Abstract
Background
Shell strength is an important trait in peanuts that impacts shell breakage and yield. Despite its significance, the genetic basis of shell strength in peanuts remains largely unknown, and the current methods for rating this trait are qualitative and subjective. This study aimed to investigate the genetics of shell strength using a segregating recombinant-inbred-line (RIL) population derived from the hard-shelled cultivar ‘Hanoch’ and the soft-shelled cultivar ‘Harari’.
Results
Initially, a quantitative method was developed using a texture analyzer, focusing on the proximal part of isolated shells with a P/5 punching probe. This method revealed significant differences between Hanoch and Harari. Shell strength was then measured in 235 RILs across two distinct environments, revealing a normal distribution with some RILs exhibiting shell strength values beyond those of the parental lines, indicating transgressive segregation. Analysis of variance indicated significant effects for the RILs, with no effects of block or year, and a broad-sense heritability estimate of 0.675, indicating a substantial genetic component. Using an existing genetic map, we identified three QTLs for shell strength, with one major QTL (qSSB02) explaining 18.7% of the phenotypic variation. The allelic status of qSSB02 corresponded significantly with cultivar designation for in-shell or shelled types over four decades of Israeli peanut breeding. Physical and compositional analyses revealed that Hanoch has a higher shell density than Harari, rather than any difference in shell thickness, and is associated with increased levels of lignin, cellulose, and crude fiber.
Conclusions
These findings provide valuable insights into the genetic and compositional factors that influence shell strength in peanut, laying a foundation for marker-assisted selection in breeding programs focused on improving pod hardness in peanuts.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05727-9.
Keywords: Peanut shell, Genetic mapping, Shell strength, QTL, Virginia-type peanut
Background
Peanut (Arachis hypogaea L.; 2n = 4x = 40) is a distinctive leguminous plant with pods that develop underground. Upon flowering, the fertilized ovules are buried in the soil to facilitate pod maturation. Below ground, the pod grows rapidly, forming a large shell wall or pericarp [1]. Initially, a succulent pericarp occupies the major part of the pod volume and serves as a temporary nutrient source for the developing seed [2]. As development progresses, the mesocarp becomes greatly reduced in size, while the exocarp undergoes lignification and stiffening [1]. Upon maturation, the pericarp, recognized as the iconic peanut shell, accounts for approximately 30% of the total pod weight. This pericarp is comprised of biopolymers, such as cellulose and lignin, and proteins, along with minerals and other bioactive compounds [3, 4].
The pod shell plays an important role in defending against diseases and insect pests during underground growth and after harvest [5, 6]. In addition, its mechanical strength reduces the pod’s susceptibility to mechanical damage during harvest, transport, and storage, which significantly influences yield and economic returns. This issue is particularly significant for the ‘in-shell’ peanut industry, in which whole pods are roasted and marketed. In that industry, the strength of the shell is vital for mitigating shell cracking, to ensure a high-grade marketable product and optimize growers’ revenues.
Shell strength (SS) in peanuts has a notable genetic component evident by consistent differences between varieties [7–10]. While the recognition of the genetic basis for SS has been an important factor in the development of improved peanut cultivars, few reports have addressed the genetic and molecular mechanisms that govern peanut pod-shell development, in general, and SS, in particular. Molecular mechanisms underlying peanut-shell development have been described in a few reports and shown to involve complex regulatory pathways associated with cell-wall synthesis and deposition [11–13]. Calcium-signaling pathways are key determinant in shell development, as calcium deficiency alter the expression of genes related to SS [14]. Understanding of these regulatory pathways offer insights into the possible enhancement of peanut SS through molecular breeding.
In addition, marker-assisted selection (MAS) using SS linked molecular markers may accelerate breeding processes and aid the development of cultivars that are resilient to mechanical damage and have improved shelf lives. A previous study of 320 peanut accessions led to the identification of significant phenotypic variations in peanut shell traits, including SS, and found 10 significant SNPs related to these traits, situated on the B genome [10]. Another study identified significant QTLs linked to pod-shell thickness, which, according to the authors, is closely correlated with SS [15]. Two major QTLs have been identified and mapped on chromosomes A08 and B08, explaining 31.1–32.3% and 16.7–16.8% of the phenotypic variation, respectively [15].
In this study, we investigated the genetics of SS in ‘Virginia’ marketing-type peanut cultivars and a recombinant-inbred-line (RIL) population. Virginia-type peanuts are primarily used in the in-shell industry, but some are also grown for the shelled-peanut market. Initially, quantitative and objective method was developed for measuring the mechanical strength of isolated shells using a texture analyzer. This method was used to assess SS among 235 segregating RILs and to identify new molecular markers associated with SS. In addition, examinations of shell thickness, density, and composition, conducted among isolated shells, reveal that shell density, rather than shell thickness, is the primary determinant of SS in this genetic background. The discussed results offer new insights into the factors influencing SS in peanuts and provide a foundation for MAS breeding of Virginia-type peanuts.
Methods
Plant material and growing conditions
The genetic analysis was based on an advanced RIL population (F7:12) that was developed from a cross between two closely related Virginia-type cultivars, ‘Hanoch’ and ‘Harari’ [16]. Hanoch is a late-maturing cultivar with long, smooth, hard pods. These qualities make it popular in the European in-shell peanut market. Harari is an early-maturing cultivar, with a chubby, reticulated, soft pod, which is primarily grown for the local shelled-peanut market. Harari has a bunch-type growth habit; whereas Hanoch has a spreading growth habit.
Two hundred and thirty-five RILs were planted in two environments: one in 2022 in Halutza, which is in the western Negev region of southern Israel (31°22"32.1 N, 34°46’81.5"E) and the other in 2023 in Kefar Masrik, in the western Galilee region of northern Israel (32°53’54.4"N 35°08’44.0"E). The western Negev location is characterized by a fine sandy-loam soil and the western Galilee site is characterized by heavy black soil. Experiments were imbedded within commercial peanut growing plots in a completely randomized block design with two replicates. Each line was planted manually in a double row in a 4-m-long bed with 90 cm gaps between rows and 40 cm between plants within each row (total of 20 plants/plot). The parental genotypes were grown randomly as control plots within the experimental field, with three replicates per block. Fields were maintained under full-irrigation conditions and all other agronomic practices were like those used in commercial plots. After harvest, the plants were left to naturally dry in the field until the seeds reached a moisture content of 12%.
Calibration of a method for measurement of the mechanical strength of shells
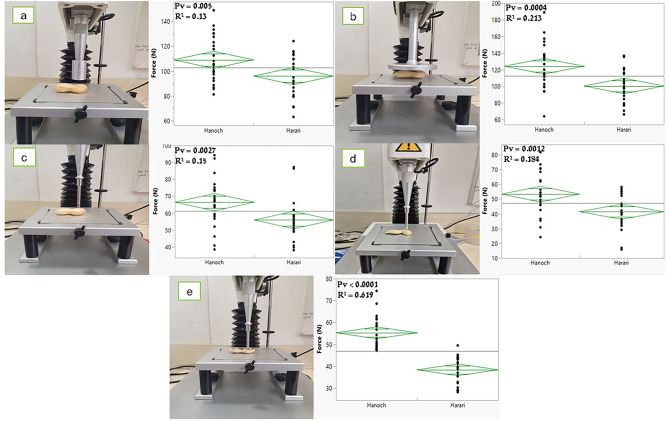
Approximately 200 sound and fully matured pods were randomly selected from each “plot” (line × block × environment). The pods were stored under the same conditions until examination. About 25 super-giant–sized pods (5–7 pods per ounce) free of stains or infections (moth eggs, mold, or rot) were selected for further analysis. The mechanical strength of the pods’ shells was examined using the TA1 texture analyzer (Stable Micro Systems) and compared with the two parental lines, Hanoch and Harari. Several combinations were investigated, using different sensors (P/20, P/75, and P/5), base plates (one heavy duty platform with 2 cm diameter hole in the middle, and another one without a hole) and variations in the positioning of the sample holder on the base plate. Additionally, we compared the results obtained when entire pods or empty shells were examined. All measurements were collected under identical test conditions, involving a 50-N load cell compressor and a running speed of 20 mm/min. The proximal portion of the pod was tested due to phenotypic instability in the distal part within this specific genetic context: In Hanoch, the distal half develops at a later point in time; whereas in Harari, both pod parts develop simultaneously. In the treatment of the empty shells, the shell was placed on the device with both the convex (Fig. 1d) and the concave (Fig. 1e) sides facing the sensor. The concave side was used to minimize the “dome” effect that may result with a non-direct resistance of the shell. The deformation-force curve was constructed using Exponent Connect software, from which the parameter peak height was extracted (force; Newton) and used as the value for SS.
Fig. 1.
Differences between the two parental lines, ‘Hanoch’ and ‘Harari’, in terms of shell mechanical strength (force; N), as measured by a texture analyzer. (a) Entire pod and p/20 probe. (b) Entire pod and p/75 probe. (c) Entire pod and p/5 probe. (d) Isolated the outer shell and p/5 probe. (e) Isolated the inner shell and p/5
In parallel with the quantitative testing, a qualitative assessment was also conducted by manually shelling an additional 25 pods from each parental line and assigning a score of 1–5 for shell strength (blind test). The correlation between the mechanical and the manual tests was calculated. Also, to initially inspect the connection between the measured SS and the actual pod breakage levels in commercially harvested plots, samples of pods were taken from four genotypes (Hanoch, Iddo, 22, 23) and two commercial plots (Ora, Shahen) in Western Negev, Israel. Around 3 kg of commercially harvested Super Giant pods (5–7 pods per ounce) were sampled. The pod breakage level was measured by counting the number of cracked pods in 100 pod sample.
Genetic analysis and trait mapping
In the subsequent step, the calibration method was applied to the 235 RILs, to conduct a genetic analysis and map the SS trait. To examine whether the mean force values (N) were normally distributed, the Shapiro-Wilk test was used in JMP. An ANOVA test was performed to examine the effects of the lines and blocks on shell strength. Heritability estimates were calculated by partitioning the general variance among lines into fixed effects (e.g., between parents and between the parents’ lines and the population mean) and random effects (differences within and between lines). Heritability was calculated as the variance component within and between lines. Genetic correlations were computed between shell strength and the other traits previously examined among this population and are presented in our database [17, 18]. The traits were maturity index, pod yield, harvest index, 50-pod weight (50PW), 50-seed weight (50SW) and shelling percentage. In addition, the correlation between SS and another important trait, 50-pod shell weight (50SHW), which was not previously reported, was calculated as well.
Additional categorical traits were recorded in this population, including branching habit (spreading or bunch), pod constriction (deep, moderate, or no constriction), and pod reticulation (rough or smooth). The correlations between SS indices and the quantitative traits were evaluated using the best linear unbiased predictors. An ANOVA test was utilized to determine the phenotypic effect of the qualitative traits on the shell-strength indices.
All SNP genotyping and genetic-map-construction procedures used in this study were described in detail in a previous report [18] and are summarized here. SNP genotyping was performed using the Affymetrix Axiom_Arachis2 SNP-array comprising 47,837 SNPs [19, 20]. Genotypic data were analyzed using the Axiom Analysis Suite 3.1, as previously described [21]. Loci positions were confirmed as previously described [22], with a few modifications [BLASTN (e-value < 1 × 10− 18) and mismatch of less than 2]. Generated genetic linkage groups (LGs) were assigned to the pseudo-molecules of the tetraploid A. hypogaea cv. Tifrunner [23].
Mapping of SS [represented by average force (N)] on the 235 RILs was performed using MapQTL v6 [24]. Interval mapping, using the maximum likelihood algorithm with an LOD (logarithm of the odds) score of 2.5 and 1000 permutations, was used to confirm the presence of a putative QTL at a 95% significance level. QTLs that explained more than 10% of the phenotypic variation were defined as major QTLs, according to the criteria set out by Collard et al. [25]. The QTL naming follows the terminology of q as QTL, followed by an abbreviation of the shell-strength trait (SS), ending with the respective chromosome.
Validation of qSSB02, qSSB03, and qSSA05 in a collection of historic peanut cultivars
The allelic situation of qSSB02, qSSB03, and qSSA05 was determined in a series of historic Virginia-type cultivars that represents more than 40 years of peanut breeding in Israel. For this investigation, leaves were collected from five seedlings of each cultivar. DNA was extracted using the GenElute™ Plant Genomic DNA Miniprep Kit method. One set of primers was designed for each QTL, based on the genomic sequence of the AX-147213175, AX-176800560, and AX-176810858 markers for qSSB02, qSSB03, and qSSA05, respectively. The primers were as follows: for qSSB02, F: 5′- CCT CTT AAG AGT GAT GTC AGA ATC − 3′ and R: 5′- GAT GTG ATG TTA TGG GGA ACA AAA G -3′; for qSSB03, F: 5′- GCG ACC CGA GGG CGT G -3′ and R: 5′- CCC GCG ACA TTA AGA CCT AAA A -3′; and for qSSA05, F: 5′- CTC TCA TTC TTT TTG CTT CTC − 3′ and R: 5′- CGG CAC AAC TTC GAA GAG ATT C -3′. The PCR conditions included 5 min of initial denaturation at 95 C; 36 cycles of denaturation (30 s at 95 C) and annealing (30 s at 62 C, 60.5 C, and 62 C for qSSB02, qSSB03, and qSSA05); extension (90 s at 72 C); and a final extension at 72 C for 5 min. Hy-Taq Ready Mix (2x)-Hylabs®) was used to amplify the PCR product.
In qSSB02, the PCR product from each parental line was polymorphic due to a 230-bp indel that exists between the two lines. Therefore, for this QTL, the PCR products were run on an 1% agarose gel. The PCR products of qSSB03 and qSSA05 were sent for Sanger sequencing at HyLabs Ltd., Israel.
Thickness, density, and compositional characterization of isolated shells
Three replicates of 300 g of peanut shells were collected from cultivars Hanoch and Harari grown in the Halutza plot (2022). The shells were cut in half and only the proximal half was retained for physical and general shell-composition analysis. Shell discs of 7-mm diameter were prepared using a cork-borer (CBSTO6-VWR). Each isolated disc was measured for shell thickness using a caliper (ABSOLUTE AOS Series 500, Mitutoyo). Subsequently, 20 discs were bulked together and weighed using an analytical scale to determine density (weight/area) values.
Compositional characterization was conducted at the Field Service Laboratory in Neve-Ya’ar Center, Israel, using an ANKOM 200 Fiber Analyzer. Three replicates of 150 g of peanut shells were examined. The relative amounts of crude fiber, lignin, hemicellulose, and cellulose were measured.
Gene ontology analysis
Gene ontology (GO) annotations for all protein coding genes were downloaded from the Legume information system database (https://mines.legumeinfo.org/arachismine/begin.do). Enrichment analysis was generated with Blast2Go [version 5.2.5] using Fisher’s exact test. The test was performed for each of the two gene sets separately, the GO annotations of all A. hypogaea [genome build 1] protein coding genes served as background vs. GO annotations of proteins within the major QTL. GO annotation with p-value < 0.05 was considered significant.
Results
Calibration of a method for measuring the mechanical strength of shells
Mature pods were sampled from two Virginia-type cultivars differing in SS (i.e., Hanoch and Harari) and the mechanical strength of their shells was compared using a texture analyzer. Five combinations of sensor configurations and sample preparations were tested. The results are presented in Fig. 1. Among the five methods investigated, the combination of employing the p/5 probe and using isolated shells and a plate with a hole on the center revealed the most significant difference in SS between Hanoch and Harari (prob[t] < 0.0001; R2 = 0.62). Consequently, this method was chosen as the preferred approach for measuring SS in other experiments within this study.
Initially, to validate its reliability, the mechanical method was compared with a qualitative assessment, where pods from each parental line were manually shelled and assigned a shell strength score (blind test). The correlation between the mechanical test values and the manual test scores was found to be very high and significant (R = 0.856; Supplementary Fig. 1). Additionally, the relationship between the selected mechanical method for measuring SS and actual pod breakage levels in several commercially harvested plots was examined. A significant negative correlation (-0.72) was observed between the mechanical method and the level of pod breakage in the field (Supplementary Fig. 2), indicating the method’s effectiveness in predicting pod damage resulting from external forces.
Phenotyping of the parents and the RIL population
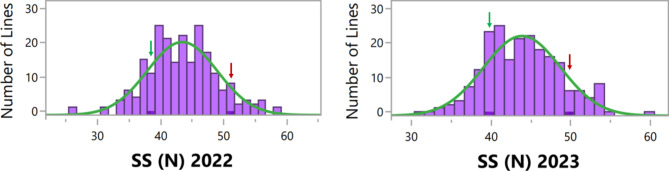
SS was examined in a RIL population derived from a cross between Hanoch and Harari. Samples were collected from field experiments conducted in two different environments. SS was found to be normally distributed among the RIL population in both environments (Fig. 2; Table 1). SS values were collected from the two parental lines as well. As expected from the calibration trial, a highly significant difference in N-force was found between the parental lines (P = < 0.0001), with averages of 52.06 N and 38.70 N for Hanoch and Harari, respectively. Parental values were within the range of the RIL population, with some RILs exhibiting SS values beyond the parental values at each end of the curve in both years, suggesting transgressive segregation (Fig. 2). ANOVA revealed a significant effect for the RILs, but no significant effects for the block, year, or RIL × year interaction (Table 2). Therefore, QTL analysis was performed using the average values for both years. The broad-sense heritability for SS was 0.675, indicating that this trait has a moderate-to-high genetic component.
Fig. 2.
Phenotypic distribution of SS in two different environments, represented by consecutive years (2022 and 2023). The y-axis corresponds to the RIL population density and the x-axis shows the amount of force (N) that the shells could withstand, based on the average values of two replicates. Arrows indicate the N-force values for ‘Hanoch’ (red) and ‘Harari’ (green). A normal distribution curve is indicated in green
Table 1.
Summary statistics of SS among parents and RILs in two field experiments
| Parents | RILs | |||||||
|---|---|---|---|---|---|---|---|---|
| Variables | Hanoch | Harari | Student’s t- test | Mean ± SD | Minimum | Maximum | Sig. of A-D testa | |
| 2022 | SS (N) | 52.29 | 38.73 | 0.0002 | 45.51 ± 7.67 | 25.59 | 58.92 | 0.384 |
| 2023 | SS (N) | 51.83 | 38.66 | < 0.0001 | 45.25 ± 7.28 | 31.32 | 60.383 | 0.225 |
| Two-year avg. | SS (N) | 52.06 | 38.70 | < 0.0001 | 45.38 ± 7.22 | 29.09 | 59.27 | 0.426 |
aThe Anderson-Darling test was used to check whether the samples represented a population with a normal distribution
Table 2.
Analysis of variance and heritability for SS in the Hanoch × Harari RIL population over two years. Block [Year] indicates the nested effect of the blocks within each year
| Trait | Variables | df | Mean square | F-ratio | P-value | H(b)2 a |
|---|---|---|---|---|---|---|
| SS | Block [Year] | 2 | 23.25 | 2.3841 | 0.933 | 0.675 |
| Year | 1 | 33.85 | 3.47 | 0.631 | ||
| RIL | 234 | 95.51 | 9.79 | < 0.0001 | ||
| RIL × Year | 220 | 11.47 | 1.17 | 0.775 | ||
| Error | 454 | 9.75 |
a Broad-sense heritability
Pearson correlations were calculated between the average SS values and a set of agricultural and marketing-related traits. (Table 3). SS was significantly negatively correlated with maturity index and shelling percentage. In contrast, SS was significantly and positively correlated with 50SW, 50PW, and 50SHW. Small, but significant negative correlations were found between SS and pod yield, and between SS and harvest index. The effects of the categorical traits on SS were examined using ANOVA (Table 4). Pod reticulation was found to have a significant, but small effect on SS, with lines with smooth shells exhibiting higher SS values than lines with rough shells. Neither branching habit nor pod constriction was found to have any significant effect on SS.
Table 3.
Summary of Pearson correlations and probability figures for SS and quantitative traits in the Hanoch × Harari RIL population
| Trait | Correlation | Probability |
|---|---|---|
| Maturity index (%) | -0.2311 | 0.0003 |
| Pod yield (g) | -0.1409 | 0.0302 |
| Harvest index | -0.1514 | 0.0197 |
| 50PW (g) | 0.4207 | < 0.0001 |
| 50SW (g) | 0.1965 | 0.0024 |
| Shelling (seed) percentage (%) | -0.4197 | < 0.0001 |
| 50SHW (g) | 0.5412 | < 0.0001 |
50PW, 50-pod weight; 50SW, 50-seed weight; 50SHW, 50-pod shell weight
Table 4.
Summary of F-test results for the effects of categorical traits on SS among the Hanoch × Harari RIL population
| Trait | F-ratio | P-value |
|---|---|---|
| Branching habit | 0.0005 | 0.9825 |
| Pod constriction | 1.32 | 0.2692 |
| Pod reticulation | 4.169 | 0.0423 |
QTL identification for SS
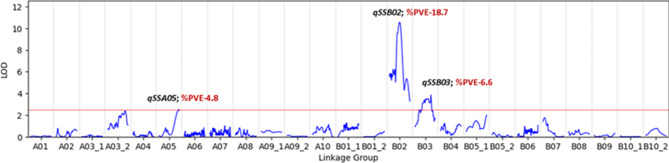
QTL mapping of the SS resulted in the identification of three QTLs with LOD scores ranging from 2.51 to 10.57 (Fig. 3; Table 5). One major QTL, qSSB02, was observed on LG B02 between AX-176816518_B2-AX-176804012_B2, spanning 15.41 Mbp, with a %PVE value of 18.7. Another minor QTL, qSSB03, was observed on LG B03 between AX-176796644_B3-147243220_B3, spanning 0.21 Mbp, with a %PVE value of 6.6. In these two QTLs, the allele that provided higher SS came from the Hanoch parental line. The third observed QTL, qSSA05, was found on LG A05 between AX-176802416_A5-AX-176809615_A5, spanning 0.09 Mbp, with a %PVE value of 4.8. In this QTL, the ‘Harari’ parent (with a weaker shell) contributed the allele for higher SS.
Fig. 3.
Whole-genome QTL analysis for the SS trait among the Hanoch × Harari RIL population. % PVE, percentage of phenotypic variance explained. %PVE was derived from the marker with the highest value within each QTL, as follows: qSSB02 - marker AX-147,213,175; qSSB03 - marker AX-176,800,560; and qSSA05 - marker: AX-176,810,858
Table 5.
QTLs identified for SS in the Hanoch × Harari RIL population
| Trait | Year | QTL | LGa | Position (cM) | Marker | Physical position (marker; Mbp) | Flanking markers | Physical position range (Mbp) | LOD | PVEb (%) | ADDc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SS | 22/23 |
qSS B02 |
B02 | 21.62 | AX-147,213,175 | 9.84 |
AX-176816518_B2- AX-176804012_B2 |
3.88–19.29 | 10.57 | 18.7 | 2.88 |
| SS | 22/23 |
qSS B03 |
B03 | 12.64 | AX-176,800,560 | 4.51 |
AX- 147243220_B3- AX- 176796644_B3 |
4.46–4.67 | 3.48 | 6.6 | 1.30 |
| SS | 22/23 |
qSS A05 |
A05 | 49.536 | AX-176,810,858 | 115.78 |
AX-176802416_A5- AX-176809615_A5 |
115.72-115.81 | 2.51 | 4.8 | -1.11 |
aLG, linkage group; bPVE, phenotypic variance explained; cADD, additive effect (negative values correspond to the ‘Harari’ parental line). 22/23, average value for 2022 and 2023
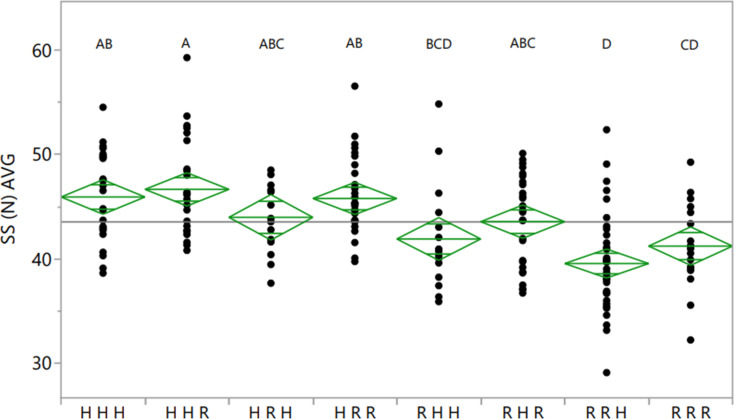
The genetic relationship between the three QTLs was analyzed by using ANOVA to compare different QTL combinations in the RIL population (Fig. 4). A general additive effect was found. The major effect of qSSB02 was observed throughout the analysis (the first four vs. the last four genotypic groups in Fig. 4). In addition, the HHR combination (i.e., Hanoch-like allele at qSSB02 and qSSB03 and Harari-like allele at qSSA05; Fig. 4), which had a positive effect in all loci, had the highest SS values; whereas the RRH genotype, which had a negative effect in all three loci, had the lowest SS value. The HHR combination (with a Harari allele on A05) had higher SS values than HHH (with a Hanoch allele at all three loci), showing again the positive effect of ‘Harari’ at qSSA05, although this difference was not statistically significant.
Fig. 4.
Marker combination analysis of three QTLs among the Hanoch × Harari RIL population. H, Hanoch allele; R, Harari allele. In each column, the left-most position of H/R is for qSSB02 (marker AX-147213175), the middle position is for qSSB03 (marker AX-176800560), and the right-most position is for qSSA05 (marker AX-176810858). Groups that are labelled with different letters are statistically different at p(F) < 0.05
The allelic status of selected markers from the three SS QTLs was evaluated in a group of 19 breeding lines and commercial varieties, spanning over 40 years of peanut breeding history in Israel (Table 6; Fig. 5). An almost perfect match was observed between the major QTL, qSSB02, and the market designation of the varieties (i.e., in-shell or shelled). Specifically, in 17 of the 19 examined cultivars, the marker accurately predicted the final market designation. In the other two QTLs, qSSB03 and qSSA05, the association between the selected marker and the market designation was less strong (13/19 and 15/19, respectively).
Table 6.
The allelic status of representative markers from the three QTLs identified for SS in a set of 19 historical peanut breeding line and commercial varieties. Highlighted in bold are genotypes with discrepancy between the allelic situation of B02 and the target market
| Line | AX-147,213,175 (B02) | AX-176,800,560 (B03) | AX-176,810,858 (A05) | Target market |
|---|---|---|---|---|
| Hanoch | 400 bp | C | A | in-shell |
| Harari | 640 bp | T | G | Shelled |
| Mona | 640 bp | T | G | Shelled |
| Orit | 400 bp | T | A | in-shell |
| Iddo | 400 bp | C | A | in-shell |
| IS-23 | 400 bp | T | G | in-shell |
| IS-22 | 400 bp | C | G | in-shell |
| Einat | 640 bp | T | G | Shelled |
| Hila | 640 bp | T | G | Shelled |
| Shulamit | 400 bp | T | A | in-shell |
| Shosh | 640 bp | T | G | Shelled |
| Rabin\ IS-57 | 400 bp | T | A | in-shell |
| 205 | 400 bp | C | A | in-shell |
| B65 | 400 bp | C | A | in-shell |
| GK7 | 400 bp | T | G | Shelled |
| IS-52 | 640 bp | T | G | Shelled |
| A76 | 640 bp | T | G | Shelled |
| IS-51 | 640 bp | T | G | Shelled |
| A89 | 400 bp | C | A | Shelled |
Fig. 5.
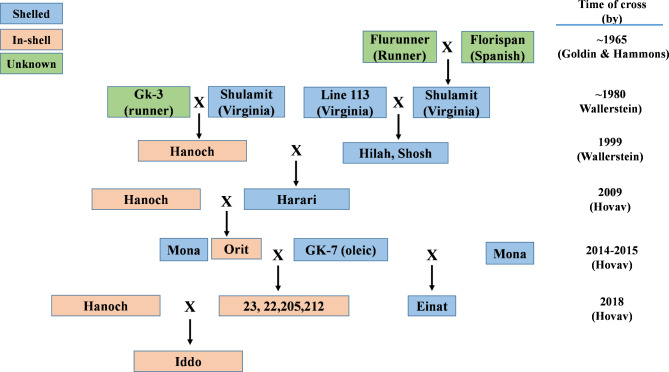
The history of peanut breeding in Israel. Each level represents a hybridization that was performed by s breeder (the name of the breeder and the year are presented on the right column). Historic cultivars are color-coded according to their main target market; blue, red, and green represent shelled, in-shell, and unknown marketing types, respectively
Thickness, density, and compositional characterization of shells
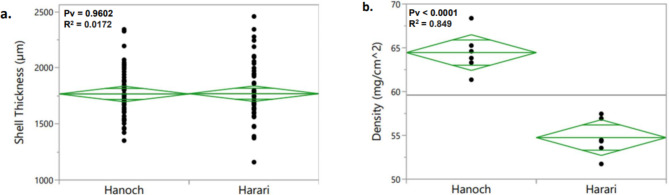
Physical and general-composition analyses were performed to better understand the differences in SS between the parental lines, Hanoch and Harari. Shell discs isolated from the two genotypes were extracted and their thickness and density were measured (Fig. 6). No significant difference was found between Hanoch and Harari in terms of pod thickness (Fig. 6a). In contrast, a significant difference was found in pod density; Hanoch had significantly greater pod density than Harari (prob[t] < 0.0001; R2 = 0.849; Fig. 6b).
Fig. 6.
Differences between Hanoch and Harari in terms of (a) shell thickness and (b) shell density
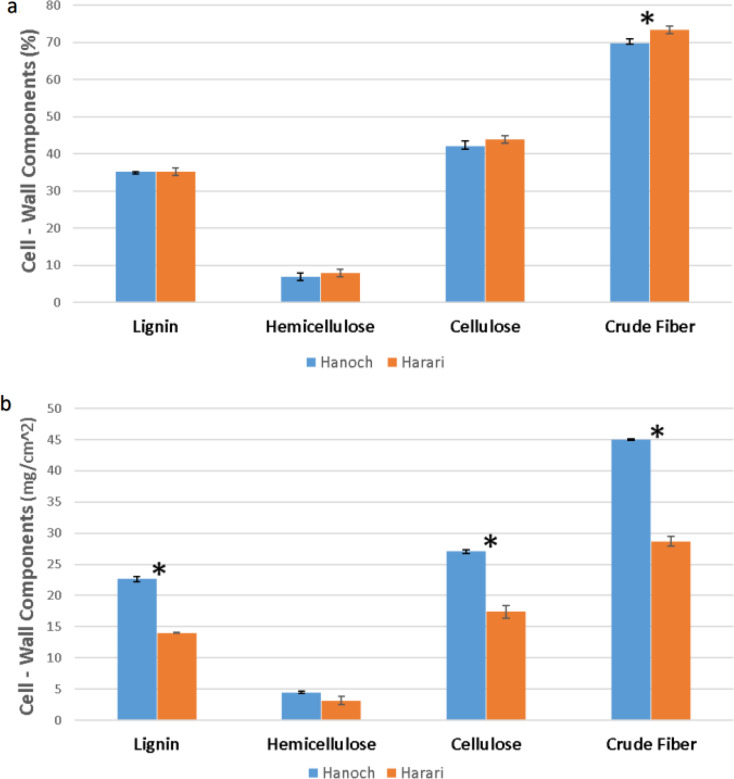
To compare the levels of cell-wall components between Hanoch and Harari, a general cell-wall composition analysis was conducted (Fig. 7). Based on shell weight (Fig. 7a), no significant differences were found between the genotypes in terms of lignin, hemicellulose, or cellulose level. However, Harari had a slightly, but significantly higher level of crude fiber. However, when the results were calculate based on shell density instead of shell weight, significantly higher levels of lignin, cellulose, and crude fiber amount per density were found in Hanoch (Fig. 7b).
Fig. 7.
Differences between Hanoch and Harari in terms of cell-wall components: (a) based on shell weight and (b) shell density. *Significant at p < 0.05
Functional annotation of the qSSB02 QTL region
To identify genes and genetic pathways potentially associated with SS, 360 genes within the qSSB02 QTL region were extracted from the Tifrunner reference genome annotation and analyzed against the entire gene set in the genome. Gene Ontology (GO) annotation showed that the majority of enriched genes had specific functional assignments: in the cellular component category, terms included cell periphery, external encapsulating structure, and cell wall; in the molecular function category, relevant terms included binding, oxidoreductase activity, and pectinesterase activity; and in the biological process category, terms included response to stimulus stress, defense, cell wall organization, and cell wall modification (Fig. 8).
Fig. 8.
Enrichment analysis of GO terms in the qSSB02 QTL region. Three component were analyzed: (a) Biological processes; (b) Molecular function; (c) Cellular component
Discussion
Yield loss due to weak pod shells is a significant concern in the Virginia-type in-shell peanut market. Therefore, there is great interest in developing new varieties with stronger SS. While SS of peanuts is known to have a genetic basis, the specific genetic mechanisms controlling it remain largely unknown. Most genetic studies of peanut have traditionally focused on traits like disease resistance, yield, and seed quality, often overlooking the important issue of shell mechanical strength. In this study, we aimed to explore the genetics of SS using a RIL population that segregates for this trait. This study represents the first attempt to genetically analyze and map shell strength in a biparental genetic structure in peanut. By utilizing closely related, elite varieties of Virginia-type peanuts as parents, we ensured that our findings would have direct relevance and significance for breeding efforts.
SS was found to be a stable trait within the inspected genetic background, as indicated by its relatively high heritability estimate (0.675) and significant differences and high R2 values among lines (averaging 0.38), as well as consistency across different environments and blocks. Similar to other pod characteristics like pod weight, seed number per pod, and pod constriction [22, 26, 27], SS showed relatively high heritability and minimal sensitivity to environmental conditions.
Our comprehensive genetic analysis identified three significant and consistent QTLs associated with SS, with a major QTL located on chromosome B02 explaining an average of 18.7% of the total phenotypic variation. This major QTL on B02 was very stable among environments (data is not shown), and it is reported here for the first time in relation to SS of peanut. In a genome-wide association study involving 320 peanut genotypes, Cui et al. [10] identified another QTL for SS on chromosome B08 that explained 12.5% of the total variation. Interestingly, in our previous study [18], when SS was manually ranked (on a scale of 0–2) within the same genetic population, neither the qSSB02 QTL nor any of the other QTLs were found to be significant. This underscores the importance of robust quantitative and subjective phenotyping methods to accurately identify QTLs with significant effects. Unfortunately, the qSSB02 QTL identified in our study is large in size (spanning 15.41 Mbp), due to sparse genetic markers and low recombination rates in this genomic region, which complicates the identification of promising candidate genes. Nonetheless, this QTL holds potential as a valuable marker for MAS breeding programs aimed at enhancing SS in Virginia-type peanuts.
Analysis of the allelic status of qSSB02 among a group of historical breeding lines and commercial varieties in Israel revealed an almost perfect match with market designation (i.e., in-shell or shelled). This suggests that indirect selection of qSSB02 has played a significant role in shaping Israel’s in-shell and shelled peanut industries. A review of Israeli peanut-breeding history (Fig. 5) revealed an interesting finding. All of the original American lines used as a source germplasm for the local breeding program (e.g., Fluranner and Florispan in 1965 by Goldin, GK3 by Wallerstein in 1980, and GK7-OL by Hovav in 2014) carried the Hanoch-like allele at qSSB02, despite not being Virginia-type varieties. Additionally, Line 113, a Virginia-type used by Yitzhak Wallerstein in the 1980s, a line of unknown origin, is probably the donor of the ‘Harari’-like undesirable allele. Therefore, we infer that the Hanoch-like allele of qSSB02, which is associated with the positive effect, represents the original condition; whereas the Harari-like allele associated with the negative effect was introduced later. This deduction was confirmed by a screen of the allelic status of qSSB02 in a collection of 64 American peanut genotypes [28], in which the Hanoch-like allele was found to be present across all four peanut marketing types (Virginia, Runner, Spanish, and Valencia); whereas the Harari-like allele was found to be present in a small subset of genotypes that is predominated by Virginia-type genotypes, such as N080820IJCT, ‘Gregory’, and ‘Jenkins Jumbo’.
An interesting finding from the QTL analysis concerns the role of the qSSA05 locus from Harari in influencing the SS phenotype. Generally, lines carrying the Hanoch genotype at the qSSB02 and qSSB03 loci, as well as the Harari genotype at qSSA05, exhibited the highest SS levels in the population. However, the difference between those lines and lines with the Hanoch genotype at all three QTLs was not statistically significant. Some lines with this HHR combination, such as HH115, consistently showed exceptionally high SS values across different environments and blocks. On the other hand, the HHR combination did not guarantee high SS, as seen in breeding line IS-22, which also possesses this combination but displays low SS (not shown). IS-22 has been excluded from commercial cultivation due to its tendency for pod breakage and seed-shattering. Together, these three QTLs contributed about 46% of the genetic variation (considering heritability rates). This suggests that there are likely additional genetic factors influencing SS that were not captured in our analysis. Nonetheless, our findings indicate that incorporating qSSA05 into MAS strategies could potentially improve SS in current peanut cultivars.
Another important finding of the current study is that the higher SS levels in Hanoch as compared to Harari are associated with shell density rather than shell thickness. This finding is substantially different from those of other studies that have reported a significant association between shell thickness and SS in peanut [10, 15, 29], as well as in other crops [30, 31]. The higher levels of shell density are evidenced by the significantly negative correlation of SS with shelling percentage and the significantly positive correlation between SS and 50SW, 50PW, and 50SHW. In peanut, similar correlations were previously found between shell thickness and pod size and between shell thickness and shelling percentage [29, 32], indicating that these relationships are not unique to shell density. Furthermore, our composition analysis revealed that this additional shell weight may not arise from the accumulation of a specific substance, but rather from the accumulation of lignin, cellulose, and crude fiber within this structure. Interestingly, we identified a strong negative association between SS and pod reticulation, with pods with smoother shells (lower pod reticulation) exhibiting higher SS levels. This suggests that both SS and pod reticulation may be regulated by the deposition of cell-wall components embedded within the “bowls” of the peanut shell. This process likely contributes to the smoother appearance of the shell without significantly affecting its overall thickness, as measured from one side to another. Several GO terms that were significant in the enrichment analysis of the qSSB02 QTL, such as cell periphery, external encapsulating structure, and cell wall (cellular component category) and cell wall organization/modification (biological process category) support this hypothesis.
Another important parameter influencing the plant biomechanics is water content. Therefore, the QTL could be potentially also related with water metabolism during maturing, not the “direct” shell strength QTLs. This assumption is unlikely to be the reason in our system, since the plants in our study were allowed to dry in the sun, and threshing was not conducted until the average seed moisture content is below 12%. While we did not directly measure shell water content, previous studies indicate a correlation between the water content of peanut seeds and shells [33]. Given that our shell material was dry at the time of harvest and stored under very dry conditions, we believe this mitigates concerns about water content affecting our results. Moreover, the high heritability rates and the presence of redundant QTLs across both years demonstrate the robustness of the findings regardless varying experimental and storage conditions. Nevertheless, we cannot completely rule out water content as a potential contributing factor, highlighting the need for further investigation in future studies.
Conclusions
Our study highlights the critical importance of SS in addressing yield losses due to weak pod shells in the Virginia-type in-shell peanut market. Through genetic analysis using a RIL population, we identified QTLs that are significantly associated with SS, underscoring the complex genetic basis of SS in peanuts, which is influenced by both major and minor QTLs across different genetic backgrounds. Our results suggest that incorporating qSSA05 into MAS programs could potentially enhance peanut SS. In addition, this study reveals insights into the composition of peanut shells, indicating that higher SS may be associated with shell density rather than thickness and is influenced by the accumulation of lignin, cellulose, and crude fiber. Understanding these genetic and compositional issues is crucial for the development of new peanut varieties with improved shell strength, to address the industry’s pressing concerns regarding pod integrity and market preferences.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Yaham LTD, Gadash Halutza and Gadash Masrik for their field and technical support.
Abbreviations
- SS
Shell strength
- RIL
Recombinant inbred line
- QTL
Quantitative trait loci
- %PVE
Percentage of phenotypic variation explained
- LG
Linkage group
- LOD
Logarithm of the odds
- MAS
Marker-assisted selection
- 50PW
50-pod weight
- 50SW
50-seed weight
- 50SHW
50-pod shell weight
Author contributions
G.B.I investigation and writing of original draft; S.K., W.M., AH and Y.L. investigation; K.G., S.G. review and editing; R.H. conceptualization, supervision and writing of original draft.
Funding
This study was funded by the Israeli Ministry of Agriculture Chief Scientists Fund (grant number 20-01-0283).
Data availability
Raw data used for the QTL analysis can be found in Supplementary Table 1 (Table S1).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Periasamy K, Sampoornam C. The morphology and anatomy of ovule and fruit development in Arachis hypogaea L. Ann Bot. 1984;53:399–411. [Google Scholar]
- 2.Boote KJ. Growth stages of peanut (Arachis hypogaea L). Peanut Sci. 1982;9:35–40. [Google Scholar]
- 3.Rico X, Gullón B, Alonso JL, Parajó JC, Yáñez R. Valorization of peanut shells: manufacture of bioactive oligosaccharides. Carbohydr Polym. 2018;183:21–8. [DOI] [PubMed] [Google Scholar]
- 4.Adhikari B, Dhungana SK, Ali MW, Adhikari A, Kim ID, Shin DH. Antioxidant activities, polyphenol, flavonoid, and amino acid contents in peanut shell. J Saudi Soc Agric Sci. 2019;18:437–42. [Google Scholar]
- 5.Perea-Moreno MA, Manzano-Agugliaro F, Hernandez-Escobedo Q, Perea-Moreno AJ. Peanut shell for energy: properties and its potential to respect the environment. Sustainability. 2018;10:3254. [Google Scholar]
- 6.Wee JH, Moon JH, Eun JB, Chung JH, Kim YG, Park KH. Isolation and identification of antioxidants from peanut shells and the relationship between structure and antioxidant activity. Food Sci Biotech. 2007;16:116–22. [Google Scholar]
- 7.Kotzamanidis C. Correlation studies of 21 traits in F2 generation of groundnut (Arachis hypogaea L). Pak J Biol Sci. 2006;9:929–34. [Google Scholar]
- 8.Kotzamanidis C. Classification and evaluation of Greek groundnut (Arachis hypogaea L.) using 17 main agronomic and quality traits. Pak J Biol Sci. 2006;9:1000–5. [Google Scholar]
- 9.Muhammad AI, Isiaka M, Fagge AA, Attanda ML, Lawan I, Dangora ND. Some engineering properties of three varieties of groundnut pods and kernels. Arid Zone J Eng Technol Environ. 2015;11:62–76. [Google Scholar]
- 10.Cui K, Qi F, Sun Z, Feng J, Huang B, Dong W, Zhang X. Genome-wide association study of physical and microstructure-related traits in peanut shell. Plant Genet Resour. 2021;19:394–404. [Google Scholar]
- 11.Clevenger J, Chu Y, Scheffler B, Ozias-Akins P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front Plant Sci. 2016;7:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Liang X, Lu Q, Li H, Liu H, Li S, et al. Global transcriptome analysis of subterranean pod and seed in peanut (Arachis hypogaea L.) unravels the complexity of fruit development under dark condition. Sci Rep. 2020;10:13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta K, Gupta S, Faigenboim-Doron A, Patil AS, Levy Y, Carrus SC, Hovav R. Deep transcriptomic study reveals the role of cell wall biosynthesis and organization networks in the developing shell of peanut pod. BMC Plant Biol. 2021;21:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Wang J, Tang Z, Guo F, Zhang Y, Zhang J, Meng J, Zheng L, Wan S, Li X. Transcriptome of peanut kernel and shell reveals the mechanism of calcium on peanut pod development. Sci Rep. 2020;10:15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Zheng Z, Sun Z, Qi F, Wang J, Wang M, Dong W, Cui K, Zhao M, Wang X, Zhang M, Wu X, Wu Y, Luo D, Huang B, Zhang Z, Cao G, Zhang X. Identification of two major QTLs for pod shell thickness in peanut (Arachis hypogaea L.) using BSA-seq analysis. BMC Genomics. 2024;25:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patil AS, Hedvat I, Levy Y, Galili S, Hovav R. Genotype-by-environment effects on the performance of recombinant inbred lines of Virginia-type peanut. Euphytica. 2018;214:83. [Google Scholar]
- 17.Agmon S, Kunta S, Dafny Yelin M, Moy J, Ibdah M, Harel A, Rabinovitch O, Levy Y, Hovav R. Mapping of stem rot resistance in peanut indicates significant effect for plant architecture locus. Crop Sci. 2022;62:2197–211. [Google Scholar]
- 18.Kunta S, Agmon S, Chedvat I, Levy Y, Chu Y, Ozias-Akins P, Hovav R. Identification of consistent QTL for time to maturation in Virginia-type peanut (Arachis hypogaea L). BMC Plant Biol. 2021;21:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clevenger JP, Korani W, Ozias-Akins P, Jackson S. Haplotype-based genotyping in Polyploids. Front Plant Sci. 2018;9:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korani W, Clevenger JP, Chu Y, Ozias-Akins P. Machine learning as an effective method for identifying true single nucleotide polymorphisms in polyploid plants. Plant Genome. 2019;12:180023. [DOI] [PubMed] [Google Scholar]
- 21.Patil AS, Popovsky S, Levy Y, Chu Y, Clevenger J, Ozias-Akins P, Hovav R. Genetic insight and mapping of the pod constriction trait in Virginia-type peanut. BMC Genet. 2018;19:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavarro C, Chu Y, Holbrook C, Isleib T, Bertioli D, Hovav R, Butts C, Lamb M, Sorensen R, Jackson SA, Ozias-Akins P. Pod and seed trait QTL identification to assist breeding for peanut market preferences. G3. (Bethesda). 2020;10:2297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertioli DJ, Jenkins J, Clevenger J, Dudchenko O, Gao D, Seijo G, et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet. 2019;51:877–84. [DOI] [PubMed] [Google Scholar]
- 24.Ooijen JW. Van. MapQTL 6. Genome. 2009.
- 25.Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–96. [Google Scholar]
- 26.Luo H, Guo J, Ren X, Chen W, Huang L, Zhou X, Chen Y, Liu N, Xiong F, Lei Y, Liao B, Jiang H. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L). Theor Appl Genet. 2018;131:267–82. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Wang Z, Ren X, Huang L, Guo J, Zhao J, Zhou X, Yan L, Luo H, Liu N, Chen W, Wan L, Lei Y, Liao B, Huai D, Jiang H. Identification of major QTL for seed number per pod on chromosome A05 of tetraploid peanut (Arachis hypogaea L). Crop J. 2019;7:238–48. [Google Scholar]
- 28.Clevenger J, Chu Y, Chavarro C, Agarwal G, Bertioli DJ, Leal-Bertioli SCM, Pandey MK, Vaughn J, Abernathy B, Barkley NA, Hovav R, Burow M, Nayak SN, Chitikineni A, Isleib TG, Holbrook CC, Jackson SA, Varshney RK, Ozias-Akins P. Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol Plant. 2017;10:309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil S. Genetic studies on hybridization in groundnut (Arachis hypogaea L). Madras Agric J. 1972;10:45–52. [Google Scholar]
- 30.Kacal M, Koyuncu MA. Cracking characteristics and kernel extraction quality of hazelnuts: effects of compression speed and positions. Int J Food Prop. 2017;20:1664–74. [Google Scholar]
- 31.Koyuncu MA, Ekinci K, Savran E. Cracking characteristics of walnut. Biosyst Eng. 2004;87:305–11. [Google Scholar]
- 32.Godoy IJ, Norden AJ. Shell and seed size relationships in peanuts. Peanut Sci. 1981;8:21–4. [Google Scholar]
- 33.Xu X, Sun Y, Yin, Xue Y, Ma F, Song C, Yin H, Zhao L. A free-space-based model for predicting peanut moisture content during natural drying. J Food Qual. 2022; 9620349.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data used for the QTL analysis can be found in Supplementary Table 1 (Table S1).