the vegan package in R (Version 4.3.2). We used the Kruskal-Wallis test with Bonferroni correction for multiple post-hoc pairwise comparisons in R (Version 4.3.2) to compare the differences in the carbohydrate metabolic pathways as delineated by the KEGG database among the four regional populations.
To ascertain the influence of environmental factors on the composition of 16S rRNA-based gut microbiota species-sample data, we performed a Detrended Correspondence Analysis (DCA). This analysis indicated that the first gradient length was 5.173. Based on this outcome, we proceeded with Canonical Correspondence Analysis (CCA), utilizing the vegan package in R (Version 4.3.2), to examine how the ratios of available protein, lipid, and SC.TNC (the ratio of neutral detergent fibre supply (NDF) to total carbohydrate supply (TNC + NDF)) impact the composition of the 16S rRNA-based gut microbiota.
Results
Regional diets
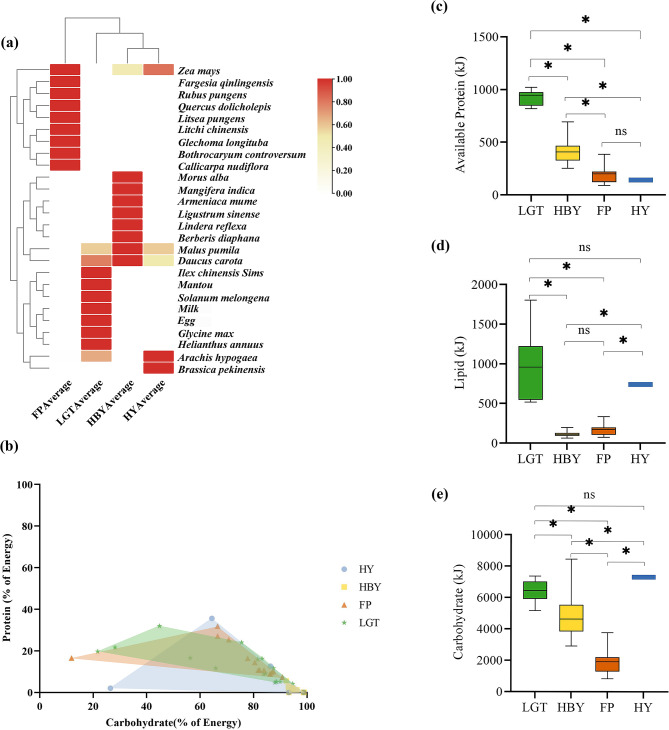
In this study, we collected food intake data from the FP, LGT, HBY and HY populations of golden snub-nosed monkeys (Fig. 1a). The FP population takes 10 food items: Zea mays, Bothrocaryum controversum, Quercus dolicholepis, Fargesia qinlingensis, Glechoma longituba, Litchi chinensis, Arachis hypogaea, Rubus pungens, Callicarpa nudiflora and Litsea pungens. The LGT population takes 9 food items: Ilex chinensis Sims, Malus pumila, Solanum melongena, Daucus carota, A. hypogaea, milk, mantou, Glycine max, Helianthus annuus and egg. The HBY population takes 9 food items: M. pumila, D. carota, Z. mays, Ligustrum sinense, Armeniaca mume, Berberis diaphana, Lindera reflexa, Morus alba and Mangifera indica. The HY population takes 5 food items: M. pumila, Z. mays, A. hypogaea, D. carota, Brassica pekinensis.
Fig. 1.
Food component, nutrient space and macronutrient intake of golden snub-nosed monkey from different regions. (a) Heat map of food intake quality of golden snub-nosed monkeys in different regions. The darker colour indicates a higher proportion of such food intake in this area compared to other areas. (b) Nutrient space of golden snub-nosed monkey populations in different regions. The figure’s points show the percentage of macronutrient energy from each food item relative to total nutrient supply. The x-axis measures carbohydrate energy’s share of total macronutrients, and the y-axis does the same for available protein. Closed shapes indicate the nutrient space for different monkey populations: blue dots for HY, yellow squares for HBY, red triangles for FP, and green pentagrams for LGT. (c) The comparison of available protein intake. (d) The comparison of carbohydrate intake. (e) The comparison of lipid intake. The asterisk (*) indicates statistical significance at the P < 0.05 (Kruskal-Wallis test)
PCA was conducted using data on the quality of food intake within each of the four populations. We discovered that food intake among golden snub-nosed monkeys significantly varied between all four subgroups (PERMANOVA, P < 0.05, Table 1; Fig. 1a).
Table 1.
Tests of intergroup differences in the quality of food intake of populations of golden snub-nosed monkeys in different regions (PERMANOVA)
| Group | F-Model | R 2 | P value | P adjusted |
|---|---|---|---|---|
| LGT vs. HBY | 53.867 | 0.658 | 0.001 | 0.001 |
| LGT vs. FP | 116.038 | 0.779 | 0.001 | 0.001 |
| LGT vs. HY | 90.021 | 0.849 | 0.002 | 0.002 |
| HBY vs. FP | 69.214 | 0.640 | 0.001 | 0.001 |
| HBY vs. HY | 9.892 | 0.310 | 0.001 | 0.001 |
| FP vs. HY | 24.022 | 0.471 | 0.001 | 0.001 |
Nutritional properties
To measure whether there were differences in nutrient space for all food intake consumed in all four populations (Fig. 1b) we used PCA. We found significant variation in nutrient space between the FP and HBY and the LGT and HBY populations (PERMANOVA, P < 0.05, Table 2).
Table 2.
Tests of intergroup differences in nutrient space of golden snub-nosed monkey populations in different regions (PERMANOVA)
| Group | R 2 | P value | P adjusted |
|---|---|---|---|
| LGT vs. HBY | 0.258 | 0.01 | 0.030 |
| LGT vs. FP | 0.023 | 0.422 | 0.422 |
| LGT vs. HY | 0.070 | 0.276 | 0.331 |
| HBY vs. FP | 0.215 | 0.004 | 0.024 |
| HBY vs. HY | 0.156 | 0.254 | 0.331 |
| FP vs. HY | 0.094 | 0.060 | 0.120 |
The energy (kJ) measurements for macronutrients showed that in the LGT population, available protein intake provided 924.76 ± 68.30 kJ (M ± SE), lipid intake provided 970.36 ± 425.16 kJ, and carbohydrate intake provided 6411.99 ± 691.74 kJ. In the HBY population, available protein intake provided 401.13 ± 103.85 kJ, lipid intake provided 115.78 ± 36.38 kJ, and carbohydrate intake provided 4824.43 ± 1337.66 kJ. For the FP population, available protein intake provided 193.10 ± 74.13 kJ, lipid intake provided 169.35 ± 65.86 kJ, and carbohydrate intake provided 1914.54 ± 707.12 kJ. The HY population as a whole exhibited an average intake of available protein providing 140.94 kJ, lipid providing 738.94 kJ, and carbohydrate providing 7310.85 kJ. Available protein energy supply decreased sequentially from the LGT, HBY, FP, and HY population. Lipid energy supply decreased sequentially from the LGT, HY, FP, and HBY population. Carbohydrate energy supply decreased sequentially from the HY, LGT, HBY, and FP population. Data for macronutrient energy in each population are shown in Fig. 1c, d, e).
Microbial compositions and functional genes
We used both Shannon and Chao1 indexes to estimate diversity and taxon richness of the gut microbiota of individuals sampled from each population, respectively. Differences in Shannon and Chao1 indexes between populations in different regions are shown in Supplementary Fig. 1. We found that taxon richness mostly decreased in the order of LGT, HBY, HY and FP. The diversity of gut microbiota was decreasing in the order of HBY, LGT, HY, and FP.
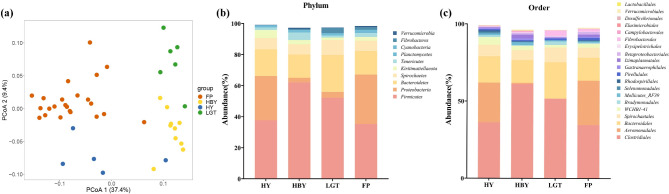
The OTU level data were used to perform PCoA, which showed significant differences between populations for the first and second principal components (Fig. 2a) (PERMANOVA, P < 0.05, Table 3).
Fig. 2.
Differences in the gut microbiota of golden snub-nosed monkeys from different regions. (a) PCoA based on OTU level gut microbiota data of golden snub-nosed monkey from different regions. Analysing regional differences in dominant bacterial populations. Relative abundance of dominant phylum (b) and order (c) in four regions based on 16S rRNA gene pools
Table 3.
Tests of intergroup differences in gut microbiota of golden snub-nosed monkey populations in different regions (PERMANOVA)
| Group | R 2 | P value | P adjusted |
|---|---|---|---|
| LGT vs. HBY | 0.657 | 0.001 | 0.001 |
| LGT vs. FP | 0.967 | 0.001 | 0.001 |
| LGT vs. HY | 0.827 | 0.030 | 0.007 |
| HBY vs. FP | 0.931 | 0.001 | 0.001 |
| HBY vs. HY | 0.802 | 0.001 | 0.001 |
| FP vs. HY | 0.702 | 0.001 | 0.001 |
To further explore variation in the golden snub-nosed monkey gut microbiota among the four populations, we performed an analysis of the composition of the gut microbiota. 16S rRNA sequencing for species annotation of the gut microbiota showed that the majority of OTUs could be classified at the phylum taxonomic level (94.5%) (Fig. 2b), the order taxonomic level (91.8%) (Fig. 2c), and the genus taxonomic level (91.2%). In total, 42 taxa were detected in the four regional populations, of which the top10 identifiable dominant phylum accounted for up to 97% of the total abundance ratio (abundance of the dominant phylum except for the total phylum abundance). Firmicutes, Proteobacteria, Bacteroidetes, Spirochaetes, Kiritimatiellaeota, Tenericutes, Planctomycetes, Cyanobacteria, Fibrobacteres, Verrucomicrobia, Euryarchaeota, Actinobacteria as dominant phyla in different regional populations. The results of the ratio of Firmicutes to Bacteroidetes (F/B) showed that HBY population was the highest at 4.11, FP population was the second highest at 2.32, HY population was the next highest at 2.20, and LGT population was the last at 2.19 and the lowest. A total of 154 Orders were annotated at the order level, and we studied the 20 orders with the highest abundance. Of these, 14 species showed enrichment among populations in all four regions, with higher abundance of Clostridiales, Bacteroidales, WCHB1-41, and Mollicutes_RF39. In particular, the abundance of Aeromonadales in the LGT population was lower than the abundance of the other populations, while the Bradymonadales was more abundant than the other populations. In the HY population we observed lower abundance of Methanomassiliicoccales compared to other populations. A total of 229 families were annotated at the family level, with Ruminococcaceae, Muribaculaceae, Lachnospiraceae, Rikenellaceae and Christensenellaceae being enriched in all four regional populations. In addition, we note that the abundance of Succinivibrionaceae is lower in the LGT population compared to the other populations, and that Acidobacteriaceae Subgroup 1 has a higher abundance compared to the other populations.
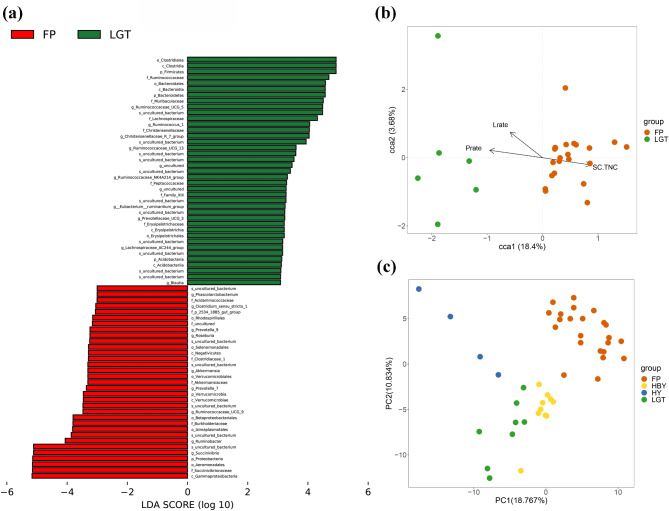
Cause the FP and LGT populations share similarities in their nutrient space, but significant differences exist in the quality of food intake, the energy supply from macronutrients, and the composition of the gut microbiota. To more effectively identify the specific gut microbiota responsible for the differences between the FP and LGT populations, we utilized LEfSe analysis to detect significant variations in gut microbiota composition between these two groups. The results, as depicted in Fig. 3a, indicate that certain microbial groups exhibit significant differences in abundance between the two populations. Specifically, the Christensenellaceae_R_7_group, Ruminococcaceae_NK4A214_group, Ruminococcaceae_UCG_13, Ruminococcaceae_UCG_5, and the Ruminococcus_1 genus is found to be more abundant in the LGT population. In contrast, the Rikenellaceae_RC9_gut_group, Ruminobacter, Ruminococcaceae_UCG_9, and Succinivibrio genus show a higher abundance in the FP population, marking a notable distinction between the FP and LGT populations in terms of their gut microbiota composition (Fig. 3a).
Fig. 3.
Differential gut microbiota, environmental influences and functional differences of golden snub-nosed monkey from different regions. (a) Differential gut microbiotas between FP and LGT populations of golden snub-nosed monkey. The groups of gut microbiotas shown are those that differ in different groupings, and the length of the bar graph indicates the magnitude of the differences. (b) Nutritional effects on microbial communities in FP and LGT. Each sample is represented by a point, while arrows emanating from the origin represent different environmental factors. The length of the arrows indicates the strength of the influence of each environmental factor on community variation; longer arrows signify greater impact. The angle between the arrows reflects their correlation: acute angles indicate positive correlation between two environmental factors, while obtuse angles indicate negative correlation. (c) PCA based on gut microbiota CAZy-family data of golden snub-nosed monkey from different regions
The results of the CCA indicate that the percentage of energy intake from available protein, the percentage of energy intake from lipid, and SC.TNC are associated with the structure of the gut microbial community. We found that the SC.TNC influence factor can well separate FP from LGT population (Fig. 3b). This indicates that carbohydrate macronutrient intake may plays an important role in the differentiation of gut microbiota in FP and LGT populations.
To estimate functional differences in the gut microbiota across the four populations, we used macro genomic sequencing methods to compare the annotation results of the CAZy database (Fig. 3c). PERMANOVA showed significant differences among all four populations. We noticed that the abundance of CAZy enzymes that dominate carbohydrate digestion were significantly different between FP and LGT populations. Previous studies have shown that carbohydrate metabolism is the most important secondary metabolic pathway in golden snub-nosed monkey populations [32], the significant difference result suggesting that carbohydrate intake has an important role in the composition and function of the gut microbiota in this study.
Upon meticulous analysis of the KEGG database annotations pertaining to carbohydrate metabolic pathways (Supplementary Fig. 2), we discerned an absence of significant variation across regional populations in the Citrate cycle (TCA cycle), Pyruvate metabolism, and Butanoate metabolism—pathways integral to cellular energy harnessing [33]. Furthermore, our result revealed a congruence between the relative abundance of Starch and sucrose metabolism, and Fructose and mannose metabolism pathways in distinct regional populations and the macronutrient intake and food components characteristic of those regions.
The hub OTUs fluctuated with the energy based on WGCNA analysis
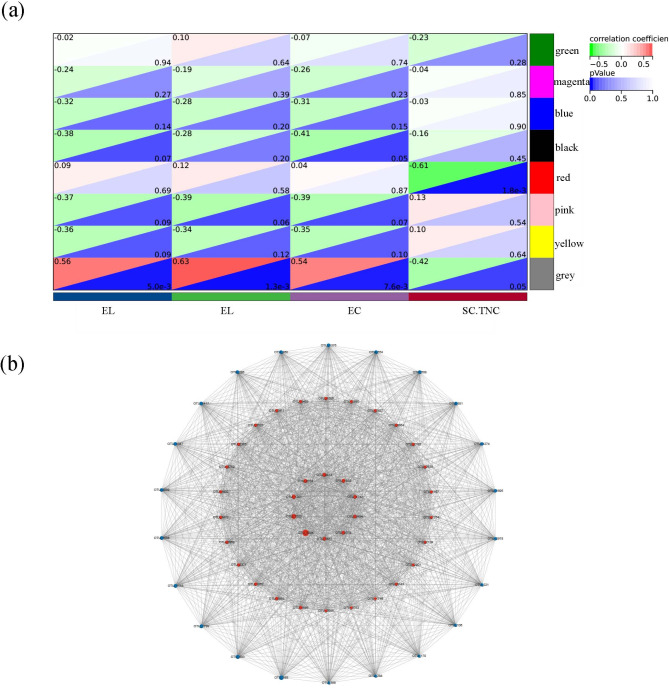
We selected the OTUs present in more than 33% of the samples in the FP and LGT subgroups, and then employed a one-step network building approach to construct the network. The network type is set to sign, and the soft threshold is set to 14. The adjacency matrix is defined based on criteria reflecting approximate scale-free topology, with a minimum module size of 60. The module detection sensitivity DeepSplit of 3. And modules correlated at 0.25 or above will be merged. Clustering results reveal that a total of 1855 OTUs are resolved into 8 different modules, where grey modules are unclassified. The correlation between module eigenvalue and trait was calculated. The module-trait relationship heatmap demonstrated the correlation coefficient between module eigenvalues and traits (Fig. 4a). 116 OTUs in the red module were significantly associated with the SC.TNC (P < 0.05).
Fig. 4.
Identification of key module and hub OTUs based on WGCNA. (a) Correlation between module eigenvalues and traits of golden snub-nosed monkey. Depth of colour corresponds to depth of correlation and P value of each module. (b) Network graph of the hub OTUs. Each node represented the OTUs who’s between centrality value was in the top 50%, and its colour represented the corresponding module, the size of each node represented the betweenness centrality value, the size of each line thickness represented the weight value between nodes (OTUs). Visualization of the complete weighted networks comprising 58 candidate hub OTUs associated with SC.TNC. Among them, 36 red nodes represent unique bacterial species in the LGT population, while 22 blue nodes denote species shared between the LGT and FP populations
We select the top 50% of OTUs based on them between centrality in the red module of WGCNA and construct a network graph in Cytoscape (Fig. 4b). 36 OTUs were exclusively found in the LGT population, while 22 OTUs were found in both the LGT and FP populations. In the red module, OTU_588, OTU_221, OTU_45, OTU_497, OTU_34, and OTU_12 was identified as candidate central OTUs (with thresholds of module membership > 0.6 and genes significance > 0.3). They belong to the following taxa: Lachnospiraceae (OTU_588), Lachnospiraceae_UCG-1 (OTU_221), Clostridiales_vadinBB60_group (OTU_497), Christensenellaceae_R-7_group (OTU_34), Rikenellaceae_RC9_gut_group (OTU_45), and Fibrobacter (OTU_12).
Discussion
Our findings indicate that the differences in food items and nutrient space among golden snub-nosed monkey populations from different regions do not fully correspond to the variations in macronutrient intake observed. Despite similarities or differences in food components among two regional populations, these do not necessarily reflect the nutrient spaces. Furthermore, the nutrient space itself does not fully represent the levels of macronutrient intake. We have observed a similar situation of unequal food intake differences and macronutrient intake differences in another generalist primate, the rhesus monkey, which is relatively specialized in macronutrient intake, despite significant differences in the food items of consumed by rhesus monkeys [34, 35]. This discrepancy may arise from environmental differences among populations. Even with the presence of similar or dissimilar food components, the varying or identical energy requirements of animals drive differences in macronutrient intake [36]. The phenomenon underscores the importance of employing a multidimensional approach, considering both food component and macronutrient intake, to interpret variation in animal dietary patterns [37].
Our results on the alpha diversity indices of the microbiota indicate that the richness of the gut microbiota decreased in the order of LGT, HBY, FP, and HY populations; and the diversity of the gut microbiota decreased in the order of HBY, LGT, HY, and FP populations (Supplementary Fig. 1). There are complex similarities or differences in gut microbiota community diversity among different populations. In light of this, placing the discussion of macronutrient intake’s effects on gut microbiota within a multidimensional context of food component and macronutrient intake will be beneficial for understanding the origins of gut microbiota community differences [21].
The ratio of Firmicutes to Bacteroidetes (F/B) indicates differences in the gut microbiota’s ability to intake nutrients [38]. Populations with higher crude fibre content in their nutrient intake tend to have higher F/B values in their gut microbiota [39]. The higher F/B ratio in FP and HBY populations may be attributed to their greater consumption of natural food compared to other populations, leading to increased intake of crude fibre (Fig. 1). This phenomenon illustrates that the F/B ratio in the gut microbiota correlate with the dietary patterns of populations in different regions. Similar to previous studies, we found that the enriched bacterial orders were found in populations from different regions with similar functions in assisting host digestion of nutrients [40] (Fig. 2). Abundance of Clostridium was positively correlated with crude fat apparent digestibility with fibre metabolism, especially crude fibre and acid detergent fibre apparent digestibility [41, 42]. Bacteroidales plays a role in cellulolytic and in the digestion of complex glycans [43, 44]. WCHB1-41 bacteria degrade mucins and convert them into short-chain fatty acids (SCFAs), which provide nutrients for other bacteria and cells [45]. Mollicutes_RF39 bacterial abundance was affected by polysaccharides such as oat β-glucan and chicory inulin [46]. Aeromonadales, found in lower abundance in LGT population, was shown to be one of the causative agents of intestinal inflammation [47], whereas Bradymonadales, found in higher abundance, was shown to be a predatory probiotic [48, 49]. We suggest that the increased abundance of Bradymonadales bacteria in the LGT population may have suppressed the abundance of harmful bacteria similar to Aeromonadales.
At the family level, gut microbiota found to be enriched in different populations are equally indicative of nutrient processing. Ruminococcaceae degrade cellulose polysaccharides and produce beneficial metabolites [50]. Muribaculaceae, widely present in gut microbiota, digest carbohydrates, induce lipolysis, and enhance insulin sensitivity [51]. Lachnospiraceae ferment dietary fibres to promote healthy gut and immune function [52]. Rikenellaceae and Christensenellaceae are associated with lipid metabolism [53]. In our study, Succinivibrionaceae exhibited lower abundance in the LGT population compared to other populations, while A. Subgroup1 showed higher abundance. Intriguingly, both bacteria are considered to be associated with nitrogen element absorption. We hypothesize that the bacterial population responsible for digesting proteins and utilizing nitrogen elements has undergone changes in the LGT population compared to other populations. The differences in these bacterial populations, which perform similar functions, may be attributed to variations in geographical environments [54]. Similarly, we found a higher abundance of Prevotellaceae in the HY population, which usually indicates a higher capacity of the host to digest simple carbohydrates [55]. We also observed the lowest abundance of Akkermansia in this population, and high abundance of this genus is usually negatively correlated with overall macronutrient intake [56]. The results of gut microbiota abundance coincide with the results that the HY population has the simplest food component and the highest macronutrient intake level compared with other populations.
We conducted a literature review of the functions of the gut microbiota resulting from the LEfSe analyses (Fig. 3a). We found that the differentially significant gut microbiota of LGT populations were strongly associated with carbohydrate digestion and were more abundant. For example, the abundance of the Christensenellaceae_R 7_group was positively correlated with the products acetate and butyrate associated with carbohydrate digestion [57]. Ruminococcaceae_NK4A214_group produces energy by fermenting dietary fibre [58]. Research on the genera Ruminococcaceae_UCG_13, Ruminococcaceae_UCG_5, and Ruminococcus_1 has demonstrated that Ruminococcaceae are involved in the breakdown of cellulose and starch, and they can produce products such as acetate that is associated with carbohydrate digestion [32, 59]. The gut microbiota results matched our macronutrient intake results, with the LGT populations having a higher carbohydrate intake than the FP populations.
Building upon previous research, we further elucidated the impact of carbohydrate energy intake on gut microbiota, utilizing the SC.TNC as a determinant of the proportion of energy derived from fibrous foods relative to the overall carbohydrate energy supply. We employed the WGCNA model to identify relevant gut microbial taxa and calculated the Chao1 index to assess the diversity across various populations. It was discovered that there existed a discrepancy between the FP and LGT populations (P < 0.05). Specifically, the Chao1 index of the LGT population was higher than that of the FP population, aligning with the nutritional findings indicating lower fibre intake in the LGT population. Concurrently, we observed no significant difference in the Chao1 index between the HBY and HY populations, which could be attributed to the similarity in the ratio of fibre to total food component between the two populations. These outcomes suggest that alterations in macronutrient intake resulting from changes in food consumption are among the factors influencing the composition and functionality of the gut microbiota in golden snub-nosed monkeys.
Some studies suggest that host evolution has a greater impact on gut microbiota than diet [60], they often only consider the types of food consumed when examining dietary influence. This study argues that such an approach is insufficient. This study has discovered that variations within the gut microbiota correspond to alterations in the host’s macronutrient intake. We propose that a comprehensive analysis, which takes into account various dimensions of food components and macronutrient intake, yields a more accurate understanding. This view is supported by studies observing the relationship between diet and gut microbiota across different species, such as rhesus monkeys, giant pandas, and others [21].
It is also noteworthy that the candidate central OTUs associated with carbohydrate digestion common to both the LGT and FP populations, are also widespread in other species and have important digestive functions. For instance, in humans Lachnospiraceae (order Clostridium) has a positive association with the digestion of beta-carotene, vitamin E, and accessible fat, while displaying a negative association with digestion of meat, total protein, and cholesterol [55]. Additionally, Lachnospiraceae (order Clostridium) is strongly influenced by non-starch polysaccharides [61]. In yaks, the C. vadinBB60 group was identified as enriched across multiple regions and seasons [45]. Additionally, the presence of Rikenellaceae_RC9_gut_group is associated with enhanced rumen function, promoting a more favourable environment for nutrient absorption and digestion [62]. The Christensenellaceae-R-7 group in chickens is linked to nutrient intake efficiency, propionate concentration, and lipid metabolism [63]. Fibrobacter plays a vital role in the metabolism and function of the microbiota of ruminants [64]. The similar to the patterns observed in this study, human gut microbiota research has identified a common “core microbiome” at the genus level across individuals, despite variations in the composition of gut microbiota among individuals [10]. This leads to the hypothesis that, regardless of feeding trait, the composition of the core gut microbiota performing digestive and absorptive functions will be similar if animals meet the same trend of macronutrient intake, whereas specific differences in food components and macronutrient intake will drive fluctuations in the composition and function of the non-core gut microbiota performing digestive and absorptive functions. The KEGG functional annotation outcomes corroborate our hypothesis. Although the relative abundance of the ‘Starch and sucrose metabolism’ and ‘Fructose and mannose metabolism’ pathways in populations from diverse geographical regions reflects a pattern of variation consistent with the changes in food component and macronutrient intake characteristic of those populations, the relative abundance of the core metabolic pathways associated with nutrient metabolism, specifically the Citrate cycle (TCA cycle) [65] and Pyruvate metabolism, exhibits no significant variation among these geographically distinct populations [66]. This observation implies an underlying uniformity in the central metabolic processes across populations, irrespective of their food component and macronutrient intake. This hypothesis helps explain why the gut microbiota is adaptive to dietary changes.
Conclusions
This study demonstrated some key results: First, although various regional populations of golden snub-nosed monkeys exhibited differences across different dietary dimensions (food component, nutrient space, and macronutrient intake), however, these disparities couldn’t be directly compared across dimensions. 2. Regional food intake differences led to shifts in macronutrient intake, thereby influencing the gut microbiota patterns observed in golden snub-nosed monkeys from various regions. 3. While the gut microbiota composition of golden snub-nosed monkeys varied notably among different regions, the core bacteria for digestive functions remained stable and enriched across different populations and taxonomic orders. The findings underscore the critical importance of considering various dietary dimensions when examining the adaptive responses to dietary alterations in generalist. Concurrently, the study reveals patterns of change within the gut microbiota that are pivotal for comprehending the mechanisms by which these microbial communities adjust to the ecological pressures induced by dietary shifts in generalist species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Y.L., Y.Y., P.Z., G.D., B.L and S.G. conceived the study and designed the experiments. G.F, H.W., X.W. and G.G. conducted the experiment, collected the data. Y.L., Y.Y., H.W., Y.M. and Y.Y. analyzed data, Y.L., Y.Y., D.D., J.J. and L.H. wrote the manuscript. All authors contributed to discussing the results and editing the manuscript and to the drafts and gave final approval for publication.
Funding
This work was supported by the Major International Joint Research Program of Natural Science Foundation of China under Grant (32220103002); National Natural Science Foundation of General Project under Grant (32370534); Natural Science Foundation of China under Grant (32371563); and Innovation Support Plan of Shaanxi Province under Grant S2021-ZC GHID-0013.
Data availability
The 16S rRNA sequence data supporting this study’s findings have been deposited at NCBI (BioProject:SUB14819847).
Declarations
Ethics approval
The animal study was reviewed and approved by the Ethics Committee of Northwest University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Slatyer RA, Hirst M, Sexton JP. Niche breadth predicts geographical range size: a general ecological pattern. Eco Lett. 2013;16(8):1104–14. 10.1111/ele.12140 [DOI] [PubMed] [Google Scholar]
- 2.Brown JH. On the relationship between abundance and distribution of species. AM NAT. 1984;124(2):255–79. 10.7208/9780226115504-035
- 3.Hardy NB, Kaczvinsky C, Bird G, Normark BB. What we don’t know about diet-breadth evolution in herbivorous insects. Annu Rev Ecol Evol S. 2020;51(1):103–22. 10.1146/annurev-ecolsys-011720-023322 [Google Scholar]
- 4.Hahn PG, Cammarano JH. Environmental context and herbivore traits mediate the strength of associational effects in a meta-analysis of crop diversity. J Appl Ecol. 2023;60(5):875–85. 10.1111/1365-2664.14382 [Google Scholar]
- 5.Hou R, Chapman CA, Jay O, Guo ST, Li BG, Raubenheimer D. Cold and hungry: combined effects of low temperature and resource scarcity on an edge-of‐range temperate primate, the golden snub‐nose monkey. Ecography. 2020;43(11):1672–82. 10.1111/ecog.05295 [Google Scholar]
- 6.Cui ZW, Wang ZL, Zhang SQ, Wang BS, Lu JQ, Raubenheimer D. Living near the limits: effects of interannual variation in food availability on diet and reproduction in a temperate primate, the Taihangshan macaque (Macaca mulatta tcheliensis). Am J Primatol. 2020;82(1):e23080. 10.1002/ajp.23080 [DOI] [PubMed] [Google Scholar]
- 7.Raubenheimer D, Hou R, Dong YL, Ren CR, Cui ZW. Towards an integrated understanding of dietary phenotypes. Philos T R Soc B. 2023;378(1891):20220545. 10.1098/rstb.2022.0545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Eco Lett. 2014;17(10):1238–46. 10.1111/ele.12329 [DOI] [PubMed] [Google Scholar]
- 9.Greene LK, Williams CV, Junge RE, Mahefarisoa KL, Rajaonarivelo T, Rakotondrainibe H, et al. A role for gut microbiota in host niche differentiation. ISME J. 2020;14(7):1675–87. 10.1038/s41396-020-0640-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amato KR, Carmody RN. Gut microbial intersections with human ecology and evolution. Annu Rev Anthropol. 2023;52:295–311. 10.1146/annurev-anthro-052721-085122 [Google Scholar]
- 11.Moore BD, Foley WJ, Wallis IR, Cowling A, Handasyde KA. Eucalyptus foliar chemistry explains selective feeding by koalas. Biol Lett. 2005;1(1):64–7. 10.1098/rsbl.2004.0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Schaik CP, Brockman DK. Seasonality in primates: studies of living and extinct human and non-human primates. Cambridge University Press; 2005. 10.1017/CBO9780511542343.002
- 13.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baniel A, Amato KR, Beehner JC, Bergman TJ, Mercer A, Perlman RF, et al. Seasonal shifts in the gut microbiome indicate plastic responses to diet in wild geladas. Microbiome. 2021;9(1):26. 10.1186/s40168-020-00977-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu WW, Wang Q, Song JJ, Xin JW, Zhang SS, Lei YH, et al. Comparison of gut microbiota of yaks from different geographical regions. Front Microbiol. 2021;12:666940. 10.3389/fmicb.2021.666940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han LL, Xue WC, Cao HW, Chen XY, Qi FS, Ma T, et al. Comparison of rumen fermentation parameters and microbiota of yaks from different altitude regions in Tibet, China. Front Microbiol. 2022;12:807512. 10.3389/fmicb.2021.807512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YH, Yan YJ, Fu HG, Jin SY, He SJ, Wang Z, et al. Does diet or macronutrients intake drive the structure and function of gut microbiota? Front Microbiol. 2023;14:1126189. 10.3389/fmicb.2023.1126189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raubenheimer D. Toward a quantitative nutritional ecology: the right-angled mixture triangle. Ecol Monogr. 2011;81(3):407–27. 10.1890/10-1707.1 [Google Scholar]
- 19.Amato KR, Leigh SR, Kent A, Mackie RI, Yeoman CJ, Stumpf RM, et al. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb Ecol. 2015;69(2):434–43. 10.1007/s00248-014-0554-7 [DOI] [PubMed] [Google Scholar]
- 20.Koch F, Ganzhorn JU, Rothman JM, Chapman CA, Fichtel C. Sex and seasonal differences in diet and nutrient intake in Verreaux’s sifakas (Propithecus verreauxi). Am J Primatol. 2017;79(4):1–10. 10.1002/ajp.22595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui ZW, Holmes AJ, Zhang WJ, Hu DL, Shao Q, Wang ZL, et al. Seasonal diet and microbiome shifts in wild rhesus macaques are better correlated at the level of nutrient components than food items. Integr Zool. 2022;17(6):1147–61. 10.1111/1749-4877.12601 [DOI] [PubMed] [Google Scholar]
- 22.Rothman JM, Plumptre AJ, Dierenfeld ES, Pell AN. Nutritional composition of the diet of the gorilla (Gorilla beringei): a comparison between two montane habitats. J Trop Ecol. 2007;23:673–82. 10.1017/S0266467407004555 [Google Scholar]
- 23.Ryan AM, Chapman CA, Rothman JM. How do differences in species and part consumption affect diet nutrient concentrations? A test with red colobus monkeys in Kibale National Park, Uganda. Afr J Ecol. 2013;51(1):1–10. 10.1111/j.1365-2028.2012.01346.x [Google Scholar]
- 24.Ren BP, Li BG, Li M, Wei FW. Inter-population variation of diets of golden snub-nosed monkeys (Rhinopithecus roxellana) in China. ATS. 2010;30(4):357–64. 10.16829/j.slxb.2010.04.001 [Google Scholar]
- 25.Hou R, He SJ, Wu F, Chapman CA, Pan R, Garber PA, et al. Seasonal variation in diet and nutrition of the northern-most population of Rhinopithecus roxellana. Am J Primatol. 2018;80(4):e22755. 10.1002/ajp.22755 [DOI] [PubMed] [Google Scholar]
- 26.Van Soest PJ. Nutritional ecology of the ruminant. Cornell university press. 1994.
- 27.Rothman JM, Chapman CA, Soest PJV. Methods in primate nutritional ecology: a user’s guide. Int J Primatol. 2012;33(3):542–66. 10.1007/s10764-011-9568-x [Google Scholar]
- 28.Guo ST, Hou R, Garber PA, Raubenheimer D, Righini N, Ji WH, et al. Nutrient-specific compensation for seasonal cold stress in a free‐ranging temperate colobine monkey. FUNCT ECOL. 2018;32(9):2170–80. 10.1111/1365-2435.13134 [Google Scholar]
- 29.Conklin-Brittain NL, Knott CD, Wrangham RW. Energy intake by wild chimpanzees and orangutans: methodological considerations and a preliminary comparison. Feeding Ecol apes Other primates. 2006;48:445–71. [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci Usa. 2011;108(1):4516–22. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YT, Yang XY, Zhang MY, Pan HJ. Comparative analysis of gut microbiota between wild and captive golden snub-nosed monkeys. Animals-Basel. 2023;13(10):1625. 10.3390/ani13101625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui ZW, Wang ZL, Shao Q, Raubenheimer D, Lu JQ. Macronutrient signature of dietary generalism in an ecologically diverse primate in the wild. Behav Ecol. 2018;29(4):804–13. 10.1093/beheco/ary003 [Google Scholar]
- 35.Cui ZW, Shao Q, Grueter CC, Wang ZL, Lu JQ, Raubenheimer D. Dietary diversity of an ecological and macronutritional generalist primate in a harsh high-latitude habitat, the Taihangshan macaque (Macaca mulatta tcheliensis). Am J Primatol. 2019;81(4):e22965. 10.1002/ajp.22965 [DOI] [PubMed] [Google Scholar]
- 36.Raubenheimer D, Simpson SJ. Eat like the animals: what nature teaches us about the science of healthy eating. New York: HarperCollins; 2020. 10.1093/emph/eoab024 [Google Scholar]
- 37.Machovsky-Capuska GE, Senior AM, Simpson SJ, Raubenheimer D. The multidimensional nutritional niche. Trends Ecol Evol. 2016;31(5):355–65. 10.1016/j.tree.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 38.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–51. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 40.Kuthyar S, Watson K, Huang S, Brent LJ, Platt M, Horvath J, et al. Limited microbiome differences in captive and semi-wild primate populations consuming similar diets. Fems Microbiol Ecol. 2022;98(10):fiac098. 10.1093/femsec/fiac098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varel VH, Richardson AJ, Stewart CS. Degradation of barley straw, ryegrass, and alfalfa cell walls by Clostridium Longisporum and Ruminococcus albus. Appl Environ Microb. 1989;55(12):3080–4. 10.1128/aem.55.12.3080-3084.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M. Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. Mbio. 2014;5(5):e01530–14. 10.1128/mbio.01530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J Biol Chem. 2009;284(37):24673–7. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda S, Hongoh Y, Sato T, Ohkuma M. Complex coevolutionary history of symbiotic Bacteroidales bacteria of various protists in the gut of termites. Bmc Evol Biol. 2009;9:1–12. 10.1186/1471-2148-9-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo N, Wu QF, Shi FY, Niu JH, Zhang T, Degen AA, et al. Seasonal dynamics of diet-gut microbiota interaction in adaptation of yaks to life at high altitude. Npj Biofilms Microbi. 2021;7(1):38. 10.1038/s41522-021-00207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng WY, Lam KL, Li X, Kong APS, Cheung PCK. Circadian disruption-induced metabolic syndrome in mice is ameliorated by oat β-glucan mediated by gut microbiota. Carbohyd Polym. 2021;267:118216. 10.1016/j.carbpol.2021.118216 [DOI] [PubMed] [Google Scholar]
- 47.Zhang JH, Meng H, Kong XC, Cheng XY, Ma T, He H, et al. Combined effects of polyethylene and organic contaminant on zebrafish (Danio rerio): accumulation of 9-nitroanthracene, biomarkers, and intestinal microbiota. Environ Pollut. 2021;277:116767. 10.1016/j.envpol.2021.116767 [DOI] [PubMed] [Google Scholar]
- 48.Wang S, Mu D, Du ZJ. Persicimonas caeni gen. nov., sp. nov., the representative of a novel wide-ranging predatory Taxon in Bradymonadales. Front Microbiol. 2020;11:698. 10.3389/fmicb.2020.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Chiu S, Wang S, Liao Y, Chang H, Ballantyne R, et al. Dietary SYNSEA probiotic improves the growth of white shrimp, Litopenaeus vannamei, and reduces the risk of Vibrio infection via improving immunity and intestinal microbiota of shrimp. Fish Shellfish Immun. 2022;127:482–91. 10.1016/j.fsi.2022.06.071 [DOI] [PubMed] [Google Scholar]
- 50.Ma YH, Deng XT, Yang X, Wang JK, Li T, Hua GY, et al. Characteristics of bacterial microbiota in different intestinal segments of Aohan fine-wool sheep. Front Microbiol. 2022;13:874536. 10.3389/fmicb.2022.874536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ou YJ, Guo YH, Chen MR, Lu XD, Guo ZB, Zheng BD. Gut microbiome–serum metabolic profiles: insight into the hypoglycemic effect of Porphyra haitanensis glycoprotein on hyperglycemic mice. Food Funct. 2023;14(17):7977–91. 10.1039/D3FO02040A [DOI] [PubMed] [Google Scholar]
- 52.Zaplana T, Miele S, Tolonen AC. Lachnospiraceae are emerging industrial biocatalysts and biotherapeutics. Front Bioeng Biotech. 2024;11:1324396. 10.3389/fbioe.2023.1324396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebastià C, Folch JM, Ballester M, Estellé J, Passols M, Muñoz M, et al. Interrelation between gut microbiota, SCFA, and fatty acid composition in pigs. Msystems. 2023;99(1):e0104923. 10.1128/msystems.01049-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang LW, Luo W, Dai QL, Zhou H, Wei W, Tang JF, et al. Giant pandas’ staple food bamboo phyllosphere fungal community and its influencing factors. Front Microbiol. 2022;13:1009588. 10.3389/fmicb.2022.1009588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Angelis M, Ferrocino I, Calabrese FM, De Filippis F, Cavallo N, Siragusa S, et al. Diet influences the functions of the human intestinal microbiome. Sci Rep-Uk. 2020;10(1):4247. 10.1038/s41598-020-61192-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. P Natl Acad Sci Usa. 2013;110(22):9066–71. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebeyew K, Chen K, Wassie T, Azad MAK, He JH, Jiang WM, et al. Dietary amylose/amylopectin ratio modulates cecal microbiota and metabolites in weaned goats. Front Nutr. 2021;8:774766. 10.3389/fnut.2021.774766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu CS, Ding LM, Jiang CX, Ma CF, Liu BT, Li DL, et al. Effects of management, dietary intake, and genotype on rumen morphology, fermentation, and microbiota, and on meat quality in yaks and cattle. Front Nutr. 2021;8:755255. 10.3389/fnut.2021.755255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang XB, Wu XY, Shang YQ, Gao Y, Li Y, Wei QG, et al. High-altitude drives the convergent evolution of alpha diversity and indicator microbiota in the gut microbiomes of ungulates. Front Microbiol. 2022;13:953234. 10.3389/fmicb.2022.953234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amato KR, Jeyakumar T, Poinar H, Gros P. Shifting climates, foods, and diseases: the human microbiome through evolution. BioEssays. 2019;41(10). 10.1002/bies.201900034. Article 1900034. [DOI] [PubMed]
- 61.Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. Isme J. 2014;8(11):2218–30. 10.1038/ismej.2014.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang C, Ge F, Yao XX, Guo X, Bao PJ, Ma XM, et al. Microbiome and metabolomics reveal the effects of different feeding systems on the growth and ruminal development of yaks. Front Microbiol. 2021;12:682989. 10.3389/fmicb.2021.682989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He ZX, Liu RR, Wang MJ, Wang Q, Zheng JM, Ding JQ, et al. Combined effect of microbially derived cecal SCFA and host genetics on feed efficiency in broiler chickens. Microbiome. 2023;11(1):198. 10.1186/s40168-023-01627-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizrahi I, Wallace RJ, Moraïs S. The rumen microbiome: balancing food security and environmental impacts. Nat Rev Microbiol. 2021;19(9):553–66. 10.1038/s41579-021-00543-6 [DOI] [PubMed] [Google Scholar]
- 65.Buchanan BB, Arnon DI. A reverse KREBS cycle in photosynthesis: consensus at last. Photosynth Res. 1990;24(1):47–53. 10.1007/BF00032643 [DOI] [PubMed] [Google Scholar]
- 66.Su YB, Peng B, Li H, Cheng ZX, Zhang TT, Zhu JX et al. Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. P Natl Acad Sci USA. 2018;115(7):E1578–87. 10.1073/pnas.1714645115 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequence data supporting this study’s findings have been deposited at NCBI (BioProject:SUB14819847).