Abstract
Background
Acinetobacter baumannii is a gram-negative, opportunistic pathogen, that is responsible for a wide variety of infections and is a significant cause of hospital-acquired infections. A. baumannii is listed by the World Health Organization (WHO) as a critical priority pathogen because of its high level of antibiotic resistance and the urgent need for alternative treatment solutions. To address this challenge, bacteriophages have been used to combat bacterial infections for more than a century, and phage research has regained interest in recent years due to antimicrobial resistance (AMR). However, although the vast majority of deaths from the AMR crisis will occur in developing countries in Africa and Asia, few phages’ studies have been conducted in these regions. In this study, we present a comprehensive characterization of the bacteriophages vAbBal23 and vAbAbd25, actives against extremely drug-resistant (XDR) A. baumannii.
Methods
Phages were isolated from environmental wastewaters in Dakar, Senegal. The host-range, thermal and pH stabilities, infection kinetics, one step growth assay, antibiofilm activity assay, sequencing, and genomic analysis, were performed to characterize the isolated phages.
Results
Comparative genomic and phylogenetic analyses revealed that vAbBal23 and vAbAbd25 belong to the Caudoviricetes class, Autographiviridae family and Friunavirus genus. Both phages demonstrated activity against strains with capsular type KL230. They were stable over a wide pH range (pH 3 to 9) and at temperatures ranging from 25 °C to 40 °C. Additionally, the phages exhibited notable activity against both planktonic and biofilm cells of targeted extremely drug resistant A. baumannii. The results presented here indicate the lytic nature of vAbBal23 and vAbAbd25. This is further supported by the absence of genes encoding toxins, resistance genes and bacterial virulence factors, highlighting their potential for future phage applications.
Conclusion
Phages vAbBal23 and vAbAbd25 are promising biological agents that can infect A. baumannii, making them suitable candidates for use in phage therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03608-7.
Keywords: Bacteriophage, vAbBal23, vAbAbd25, Characterization, Acinetobacter baumannii, Antimicrobial resistance
Introduction
Acinetobacter baumannii is a gram-negative opportunistic pathogen that is primarily responsible for a variety of infections, including pneumonia, urinary tract infections, ventilator-associated pneumonia, and bacteremia. This bacterium is widely distributed in diverse environments such as water, soil, food, activated sludge, and the human body [1, 2], A. baumannii poses a significant challenge as a nosocomial pathogen, contributing to multidrug-resistant (MDR) infections and representing approximately 20% of infections in intensive care units worldwide [3]. Unfortunately, over 80% of A. baumannii strains exhibit resistance to antibiotics, including β-lactams, carbapenems, aminosids and cyclins, owing to the presence of multiple genetic elements encoding antimicrobial resistance [4, 5]. Despite the use of carbapenems for treating A. baumannii infections, a recent surge in carbapenem resistance represents an important challenge in treatment [6]. One of the pivotal virulence characteristics of this bacterium is its ability to form biofilms, which are communities of bacteria encased within a self-produced extracellular matrix [7] that adhere to either biotic or abiotic surfaces [8]. Within biofilms, bacteria exhibit high resistance to antibiotics and disinfectants [9], making their inhibition or eradication challenging in various settings, including husbandry, the food industry, and clinical environments [10]. Additionally, antibiotic resistance facilitates bacterial dissemination, potentially exacerbating the prevalence of healthcare associated infections caused by these bacteria [11]. According to the World Health Organization, A. baumannii ranks among the most dangerous pathogens, underscoring the urgent need for novel therapeutic strategies [12]. Given the global rise in antibiotic resistance, bacteriophage therapy has reemerged as a promising avenue of investigation [13].
Bacteriophages are viruses capable of targeting and eliminating specific bacteria. Bacteriophages are ubiquitous in environments containing their host bacteria, and play pivotal roles in various biological processes [14]. While they were employed for treating various diseases until the 1940s, the advent of antibiotics in 1941 led to a decline in the use of bacteriophage therapy, although it persisted as a treatment modality in Soviet-aligned countries [15]. The key characteristics of phages include their specificity and ability to replicate within the host and at the site of infection without adverse effects [16]. In recent decades, a growing body of evidence from several studies has demonstrated the efficacy of phage therapy in treating drug-resistant bacterial infections [17], owing to its bacteriolytic activity and ability to eradicate bacterial cells within biofilms [18].
This study explores the properties of phages isolated from environmental wastewaters in Dakar, Senegal that target extremely drug resistant (XDR) A. baumannii isolates. The biological properties of the potential therapeutic phages, including plaque morphology, host range, stability, burst size, and planktonic and biofilm activity, were evaluated. In addition, the whole-genome annotation of phages was investigated, which provides vital information for further therapeutic development and applications.
Materials and methods
Collection and identification of isolates
In this study, all 13 XDR A. baumannii strains from hospital acquired infections were collected from the biobanks of two routine laboratories at tertiary hospitals in Dakar. Conventional phenotypic identification via the disk diffusion method confirmed all A. baumannii isolates as XDR, according to the criteria defined by Magiorakos et al. [19]. The K locus types (KL) were determined through whole genome sequencing (WGS) and subsequent bioinformatic analysis using Kaptive 2.0 [20]. The bacterial cultures were grown in Luria Bertani (LB) broth or on agar (Difco Laboratories, Detroit, MI, USA) at 37 °C. Bacterial growth was monitored turbidimetrically by measuring the optical density at 600 nm (OD600), with an OD unit of 1.0 corresponding to 3 × 108 cells/mL.
Isolation, purification, and host range of bacteriophages
Bacteriophages specific to A. baumannii were isolated from environmental wastewater sources in Dakar, Senegal. The presence of phages was initially assessed using the double layer method, as outlined by Kusradze et al. [21]. In summary, a 1 mL wastewater sample, was centrifuged and filtered through a 0.22-µm-pore-size membrane (Millex®, syringe filter). After, 10 µL of the filtered sample was added on an overnight bacterial solution and inoculated into 2.5 mL of melted nutrient agar medium (0.7%). The resulting mixture was poured onto a plate containing 1.5% nutrient agar, creating two-layer plates. After the top layer solidified, the plates were incubated at 37 °C for 24 h. The formation of plaques on the plate indicates bacterial susceptibility to the phage. Single-plaque isolation was subsequently conducted for phage purification. The host range of the phages was evaluated through a spot test by testing a serial dilution of the phage stock (1010 PFU/mL) against 13 clinical XDR isolates and determining the efficiency of plating (EOP).
Effects of temperature and pH on the phage stability
To evaluate the thermal stability of the phages, 100 µL of phage solution (1 × 1010 PFU/mL) was incubated at temperatures ranging from 25 °C to 70 °C for 1 h. Following incubation, 50 µL samples were collected for phage titration. These samples were then subjected to 10-fold dilution, and 4 µL of each dilution was spotted onto LB agar plates containing host bacteria, followed by overnight incubation. The plates were subsequently examined to determine phage titers. For the assessment of pH stability, 10-fold diluted aliquots were prepared after 1 h of incubation in SM buffer at various pH values (3, 5, 9, and 12), with pH 7.5 serving as the control. The phage titers were then determined. All experiments were conducted in triplicate biological assays, with each assay comprising triplicate plaque assays.
In vitro activity of phages against XDR A. baumannii
The in vitro bacteriolytic activity of the phages against their respective bacterial hosts was evaluated at various multiplicity of infection (MOIs) values, following the method described by Carlson [22]. Overnight bacterial cultures were diluted to an optical density at 600 nm (OD600) of 0.2 in fresh LB medium and incubated at 37 °C until they reached the mid-log phase. The microplates were then prepared by adding bacterial suspensions and phages at different MOIs (1, 10− 1, 10− 3), followed by incubation at 37 °C with agitation. The control group consisted of A. baumannii isolates incubated with LB medium without phages. Bacterial growth was monitored by measuring the OD600 every 10 min using a spectrophotometer for 16 h. Each experiment was independently conducted in triplicate, with triplicate assays performed for each replicate.
For one-step growth experiments, a previously described method was employed [23]. Briefly, following phage adsorption at room temperature for 5 min and centrifugation at 10,000 g for 30 s, the supernatant was removed. The pelleted cells were resuspended in 20 mL of preheated (37 °C) LB broth and centrifuged at 10,000 g for 30 s (repeated 3 times). The final resuspended pellet was then incubated at 37 °C. Samples were collected at 5 min intervals, and phage titers were immediately determined. Each experiment was replicated three times.
Biofilm formation and the antibacterial activity of the phage against biofilms
The antibiofilm activities of phages vAbBal23 and vAbAbd25 were experimentally assessed at two different MOIs (10 and 1). To evaluate the effects of the phages on the biofilm formation of the host strain AB12, we conducted inhibition experiments on bacterial growth in 96-well plates and the OD600 values of each well were measured at 24 h post bacterial inoculation. We investigated the effects of phages on pre-existing biofilms formed by two XDR strains using a method described previously by Zaki et al. [24]. Overnight cultures of each strain were diluted 1:10 in fresh LB medium and incubated at 37 °C until an OD600 of 0.2 was reached. Subsequently, 100 µL of each culture was transferred to individual wells of a 96-well microtiter plate and incubated at 37 °C for 24 h to allow biofilm formation. Following incubation, the plates were emptied and cleaned three times with phosphate buffer saline (PBS). Each well was then treated with either 100 µL of the phage solution in SM buffer (pH 7.4) at various MOIs (1 and 10) or supplemented with LB medium. The plates were further incubated for 24 h at 37 °C. Following the incubation period, the wells were emptied and cleaned twice with sterilized PBS. Subsequently, each well was stained with 150 µL of 1% crystal violet. After 15 min of staining at room temperature, the wells were rinsed with sterile water, and solubilized in 150 µL of 100% ethanol for 10 min. A volume of 100 µL from each well was then transferred to a fresh 96-well plate. The reduction in biomass was determined by measuring the difference in absorbance at 600 nm between the control (untreated) and phage-treated wells. Each experiment was replicated at least three times. For quantification, after incubation of the plates for 24 h, the supernatants were collected for planktonic cell counting and adherent cells were scraped from the surface of the well using pipette tips and suspended in PBS for biofilm cell counting. Serial dilutions of suspended cells were then plated for bacterial counting. Each experiment was replicated three times.
Genomic DNA extraction
Genomic DNA (gDNA) was isolated from high-titer stocks (> 1010 PFU/mL). Initially, 1 mL of phage lysate was treated with 10 µL of DNase I (20 U) and 4 µL of RNase A (20 mg/mL), followed by incubation for 30 min at 37 °C. DNA extraction was then performed using the phenol-chloroform method [25]. Proteins were removed and phage DNA was purified by two cycles of phenol/chloroform/isoamyl alcohol extractions and DNA was precipitated with isopropanol. After washing in 70% ethanol, the pellets were resuspended in 30 µL of water. Nucleic acid concentrations were quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific) to assess sample quality.
Genome sequencing and bioinformatic analysis of sequencing data
For library preparation, 1 ng of DNA was utilized with Nextera XT DNA library preparation kits (Illumina®, San Diego, CA, USA), following the manufacturer’s protocol. WGS was conducted on Illumina® iSeq100 sequencers, with a 300-cycle i1 Reagent V2 Kit (Illumina®, San Diego, CA, USA). Quality control of the reads was performed using FastQC v0.12.1 [26], followed by adaptor trimming with trim-galore v0.6.10 [27]. The trimmed paired-end reads were de novo assembled using SPAdes v.3.15.5 [28] in careful mode. Coverage per contig and assembly validation were assessed using BBMap v 35.85 with default parameters [29], and contaminating host DNA was manually removed. The input genome, sorted and indexed using Samtools v1.18 [30], and assembly error corrections were performed using Pilon v1.24 with default parameters [31]. The predicted coding sequences (CDS), transfer RNAs (tRNAs), transfer-messenger RNAs (tmRNAs), virulence factors (VFs), antimicrobial resistance genes (AMRs), clustered regularly interspaced short palindromic repeats (CRISPRs), and functional annotations for CDSs were identified using Pharokka v1.3.0 with default parameters [32]. Circular genome visualization was performed using the option pharokka_plotter. The whole genome sequences of phages underwent phylogenetic analyses utilizing the Virus Classification and Tree building Online Resource (VICTOR) (https://ggdc.dsmz.de/victor.php, accessed on 23 April 2024). Phage sequences were sourced from the NCBI nucleotide database. Pairwise comparisons of the nucleotide sequences were performed using the Genome-BLAST Distance Phylogeny (GBDP) method, with the recommended settings for prokaryotic viruses [33]. In accordance with ICTV recommendations, the phage taxonomy was refined using VIRIDIC (http://rhea.icbm.uni-oldenburg.de/VIRIDIC/, accessed on 23 April 2024), which employs the virus intergenomic distance calculation method under the BLASTn default settings [34]. Double-stranded DNA (dsDNA) bacteriophages are classified into the same species if they share an average nucleotide identity (ANI) of 95% or greater. Phages with an ANI of at least 70% across the entire genome are grouped within the same genus [34, 35].
Results
Antimicrobial susceptibility testing
The A. baumannii strains, which were classified as sensitive or resistant, presented varying levels of susceptibility to the tested antibiotics, and a resistance phenotype was determined. The bacterial strains showed resistance major antibiotics such as imipenem, tobramycin, tetracyclin and ciprofloxacin. All the isolates were characterized as XDR (Supplementary Table 1).
Phage isolation, purification and host range
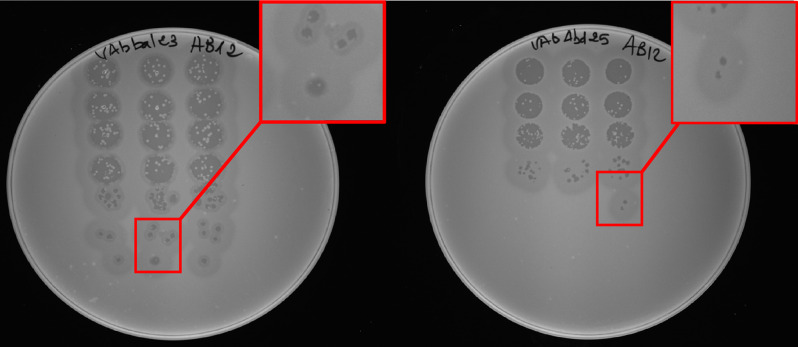
Two phages vAbBal23 and vAbAbd25 were isolated from environmental wastewater samples, using XDR A. baumannii AB12 as the host strain. Strain AB12 belong to the sequence type 164 (ST164) and has the capsular type KL230. A spot test of the serial diluted phages on a bacterial lawn of strain AB12 was done and phage vAbBal23 formed clear plaques 1–2 mm in diameter with a halo, whereas phage vAbAbd25 formed clear plaques < 1 mm in diameter without a halo (Fig. 1). Host range analysis showed that among the 7 different KL types, both phages could infect only two strains with the same KL type (KL230) (Supplementary Table 2).
Fig. 1.
Plaque morphology of phage vAbBal23 (clear plaque 2 mm with a halo) and vAbAbd25 (clear plaque, < 1 mm without a halo) on isolate AB12
Physicochemical stability of the phages
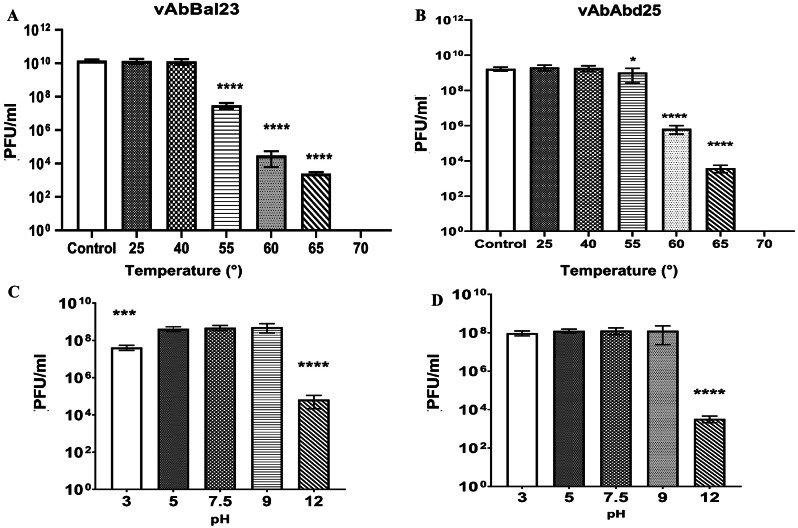
We assessed the ability of phage particles to remain infectious across a range of temperatures and nonneutral pH solutions. Each experiment was conducted in triplicate repetitions and p values (“*” for p-value < 0.05, “**” for p-value < 0.01, “***” for p-value < 0.001, and “****” for p-value < 0.0001) were used for statistical and quantitative analysis. The temperature stability tests revealed that after one hour of exposure, both phages remained fully infectious at temperatures up to 40 °C, but they lost titer at higher temperatures. A thermal transition was observed at a midpoint of approximately 55 °C (p < 0.0001 for vAbBal23, p = 0.0319 for vAbAbd25), with a subsequent decrease in phage viability up to 65 °C (p < 0.0001), and no viable phage was observed at 70 °C (p < 0.0001) (Fig. 2A and B). The optimal pH values for maintaining phage activity ranged from 5 to 9. However, the titer of phages was significantly reduced when they were exposed to an alkaline buffer solution at pH 12 (p < 0.0001). Additionally, the two phages showed different tolerances to acidic environments; vAbAbd25 remained stable at pH 3, whereas vAbBal23 showed a slight decrease in phage viability (p < 0.0001) after being exposed to a pH 3 buffer for one hour (Fig. 2C and D).
Fig. 2.
Stability of phages vAbBal23 and vAbAbd25 under various conditions. (A) and (B) Effects of temperature on the stability of phages. The phage was incubated at different temperatures for 1 h. (C) and (D) Effects of pH on the stability of phages vAbBal23 and vAbAbd25. The phage was incubated for 1 h at different pH values. The experiments were independently performed in triplicate with triplicate assay. The data are presented as the means ± standard errors from three replicates. “ns” means no significant difference, “*” means p < 0.05, “**” means p < 0.01, “***” means p < 0.001, “****” means p < 0.0001
Infection dynamics and one step growth assay
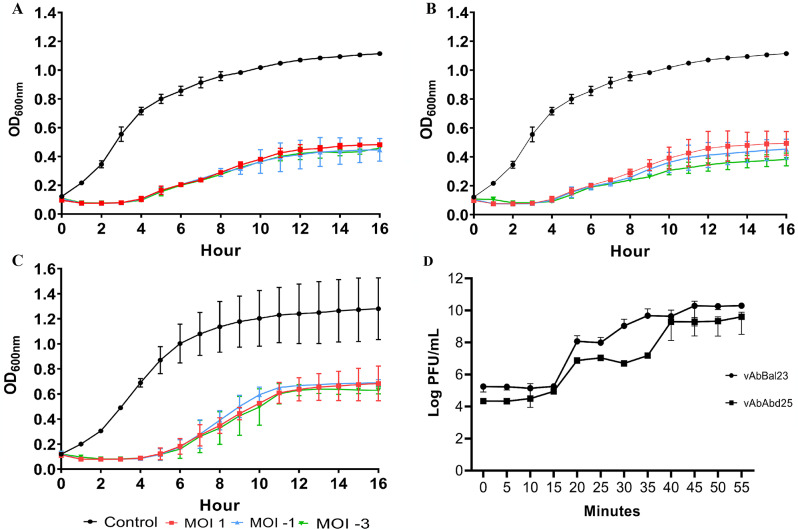
The growth curves of the host strain AB12 were monitored after treatment with vAbBal23 and vAbAbd25 by measuring changes in the OD600 nm over a 16 h incubation period. Comparisons were made between untreated bacterial growth and growth in the presence of phage lysate at different MOIs (1, 10− 1, 10− 3). The results revealed a steady increase in growth for uninfected XDR A. baumannii isolates, whereas treatment with phages significantly inhibited bacterial growth. Both phages showed similar bactericidal activity, with phage vAbBal23 treatment resulting in a 70% reduction in bacterial growth and vAbAbd25 treatment showing a 71.7% of reduction. Treatment with a combination of both phages resulted in a slight increase in bactericidal reduction to 75.8%. No significant differences were observed with different MOI treatments (Fig. 3A, B and C).
Fig. 3.
In vitro planktonic cell lysis assay evaluating phage A) vAbBal23 against strain AB12, B) vAbAbd25 against strain AB12 and C) a combination of phages vAbBal23-vAbAbd25 against strain AB12. Each phage was studied at multiplicities of infection (MOIs) 1, 10− 1, and 10− 3. The data represent the means and standard errors from three biological and technical triplicate experiments for single phage treatment and triplicate experiments for phage cocktail treatment. D) One-step growth curve of phages vAbBal23 and vAbAbd25 on AB12. The data points indicate the PFUs/mL at different time points. Each data point represents the mean of three independent measurements
According to the one-step growth assay, the latent period for both phages was approximately 15 min, defined as the time between adsorption and the start of the initial burst. The estimated burst size, the average number of new phage particles released per infected cell, was 2 103 and 270 PFU/cell for vAbBal23 and vAbAbd25, respectively (Fig. 3D).
Biofilm assay
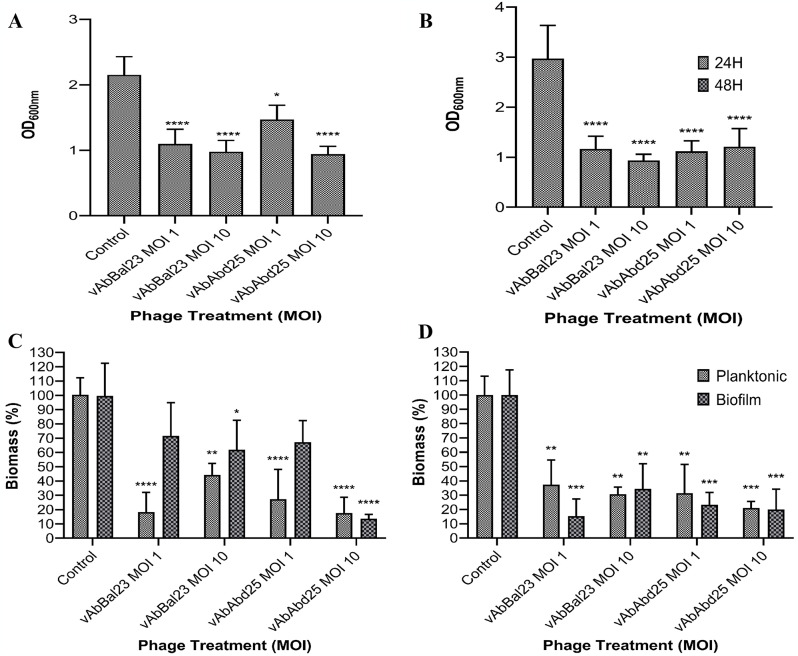
Both phages, at all assayed MOIs, significantly succeeded in subverting biofilms (p < 0.05). In co-cultures set-up at MOI 1, significant losses (p < 0,05) of approximately 70% and 71.7% biomass were observed after 24 h for vAbBal23 and vAbAbd25 on AB12 (Fig. 4A and B). For the viable count assay, the results demonstrated significant reductions (p < 0.05) in both planktonic and biofilm cells for the phages vAbBal23 and vAbAbd25 (Fig. 4C and D).
Fig. 4.
Efficacity of phages vAbBal23 and vAbAbd25 to reduce preformed biofilms for strains (A) AB12, (B) AB13 and quantification of planktonic and biofilm (C) AB12, (D) AB13 after phage treatments. The biofilm biomass and bacterial cell viability were determined by the crystal violet assay and the colony counting method, respectively. The data are illustrated in a violin plot derived from a biological triplicate and technical triplicate experiments. The statistical significance is denoted as “*” for p-value < 0.05, “**” for p-value < 0.01, “***” for p-value < 0.001, and “****” for p-value < 0.0001
Genomic characterization
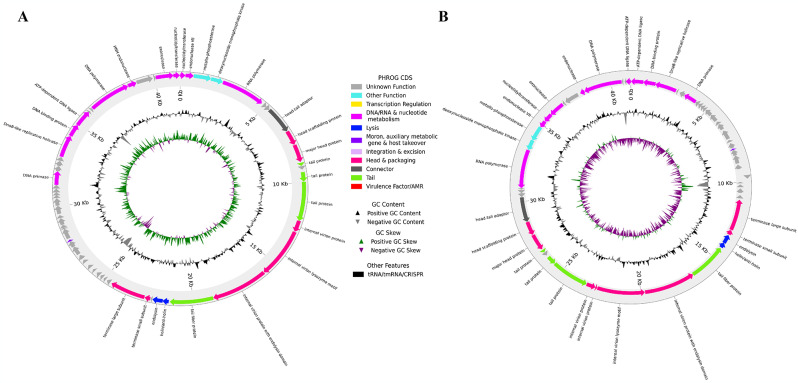
The genomes of phages vAbBal23 and vAbAbd25 were sequenced and duly assembled, resulting in contigs of 41,386 bp and 41,193 bp genome size, respectively. The GC content of the vAbBal23 phage genome is 39.2%, whereas that of the vAbAbd25 phage genome is 39.3%. The genome of phage vAbBal23 encodes 60 protein-coding genes (CDSs), while phage vAbAbd25 encodes 58 CDSs. A comparison of the annotated CDSs in both phage genomes revealed that, 32 CDSs in vAbBal23 and 28 CDSs in vAbAbd25 are predicted to be hypothetical proteins or proteins of unknown function. Both genomes contain known proteins relevant to the phage tail, head, packaging, DNA metabolism-related proteins, and host lysis proteins (endolysin, holin/anti-holin). No genes related to toxins, virulence factors, antibiotic resistance, or integrase enzymes were detected among the CDSs with predicted functions in phage genomes. Circular maps of the annotated genomes of phages vAbBal23 and vAbAbd25 are displayed in Fig. 5, and a detailed description of the CDS is available in Supplementary Tables 3 and 4.
Fig. 5.
Predicted coding sequences (CDSs) of phages (A) vAbBal23 and (B) vAbAbd25 with transfer RNAs (tRNAs), transfer-messenger RNAs (tmRNAs), virulence factors (VFs), antimicrobial resistance genes (AMRs), clustered regularly interspaced short palindromic repeats (CRISPRs) and functional annotation of the CDSs
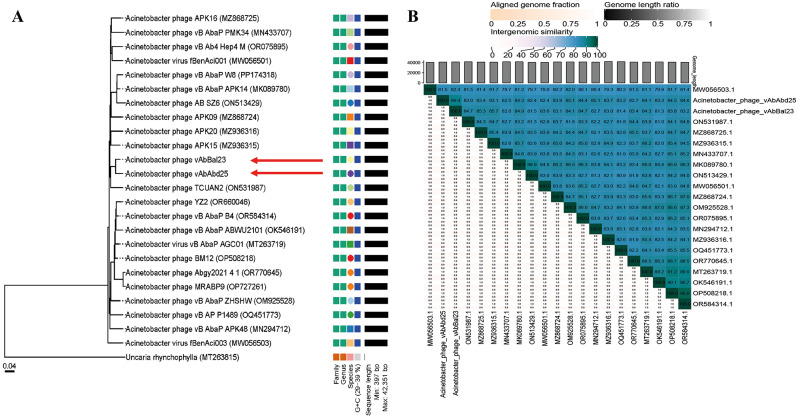
The phylogenetic tree generated by the Virus Classification and Tree Building Online Resource (VICTOR) serves as an effective tool for viral classification. In the VICTOR phylogenetic tree, phages vAbBal23 and vAbAbd25, along with their homologous sequences, were compared with other phages classified in the ICTV classification. The analysis revealed that vAbBal23 and vAbAbd25 resemble each other and cluster together with phages belonging to the unclassified Friunavirus group (Fig. 6A), which is consistent with previous findings. The hosts of these unclassified Friunavirus phages are from the Acinetobacter genus. Average nucleotide identity (ANI) analysis of phages vAbBal23 and vAbAbd25 with closely related phages shows that both phages represent new species in the genus Friunavirus (ANI < 95%) and propose the name “Friunavirus sninA” and “Friunavirus sninB” respectively (Fig. 6B).
Fig. 6.
Phylogenetic and comparative genomic analyses of the phages vAbBal23 and vAbAbd25. (A) Phylogenetic tree of vAbBal23 and vAbAbd25 generated by VICTOR using the whole-genome sequences of phage homologs in BLASTn. (B) Percentage sequence similarity between phages calculated using VIDIRIC
Discussion
An infection caused by a bacterial strain resistant to all available antibiotics can represent a fatal outcome for a patient. Each year, approximately 700,000 individuals die from untreatable bacterial infections [36]. Phage therapy has the potential to serve not only as an exceptional clinical option to prevent such fatalities but also as a standard treatment if certain challenges are addressed. One major challenge is the highly specific nature of phages, which can infect only a small percentage of bacterial strains.
In this study, we isolated two Acinetobacter phages, vAbBal23 and vAbAbd25, that demonstrated a narrow host range, which is a typical feature of Acinetobacter phages [37, 38]. Both phages showed significant bactericidal efficacy, reducing bacterial populations by up to 75.8%. Phages targeting A. baumannii are relatively specific to the Acinetobacter genus [39]. This limited host range is advantageous as it minimizes the elimination of other bacterial species, preserving the microbiota [40]. However, it can represent a challenge in identifying phages that specifically match the bacteria’s K locus for each infection [41]. Further characterization of Acinetobacter phages vAbBal23 and vAbAbd25 focused on their potential application as antimicrobial therapeutics.
Phage attachment to the bacterial surface represents the initial stage of the phage life cycle. Furthermore, alterations in environmental pH and temperature conditions, where the phage resides, are recognized as crucial factors influencing the phage life cycle [42]. The phages vAbBal23 and vAbAbd25 exhibit high stability across a temperature range from 25 °C to 40 °C, notably showing sustained viability at the typical human body temperature of 37 °C. Additionally, the phages demonstrated exceptional stability across a wide range of temperatures and pH values, making them suitable for pharmaceutical formulation and therapeutic use. Its resilience in both acidic and alkaline environments (pH 3–9) also allows for oral administration without compromising its viability in the gastrointestinal tract [43].The resilience of these phages to pH and temperature, is a pivotal factor determining their suitability for therapeutic applications, particularly considering the various routes of administration available [44].
The one-step growth curve revealed that both phages had a short latent period of 15 min. However, vAbBal23 had a larger burst size (2,103 PFU per infected cell), whereas vAbAbd25 had a smaller burst size (270 PFU per infected cell).Studies have indicated that phages with a long latent period may experience prolonged and less efficient growth and replication, whereas those with a short latency can replicate more rapidly and efficiently release progeny phages [45]. Additionally, phages with a large burst size are regarded as more virulent because they can quickly and efficiently eradicate bacterial infections [46].
In this study, we also explored the impact of phages vAbBal23 and vAbAbd25 on reducing the biomass of A. baumannii biofilm through disruption assays. Biofilms are bacterial communities that exhibit strong resistance to antibiotics and are often associated with chronic infections [47, 48]. Several factors contribute to the heightened antimicrobial resistance of biofilm-associated microorganisms, including the extracellular matrix, which forms a physical barrier limiting the diffusion of antimicrobial agents [49]. Furthermore, the depletion of nutrients and oxygen within the biofilms can lead bacteria to enter a stationary phase, reducing their susceptibility to antimicrobial agents [50]. The ability of bacteria to form biofilms on various surfaces increases the risk of contamination, especially in settings such as healthcare facilities and food processing industries [51]. Despite efforts, devising effective strategies to eradicate biofilms remains challenging, and suitable agents for controlling bacterial biofilms are currently lacking [52]. The impact of the isolated phages on two XDR A. baumannii isolates embedded in biofilms was assessed. The isolated lytic phage effectively reduced the biofilm content of these isolates by up to 68% after 24 h of treatment and therefore has the potential to be successfully used as a biofilm treatment agent. To improve phage effectiveness and more efficiently eradicate bacterial biofilms, bacteriophage cocktails, comprising multiple phages with varying host ranges and modes of action offer promising approaches and can expand the host range, and prevent the formation of phage-resistant mutants [22, 53]. Additionally, phages can be engineered and combined with antibiotics to enhance antimicrobial activity [54]. For example, Grygorcewicz et al. recently demonstrated that a cocktail of A. baumannii phages, when combined with antibiotics, exhibited potent lytic activity in eradicating biofilms in human urine [55].
A critical aspect of this characterization involved the analysis of the phages genomes. This comprehensive analysis aimed to ensure the absence of resistance genes, virulence factors and the inability of the virus to undergo lysogenic conversion, as either of these traits would render it unsuitable for therapeutic use [56]. WGS analysis revealed that either of the two phages carried virulence, antibiotic resistance, or bacterial toxin-related genes. Phages with these characteristics are considered to have a low risk of promoting antibiotic resistance, making them suitable candidates for treating bacterial infections [57]. Additionally, we confirmed that both phages are indeed lytic phages with no typical prophage conversion genes, such as integrase or repressor genes. However, approximately half of the predicted gene products remain unidentified. The successful utilization of other phages belonging to the Friunavirus genus has been reported in phage therapy trials [58, 59] and supports the appropriateness of the characterized phages in this study for potential application in phage therapy.
An area for extending this study involves testing the combination of multiple lytic phages with good host ranges and promising antibacterial activity or a combination of phages with antibiotics to assess their effects on MDR and biofilm-producing A. baumannii. Also, future research may explore phage-derived enzymes as potential biological agents to target A. baumannii and its biofilms. Therefore, the establishment of large phage banks containing thoroughly characterized phages is crucial for the development of phage therapy in Senegal.
Conclusion
This study suggests that the phages vAbBal23 and vAbAbd25 exhibit good heat tolerance and a wide range of pH stabilities. They are effective against XDR A. baumannii in both plankonic cells and biofilms. Detailed genomic annotation excluded the presence of toxin-coding genes, antibiotic resistance determinants, and other bacterial virulence factors. The results presented here support the potential of the phages vAbBal23 and vAbAbd25 as therapeutic agents against XDR resistant A. baumannii infections.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Pr Amadou Diop and Pr Assane Dieng for providing the strains used in this study. The authors also thank Maïmouna Mbanne Diouf for technical assistance with Sequencing and all the members of the Pole of Microbiology, Pasteur Institute of Dakar for their valuable assistance.
Abbreviations
- AMR
Antimicrobial resistance
- ANI
Average nucleotide identity
- CRISPRs
Clustered regularly interspaced short palindromic repeats
- EOP
Efficiency of plating
- ICTV
International Committee on Taxonomy of Viruses
- LB
Luria Bertani
- MDR
Multidrug-resistant
- MOI
Multiplicity of infection
- OD
Optical density
- PBS
Phosphate buffer saline
- PFU
Plaque forming unit
- tRNA
Transfer RNA
- tmRNA
Transfer-messenger RNA
- VFs
Virulence factors
- VICTOR
Virus Classification and Tree Building Online Resource
- WHO
World Health Organization
- WGS
Whole genome sequencing
- XDR
Extremely drug-resistant
Author contributions
I.N. designed the study, isolated and characterized the phages, performed the data and bioinformatic analyses and drafted the manuscript. L.D. supervised the study. O.S. assisted with the environmental samples collection for phage isolation. M.M.D. assisted with sequencing. M.M.D., A.C., B.S.B., C.F., Y.D., N.D. provided resources to perform the study. L.D., O.S., M.M.D., A.C., B.S.B., C.F., Y.D., N.D., G.C.M., A.S. have substantively reviewed and edited the manuscript. G.C.M. and A.S. participated in the design, supervised, administrated the study and carried out the final validation of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by GCDM at the Institut de Recherche pour le Développement (IRD), France.
Data availability
The genome assemblies and annotations were deposited in the European Nucleotide Archive (ENA) under the project number PRJEB75249 and SRA and assembly accession numbers ERR12951254 and ERR12951255 for vAbBal23 and vAbAbd25 respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a Century of challenges. Clin Microbiol Rev. 2017;30(1):409–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes LC, Visca P, Towner KJ. Acinetobacter Baumannii: Evolution of a Global Pathogen. Pathog Dis. 2014;71(3):292–301. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y-T, Tsao S-M, Hsueh P-R. Clinical Outcomes of Tigecycline Alone or in Combination with Other Antimicrobial Agents for the Treatment of Patients with Healthcare-Associated Multidrug-Resistant Acinetobacter Baumannii Infections. Eur J Clin Microbiol Infect Dis. 2013;32:1211–20. [DOI] [PubMed] [Google Scholar]
- 4.Pendleton JN, Gorman SP, Gilmore BF. Clinical Relevance of the Eskape Pathogens. Expert Rev Anti Infect Ther. 2013;11:297–308. [DOI] [PubMed] [Google Scholar]
- 5.Wong DW, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna BM, Spellberg B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin Microbiol Rev. 2016;30:409–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris FC, Dexter C, Kostoulias X, Uddin MI, Peleg AY. The Mechanisms of Disease Caused by Acinetobacter Baumannii. Front Microbiol. 2019;10. [DOI] [PMC free article] [PubMed]
- 7.Tzu L, Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat Rev Microbiol. 2004;2:95–108. [DOI] [PubMed] [Google Scholar]
- 8.Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. Biology of Acinetobacter Baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front cell infect microbiol. 2017;7:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatesan N, Perumal G, Doble M. Bacterial Resistance in Biofilm-Associated Bacteria. Future Microbiol. 2015;10 11:1743–50. [DOI] [PubMed] [Google Scholar]
- 10.Penesyan A, Gillings MR, Paulsen IT. Antibiotic Discovery: Combatting Bacterial Resistance in Cells and in Biofilm Communities. Molecules. 2015;20:5286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung CYJ, Weitz JS. Modeling the Synergistic Elimination of Bacteria by Phage and the Innate Immune System. J Theor Biol. 2017;429:241–52. [DOI] [PubMed] [Google Scholar]
- 12.Tacconelli E. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development. 2017.
- 13.Lyon J. Phage Therapy’s Role in Combating Antibiotic-Resistant Pathogens. JAMA. 2017;318 18:1746–8. [DOI] [PubMed] [Google Scholar]
- 14.Kakasis A, Panitsa G. Bacteriophage Therapy as an Alternative Treatment for Human Infections. A Comprehensive Review. Int J Antimicrob Agents. 2019;53(1):16–21. [DOI] [PubMed] [Google Scholar]
- 15.Theuretzbacher U, Outterson K, Engel A, Karlén A. The Global Preclinical Antibacterial Pipeline. Nat Rev Microbiol. 2020;18(5):275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viertel TM, Ritter K, Horz H-P. Viruses Versus Bacteria—Novel Approaches to Phage Therapy as a Tool against Multidrug-Resistant Pathogens. J Antimicrob Chemother. 2014;69(9):2326–36. [DOI] [PubMed] [Google Scholar]
- 17.García-Quintanilla M, Pulido MR, López-Rojas R, Pachón J, McConnell MJ. Emerging Therapies for Multidrug Resistant Acinetobacter Baumannii. Trends Microbiol. 2013;21(3):157–63. [DOI] [PubMed] [Google Scholar]
- 18.Sulakvelidze A, Alavidze Z, Morris JG Jr. Bacteriophage Therapy. Antimicrob Agents Chemother. 2001;45(3):649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magiorakos A, Srinivasan A, Carey RB, Carmeli Y, Falagas M, Giske C, Harbarth S, Hindler J, Kahlmeter G, Olsson-Liljequist B. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin Microbiol Infect. 2012;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 20.Lam MM, Wick RR, Judd LM, Holt KE, Wyres KL. Kaptive 2.0: Updated Capsule and Lipopolysaccharide Locus Typing for the Klebsiella Pneumoniae Species Complex. Microb genom. 2022;8(3). [DOI] [PMC free article] [PubMed]
- 21.Kusradze I, Karumidze N, Rigvava S, Dvalidze T, Katsitadze M, Amiranashvili I, Goderdzishvili M. Characterization and Testing the Efficiency of Acinetobacter Baumannii Phage Vb-Gec_Ab-M-G7 as an Antibacterial Agent. Front Microbiol. 2016;7:1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson K. Working with Bacteriophages: Common Techniques and Methodological Approaches. CRC press Boca Raton, FL; 2005.
- 23.Goodridge L, Gallaccio A, Griffiths MW, Morphological. Host Range, and Genetic Characterization of Two Coliphages. Appl Environ Microbiol. 2003;69(9):5364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaki BM, Fahmy NA, Aziz RK, Samir R, El-Shibiny A. Characterization and Comprehensive Genome Analysis of Novel Bacteriophage, Vb_Kpn_Zckp20p, with Lytic and Anti-Biofilm Potential against Clinical Multidrug-Resistant Klebsiella Pneumoniae. Front Cell Infect Microbiol. 2023;13:1077995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. Identification of Associated Proteins by Coimmunoprecipitation. Cold Spring Harb Prot. 2006;2006(1):pdb. prot3898. [DOI] [PubMed]
- 26.Andrews S. Babraham Bioinformatics-Fastqc a Quality Control Tool for High Throughput Sequence Data. URL: https://www.bioinformaticsbabrahamacuk/projects/fastqc. 2010.
- 27.Krueger F. Trim Galore!: A Wrapper around Cutadapt and Fastqc to Consistently Apply Adapter and Quality Trimming to Fastq Files, with Extra Functionality for Rrbs Data. Github. 2015. Available from : https://github.com/FelixKrueger/TrimGalore.
- 28.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using Spades De Novo Assembler. Curr prot bioinf. 2020;70(1):e102. [DOI] [PubMed] [Google Scholar]
- 29.Bushnell B. Bbmap Short-Read Aligner, and Other Bioinformatics Tools. Berkeley: University of California. 2015.
- 30.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM. Twelve Years of Samtools and Bcftools. Gigascience. 2021;10(2):giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE. 2014;9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald P-J, Vreugde S. Pharokka: A Fast Scalable Bacteriophage Annotation Tool. Bioinf. 2023;39(1):btac776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff JP, Göker M, Victor. Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics. 2017;33(21):3396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moraru C, Varsani A, Kropinski AM. Viridic—a Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses. 2020;12(11):1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S. Changes to Virus Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol. 2019;164(9):2417–29. [DOI] [PubMed] [Google Scholar]
- 36.Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens. 2021;10(10):1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merabishvili M, Vandenheuvel D, Kropinski AM, Mast J, De Vos D, Verbeken G, Noben J-P, Lavigne R, Vaneechoutte M, Pirnay J-P. Characterization of Newly Isolated Lytic Bacteriophages Active against Acinetobacter Baumannii. PLoS ONE. 2014;9(8):e104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soontarach R, Srimanote P, Enright MC, Blundell-Hunter G, Dorman MJ, Thomson NR, Taylor PW, Voravuthikunchai SP. Isolation and Characterisation of Bacteriophage Selective for Key Acinetobacter Baumannii Capsule Chemotypes. Pharmaceuticals. 2022;15(4):443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghajavand H, Esfahani BN, Havaei A, Fazeli H, Jafari R, Moghim S. Isolation of Bacteriophages against Multidrug Resistant Acinetobacter Baumannii. Res Pharm Sci. 2017;12(5):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyman P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals. 2019;12(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leptihn S, Loh B, Complexity. Challenges and Costs of Implementing Phage Therapy. Future Medicine; 2022. pp. 643–6. [DOI] [PubMed]
- 42.Tomat D, Aquili V, Casabonne C, Quiberoni A. Influence of Physicochemical Factors on Adsorption of Ten Shigella Flexneri Phages. Viruses. 2022;14(12):2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Atrees DM, El-Kased RF, Abbas AM, Yassien MA. Characterization and Anti-Biofilm Activity of Bacteriophages against Urinary Tract Enterococcus Faecalis Isolates. Sci Rep. 2022;12(1):13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernández L, Gutiérrez D, García P, Rodríguez A. The Perfect Bacteriophage for Therapeutic Applications-a Quick Guide. Antibiot (Basel). 2019;8(3). [DOI] [PMC free article] [PubMed]
- 45.Abedon ST. Selection for Bacteriophage Latent Period Length by Bacterial Density: A Theoretical Examination. Microb Ecol. 1989;18:79–88. [DOI] [PubMed] [Google Scholar]
- 46.Hyman P, Abedon ST. Bacteriophage Host Range and Bacterial Resistance. Adv Appl Microbiol. 2010;70:217–48. [DOI] [PubMed] [Google Scholar]
- 47.Khalifa L, Brosh Y, Gelman D, Coppenhagen-Glazer S, Beyth S, Poradosu-Cohen R, Que Y-A, Beyth N, Hazan R. Targeting Enterococcus Faecalis Biofilms with Phage Therapy. Ap environ microbiol. 2015;81(8):2696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, Ellwood R, Giacaman RA, Herrera D, Herzberg MC. Role of Microbial Biofilms in the Maintenance of Oral Health and in the Development of Dental Caries and Periodontal Diseases. Consensus Report of Group 1 of the Joint Efp/Orca Workshop on the Boundaries between Caries and Periodontal Disease. J Clin Periodontol. 2017;44:S5–11. [DOI] [PubMed] [Google Scholar]
- 49.Singh S, Singh SK, Chowdhury I, Singh R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. open microbiol journ. 2017;11:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bamford NC, MacPhee CE, Stanley-Wall NR. Microbial Primer: An Introduction to Biofilms–What They Are, Why They Form and Their Impact on Built and Natural Environments. Microbiol. 2023;169(8):001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Ye C, Soteyome T, Zhao X, Xia J, Xu W, Mao Y, Peng R, Chen J, Xu Z. Inhibitory Effects of Two Types of Food Additives on Biofilm Formation by Foodborne Pathogens. Microbiol Open. 2019;8(9):e00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joshi RV, Gunawan C, Mann R. We Are One: Multispecies Metabolism of a Biofilm Consortium and Their Treatment Strategies. Front Microbiol. 2021;12:635432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fong SA, Drilling A, Morales S, Cornet ME, Woodworth BA, Fokkens WJ, Psaltis AJ, Vreugde S, Wormald P-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas Aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front cell infect microbiol. 2017;7:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang C, Yu X, Guo W, Guo C, Guo X, Li Q, Zhu Y. Bacteriophage-Mediated Control of Biofilm: A Promising New Dawn for the Future. Front Microbiol. 2022;13:825828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grygorcewicz B, Wojciuk B, Roszak M, Łubowska N, Błażejczak P, Jursa-Kulesza J, Rakoczy R, Masiuk H, Dołęgowska B. Environmental Phage-Based Cocktail and Antibiotic Combination Effects on Acinetobacter Baumannii Biofilm in a Human Urine Model. Microb Drug Resist. 2021;27(1):25–35. [DOI] [PubMed] [Google Scholar]
- 56.Al-Anany AM, Fatima R, Hynes AP. Temperate Phage-Antibiotic Synergy Eradicates Bacteria through Depletion of Lysogens. Cell Rep. 2021;35(8). [DOI] [PubMed]
- 57.Luong T, Salabarria A-C, Roach DR. Phage Therapy in the Resistance Era: Where Do We Stand and Where Are We Going? Clin Ther. 2020;42(9):1659–80. [DOI] [PubMed] [Google Scholar]
- 58.Wu M, Hu K, Xie Y, Liu Y, Mu D, Guo H, Zhang Z, Zhang Y, Chang D, Shi Y. A Novel Phage Pd-6a3, and Its Endolysin Ply6a3, with Extended Lytic Activity against Acinetobacter Baumannii. Front Microbiol. 2019;9:3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho YH, Tseng CC, Wang LS, Chen YT, Ho GJ, Lin TY, Wang LY, Chen LK. Application of Bacteriophage-Containing Aerosol against Nosocomial Transmission of Carbapenem-Resistant Acinetobacter Baumannii in an Intensive Care Unit. PLoS ONE. 2016;11(12):e0168380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assemblies and annotations were deposited in the European Nucleotide Archive (ENA) under the project number PRJEB75249 and SRA and assembly accession numbers ERR12951254 and ERR12951255 for vAbBal23 and vAbAbd25 respectively.