Abstract
Background
The Mendelian Disorders of Cornification (MeDOC) comprise a large number of disorders that present with either localised (palmoplantar keratoderma, PPK) or generalised (ichthyoses) signs. The MeDOC are highly heterogenic in terms of genetics and phenotype. Consequently, diagnostic process is challenging and before implementation of the next generation sequencing, was mostly symptomatic, not causal, which limited research on those diseases. The aim of the study was to genetically characterise a cohort of 265 Polish patients with MeDOC and to get insight into the skin lesions using transcriptome and lipid profile analyses.
Results
We detected causal variants in 85% (226/265) patients. In addition to the primary gene defect, a pathogenic variant in another gene involved in MeDOC pathology was identified in 23 cases. We found 150 distinct variants in 33 genes, including 32 novel and 16 recurrent (present in > 5 alleles). In 43 alleles large rearrangements were detected, including deletions in the STS, SPINK5, CERS3 and recurrent duplication of exons 10–14 in TGM1. The RNA analysis using samples collected from 18 MeDOC patients and 22 controls identified 1377 differentially expressed genes - DEG. The gene ontology analysis revealed that 114 biological processes were upregulated in the MeDOC group, including i.e. epithelial cell differentiation, lipid metabolic process; homeostasis; regulation of water loss via skin; peptide cross-linking. The DEG between TGM1 and ALOX12B patients, showed that RNA profile is highly similar, though fatty acid profile in epidermal scrapings of those patients showed differences e.g. for the very long chain fatty acids (VLCFAs; FAs ≥ C20), the very long-chain monounsaturated fatty acids (VLC-MUFAs, FAs ≥ C20:1) and the n6 polyunsaturated fatty acids (n6 PUFAs).
Conclusion
Our results show that NGS-based analysis is an effective MeDOC diagnostic tool. The Polish MeDOC patients are heterogenic, however recurrent variants are present. The novel variants and high number of TGM1 and SPINK5 copy number variations give further insight into molecular pathology of MeDOC. We show that secondary variants in MeDOC-related genes are present in a significant group of patients, which should be further investigated in the context of phenotype modifiers. Finally, we provide novel RNA and lipid data that characterise molecularly MeDOC epidermis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13023-024-03395-4.
Keywords: Genodermatoses, MeDOC, PPK, Ichthyoses, RNAseq, Genes, Variants, Secondary findings
Background
The human epidermis is a stratified epithelium composed of morphologically distinct cellular layers. In the basal layer, keratinocytes divide and subsequently initiate differentiation, which involves morphological and physiological changes. Finally, in the upper layers of the epidermis, they undergo cornification, a multistep process, during which programmed cell death is initiated and live keratinocytes are transformed into apoptotic corneocytes. The dead cells, surrounded by lipid monolayers (corneocyte lipid envelopes), are embedded in lipid-protein matrix forming a very regular structure of the epidermal barrier. On the molecular level, the cornification process involves synthesis of several structural proteins, enzymes and lipids [1]. The disturbance of keratinocyte differentiation, faulty intercellular adhesion as well as quality and quantity changes in protein/lipid content lead to homeostasis disturbance and may have clinical implications. So far, pathogenic variants in over 100 genes are known to be causative for epidermal barrier dysfunction, commonly referred to Mendelian Disorders of Cornification (MeDOC).
The MeDOC comprise a large number of non-syndromic (skin-limited) and syndromic disorders (skin and other organs affected). The disruption of cornification is manifested clinically by either localised (palmoplantar keratoderma, PPK) or generalised (Ichthyoses) hyperkeratosis in the form of scaling and keratoderma often accompanied by collodion membrane, erythema, blisters, erosions, atopy, infections, and other symptoms [2, 3].
The phenotypes of patients have many similarities, like scaling and erythema. However, they can vary significantly even within the same subtype, regardless of the underlying causative gene defects [4]. This means that a clinical diagnosis cannot be easily correlated with a specific molecular defect. Therefore, before genetic analyses became available, complex laboratory tests, such as histology of skin biopsies by electron and light microscopy, immunohistochemical staining, enzymatic activity and biochemical tests were necessary to specify the diagnosis. Consequently, the diagnosis was mostly symptomatic, not causal. This aspect had further implications on research limiting the possibility of comparative characterization and therapeutic research in patients with different causes of MeDOC.
The aim of the study was to genetically characterise a cohort of 265 Polish patients with cornification disorders. We describe novel pathogenic variants and provide unique data about incidental findings in MeDOC patients, which may influence future genotype-phenotype studies. Additionally, we present the results of transcriptome and lipid analysis in skin biopsies and scrapings, to further characterise skin lesions.
Methods
Patients
The group of 265 patients with clinical symptoms of MeDOC were enrolled for molecular analysis. The patients were referred by clinical geneticist and dermatologists across Poland in the years 2016–2022.
Each patient gave informed consent to participate in the study. The analysis was performed in the Department of Medical Genetics and Institute of Mother and Child (IMiD, 253 cases) and in the MedGen Medical Center (12 cases).
In the years of 2016–2020 as part of the project “Change in global gene expression versus keratin and lipid profile in rare skin disorders from the group of ichthyosis. (2014/13/D/NZ5/03304 )” the patients were asked to give consent for a skin biopsy to perform functional studies. Skin biopsies and scrapings of 18 MeDOC patients were taken (Additional file 1, part a). The 22 control skin samples were collected from persons without clinical signs of cornification disorders during surgical procedures performed for other medical indications. In the case of 10 patients with ALOX12B (n = 5) and TGM1 (n = 5), from whom the biopsies were taken, we obtained detailed clinical questionnaire (Additional file 1, part b). These patients manifest typical features of ARCI (autosomal recessive congenital ichthyosis), although pain, keratosis pilaris, nail changes and sparse hair were more prevalent in TGM1 patients.
Genotyping
The Next Generation sequencing (NGS) panel
In the majority of patients (245 cases) genotyping was performed using a customized NGS panel covering all coding exons of 60 genes: AAGAB, ABCA12, ABHD5, ADAM10, ALDH3A2, ALOX12B, ALOXE3, AP1S1, AQP5, CDSN, CLDN1, CSTA, CTSC, CYP4F22, DSG1, DSP, EBP, ENPP1, ERCC2, ERCC3, FERMT1, FLG, GJA1, GJB2, GJB3, GJB4, GTF2H5, HOXC13, JUP, KANK2, KRT1, KRT10, KRT16, KRT17, KRT2, KRT9, LIPN, LOR, MBTPS2, MPLKIP, NIPAL4, NSDHL, PEX7, PHYH, PKP1, PNPLA1, POFUT1, POGLUT1, POMP, SERPINB7, SLC27A4, SLURP1, SNAP29, SPINK5, ST14, STS, SUMF1, TGM1, TRPV3, VPS33B. The panels were supplied by Roche and Agilent. The libraries were made using the KAPA Library Preparation Kit and SureSelect Kit. The MiSeq sequencer (Illumina) was used to sequence the samples. The bioinformatic analysis was performed using pipeline based on Ensembl Variant Effect Predictor (VEP). The reads were aligned against the GRCh38 human genome assembly. The following databases were used for variant annotations: the SNPdb (NCBI), Ensembl, OMIM, GnomAD, ClinVar, and HGMD Professional. The Copy Number Variation (CNV) analysis was performed using CODEX, CoNIFER, cn.mops, ExomeDepth and XHMM algorithms.
The clinical significance of the variants was assessed according to ACMG [5] using Varsome (https://varsome.com/) and Franklin (https://franklin.genoox.com) online software packages.
Verification of NGS results
All variants detected by NGS were verified as described below
Whenever possible, biparental origin of the variants was verified by testing the parents (n = 122 families, data not shown).
Single nucleotide variants: Sanger sequencing
The presence of all pathogenic and likely pathogenic variants detected using NGS analysis was confirmed by Sanger sequencing. The primers were designed using primer3 software (https://primer3.ut.ee/) and are available upon requests together with PCR conditions.
Rare FLG (filaggrin) variants confirmation (apart from p.Arg501Ter and p.Ser761CysfsTer36) was performed in the Departments of Dermatology and Clinical Genetics at the Maastricht University Medical Centre + as described before [6].
Copy number variation
The TGM1 duplication was confirmed by qPCR using TaqMan Copy Number Assay (ThermoFisher Scientific). The probes Hs01865876_cn (intron 12-exon 13 boundary, Chr.14:24249114–24263210 on Build GRCh38) and Hs03052754_cn (exon 14, Chr.14:24249114–24263210 on Build GRCh38) were used to analyse the TGM1 gene, the control probe was TaqMan™ Copy Number Reference Assay, human, TERT (ThermoFisher Scientific). Reactions were performed according to the manufacturer’s recommendations using the CFX Opus 96 Real-Time PCR System (BioRad).
The presence of deletions was confirmed by analysis of the parental samples using the NGS gene panel or, in case of STS, by Multiplex Ligation-dependent Probe Amplification (MLPA), see below.
MLPA analysis of the STS gene
The STS gene analysis was performed using MLPA in 9 patients with the suspicion of X-linked ichthyosis. It was also used to confirm deletions that were detected by NGS. The MLPA was performed using The SALSA MLPA Probemix P160 STS (MRC Holland) on Sequencer ABI 3500. The results were analysed using Coffalyser.Net™ (MRC Holland).
Analysis of the most common variants in the FLG gene
In 11 cases, analysis of the two most common pathogenic variants p.Arg501Ter and p.Ser761CysfsTer36 in FLG was performed by Sanger sequencing using primers and PCR conditions as described by Sandilands et al. [7].
Functional studies
Skin biopsy and scrapings
The 3 mm skin biopsies were taken from the patients and controls for RNA sequencing, while for lipid analysis, only the scrapings were collected mechanically using a scalpel. The biopsies and scrapings were snap frozen immediately. The epidermis was detached mechanically in a cryotome prior to further studies. All samples were stored at -80 C.
RNA sequencing
The RNA isolation and sequencing were performed as we described previously [8]. Briefly, RNA was isolated from homogenized epidermis with RNeasy Micro Kit (Qiagen) and quantified on Agilent 2100 Bioanalyzer using an RNA 6000 Pico Kit (Agilent Technologies, Ltd.). The polyA enriched RNA libraries were prepared using the QuantSeq 3’ mRNA-Seq Library Prep Kit according to the manufacturer’s protocol (Lexogen GmbH, Vienna, Austria) and the libraries were single-end sequenced (75 bp) on a HiSeq 1500 (Illumina, San Diego, CA 92122 USA). Reads were aligned to the reference human genome GRCh38 (Ensembl database) using STAR aligner and counted using HT seq. Genes with exceptionally low expression (less than 5 reads across all samples) were excluded from further analysis. Linear models implemented in the edgeR package were used for normalization and differential expression analysis. The adjusted p-value (FDR) < 0.05 and |logFC| > 2 were used to determine the list of differentially expressed genes. An enrichment test implemented in the systemPipeR R package was used for gene ontology (GO) enrichment analysis. The genes that were considered in this type of analysis were those with |logFC| > 1.5 and an adjusted p-value (FDR) < 0.05. The returned corresponding Bonferroni adjusted p-value < 0.05 was used as the significance threshold for each GO. The R version 3.6.2 was used for all statistical analysis. The genes considered for GO analysis, were also included in Reactome’s Pathway Analysis based on hypergeometric distribution for pathway over-representation analysis. The Reactome version 87 on 27/12/2023 was used.
Fatty acid (FA) analysis
Total lipids were extracted according to the Folch method [9]. Subsequently, the lipid extracts were dried under a stream of nitrogen and hydrolyzed with 0.5 M KOH at 90 °C for 3 h. The solution was acidified and 1mL of water was added. The unesterified fatty acids (FAs) were extracted with n-hexane (3 × 1 mL) and the solvent was then completely evaporated under a stream of nitrogen. To obtain fatty acid methyl esters (FAMEs), a 10% boron trifluoride–methanol solution was added to each sample and heated at 55 °C for 90 min. Then, 1mL of water was added to the reaction mixture and FA derivatives were extracted with n‐hexane (3 × 1 mL). 19 methyleicosanoic acid was used as an internal standard. The analysis of FAs was performed by gas chromatography – mass spectrometry (GC-MS) [10].
Results
Genotyping
We detected causal variants for clinical phenotypes in 226/265 patients, which gave an overall diagnostic yield of 85%. The different detection rates were achieved, with respect to the method used (Table 1).
Table 1.
Results of genotyping performed using different diagnostic strategies
| Total number of patients | Number of patients with causative pathogenic variants identified | Number of patients with one pathogenic variant in AR inheritance or no variants at all (%) | |
|---|---|---|---|
| TOTAL: | 265 | 226 (85%) | 39 (15%) |
| NGS customized panel: | 245 | 214 (87%) | 31 (13%) |
| STS analysis by MLPA | 9 | 8 (89%) | 1 (11%) |
| FLG - two mutations test by Sanger | 11 | 4 (36%) | 7 (64%) |
In 214/245 (87%) patients analysed by NGS, the genetic cause of the disease was elucidated (Table 1 and Additional file 2). In 31/245 (13%) cases either one pathogenic/likely pathogenic variant was found (while autosomal recessive (AR) inheritance was expected) or no pathogenic variants were identified. In 9 cases, only the STS gene analysis by MLPA was performed enabling for detection of STS gene deletions in 8/9 (89%) cases. In a subgroup of 11 patients, we only performed limited genetic analysis comprising the two most common FLG variants: p.Arg501Ter and p.Ser761CysfsTer36. Two pathogenic variants were detected in 4/11 (36%) cases.
In 23 cases of 245 analysed by NGS, in addition to the primary gene defect, a pathogenic variant(s) in another gene(s) were detected (Table 2). The majority of secondary findings (17/23) were variants already reported in the ClinVar database as pathogenic (P) or likely pathogenic (LP). One variant has the conflicting status (LP vs. variants of unknown significance, VUS) and 3 are reported as VUS. In 3 cases novel variants were found, while in one patient (No #19 in Table 2), known pathogenic variants were found in two other genes.
Table 2.
The primary genotypes and secondary findings
| No (sex) | Gene and variants found causative of phenotype | Other (P, LP and VUS) findings in NGS panel (all variants were detected as heterozygotes) |
||||
|---|---|---|---|---|---|---|
| gene | allele 1 | allele 2 | gene | allele | Classification in ClinVar | |
| 1 | ALOX12B | c.1454T>C p.(Phe485Ser) | c.1562A>G p.(Tyr521Cys) | TGM1 | c.377G>A p.(Arg126His) |
known pathogenic VCV000279909.12 |
| 2 | ALOXE3 | c.700 C>T p.(Arg234Ter) |
c.700 C>T p.(Arg234Ter) |
ERCC3 | c.460C>T p.(Gln154Ter) |
known pathogenic VCV000801749.3 |
| 3 | ALOX12B | c.1562A>G p.(Tyr521Cys) | c.1562A>G p.(Tyr521Cys) | ERCC3 | c.325C>T p.(Arg109Ter) |
known pathogenic VCV000265515.32 |
| 4 | ABCA12 | c.4139A>G p.(Asn1380Ser) | c.6100A>G p.(Asn2034Asp) | FLG | c.7339C>T p.(Arg2447Ter) |
pathogenic/likely pathogenic VCV000050932.46 |
| 5 | ALOX12B | c.1265C>T p.(Pro422Leu) | c.1562A>G p.(Tyr521Cys) | FLG | c.2282_2285del p.(Ser761CysfsTer36) |
known pathogenic VCV000016320.59 |
| 6 | ALOX12B | c.1562A>G p.(Tyr521Cys) |
c.2094C>A p.(Ser698Arg) |
FLG | c.2282_2285del p.(Ser761CysfsTer36) | known pathogenic VCV000016320.59 |
| 7 | ALOXE3 | c.700C>T p.(Arg234Ter) | c.1472T>G p.(Leu491Arg) | FLG | c.5702del p.(Gly1901AlafsTer194) | NOVEL* |
| 8 | TGM1 | c.1135G>C p.(Val379Leu) |
c.(1402 + 1_1401-1)_(2225 + 1_2226-1)dup p.? |
FLG | c.2282_2285del p.(Ser761CysfsTer36) |
known pathogenic VCV000016320.59 |
| 9 | TGM1 | c.1135G>C p.(Val379Leu) |
c.1500T>A p.(Ser500Arg) |
FLG |
c.3982C >T p.(Gln1328Ter) |
NOVEL* |
| 10 | ALOX12B | c.1A>G p.(Met1?) |
c.2094C>A p.(Ser698Arg) |
FLG | c.2282_2285del p.(Ser761CysfsTer36) | known pathogenic VCV000016320.59 |
| 11 | STS | c.(?_-1)_(*1_? )del p.1Met_583Terdel | (-) | FLG | c.2282_2285del p.(Ser761CysfsTer36) | known pathogenic VCV000016320.59 |
| 12 | KRT10 | c.467G>A p.(Arg156His) | (-) | FLG | c.2282_2285del p.(Ser761CysfsTer36) | known pathogenic VCV000016320.59 |
| 13 | KRT2 | c.1459G>A p.(Glu487Lys) | (-) | ABCA12 | c.6611G>A p.(Arg2204Gln) | VUS RCV001136974.4 |
| 14 | STS | c.(?_-1)_(*1_? )del p.1Met_583Terdel | ABCA12 | c.179G>C p.(Arg60Pro) | VUS/likely pathogenic VCV000264998.5 | |
| 15 | SPINK5 | c.(?_-1)_(410 + 1_411-1)del p.? |
c.(?_-1)_(410 + 1_411-1)del p.? |
ALOXE3 | c.1432 A>C p.(Ser478Arg) | VUS RCV001127791.4 |
| 16 | KRT2 | c.566T>C p.(Phe189Ser) | (-) | ALOX12B | c.1562 A>G p.(Tyr521Cys) |
pathogenic VCV000039546.40 |
| 17 | ALOXE3 | c.700C>T p.(Arg234Ter) |
c.1889C>T p.(Pro630Leu) |
VPS33B | c.199T>C p.(Tyr67His) | NOVEL* |
| 18 | SPINK5 | c.1825C>T p.(Gln609Ter) | c.(?_-1)_(1479 + 1_1480-1)del p.? | GTF2H5 | c.49 A>T p.(Lys17Ter) |
pathogenic VCV000975159.1 |
| 19 | SLC27A4 | c.1541A>G p.(Glu514Gly) | c.1510C>T p.(Arg504Cys) | FLG | c.7339C>T p.(Arg2447Ter) |
pathogenic/likely pathogenic VCV000050932.46 |
| NIPAL4 | c.341C>A p.(Ala114Asp) |
pathogenic/likely pathogenic VCV000001731.29 |
||||
| 20 | ALOXE3 | c.700C>T p.(Arg234Ter) |
c.700C>T p.(Arg234Ter) |
DSP | c.4198C>T p.(Arg1400Ter) |
pathogenic/likely pathogenic VCV000199884.19 |
| 21 | ALOX12B | c.467_470dup p.(His158CysfsTer20) | c.1562A>G p.(Tyr521Cys) | TGM1 | c.1631A>G p.(Tyr544Cys) | VUS RCV000664924.1 |
| 22 | ALOX12B | c.1207C>T p.(His403Tyr) |
c.1790C>A p.(Ala597Glu) |
WNT10A | c.321C >A p.(Cys107Ter) |
pathogenic VCV000004461.41 |
| 23 (F) | FLG | c.1501C>T p.(Arg501Ter) | c.7339C>T p.(Arg2447Ter) | STS | c.1316A>G p.(His439Arg) |
pathogenic VCV000010556.1 |
* see Table 3 ACMG classification
Variant characteristics
In total, we detected 150 distinct variants in 415 alleles of 33 genes (Table 2, Additional file 2 and 3). The alleles with pathogenic/likely pathogenic variants in the ALOX12B gene were the mostly represented (n = 115), followed by variants in the TGM1 (n = 68), FLG (n = 39), ALOXE3 (n = 36), and STS (n = 27) genes. The number of distinct variants was also the highest in the TGM1 (n = 23) and the ALOX12B genes (n = 22). Among 150 distinct variants detected, 32 were previously undescribed and 118 known. The most common was c.1562 A > G p.(Tyr521Cys) in the ALOX12B gene, detected in 52 alleles. Eight variants were detected in over 10 alleles, and total of 51 were detected at least 2 times (Additional file 4). Importantly, 99 variants (66%) were unique. Among 32 novel ones, as many as 9 were found in ALOX12B, followed by 4 in ALOXE3, 4 in KRT1, and 3 in ABCA12 and KRT10 (Table 3).
Table 3.
The variants identified for the first time in this study
| GENE (numer of variants) |
VARIANT | Number of alleles with the variant | ACMG classification | ACMG main criteria |
|---|---|---|---|---|
| ABCA12 (n = 3) | c.6100 A>G p.(Asn2034Asp) | 1 | VUS | PM2, PP3 |
| c.6194del p.(Asn2065ThrfsTer3) | 1 | likely pathogenic | PVS1, PM2 | |
| c.758delT p.(Phe253SerfsTer27) | 1 | likely pathogenic | PVS1, PM2 | |
| ALOX12B (n = 9) | c.1454T>C p.(Phe485Ser) | 2 | VUS | PM2, PM1, PP2, PP3 |
| c.962T>A p.(Met321Lys) | 2 | VUS | PM2, PM1, PP2 | |
| c.808 A>C p.(Asn270His) | 1 | VUS | PM2, PM1, PP2, PP3 | |
| c.1154T>C p.(Val385Ala) | 1 | likely pathogenic | PM1, PP2, PM2, PP3 | |
| c.1446dupC p.(Asn483GlnfsTer2) | 1 | likely pathogenic | PVS1, PM2 | |
| c.1529 A>G p.(Glu510Gly) | 1 | VUS | PM2, PP2 | |
| c.808 A>C p.(Asn270His) | 1 | VUS | PM2, PM1, PP2, PP3 | |
| c.911T>C p.(Leu304Ser) | 1 | VUS | PM2, PM1, PP2, PP3 | |
| c.1102–2 A>T p.? | 1 | likely pathogenic | PVS1, PM2 | |
| ALOXE3 (n = 4) | c.707del p.(Leu236ArgfsTer44) | 1 | likely pathogenic | PVS1, PM2 |
| c.1472T>G p.(Leu491Arg) | 1 | VUS | PM2, PP3 | |
| c.842delG p.(Gly281ValfsTer38) | 1 | likely pathogenic | PVS1, PM2 | |
| c.984 C>A p.(Tyr328Ter) | 1 | likely pathogenic | PVS1, PM2 | |
| CERS3 (n = 1) | c.(999+1_1000-10)_(*11_? )del p.? | 2 | VUS | Uncertain (0.75) |
| DSG1 (n = 1) | c.518–2 A>G p.?* | 1 | likely pathogenic | PVS1, PM2 |
| FLG (n = 1, SF) | c.5702del p.(Gly1901AlafsTer194) | 1 | likely pathogenic | PVS1, PM2 |
| KRT1 (n = 4) | c.1430T>C p.(Leu477Pro) | 1 | VUS | PM2, PM1 |
| c.1535delT p.(Ile512ThrfsTer102)* | 1 | likely pathogenic | PVS1, PM2 | |
| c.539 A>T p.(Glu180Val) | 1 | VUS | PM2, PM1, PP3 | |
| c.551T>A p.(Ile184Asn) | 1 | VUS | PM2, PM1, PP3 | |
| KRT10 (n = 3) | c.1441_1448del p.(Gly481ArgfsTer97) | 1 | likely pathogenic | PVS1, PM2 |
| c.1554dupC p.(Ser519GlnfsTer62)* | 1 | likely pathogenic | PVS1, PM2 | |
| c.1689_1690del p.(Ser563ArgfsX17) | 1 | likely pathogenic | PVS1, PM2 | |
| SLC27A4 (n = 1) | c.1541 A>G p.(Glu514Gly) | 1 | VUS | PM2, PP3 |
| SPINK5 (n = 1) | c.(?_-1)_(410+1_411-1)del p.? | 3 | VUS | Uncertain (0.75) |
| ST14 (n = 1)** | c.2086 C>A p.(Arg696Ser)** | 1 | VUS | PM2 |
| TGM1 (n = 2) | c.1500T>A p.(Ser500Arg) | 2 | VUS | PM2 |
| c.1490 A>T p.(Glu497Val) | 1 | likely pathogenic | PP3, PM2 | |
| VPS33B (n = 1, SF) | c.199T>C p.(Tyr67His) | 1 | VUS | PM2, PP3 |
SF- variants were found as secondary findings; *phenotype details of this patient were partially published by us recently [11]; ** the variant in ST14 gene was identified in one allele only, while defects in ST14 cause autosomal recessive ichthyosis. Therefore molecular cause of MeDOC in this case is not known
Among all variants we found, the majority were SNV (single nucleotide variants), however, 43/415 (10.3%) alleles harboured large rearrangements (CNV – copy number variations) (Table 4). Deletion of the STS gene was the most common. Moreover, in 6 alleles of SPINK5, large deletions were found encompassing exons 1–16: c.(?_-1)_(1479 + 1_1480-1)del (in 3 alleles) and exons 1–5: (c.(?_-1)_(410 + 1_411-1)del (in 3 alleles, variant not reported before). Finally, one patient had a homozygous deletion of exon 12 in CERS3 gene. In nine TGM1 alleles we detected duplication of exons 10–14 (c.(1402 + 1_1401-1)_(2225 + 1_2226-1)dup).
Table 4.
The list of CNV identified in the study
| Gene | CNV | Number of alleles | Numer of patients |
|---|---|---|---|
| STS |
c.(?_-1)_(*1_? )del p.01Met_583Terdel |
26 | 26 |
| TGM1 | c.(1402 + 1_1401-1)_(2225 + 1_2226-1)dup p.? | 9 | 9 |
| SPINK5 |
c.(?_-1)_(1479 + 1_1480-1)del p.? |
3 | 3 |
| SPINK5 |
c.(?_-1)_(410 + 1_411-1)del p.? |
3 | 2 |
| CERS3 |
c.(999 + 1_1000-10)_(*11_? )del p.? |
2 | 1 |
Functional studies
The RNA samples were collected from 18 patients and from 22 anonymous controls without signs of a cornification disorder. There were 7 cases with pathogenic variants in TGM1, 5 in ALOX12B, 2 in ALOXE3 and one in LOR, SPINK5, STS and ABCA12, each. Median age of the patients was: 21.5 ± 13.5. The control group did not manifest any clinical symptoms of a cornification disorder and included: 13 samples obtained from female adults (BMI 25–33, median age: 50,5 ± 20.87), 8 samples from children (BMI = 18.5–25, median age: 6.5 ± 3.5) and 1 from adult men. The characteristics of patients and controls, including skin samples collection sites and type are given in the Additional file 1.
RNA analysis
Principal component analysis (PCA) was performed on RNA-seq expression data to verify if samples are grouping according to considered conditions (Additional file 5). The PCA revealed that MeDOC samples fall apart from healthy ones. PCA also revealed that grouping according to the skin sample collection site was not exact. In order to get insight into the expression pattern differences between epidermis of thorax and limb of the adult controls, we performed the Differentially Expressed Genes (DEG) analysis and discovered only 39 of them. We also performed DEG analysis between children and adult controls, which reached 137. Those numbers were however much smaller than the values of DEGs between patient and controls groups, which justified further analyses.
MeDOC patients vs. controls
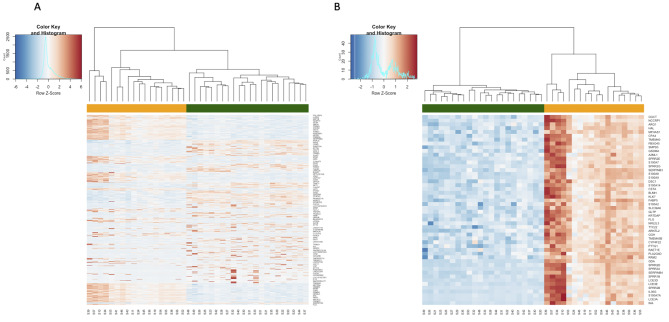
First, we performed the general analysis comprising all patients and control samples and defined 1377 DEG of which 647 were upregulated and 730 downregulated (Fig. 1). The statistical overrepresentation test using Reactome pathways has shown that the following pathways are over-represented: innate immune system, interleukin-36 pathway, keratinization, gap junction trafficking and regulation, post-translational modification: synthesis of GPI (Glycosylphosphatidylinositol) -anchored proteins, sphingolipid metabolism and neutrophil degranulation (Additional file 6). The gene ontology (GO) analysis revealed that 114 biological processes were upregulated in the MeDOC group, while 192 were downregulated. The upregulated GO Biological Processes (GO BP) include i.e. epithelial cell differentiation; epidermis development; organonitrogen compound, small molecule metabolic, lipid metabolic process and catabolic process; homeostatic process; regulation of water loss via skin; peptide cross-linking; proteolysis; secretion; cell death; oxidation-reduction process, generation of precursor metabolites and energy, transport. Conversely, the following GO BP were downregulated: locomotion; cell adhesion and motility; developmental process; cell differentiation including positive regulation of cell differentiation; extracellular structure organization; anatomical structure morphogenesis; cell-cell signalling and communication; response to endogenous stimulus; cilium organization; regulation of transport including positive regulation of transport and cation transport. See Additional file 7 for further data.
Fig. 1.
Heatmaps of DEGs patients vs. control
Total overview, B- top 50 DEGs
Considering the fact that patients with TGM1 and ALOX12B variants were mostly represented in our MeDOC group, we performed the TGM1 patients vs. controls and ALOX12B patients vs. controls analysis and identified 1061 and 926 DEGs, respectively. Next, we compared these two sets of DEGs and found that 512 DEGs were common for TGM1 and ALOX12B groups (Fig. 2). However, when ALOX12B vs. TGM1 DEGs were compared, only 8 were found to be significant, showing that the similarity between these two sets was indeed high.
Fig. 2.
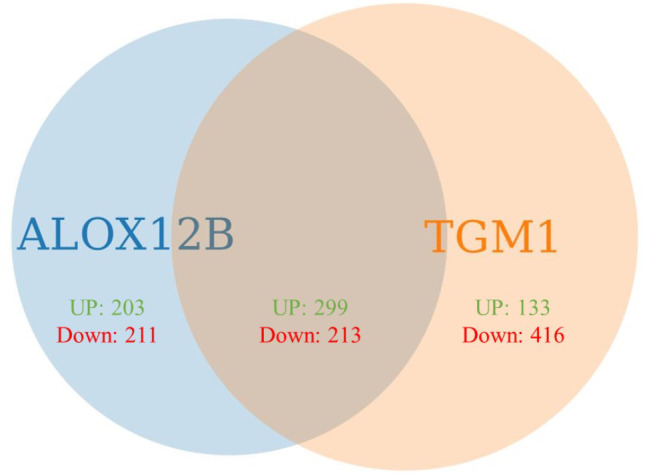
Venn Diagram showing DEGs identified in ALOX12B patients vs. controls and TGM1 patients versus control. UP –upregulated DEGs, Down – downregulated DEGs
In terms of Reactome pathways overrepresentation testing, the general pattern was similar to that observed in combined MeDOC samples vs. controls with slight differences only, e.g. “sphingolipid metabolism and gap junction trafficking and regulation” were reported for ALOX12B group, while “chemokine receptors bind chemokines” in TGM1 patients (data not shown).
Lipid analyses
We performed fatty acid (FAs) profile in epidermal scrapings of patients with ichthyosis caused by pathogenic variants in ALOX12B and TGM1. Since the sampling site seems to have an effect on the FAs profile (the FAs profile of the back differed from that of the arm), to eliminate the sample site bias we performed two distinct analyses for scrapings taken from arm and back (Tables 5 and 6).
Table 5.
Composition of selected fatty acids in skin of ichthyosis patients with ALOX12B vs. TGM1 pathogenic variants (arm)
| AGE | 29.7 ± 7.23 | 40.0 ± 14.1 | 0.479 |
|---|---|---|---|
| FATTY ACIDS | ALOX12B patients | TGM1 patients | p |
| 16:0 | 21.6 ± 6.35 | 16.8 ± 4.65 | 0.434 |
| 18:0 | 7.33 ± 1.62 | 12.1 ± 2.79 | 0.086 |
| 20:0 | 0.98 ± 0.09 | 1.52 ± 0.17 | 0.016 |
| 22:0 | 1.42 ± 0.29 | 2.62 ± 0.35 | 0.025 |
| 24:0 | 4.42 ± 0.99 | 9.40 ± 2.46 | 0.045 |
| 26:0 | 3.52 ± 1.10 | 7.53 ± 2.62 | 0.088 |
| 28:0 | 2.42 ± 1.06 | 4.49 ± 2.91 | 0.317 |
| 30:0 | 0.60 ± 0.37 | 0.88 ± 0.68 | 0.584 |
| 32:0 | 0.04 ± 0.03 | 0.09 ± 0.07 | 0.370 |
| other ECFA | 5.03 ± 1.21 | 4.07 ± 2.10 | 0.548 |
| ECFA | 47.4 ± 5.65 | 59.55 ± 0.27 | 0.063 |
| 21:0 | 0.14 ± 0.07 | 0.19 ± 0.05 | 0.476 |
| 23:0 | 0.56 ± 0.25 | 1.04 ± 0.52 | 0.243 |
| 25:0 | 1.15 ± 0.25 | 2.21 ± 0.79 | 0.104 |
| 27:0 | 0.60 ± 0.27 | 1.26 ± 0.85 | 0.270 |
| 29:0 | 0.31 ± 0.15 | 0.52 ± 0.41 | 0.443 |
| 31:0 | 0.03 ± 0.02 | 0.05 ± 0.04 | 0.574 |
| other OCFA | 3.35 ± 0.35 | 3.08 ± 0.43 | 0.485 |
| OCFA | 6.14 ± 0.87 | 8.34 ± 2.23 | 0.200 |
| other SFA | 3.75 ± 0.77 | 2.84 ± 0.40 | 0.233 |
| TOTAL SFA | 57.3 ± 5.30 | 70.7 ± 1.56 | 0.045 |
| 16:1 | 8.07 ± 1.19 | 3.94 ± 2.80 | 0.096 |
| 18:1 | 25.6 ± 4.32 | 17.9 ± 2.64 | 0.116 |
| 19:1 | 0.02 ± 0.01 | 0.04 ± 0.01 | 0.282 |
| 20:1 | 0.25 ± 0.05 | 0.47 ± 0.16 | 0.014 |
| 22:1 | 0.04 ± 0.02 | 0.12 ± 0.01 | 0.027 |
| 24:1 | 0.05 ± 0.03 | 0.19 ± 0.03 | 0.220 |
| 26:1 | 0.03 ± 0.01 | 0.08 ± 0.03 | 0.282 |
| other MUFA | 1.67 ± 0.31 | 0.70 ± 0.61 | 0.228 |
| TOTAL MUFA | 35.7 ± 4.24 | 23.4 ± 0.66 | 0.031 |
| 16:2n6 | 0.01 ± 0.004 | 0.01 ± 0.00 | 0.219 |
| LA | 6.87 ± 1.28 | 5.51 ± 1.00 | 0.300 |
| ARA | 0.03 ± 0.000 | 0.08 ± 0.06 | 0.192 |
| DGLA | 0.02 ± 0.006 | 0.06 ± 0.01 | 0.015 |
| EDA | 0.03 ± 0.02 | 0.09 ± 0.05 | 0.165 |
| AdA | 0.01 ± 0.000 | 0.02 ± 0.000 | 0,200* |
| PUFA n6 | 6.96 ± 1.29 | 5.77 ± 0.88 | 0.345 |
| ALA | 0.02 ± 0.006 | 0.03 ± 0.01 | 0.495 |
| EPA | 0.03 ± 0.02 | 0.05 ± 0.04 | 0.463 |
| DPA | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.789 |
| DHA | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.870 |
| PUFA n3 | 0.08 ± 0.01 | 0.11 ± 0.03 | 0.219 |
Value is mean ± SD. Content of FA given as a percentage (%). p–value Student’s t–test, * p–value Mann–Whitney Rank Sum Test. AdA – adrenic acid (22:4 n6); ALA – α-linolenic acid (18:3 n3); ARA – arachidonic acid (20:4 n6); DGLA –dihomo-γ-linolenic acid (20:3 n6); DHA – docosahexaenoic acid (22:6 n3); DPA – docosapentaenoic acid (22:5 n3); EDA – eicosadienoic acid (20:2 n6); EPA – eicosapentaenoic acid (20:5 n3); HDA – hexadecadienoic acid (16:2 n6), LA – linoleic acid (18:2 n6); ECFA – even chain fatty acids; MUFA – monounsaturated fatty acids. OCFA – odd chain fatty acids; PUFA – polyunsaturated fatty acids; SFA – saturated fatty acids. Boldface - major groups of fatty acid
Table 6.
Composition of selected fatty acids in skin of ichthyosis patients with ALOX12B vs. TGM1 pathogenic variants (back)
| age | 17.0 ± 2.83 | 34.0 ± 21.1 | 0.378 |
|---|---|---|---|
| FATTY ACIDS | ALOX12B patients | TGM1 patients | p |
| 16:0 | 26.9 ± 4.77 | 14.3 ± 2.28 | 0.078 |
| 18:0 | 8.06 ± 0.64 | 13.8 ± 5.61 | 0.290 |
| 20:0 | 0.80 ± 0.03 | 1.86 ± 0.86 | 0.225 |
| 22:0 | 0.85 ± 0.05 | 3.49 ± 1.10 | 0.077 |
| 24:0 | 2.96 ± 0.78 | 11.7 ± 3.01 | 0.058 |
| 26:0 | 2.25 ± 0.78 | 7.69 ± 2.99 | 0.131 |
| 28:0 | 1.53 ± 0.28 | 4.78 ± 1.45 | 0.090 |
| 30:0 | 0.33 ± 0.13 | 0.95 ± 0.08 | 0.028 |
| 32:0 | 0.05 ± 0.02 | 0.08 ± 0.04 | 0.412 |
| other ECFA | 7.39 ± 0.42 | 3.11 ± 0.52 | 0.012 |
| ECFA | 51.1 ± 2.98 | 61.7 ± 0.90 | 0.040 |
| 21:0 | 0.06 ± 0.000 | 0.37 ± 0.17 | 0.123 |
| 23:0 | 0.33 ± 0.11 | 1.74 ± 0.42 | 0.045 |
| 25:0 | 0.80 ± 0.42 | 2.74 ± 0.81 | 0.096 |
| 27:0 | 0.37 ± 0.15 | 1.23 ± 0.40 | 0.105 |
| 29:0 | 0.21 ± 0.13 | 0.51 ± 0.000 | 0.085 |
| 31:0 | 0.06 ± 0.06 | 0.08 ± 0.05 | 0.759 |
| other OCFA | 6.25 ± 1.71 | 2.90 ± 1.63 | 0.333* |
| OCFA | 8.06 ± 2.59 | 9.55 ± 3.39 | 0.671 |
| other SFA | 5.21 ± 0.02 | 1.55 ± 0.04 | 0.000 |
| TOTAL SFA | 64.4 ± 0.37 | 72.8 ± 4.26 | 0.108 |
| 16:1 | 16.1 ± 0.42 | 3.07 ± 0.30 | 0.001 |
| 18:1 | 12.7 ± 1.68 | 18.6 ± 6.12 | 0.321 |
| 19:1 | 0.01 ± 0.00 | 0.04 ± 0.01 | 0.095 |
| 20:1 | 0.14 ± 0.04 | 0.39 ± 0.21 | 0.231 |
| 22:1 | 0.02 ± 0.00 | 0.10 ± 0.08 | 0.314 |
| 24:1 | 0.03 ± 0.01 | 0.24 ± 0.23 | 0.331 |
| 26:1 | 0.03 ± 0.02 | 0.06 ± 0.04 | 0.406 |
| other MUFA | 3.25 ± 0.98 | 0.59 ± 0.10 | 0.062 |
| TOTAL MUFA | 32.3 ± 0.22 | 23.1 ± 5.95 | 0.160 |
| 16:2n6 | 0.01 ± 0.000 | 0.01 ± 0.005 | 0.423 |
| LA | 3.25 ± 0.56 | 3.85 ± 1.69 | 0.681 |
| ARA | 0.01 ± 0.000 | 0.07 ± 0.000 | 0.333* |
| DGLA | 0.02 ± 0.01 | 0.02 ± 0.01 | 1.000* |
| EDA | 0.01 ± 0.005 | 0.05 ± 0.04 | 0.267 |
| AdA | 0.01 ± 0.005 | 0.03 ± 0.02 | 0.353 |
| PUFA n6 | 3.29 ± 0.56 | 4.01 ± 1.72 | 0.633 |
| ALA | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.667* |
| EPA | 0.01 ± 0.000 | 0.02 ± 0.000 | 0.333* |
| DPA | 0.02 ± 0.01 | 0.04 ± 0.01 | 0.155 |
| DHA | 0.02 ± 0.01 | 0.04 ± 0.01 | 0.333* |
| PUFA n3 | 0.07 ± 0.01 | 0.13 ± 0.01 | 0.028 |
Value is mean ± SD. Content of FA given as a percentage (%). p–value Student’s t–test, * p–value Mann–Whitney Rank Sum Test. AdA – adrenic acid (22:4 n-6); ALA – α-linolenic acid (18:3 n-3); ARA – arachidonic acid (20:4 n-6); DGLA –dihomo-γ-linolenic acid (20:3 n-6); DHA – docosahexaenoic acid (22:6 n-3); DPA – docosapentaenoic acid (22:5 n-3); EDA – eicosadienoic acid (20:2 n-6); EPA – eicosapentaenoic acid (20:5 n-3); HDA – hexadecadienoic acid (16:2 n-6), LA – linoleic acid (18:2 n-6); ECFA – even chain fatty acids; MUFA – monounsaturated fatty acids. OCFA – odd chain fatty acids; PUFA – polyunsaturated fatty acids; SFA – saturated fatty acids. Boldface - major groups of fatty acid
The largest differences were observed for the very long chain fatty acids (fatty acids with carbon chain length ≥ C20 - FAs ≥ C20, VLCFAs; ), the very long-chain monounsaturated fatty acids (VLC-MUFAs, FAs ≥ C20:1) and the n6 polyunsaturated fatty acids (n6 PUFAs) (Table 5). Due to the small sample size, many FAs only showed an upward or downward trend (Tables 5 and 6). Differences were found in ultra-long chain fatty acids (ULCFA, FAs ≥ C28) and n3 polyunsaturated fatty acids (n3 PUFAs). Interestingly, the palmitoleic acid content was 5 times lower in the TGM1 sample compared to ALOX12B (Table 6).
Discussion
General summary
In this study, we have conducted comprehensive molecular analysis of a cohort of 265 Polish patients with cornification disorders. Novel pathogenic variants and modifying variants were detected in MeDOC patients. For part of the patients, we also preformed transcriptome analysis and described characteristics of ALOX12B vs. TGM1 deficient epidermis on mRNA and lipid levels.
In the cases analysed by NGS (245) the diagnostic yield of 87% was achieved, a value similar to the diagnostic efficacy obtained by others [12]. A higher detection rate of 89% was obtained in a small subgroup of X-linked ichthyosis patients, selected by an X-linked inheritance pattern, in whom only the STS MLPA test was performed.
The majority of patients were sent from different clinical centres across Poland and their clinical evaluation was limited or not available, making the unbiased, broad NGS-gene panel the method of choice. However, as the clinical data were not fully available, we had to trust the suspicion of MeDOC made by the referring clinician and therefore could not assess with certainty if in the 13% of unsolved cases, the right diagnostic methods were used. For example Nagtzaam et al., reported that in Dutch population in 20% of cases with clinical suspiction of X-linked ichthyosis, the genetic analysis showed variants in the other gene(s) than STS, leading to diagnosis change [13].
Based on the presented results, we can conclude that in the referred population ARCI patients were mostly represented, (126/254, 49%). In 56/126 (45%) cases, the ALOX12B gene was involved, making it the most common cause of ARCI in the tested population, while in 33/126 (26%) and 18/126 (14%) of cases the TGM1 and ALOXE3 genes were causal, respectively.
The overrepresentation of ALOX12B patients was also noticed among Czech ARCI patients, where 18/47 (38%) probands had ALOX12B pathogenic variants [14] and among Middle-Eastern ARCI patients of Muslim origin – 26% [15]. This is in contrast to several other European studies, that indicated the TGM1 variants as the ARCI leading cause, e.g. in Austria, Scandinavia, Galicia [12, 15–17]. These discrepancies could at least partially be explained by the high abundancy of recurrent ALOX12B and ALOXE3 variants in the Polish and regional (Czech) population [14]. Nevertheless, in the future, the availability of more population-specific genomic data will enable more precise estimations regarding variant frequency.
A large multicenter study on 224 ALOX12B and ALOXE3 patients of various ethnicities has shown that the p.Tyr521Cys variant accounts for 22% of all ALOX12B alleles [18]. In our group however, this variant was detected in 45% (52 of 115) of ALOX12B alleles (including the one allele found as a secondary finding). In Austrian and Czech cohorts this variant was also frequent and present in 6/14 (42%) and 10/36 (28%) of ALOX12B alleles, respectively [12, 14]. Although the origin of the p.Tyr521Cys variant is not known, our results suggest that a founder effect may have contributed to its high prevalence in Poland.
Of note, another frequent variant in ALOX12B: p.Ala597Glu was found in 11/115 alleles (9.5%) in our cohort, similarly to Czech patients (4/36 alleles – 10%), while in multicentre studies mentioned above, it was found in 12/282 (4%) alleles, confirming its regional higher prevalence [14, 18]. For ALOXE3, the most prevalent variant: p.Arg234Ter was detected in 22/37 (59%) alleles and 9/18 (50%) in Polish and Czech cohorts, respectively. Among various ethnicities this variant was present in 21% of ALOXE3 alleles. Interestingly, the p.Pro630Leu was present in our population only once, while in a multicentred and Czech cohort it was frequent (40% and 28% of ALOXE3 alleles, respectively) [14, 18].
The TGM1 results revealed, beside two well-known recurrent pathogenic variants: p.Arg126His and p.Val379Leu, an additional one: duplication of exons 10–14 (c.(1402 + 1_1401-1)_(2225 + 1_2226-1)dup), found in 9 probands. According to the HGMD database (v. Professional 2023.3), this variant was described only once in the literature before, but was detected using in silico NGS analysis in a single patient and its presence was not confirmed by DNA analysis [17]. We performed DNA quantitative tests in probands and their parents, which confirmed that the duplication was present (manuscript under preparation).
Additional variants
In 23 cases of 245 analysed by NGS (9%), we revealed additional variants in genes encoding proteins involved in epidermal barrier formation. Most of them were known pathogenic/likely pathogenic ones, however few were novel and few were VUS. There is no consensus in Poland and Europe as to the genetic testing and reporting of single allele incidental findings in autosomal recessive disorders. For example, in Poland carriership of pathogenic/likely pathogenic variants in the genes included in the phenotype-related panels are reported for the purpose of genetic counselling, while in the Netherlands often only causative variants are, and carrier status is not a part of the diagnostic result. Furthermore, there is an open question whether the single allele variants influence the phenotype. This aspect is intriguing in particular, considering that in 10/23 cases, the additional gene was FLG. This gene encodes filaggrin, the multitasking protein involved in epidermal differentiation, barrier formation and moisturising. FLG defects lead to ichthyosis vulgaris (IV), an autosomal semidominant condition with incomplete penetrance (83–96%) and variable expressivity [19, 20]. Biallelic pathogenic variants in FLG are associated with a relatively severe phenotype, however the vast majority of patients have a mild phenotype of IV and/or symptoms of atopy due to heterozygous mutation in FLG [20–24].
It has already been anecdotally observed that FLG variants exaggerate the symptoms of X-linked ichthyosis [25–29]. Moreover, in a Dutch cohort of 109 male patients with clinical suspicion of X-linked ichthyosis (XLI), FLG variants were concomitant with an STS pathogenic variant in 4% of cases [13]. Another report showed a FLG variant in a patient with ARCI caused by PNPLA1 pathogenic variants, however it is impossible to say if and how the phenotype was influenced by filaggrin defect [30]. It seems that FLG variants are not well recognized or described as incidental findings in ichthyosis types other than X-linked ichthyosis. It is safe to assume that the frequency of FLG pathogenic variants in the general European population is around 4–7% [20, 31]. However, in the available reports on ichthyosis cohorts, the presence of FLG pathogenic variants is either not reported or was not investigated.
In the remaining cases, we detected pathogenic variants in the other genes, that can be causative for different types of autosomal recessive ichthyosis (ARCI). Importantly, six of them were nonsense variants, meaning that only 50% of protein would be produced, provided the second allele is normal. As many of the ARCI subtypes share the same pathways/processes in the epidermis such an haploinsufficiency of the other protein might hypothetically have some influence on the phenotype. Such observations have already been made in other genodermatoses, e.g. epidermolysis bullosa [32–34]. Therefore further studies are needed to verify if this has clinical implications in ichthyosis.
Transcriptomics and lipidomics
The transcriptome analysis of 18 patients revealed overexpression of genes encoding proteins involved in immunity, epidermis development, differentiation and signalling. In contrary, downregulation of those involved in cell adhesion and motility, developmental process and differentiation, extracellular structure organization, signalling and communication was shown.
Recent transcriptomic analyses showed the Th17/Th22 mediated immune skewing in different clinical subtypes of ichthyosis [35, 36]. In accordance with their studies, the Th17/Th22 markers were also upregulated in our group. Six of them were among the top 50 Differentially Expressed Genes (DEG) (S100A9, S100A8, S100A7, SERPINB3, SERPINB4, S100A7A), but the overall number of upregulated genes involved in Th17/Th22 immune response included also IL36G, IL36RN, IL26, KLK10, EPN3, PI3, VNN3 and others. Importantly, despite general convergent gene expression profile, some discrepancies regarding individual genes are noticeable. For example, Kim detected IL17A/C, IL22 and IL-23R expression using RNA-seq, that was below the threshold in Malik analysis based on microarrays [36]. In our group, only IL17C and IL22RA1 (Subunit Alpha1 for IL22) were upregulated, although in IL22RA1 logFC was 1.6. This may reflect individual differences, methodological issues and heterogeneity of ichthyoses, as discussed below. Nevertheless, genes involved in cornification and barrier formation were concordantly upregulated in this and previous studies.
Furthermore, in contrast to Kim et al., who reported downregulation of lipid metabolism genes, we found upregulation of several genes involved in unsaturated fatty acid biosynthetic, oxoacid and lipid metabolic processes. However, when we compared our results with those of Kim, it turned out that of 277 genes upregulated in our group, around 40 were also upregulated in their patients and only a few were assigned as downregulated [36]. Moreover, a set of genes related to lipid biosynthesis was shown to be upregulated in TGM1-ARCI patients, as reported by Zhang et al. [37]
In order to have further insight into this issue, we compared the expression profile narrowing to two patients subgroups: ALOX12B-deficient (n = 5) and TGM1-deficient (n = 7) patients. Those subgroups were the most numerous in our group of patients. The proteins encoded by both genes are involved in production and formation of the corneocyte lipid envelope. Specifically, ALOX12B encodes arachidonate 12-lipoxygenase which is a key enzyme processing arachidonic acid (20:4n–6) during synthesis of barrier lipids, while transglutaminase 1 (TGM1) mediates the cross-linking of proteins in the corneocyte protein envelope and the attachment of the corneocyte lipid envelope [38].
Although differences between the number of upregulated and downregulated genes were shown when ALOX12B-deficient vs. controls and TGM1-deficient vs. controls DEGs sets were compared, we haven’t found DEGs between ALOX12B-deficient and TGM1-deficient patients, emphasizing that the expression pattern in those two groups are highly similar. In contrast, when we compared the fatty acids (FAs) profile in the epidermis taken from lesional skin of ALOX12B-deficient and TGM1-deficient patients, the differences in content of selected groups of FAs were visible. Recently we analysed the FAs profile in normal epidermis, which showed the differences in FA distribution with respect to age and skin location [10]. Therefore, the results of MeDOC patient analyses were narrowed with respect to collection site. We showed that the content of C16:1 in TGM1 was lower than in ALOX12B patients (inactive ALOX12B impairs processing of lipids biosynthesis). Conversely, the number of long chain FAs was higher in TGM1 patients. Though the tendency is visible, the statistical significance was achieved in only certain classes of FAs, probably due to small group sizes.
Previous studies on atopic dermatitis and the Netherton syndrome have already shown that the level of C16-18 adversely corresponds with epidermal barrier function [39, 40]. Importantly, it has been proposed, that the altered ratio of mono – vs. saturated long chain fatty acids affects production of substrates necessary for ceramide synthesis [41]. Moreover, in vitro studies provided data showing that increase in FAs and/or monounsaturated fatty acids (MUFA) content influences lipid organisation and, consequently, barrier permeability [40]. Our results also focus on this aspect showing that in ALOX12B patients, the amount of total even chain fatty acids (ECFA), odd chain fatty acids (OCFA) and saturated fatty acids (SFA) were diminished compared to the TGM1 group, with the exception of MUFA that was increased. Although detailed data on lipid composition in MeDOC skin are limited, it is well known that the ratio of main lipid types, as well as their content and organisation, vary in different ichthyosis types [42]. Whether phenotypic differences between patients with defects in ALOX12B and TGM1 genes are directly related to the different lipid levels and composition is highly intriguing. Unfortunately, due to limited data, we are currently unable to comment on this aspect. Of note, however, we are the first to publish comparative data on FA in epidermis of patients with ARCI. Hence, providing further insights into the complexity of lipid homeostasis in the ARCI epidermis, we also provoke novel (yet unanswered) questions.
Limitation of the study
We analysed large cohort of patients with different types of MeDOC, the clinical data in most cases were highly limited, though. In majority of cases the referral doctors indicated only “ichthyosis” or “keratoderma” on the referral form. Importantly, the material was referred to our laboratories by over 70 clinicians (geneticists, dermatologists, neonatologists) from over 30 different clinical canters across country. Therefore, in frame of this manuscript it was not possible to provide clinical descriptions. Importantly, MeDOCs are group of rare diseases, therefore the most valuable phenotypic evaluations, enabling the assessment of clinical nuances, would be those performed in specialized reference canters. Nevertheless, few cases of MeDOC patients presented in this article were phenotypically characterised before (as indicated in Additional file 3) [8, 11, 43].
Although the genetic basis of MeDOC are generally well established, the functional consequences of genetic defects in barrier formation genes is less recognised. Currently more tools are available for robust wide-ranging biochemical research, however there are still several limitations of such studies. Accordingly, there were also limitations of our study. Firstly, we encountered difficulties in collecting epidermal samples from healthy volunteers. We struggled to match age, sex and biopsy site in our control group. Therefore, the majority of control skin was mostly obtained during bariatric surgery from normal skin of feminine abdomen without clinical symptoms of MeDOC. Also genotyping was not performed in the control group.
Conclusions
In conclusion, our results are the first compilation of genetic analysis of a large cohort of 265 patients with inherited epidermal barrier dysfunction of Polish origin. We report novel variants, and show that copy number variations are among the major group of molecular defects not only in STS, but also in TGM1 and SPINK5 genes. This significantly broadens the knowledge about molecular pathology in these disorders. Furthermore, we provide evidence that the pathogenic variants in more than one gene are detected in several patients, which should be further investigated in the context of phenotypic modifiers. In terms of functional studies, our data add an unique characteristic of ALOX12B vs. TGM1 deficient epidermis on mRNA and lipid levels.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all the patients and their parents for participation in this study and cooperation.
Abbreviations
- ACMG
American College of Medical Genetics
- ARCI ALA
α-linolenic acid (18:3 n3)
- DEG
Differentially Expressed Genes
- ECFA
Even chain fatty acids
- FA
Fatty acids
- FDR
False Discovery Rate
- GO BP
GO Biological Processes
- GO
Gene ontology
- LogFC
log fold change
- MeDOC
Ichthyoses or Mendelian Disorders of Cornification
- MUFA
monounsaturated fatty acids
- n3 PUFAs
n3 polyunsaturated fatty acids
- n6 PUFAs
n6 polyunsaturated fatty acids
- OCFA
Odd chain fatty acids
- PCA
Principal component analysis
- PPK
Palmolpantar keratoderma ()
- PUFA
Polyunsaturated fatty acids
- SFA
Saturated fatty acids
- ULCFA
Ultra-long chain fatty acids (FAs ≥ C28)
- VLCFAs
Very long chain fatty acids (FAs ≥ C20)
- VLC-MUFAs
Very long-chain monounsaturated fatty acids (FAs ≥ C20:1)
Author contributions
Conceptualization: KWT ; data curation: ASz, JZW, MD ; writing-original draft preparation: KWT; data analysis: KWT, JS, AM, KN, AST, ASz, JZW, MD, TG; investigation and writing-review and editing: KWT, AST, BWa, AP, AMR, ASz, JZW, JC, MvG, JB, AG; patient qualification and clinical evaluation: KO, KW, AKK, BWa, AP, RŚ, NBW, LR, OS, NK, BHN, AEM, AJT, PW, IJ, AJS, JK, CK; methodology: KWT, AM, BWo, BG, AD, AG, SRZ, MD, DD, MJ, KD, BD, EO, JW, AB, JC, MvG; project administration: KWT; resources: KWT; supervision: CK, JB, AG.
Funding
The study was funded by the grant 2014/13/D/NZ5/03304 (to KWT) from National Science Center (NCN).
Data availability
The datasets supporting the conclusions of this article are available in the repository of Institute of Mother and Child in Warsaw and are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This research was performed in accordance with the local ethic committee agreement (Opinion no 27/2014 issued on 17.11.2014 in the Institute of Mother and Child, Warsaw, Poland).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lefèvre-Utile A, Braun C, Haftek M, Aubin F. Five functional aspects of the epidermal barrier. Int J Mol Sci. 2021;22:11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmuth M, Martinz V, Janecke AR, Fauth C, Schossig A, Zschocke J, et al. Inherited ichthyoses/generalized mendelian disorders of cornification. Eur J Hum Genet. 2013;21:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dev T, Mahajan VK, Sethuraman G. Hereditary Palmoplantar Keratoderma: a practical Approach to the diagnosis. Indian Dermatol Online J. 2019;10:365–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vahlquist A, Törmä H, Ichthyosis. A Road Model for skin research. Acta Derm Venereol. 2020;100:adv00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, ACMG Laboratory Quality Assurance Committee, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Leersum FS, Nagtzaam IF, van Oosterhoud CN, Ghesquiere SAI, Brandts RRHFJ, Gostyński A, et al. Improving the diagnostic yield for filaggrin: concealed mutations in the Dutch population. J Allergy Clin Immunol. 2020;145:1704–e17062. [DOI] [PubMed] [Google Scholar]
- 7.Sandilands A, Terron-Kwiatkowski A, Hull PR, O’Regan GM, Clayton TH, Watson RM, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007;39:650–4. [DOI] [PubMed] [Google Scholar]
- 8.Wertheim-Tysarowska K, Osipowicz K, Gielniewski B, Wojtaś B, Szabelska-Beręsewicz A, Zyprych-Walczak J, et al. The epidermal transcriptome analysis of a Novel c.639_642dup LORICRIN variant-delineation of the loricrin Keratoderma Pathology. Int J Mol Sci. 2023;24:9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folch J, Lees M, Sloane S, Gh. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 10.Mika A, Pakiet A, Szczygielski O, Woźniak K, Osipowicz K, Kowalewski C, et al. Fatty acid profiles in various lipid fractions in the female epidermis. Does the body site and age matter? Acta Biochim Pol. 2022;69:657–71. [DOI] [PubMed] [Google Scholar]
- 11.Pietrzak A, Wawrzycki B, Schmuth M, Wertheim-Tysarowska K. Structural and functional foot disorders in patients with genodermatoses: a single-centre, retrospective chart review. Orphanet J Rare Dis. 2022;17:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidl-Philipp M, Schatz UA, Gasslitter I, Moosbrugger-Martinz V, Blunder S, Schossig AS, et al. Spectrum of ichthyoses in an Austrian ichthyosis cohort from 2004 to 2017. J Dtsch Dermatol Ges. 2020;18:17–25. [DOI] [PubMed] [Google Scholar]
- 13.Nagtzaam IF, van Leersum FS, Kouwenberg LCM, Blok MJ, Vreeburg M, Steijlen PM, et al. STS pathogenic variants in a Dutch patient cohort clinically suspected for X-linked ichthyosis show genetic heterogeneity. Br J Dermatol. 2022;187:820–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borská R, Pinková B, Réblová K, Bučková H, Kopečková L, Němečková J, et al. Inherited ichthyoses: molecular causes of the disease in Czech patients. Orphanet J Rare Dis. 2019;14:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohamad J, Samuelov L, Malchin N, Rabinowitz T, Assaf S, Malki L, et al. Molecular epidemiology of non-syndromic autosomal recessive congenital ichthyosis in a Middle-Eastern population. Exp Dermatol. 2021;30:1290–7. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Pazos L, Ginarte M, Fachal L, Toribio J, Carracedo A, Vega A. Analysis of TGM1, ALOX12B, ALOXE3, NIPAL4 and CYP4F22 in autosomal recessive congenital ichthyosis from Galicia (NW Spain): evidence of founder effects. Br J Dermatol. 2011;165:906–11. [DOI] [PubMed] [Google Scholar]
- 17.Pigg MH, Bygum A, Gånemo A, Virtanen M, Brandrup F, Zimmer AD, et al. Spectrum of autosomal recessive congenital ichthyosis in Scandinavia: clinical characteristics and novel and recurrent mutations in 132 patients. Acta Derm Venereol. 2016;96:932–7. [DOI] [PubMed] [Google Scholar]
- 18.Hotz A, Kopp J, Bourrat E, Oji V, Komlosi K, Giehl K, et al. Meta-analysis of mutations in ALOX12B or ALOXE3 identified in a large cohort of 224 patients. Genes (Basel). 2021;12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown SJ, McLean WH. One remarkable molecule: filaggrin. J Invest Dermatol. 2012;132:751–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thyssen JP, Godoy-Gijon E, Elias PM. Ichthyosis vulgaris: the filaggrin mutation disease. Br J Dermatol. 2013;168:1155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer J, Bourrat E. Genetics of inherited ichthyoses and related diseases. Acta Derm Venereol. 2020;100(7):adv00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moosbrugger-Martinz V, Leprince C, Méchin MC, Simon M, Blunder S, Gruber R, Dubrac S. Revisiting the roles of filaggrin in atopic dermatitis. Int J Mol Sci. 2022;23:5318. [DOI] [PMC free article] [PubMed]
- 23.Hoober JK, Eggink LL. The Discovery and function of Filaggrin. Int J Mol Sci. 2022;23:1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drislane C, Irvine AD. The role of filaggrin in atopic dermatitis and allergic disease. Ann Allergy Asthma Immunol. 2020;124:36–43. [DOI] [PubMed] [Google Scholar]
- 25.Liao H, Waters AJ, Goudie DR, Aitken DA, Graham G, Smith FJ, et al. Filaggrin mutations are genetic modifying factors exacerbating X-linked ichthyosis. J Invest Dermatol. 2007;127:2795–8. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Tan L, Shen N, Lu Y, Zhang Y. Exacerbation of ichthyosis vulgaris phenotype by co-inheritance of STS and FLG mutations in a Chinese family with ichthyosis: a case report. BMC Med Genet. 2018;19:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakane Y, Yoshikawa T, Takeichi T, Kono M, Akiyama M. Dual clinical features of fine and rough scales seen in a combined ichthyosis vulgaris and X-linked recessive ichthyosis patient with atopic dermatitis. J Dermatol. 2024;51:e49–50. [DOI] [PubMed] [Google Scholar]
- 28.Ramesh R, Chen H, Kukula A, Wakeling EL, Rustin MH, McLean WH. Exacerbation of X-linked ichthyosis phenotype in a female by inheritance of filaggrin and steroid sulfatase mutations. J Dermatol Sci. 2011;64:159–62. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Si N, Liu Y, Zhang D, Wang R, Zhang Y, et al. Steroid sulfatase and filaggrin mutations in a boy with severe ichthyosis, elevated serum IgE level and moyamoya syndrome. Gene. 2017;628:103–8. [DOI] [PubMed] [Google Scholar]
- 30.Rossel SVJ, Clabbers JMK, Steijlen PM, van den Akker PC, Spuls PI, Middelkamp Hup MA, et al. Expanding the molecular and clinical spectrum of autosomal recessive congenital ichthyosis caused by pathogenic variants in NIPAL4 and PNPLA1 and evaluation of novel therapeutic interventions. J Eur Acad Dermatol Venereol. 2023;37:e1405–9. [DOI] [PubMed] [Google Scholar]
- 31.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. [DOI] [PubMed] [Google Scholar]
- 32.Padalon-Brauch G, Ben Amitai D, Vodo D, Harel A, Sarig O, et al. Digenic inheritance in epidermolysis bullosa simplex. J Invest Dermatol. 2012;132:2852–4. [DOI] [PubMed] [Google Scholar]
- 33.Floeth M, Bruckner-Tuderman L. Digenic junctional epidermolysis bullosa: mutations in COL17A1 and LAMB3 genes. Am J Hum Genet. 1999;65:1530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wertheim-Tysarowska K, Sota J, Kutkowska-Kaźmierczak A, Woźniak K, Bal J, Kowalewski C. Coexistence of KRT14 and KRT5 mutations in a Polish patient with epidermolysis bullosa simplex. Br J Dermatol. 2014;170:468–9. [DOI] [PubMed] [Google Scholar]
- 35.Malik K, He H, Huynh TN, Tran G, Mueller K, Doytcheva K, et al. Ichthyosis molecular fingerprinting shows profound TH17 skewing and a unique barrier genomic signature. J Allergy Clin Immunol. 2019;143:604–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M, Mikhaylov D, Rangel SM, Pavel AB, He H, Renert-Yuval Y, et al. Transcriptomic analysis of the Major Orphan Ichthyosis subtypes reveals Shared Immune and Barrier signatures. J Invest Dermatol. 2022;142:2363–e237418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Ericsson M, Weström S, Vahlquist A, Virtanen M, Törmä H. Patients with congenital ichthyosis and TGM1 mutations overexpress other ARCI genes in the skin: part of a barrier repair response? Exp Dermatol. 2019;28:1164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guneri D, Voegeli R, Doppler S, Zhang C, Bankousli AL, Munday MR, et al. The importance of 12R-lipoxygenase and transglutaminase activities in the hydration-dependent ex vivo maturation of corneocyte envelopes. Int J Cosmet Sci. 2019;41:563–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Smeden J, Janssens M, Kaye EC, Caspers PJ, Lavrijsen AP, Vreeken RJ, et al. The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp Dermatol. 2014;23:45–52. [DOI] [PubMed] [Google Scholar]
- 40.van Smeden J, Janssens M, Boiten WA, van Drongelen V, Furio L, Vreeken RJ, et al. Intercellular skin barrier lipid composition and organization in Netherton syndrome patients. J Invest Dermatol. 2014;134:1238–45. [DOI] [PubMed] [Google Scholar]
- 41.Rabionet M, Gorgas K, Sandhoff R. Ceramide synthesis in the epidermis. Biochim Biophys Acta. 2014;1841:422–34. [DOI] [PubMed] [Google Scholar]
- 42.Knox S, O’Boyle NM. Skin lipids in health and disease: a review. Chem Phys Lipids. 2021;236:105055. [DOI] [PubMed] [Google Scholar]
- 43.Osipowicz K, Wertheim-Tysarowska K, Kwiek B, Jankowska E, Gos M, Charzewska A, et al. Bullous diseases caused by KRT1 gene mutations: from epidermolytic hyperkeratosis to a novel variant of epidermolysis bullosa simplex. Postepy Dermatol Alergol. 2021;38:1032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in the repository of Institute of Mother and Child in Warsaw and are available from the corresponding author upon reasonable request.