Abstract
In plants, the BAX inhibitor-1 (BI-1) gene plays a crucial part in controlling cell death under stress conditions. This mechanism of Programmed Cell Death (PCD) is genetically regulated and is crucial for the elimination of unwanted or damaged cells in a controlled manner, which is essential for normal development and tissue maintenance. A study on cucumber identified and characterized five BI-1 genes: CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5. These genes share conserved domains, indicating common evolutionary history and function. Physicochemical analysis revealed their molecular weights and isoelectric points, while subcellular localization showed their presence in different cellular compartments. The phylogenetic analysis highlighted evolutionary relationships with related crops. Chromosomal distribution and synteny analysis suggested segmental or tandem duplications within the gene family. Protein-protein interaction analysis revealed extensive interactions with other cucumber proteins. Cis-regulatory elements in the promoter regions provided insights into potential functions and transcriptional regulation. miRNAs showed diverse regulatory mechanisms, including mRNA cleavage and translational inhibition. The CsBI3, CsBI4 and CsBI5 genes exhibit elevated expression levels during cold stress, suggesting their vital involvement in cucumber plant defense mechanisms. The application of chitosan oligosaccharides externally confirms their distinct expression patterns. The qRT-PCR confirms the upregulation of CsBI genes in ToLCNDV-infected plants, indicating their potential to mitigate biotic and abiotic stresses. The comprehensive genome-wide exploration provides opportunities for the development of cold-tolerant and virus-resistant cucumber variants by traditional breeding or gene.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10704-5.
Keywords: Cucumis sativus, BAX inhibitor-1, ER stress, Ca+ 2 level, Bioinformatics, Biotic stress, Abiotic stress
Introduction
The BAX Inhibitor-1 (BI-1) gene family plays a pivotal role in plant resilience to both abiotic and biotic stresses. During adverse environmental conditions like drought, salinity, or pathogen attacks, BI-1 genes are upregulated, regulating programmed cell death pathways to maintain cellular stability [1–4]. By inhibiting cell death, BI-1 proteins enhance stress tolerance and bolster plant defenses against pathogens, making them crucial regulators in plant stress responses [5]. Cucumber cultivation faces numerous biotic and abiotic challenges that result in substantial yield losses. About 80–97% yield losses were attributed to biotic and abiotic stress in cucumber [6, 7]. The BI-1 gene family in cucumber reduces yield losses by enhancing stress tolerance and regulating defense mechanisms. It modulates cell death pathways during both biotic and abiotic stress, reducing pathogen spread and tissue damage, and ultimately improving plant health and yield [2, 3].
BAX inhibitor-1 (BI-1) is a eukaryotic conserved sensor localized in the endoplasmic reticulum (ER). BI-1 is a protein that was initially discovered by screening a complementary DNA library of humans in yeast [8]. BI-1 is commonly referred to as a “cell death suppressor” as its role in mammals involves preventing BAX-induced cell death by monitoring fluctuations in Ca+ 2 within the ER [9]. BI-1 has been found involved in anti-programmed cell death activity. This mechanism Programmed Cell Death (PCD) is genetically regulated and is crucial for the elimination of unwanted or damaged cells in a controlled manner which is essential for normal development and tissue maintenance [10]. BI-1 prevents autophagy in combination with cytochrome P450 and IRE1α in mammals [11]. BI-1 is evident to overcome PCD in plants induced by different stimuli [12, 13]. It is currently unclear if there is a direct interaction between BI-1 and IRE1 that regulate the pathways involved in PCD. AtBI-1 is an additional sensor of eukaryotic ER stress that can detect changes in ER calcium stores and manage PCD [12]. Arabidopsis thaliana BI-1 homologs in plants contribute to defense against externally introduced Bax, thus validating the capability of protective property of BI-1 homologs [14–16].
Previous research indicates that plant BI-1 is linked to various environmental stress responses. For instance, overexpression of BI-1 in Arabidopsis, tobacco, rice, cabbage, corn mildew and sugarcane enhances tolerance to oxidative, salt, and drought stresses [3, 17–23]. Conversely, BI-1 deletion increases sensitivity to heat shock and ER stress in plants. BI-1 also plays a role in resistance against pathogens like Pseudomonas syringae pv. tomato DC3000 carrying AvrRPT2, Blumeria graminis f. sp. tritici, Botrytis cinerea, Chalara elegans, Puccinia striiformis, and Moniliophthora perniciosa [24]. In wheat infected with Fusarium graminearum, the wheat BI-1 gene (Triticum aestivum bax inhibitor-1) TaBI-1.1 was identified through RNA-sequence analysis [25, 26]. Understanding the BI-1 mechanism could lead to the development of plants resilient to both abiotic and biotic stresses.
Cucumis sativus L. is among the earliest cultivated plants, and an important member of the Cucurbitaceae family [27]. Pakistan as an agrarian land, takes advantage of a suitable climate that allows year-round cultivation of vegetables. Cucumber is an annual vegetable crop having bisexual flowers, it can be a creeper or climber vine and completes its life cycle in 90–120 days [28]. It is believed that cucumber originated and bred in India while China is the largest producer with 75% annual production of the world [29]. Even so, over the past twenty years, viral diseases have significantly reduced the production of vegetable crops. Cucumber green mottle mosaic virus (CGMMV), Zucchini yellow mosaic virus (ZYMV), Watermelon mosaic virus (WMV) [30], Papaya ring spot virus (PRSV), Tomato leaf curl New Delhi virus (ToLCNDV), and Squash leaf curl China virus (SLCCNV) are among the viruses that cause yellow mosaic infection in members of Cucurbitaceae family [31]. Infections caused by Begomoviruses are responsible for the yellowing, folding, curling, cupping, etc. of leaves in cucumber. Many recent reports indicated a huge loss of current crops due to Begomoviruses infections which poses a serious thought to its remedy [32–35].
Cucumber is highly vulnerable to various diseases, leading to reduced yield and commercial losses. Cucumber can be encountered by both biotic and abiotic stresses [36]. Throughout its life cycle, cucumber in agriculture is subjected to unfavorable climatic circumstances such as salt, drought, and extreme temperatures, which have an unfavorable effect on crop productivity and growth [37]. Stress from both high and low temperatures dramatically modifies the molecular response [38]. Early diagnosis of diseases is needed to prevent the crop from severe infection [39]. To overcome these viral infections, various techniques were previously used and still new strategies are evolving like immunization [40, 41].
The primary goal of this study was to enhance our comprehension of genome-wide analysis and the role of the BI-1 gene in conferring resistance against Begomoviruses. BI-1, which inhibits cell death, is present in both animals and plants [14, 42, 43]. The mRNA expression of BI-1 has been studied in different tissues of plants which boost up during aging, biotic and abiotic stresses [44, 45]. During cold stress, CsBI1 and CsBI5 genes exhibit high expression levels, indicating their crucial role in the abiotic defense mechanism in cucumber plants. Cucumber plants infected with ToLCNDV showed increased expression levels of the BI-1 genes as determined by qRT-PCR [46, 47], indicating their involvement in defending against both biotic and abiotic stresses because of BI-1 involvement in crucial cellular processes and potential contribution to resistance against various biotic and abiotic stresses, BI-1 presents a promising candidate for genome-wide analysis and comprehensive genomic research. Identifying and characterizing BI-1 on a genomic scale provides opportunities for developing novel virus-resistant and cold-tolerant varieties through gene cloning or integration using conventional breeding approaches.
Materials and methods
Database search and sequence retrieval
To identify the BI-1 gene in C. sativus, the protein sequence of the BAX1-I domain from Arabidopsis thaliana retrieved from NCBI (National Center for Biotechnology Information) was BLAST against the whole genome of C. sativus (Cucumber Chinese long v2), specifically the entry with the identifier PF01027. From the Motif Search [48] a sequence representing the BAX1-I domain was isolated. This particular sequence was utilized to search for BI-1 in the Cucurbit Genomics Database (http://cucurbitgenomics.org/blast). The retrieved amino acid sequences were subjected to the Conserved Domain Database (CDD) at https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi using the default settings. All those proteins that did not contain the conserved domain (PF01027) corresponding to the BAX1-I domain were excluded from further analysis [49].
Determination of physio-chemical properties of cucumber (Cucumis sativus) BI-1 proteins
The amino acid length, molecular weight (Mw), and isoelectric point (pI) of the BI-1 gene were estimated using the ProtParam tool [35, 50, 51]. Chromosomal position, chromosome direction, and CDS length size were all determined using the Cucurbit Genomics Database. Additionally, the WoLF PSORT online tool was used to predict the sub-cellular localization of BI-1 [52].
Multiple sequence alignment and phylogenetic analysis
The BI-1 gene’s amino acid sequences were aligned using Clustal W 2.1, and the phylogeny was investigated using MEGA XI [48, 53]. The neighbor-joining method was employed with 1000 replications of bootstrapping and partial deletion.
Cis-regulatory elements, gene structure and conserved motifs recognition
The analysis of the promoter region involved obtaining a genomic sequence of 1000 upstream base pairs. The sequences were analyzed using the PlantCare database [33, 49] to predict cis-regulatory elements. To validate these predictions, the PLACE database (https://www.dna.affrc.go.jp/PLACE/) was consulted. Subsequently, the protein sequences of BI-1 with the highest number of motifs were recognized by the MEME (Multiple EM for Motif Elicitation) program (https://meme-suite.org/meme/) [49]. To learn about the gene structure of BI-1 proteins, the Cucurbit Genomics Database was used to find their genomic and coding sequences. GSDS (Gene Structure Display Server) 2.0 was used to visualize the structure of the genes (http://gsds.gao-lab.org/) [33, 49].
Gene duplication analysis
The approximate divergence period of the cucumber (Cucumis sativus) BI-1 gene family was calculated using the Ks and Ka values. The MUSCLE algorithm in the MEGA-X program was used to align the protein sequences [49]. Then, the Ka/Ks calculator from the TBTools software was used to determine the quantity of Ka and Ks substitution rates [54]. The Ka/Ks ratios were taken into account in order to estimate the rates of molecular evolution for each pair of paralogous genes [55–57].
Transcriptomic analysis
The Cucurbit Expression Atlas Cucurbit Genomics Database (CuGenDB) was used to locate the cucumber (Cucumis sativus) transcriptome data (PRJNA80169), and TBtools was used to do the tissue-specific expression analysis of 5 CsBI genes and displayed the results as a heatmap.
To examine the precise expression profile of BI-1 in response to cold stress, the NCBI GEO database (Gene Expression Omnibus) was used. The GEO database was used to obtain RNA-sequence data of cucumber (Cucumis sativus) plants that had undergone treatment with chitosan oligosaccharide in the field (GSE224757) [58]. The Heatmap Illustrator tool in TBtools was then used to visualize the expression patterns and showed the links between the expression profiles using hierarchical clustering [59]. In addition, sequences taken from the GEO database were utilized to look at how they were expressed in response to the begomovirus [35].
Putative microRNA target sites analysis
The micro-RNA dataset for cucumber (Cucumis sativus) plants was downloaded from the Plant miRNA Encyclopedia (https://www.pmiren.com/download). By examining the coding sequence sequences, psRNA Target (https://plantgrn.noble.org/psRNATarget/analysis?function=3/) was used to search for miRNAs that target the BI-1 genes of cucumber (Cucumis sativus) [55, 60].
Chromosomal location, synteny, and dual synteny analysis of BI-1 genes
The Cucurbit Genomics Database was used to gather crucial data on the size of chromosomes and the precise placements of genes. Synteny analysis was carried out using the Advanced Circosprogramme in TBtools [61, 62]. The TBtools software’s One Step MCScanX and Dual Synteny Plot were used to create gene con/uration similarity maps. These maps show how similar several species’ gene configureurations are to one another [55, 63].
Analysis of protein-protein interaction network
By building protein interaction networks, the study examined the connections of all five cucumber (Cucumis sativus) proteins. This was done by using the Search tool in the STRING database v11.5, with various parameters and a default value of a medium confidence score of 0.400 [49, 64]. The STRING database version 11.5’s local network clusters, KEGG pathways, biological processes, chemical activities, and other functional relationships were all present in the interaction network [55, 65].
.
Physiological and bio-chemicals under virus stress
Chlorophyll content estimation
Fresh cucumber (Cucumis sativus) leaves were used for chlorophyll content estimation. Leaf extracts were dissolved in 80% acetone and the absorbance values for Chl. a, Chl. b, and total Chl [66]. were determined at 450 nm, 650 nm, and 663 nm using using a UV-Vis Spectrophotometer (AnalytikJena SPECORD-200) [67].
Peroxidase activity determination
To determine peroxidase activity, a 0.05 mol/L sodium phosphate buffer solution was prepared by diluting 0.2 mol/L buffer solution. A 0.3% H2O2 solution was prepared by diluting H2O2 with water, and a 0.2% Guaiacol solution was prepared by adding guaiacol to the buffer solution. Sample preparation involved mixing buffer solution, H2O2 solution, and plant extract, followed by absorbance measurement at 470 nm using a UV-Vis Spectrophotometer (AnalytikJena SPECORD-200) [68, 69].
Catalase activity determination
Catalase activity was quantified using a 0.05 mol/L sodium phosphate buffer solution and a 0.3% H2O2 solution. Sample preparation included mixing buffer solution, H2O2 solution, and plant extract, followed by absorbance measurement at 240 nm using a UV-Vis Spectrophotometer (AnalytikJena SPECORD-200) [66].
Superoxide dismutase (SOD) activity determination
SOD activity was quantified by adjusting the reaction mixture to a final volume of 3 mL with specific components. The mixture was incubated at room temperature, and the absorbance was measured at 560 nm using a UV-Vis Spectrophotometer (AnalytikJena SPECORD-200) [70, 71].
Effect of tomato leaf curl New Delhi virus (ToLCNDV) stress on cucumber (C. Sativus) varieties
Two cucumber (Cucumis sativus) varieties, “Oscar” and “ICSCU1652”, were obtained and cultivated in a controlled environment at the Faculty of Agriculture Sciences, University of the Punjab, Lahore. A total of 300 seeds from both varieties were planted, and after 10–12 days, plants with uniform size and good health were selected for inoculation. Sixty plants from each variety were then divided into two groups named Cu-B1 and Cu-B2, with three replicates each. Inoculation with the ToLCNDV clone (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6460037/) was carried out using a syringe. Young leaf samples were collected from both control and inoculated plants when symptoms began to appear. These collected samples were promptly frozen in liquid nitrogen and stored at -80 °C. The remaining plants were harvested for subsequent analysis of their physical characteristics, physiological attributes, and biochemical components [72].
RNA extraction and qRT-PCR analysis
The plants were collected from the Departments of Horticulture, Punjab University, Lahore with voucher HDpu7598 and submitted to Horticultural Bank Punjab University Lahore. Young leaf tissues were used to isolate total RNA, and the Pure Link RNA Mini Kit from Invitrogen (Catalogue No. 12183018 A) was employed for this purpose. The RNA samples were quantified and standardized using NanoDrop Quantification. Primers for five BI-1 genes were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Real-time expression analysis of the target genes having FPKM values were carried out using SYBR Select Master Mix (Catalogue No. 4472903), with cDNA as a template and the appropriate primers. Expression levels were quantified by calculating the 2–∆∆Ct value relative to the control, and GAPDH was employed as an internal reference. The primer sets and protocols for quantifying virus titer were used as described [73].
Statistical analysis
The data was subjected to statistical analysis using a two-way factorial Randomized Complete Block Design (RCBD). Analysis of variance (ANOVA) was utilized to assess variances, and means were differentiated using the least significant difference, with a significance level set at P < 0.05. The entire statistical analysis was carried out using the Statistix 8.1 software.
Results
Identification of BI-1 gene in C. Sativus
Six BI-1 proteins were identified. For the subsequent investigation, proteins resulting from identical gene isoforms or having truncated domains were ignored. Five different BI-1 genes were eventually identified and used as the basis for further research.
Physio-chemical properties of C. Sativus BI-1 gene
The CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5 genes encode proteins that vary in length, typically between 238 and 293 amino acids. These proteins exhibit molecular weights ranging from 26.7 to 32.4 kD, with CsBI1 being the largest among them. The isoelectric points (pI) of these proteins range from 6.56 to 8.67, reflecting a diversity in their charge properties across different pH levels. This variation in amino acid length, molecular weight, and isoelectric points suggests that each protein might have distinct functional roles and stability under varying physiological conditions. The genes CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5 are all localized in the plasma membrane. This specific localization is crucial as it plays a significant role in their functional dynamics within the cellular environment (Table 1).
Table 1.
List of all five non-reduntant proteins observed in the genome of C. Sativus
| Gene ID | Chr. No | Chromosome Location | Direction | Length Amino Acid | Isoelectric Point | Molecular weight (Mw) | Subcellular Localization | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rename | Cucumber (Chinese Long) v2 | Cucumber (Chinese Long) v3 | Start | End | Genomic | Peptide | (pI) | (KD) | |||
| CsBI1 | Csa6G061760.1 | CsaV3_6G005470.1 | 6 | 4,664,965 | 4,667,387 | F | 735 | 244 | 6.56 | 27 | Plasma Membrane |
| CsBI2 | Csa2G222100.1 | CsaV3_2G013450.1 | 2 | 10,735,003 | 10,736,757 | F | 717 | 238 | 7.74 | 26.7 | Plasma Membrane |
| CsBI3 | Csa2G223750.1 | CsaV3_2G013630.1 | 2 | 10,894,380 | 10,896,518 | R | 726 | 241 | 8.62 | 26.9 | Plasma Membrane |
| CsBI4 | Csa3G912950.1 | CsaV3_3G049850.1 | 3 | 39,597,173 | 39,599,440 | R | 882 | 293 | 7.12 | 32.4 | Plasma Membrane |
| CsBI5 | Csa3G912940.1 | CsaV3_3G049840.1 | 3 | 39,593,461 | 39,595,587 | R | 753 | 250 | 8.67 | 27.6 | Plasma Membrane |
Comparative phylogenetic analysis of BI-1 proteins
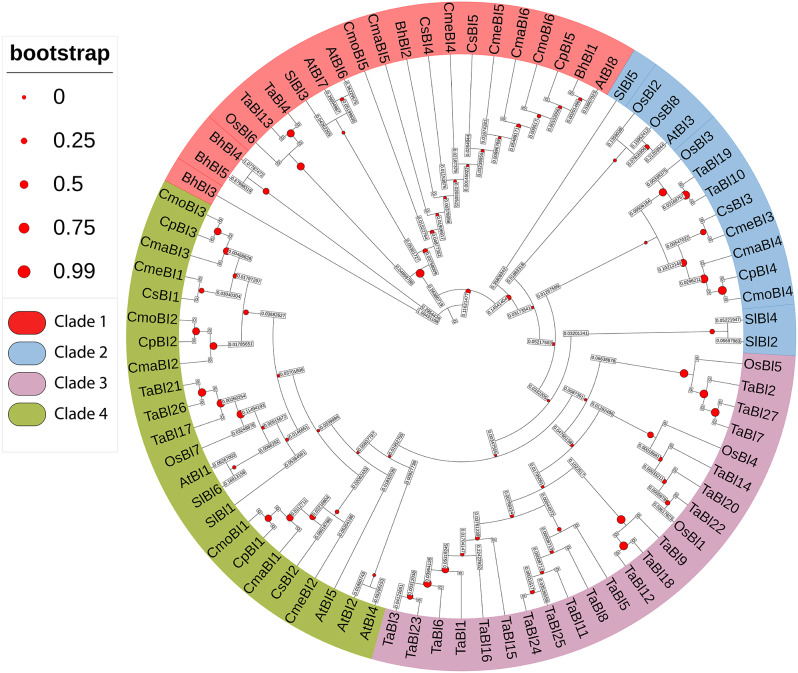
The results showed four main clades emerging from the rooted tree, with a total of 81 BI-1 genes classified into Clade 1, Clade 2, Clade 3, and Clade 4. Clade 3 and Clade 4 were the largest groups, each consisting of twenty-three BI-1 genes. Clade 1 contained twenty-one BI-1 genes, while Clade 2 was the smallest group with fourteen BI-1 genes. These groups were formed based on the presence of Arabidopsis genes, indicating the evolutionary history of different crops. The functions of all genes in each clade may be similar to the Arabidopsis gene present in that clade (Fig. 1).
Fig. 1.
The phylogenetic tree was generated based on the full-length sequences of BI-1 proteins from C. sativus (Cs), A. thaliana (At), C. melo (Cm), C. pepo (Cp), C. moschata (Cm), Oryza sativa (Os), Triticum aestivum (Ts), Benincasa hispida (Bh), Solanum lycopersicum (Sl), and C. maxima (Cm). Each clade was indicated with a specific color: Clade 1 (red), Clade 2 (blue), Clade 3 (purple), and Clade 4 (green)
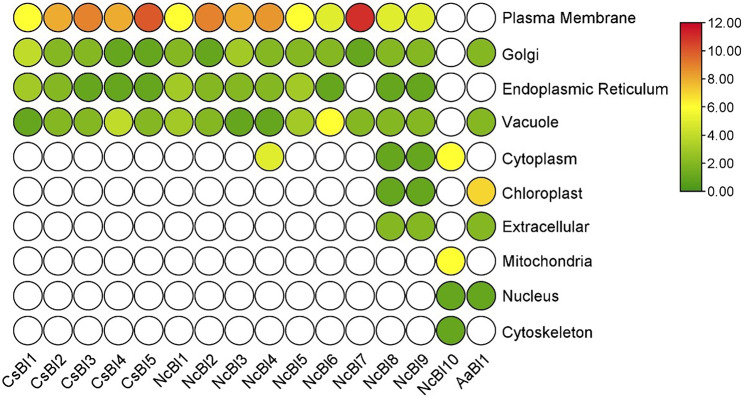
Sub-cellular localization signals in BI-1 gene of C. Sativus
Sub-cellular location of CsBI1, CsBI2, CsBI3, CsBI4 and CsBI5 genes was predicted in the plastid, endoplasmic reticulum, golgi apparatus and vacuole. CsBI5 was highly localized in the plastid. CsBI1 on the other hand, was typically located in the both plastid and golgi body. CsBI5 amount was high in the plastid as compared to golgi body, ER and vacuole. The maximum localization of cucumber and tobacco BI-1 proteins was identified in the plasma membrane and vacuoles. In contrast, leeks showed high localization in the chloroplast (Fig. 2).
Fig. 2.
Heat map showing the sub-cellular localization of C. sativus (CsBI), Allium ampeloprasum (AaBI) and Nicotiana tabacum (NtBI) proteins. Green color indicates the lowest and red color indicates the highest number of signals predicted
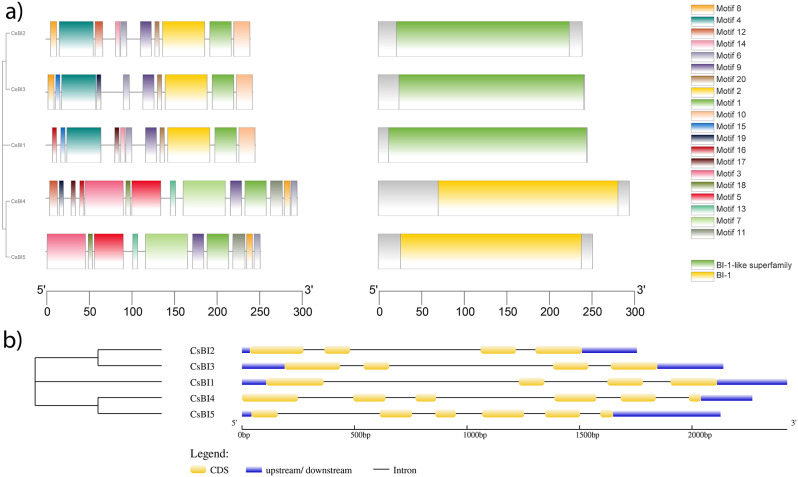
Analysis of conserved motif domains and gene structure in C. Sativus BI-1 family genes
These non-redundant BI-1 protein sequences from C. sativus exhibit highly conserved domains. All genes of C. sativus have conserved domain (Table 1S). Motif 1 was found present in all genes thus depicting it may have functional role in encoding BI-1 protein. CsBI1, CsBI2, and CsBI3 genes contain the conserved motifs (8, 4, 12, 14, 6, 9, 20, 2, 1, and 10). CsBI4 and CsBI5 also have their own set of conserved motifs (1, 15, 19, 16, 17, 3, 18, 5, 13, 7, and 11). All five BI-1 genes possess the BI-1 domain, which remains conserved across them (Table 2S). In silico analysis revealed that, number of exons varies from one to six in cucumber BI-1. CsBI, CsBI2 and CsBI3 each had 4 exons while CsBI4 and CsBI5 had 6 exons (Fig. 3a, b).
Fig. 3.
(a) Identification of conserved domains in C. sativus through NCBI CDD using the sequence of BAX1-I (b) Gene structure and phylogeny of BI-1 genes from C. sativus. Yellow bars represent exons and black lines represent introns in genetic pattern display of the CsBI1, CsBI2, CsBI3, CsBI4 and CsBI5 genes
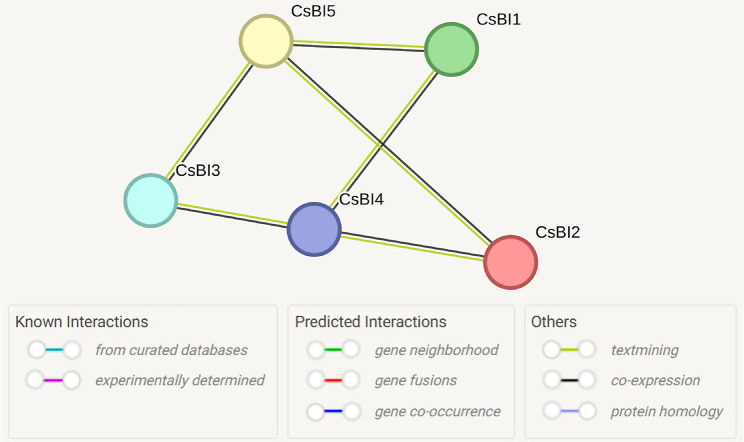
Analysis of protein-protein interaction network
The protein-protein interaction (PPI) analysis among BI-1 proteins revealed the complex networks of interactions. With a minimum required interaction score indicating low confidence set at 0.150, the protein-protein interaction (PPI) network consisted of 5 nodes and 6 edges. The average node degree was 2.4, while the average low clustering coefficient reached 0, suggesting dense interconnectivity within the network. Notably, the expected number of edges was 0, underscoring significant enrichment of protein interactions. The PPI enrichment p-value was < 1.29 e-12, emphasizing the substantial significance of the observed interactions.
BI-1 proteins CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5 demonstrated interactions with themselves within the C. sativus genome (Fig. 4). This indicated that CsBI1 proteins were linked with CsBI4 and CsBI5, while CsBI2 proteins also exhibited linkages with CsBI4 and CsBI5. Similarly, CsBI3 proteins were connected with CsBI4 and CsBI5. These interactions suggested that these proteins might have had similar functions in cucumber (Fig. 4).
Fig. 4.
Protein-protein interactions of C. sativus BI-1 proteins, predicted through the STRING database, showed that CsBI1, CsBI2, and CsBI3 proteins are all linked with CsBI4 and CsBI5
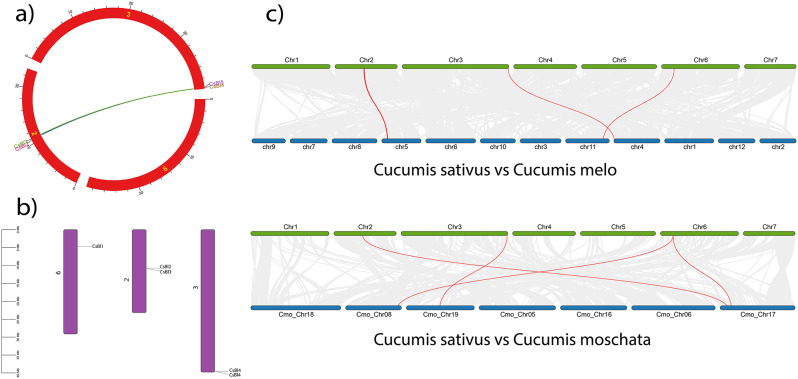
Synteny analysis and chromosomal location of C. sativusBI-1 genes
During the synteny analysis both the genes i.e., CsBI2 and CsBI3 were present on chromosome 2 linked to CsBI4 and CsBI5 genes which were present on chromosome 3. The synteny analysis has revealed the presence of tandem duplication in C. sativus. Chromosomal distribution analysis of C. sativus BI-1 genes demonstrated that CsBI1 were located on chromosome 6, CsBI2 and CsBI3 on chromosome 2, and CsBI4 and CsBI5 on chromosome 3.
The dual synteny analysis has revealed the presence of segmented duplication. All of the BI-1 genes in C. sativus have syntenic relationship with BI-1 genes in all other species C. melo and C. moschata. CsBI1, CsBI2, CsBI3, CsBI4 and CsBI5of C. sativus shows syntenic relationships with gene members of their respective groups located on different chromosomes in C. melo and C. moschata(Fig. 5a, b, c).
Fig. 5.
(a) Syntenic relationship of BI-1 genes within C. sativus genome (b) Distribution of BI-1 genes on chromosomes within C. sativus genome. Purple bars represented the chromosomes and label on the top of each bar represented the chromosome number. BI-1 genes were represented in multiple colors on the chromosome of C. sativus genome (c) Dual synteny of C. sativus BI-1 genes with C. melo. Colored horizontal bars represent chromosomes and label on top C. sativus and on bottom C. melo and C. moschata of these horizontal bars represent the chromosome number of the respective plant species. The orthologue gene pairs are highlighted by red lines and gray lines indicate collinear blocks of C. sativus with the genome of C. melo and C. moschata in the background
Evaluation of C. Sativus BI-1 genes duplication event
Generally, a Ka/Ks ratio greater than 1 indicates positive selection, a ratio approximately equal to 1 suggests neutral selection, while a ratio less than 1 implies the likelihood of purifying selection, showing minimal functional divergence exhibited by the duplicated genes [74]. The ratio of nonsynonymous mutations (Ka) to synonymous mutations (Ks) is denoted by Ka/Ks, varied from 0.092 in the CsBI2_CsBI3 pair to 0.452 in the CsBI1_CsBI5 pair. In the paralogous groups of all seven pairs in C. sativus, the Ka/Ks ratio was less than 1, recommending a low likelihood of significant functional divergence during the process of duplication, likely by virtue of purifying selection among these paralogous (Fig. 6).
Fig. 6.
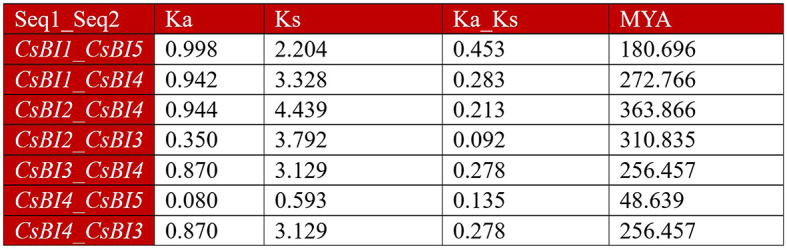
Ks and Ka values for BI-1 gene pairs of C. sativus were calculated using the TBtools Simple Ks/Ka Calculator. This evaluation provides insights into the duplication events of C. sativus BI-1 genes
Analysis of cis-regulatory elements of BI-1 genes
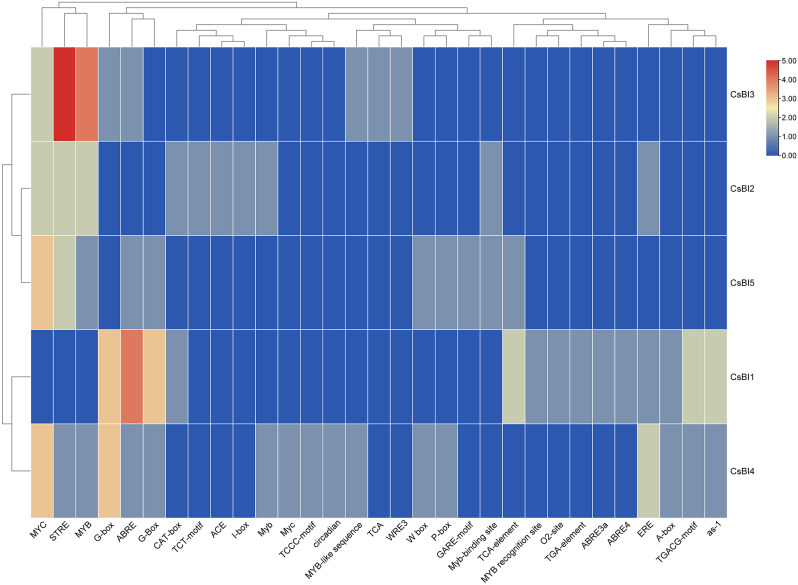
Cis-elements are crucial regulatory sequences found in the promoter regions of genes, influencing their expression in response to various environmental and physiological cues. Among the abiotic stress-responsive cis-elements, ABRE (Abscisic Acid-Responsive Element) plays a role in drought and stress responses, while ERE (Ethylene-Responsive Element) responds to ethylene signaling. MBS (MYB binding site) is involved in drought-inducibility, and DRE core (Dehydration-Responsive Element) is central to the plant’s response to dehydration stress. TCA-element is associated with salicylic acid responsiveness, LTR (Low-Temperature-Responsive Element) is activated under low-temperature stress, and ARE (Anaerobic Responsive Element) is involved in anaerobic conditions. The TCT-motif responds to copper stress, CCGTCC motif is linked to ABA and MeJA responsiveness, STRE (Stress-Responsive Element) is generally associated with various stress responses, and the CAT-box is specific to the catalase gene promoter, involved in oxidative stress responses. On the other hand, among the biotic stress-responsive cis-elements, E2Fb, a cell cycle-regulated element, may be activated during pathogen infection. The Myb-binding site is linked to the response to fungal infections, while AP-1 (Activator protein 1) plays a role in the plant’s response to pathogens. TGA-element is associated with salicylic acid signaling and defense against pathogens, while the AE-box is involved in anaerobic induction. The W box is activated in response to physical damage or wounding, and as-1 (Tobacco pathogenesis-related gene 1 element) is related to pathogen defense. The Myb motif is a general Myb binding site that can be involved in defense responses, and motif I has a role in plant defense mechanisms. Lastly, the RY-element serves as a binding site for transcription factors related to pathogen responses. These cis-elements collectively contribute to the intricate regulatory network governing plant responses to both abiotic and biotic stresses. The cis-regulatory elements (CRE) analysis showed that CsBI1 had CREs such as G-box, ABRE, G-Box, CAT-box, TCA-element, MYB recognition site, O2-site, TGA-element, ABRE3a, ABRE4, ERE, A-box, TGACG-motif and as-1 motif. CsBI2 had MYC, STRE, MYB, CAT-box, TCT-motif, ACE, I-box, Myb, Myb-binding site and ERE motif. CsBI3 contained MYC, STRE, MYB, G-box, and ABRE, MYB-like sequence, TCA and WRE3 motif. CsBI4 possessed MYC, STRE, MYB, G-box, ABRE, G-Box, Myb, Myc, TCCC-motif, and circadian, MYB-like sequence, Wbox, P-box, ERE, A-box, TGACG-motif and as-1 motif. CsBI5 had MYC, STRE, MYB, ABRE, G-Box, W box, P-box, GARE-motif, Myb-binding site and TCA element motif (Table 3S) (Fig. 7).
Fig. 7.
A heat map representing the cis-elements found in C. sativus BI-1 genes. The cis-elements are categorized into three groups: phytohormone responses, stress responses, and growth and development
Putative miRNA targets in C. Sativus
A class of short RNAs (sRNAs) known as microRNAs (miRNAs) attach to target mRNAs (messenger RNAs) at highly complementary locations to suppress the expression of certain genes. miRNAs have played a crucial role in plant life by regulating developmental programmes and carrying out responses to biotic and abiotic stimuli through their complex integration into gene expression programmes [51, 75]. Consequently, a total of 18 miRNAs were discovered, all of which targeted CsBI1, CsBI2, CsBI3, CsBI4 and CsBI5 genes. These miRNAs ranged in length from 21 to 23 amino acids. The identified miRNAs were specific and targeted individual genes, although multiple miRNAs targeted a single gene, with the exception of csa-novel-mir77, (the regulation of plant response to salt and drought stresses in an abscisic acid) which targeted two genes [76]. While most of the miRNAs caused inhibition through cleavage, two miRNAs hindered the translation process of their respective targeted genes. 3 miRNAs were targeting CsBI1, 3 miRNAs targeted CsBI2, 4 miRNAs were targeting CsBI3, 6 miRNAs were targeting CsBI4 and 2 miRNAs were targeting CsBI5 (Table 4S).
Physiological and bio-chemicals under virus stress
Impact of virus infection on chlorophyll content antioxidant activity
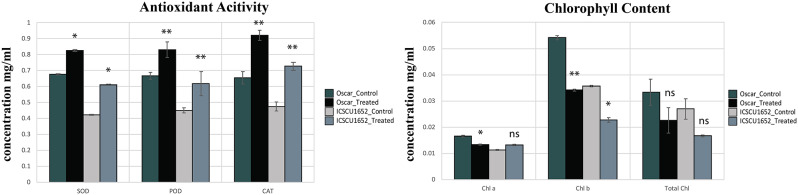
When it comes to chlorophyll concentration between two varieties of cucumber, virus inoculated plants possessed significantly lower concentration of Chl a, Chl b and total Chl. as compared to control group. However, chlorophyll concentration was found lower in ICS CU-1652 than the other variety. While, the plants with virus inoculum resulted in a substantial increase in the antioxidant activity of both varieties. However; the antioxidant level was higher in the “Oscar” variety of cucumber (Fig. 8).
Fig. 8.
Graphical representation of the effect of virus infection on chlorophyll content (concentration mg/ml) and antioxidant activity (unit mg/protein) in both plant varieties (Less than 0.05=*, Less than 0.01=**, ns = non significant). Graph was constructed with R programming language. The graphs illustrate how virus infection impacts these physiological parameters, highlighting differences between the two varieties
Validation of gene expression of BI-1 in response to ToLCNDV by RT-qPCR
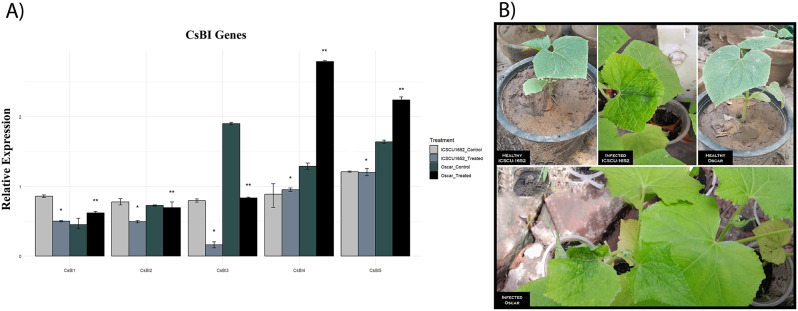
The variations in BI-1 gene expression in two varieties of cucumber were analyzed using real time quantitative PCR after 15 days of infection. After infection with ToLCNDV, expression changes of 5 BI-1 genes in both varieties of cucumber were observed. The expression patterns of all members of the BI-1 gene family were notably different. The CsBI1 gene was found to be up-regulated in response to Begomovirus infection in the “Oscar” variety, as compared to the “ICSCU1652” variety. CsBI2 showed a slight upregulation compared to the CsBI1 gene. On the other hand, CsBI3, CsBI4, and CsBI5 exhibited a high level of upregulation in the “Oscar” variety under Begomovirus infection. Overall, the results demonstrate that all members of the CsBI gene family were significantly upregulated (Fig. 9a, b).
Fig. 9.
(a) Quantitative real-time PCR analysis of the relative expression pattern of BI-1 gene family in cucumber leaf after inoculation of ToLCNDV (Less than 0.05=*, Less than 0.01=**, ns = non significant). Graph was constructed with R programming language (b) Representation of healthy and infected plants of both varieties
Transcriptomic analysis of C. Sativus BI-1 genes
Tissue-specific expression analysis
Using the RNA-seq data on the CuGenDB, the tissue-specific expression of the five CsBI genes was summarized. Some genes, such as CsBI1 and CsBI2, which were extensively expressed in each tissue, suggested that these BI-1 genes are crucial for cucumber development. Few tissues had modest levels of CsBI5 expression. Different tissues had variable levels of CsBI4 expression; for instance, tendril base, tendril, and male flowers had relatively high levels, whereas other tissues had lower levels. These showed that the CsBI genes had various functions during cucumber growth (Fig. 10).
Fig. 10.
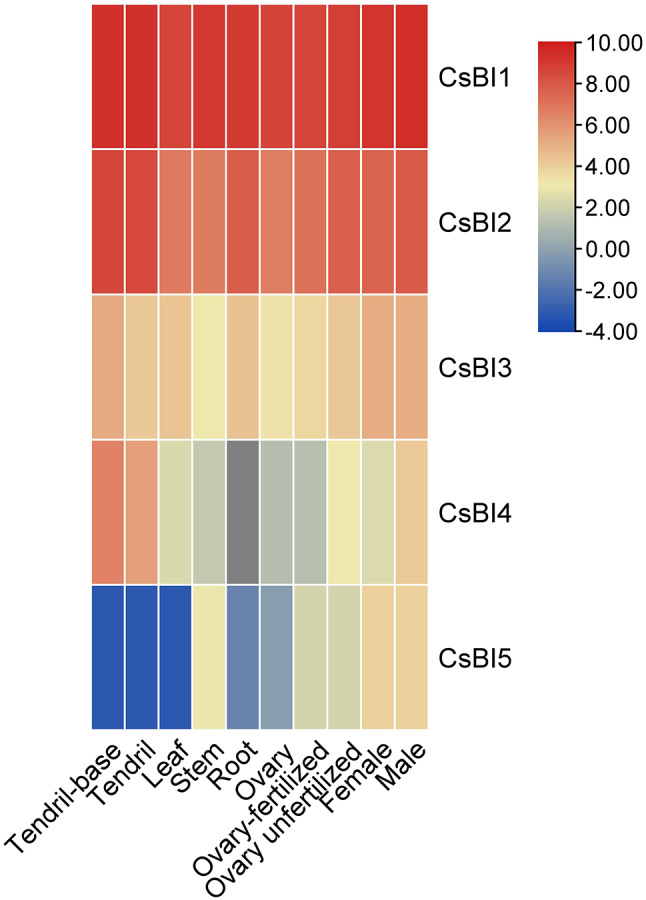
Expression of the CsBI genes in different cucumber tissues. Based on publicly available transcriptome data (PRJNA80169), the transcriptional levels of CsBI genes in ten tissues or organs of cucumber Chinese long V2 were examined. Using a heatmap and a log2RPKM value, the expression of CsBI genes across the entire genome was displayed. From blue to red, the color scale depicted escalating expression levels
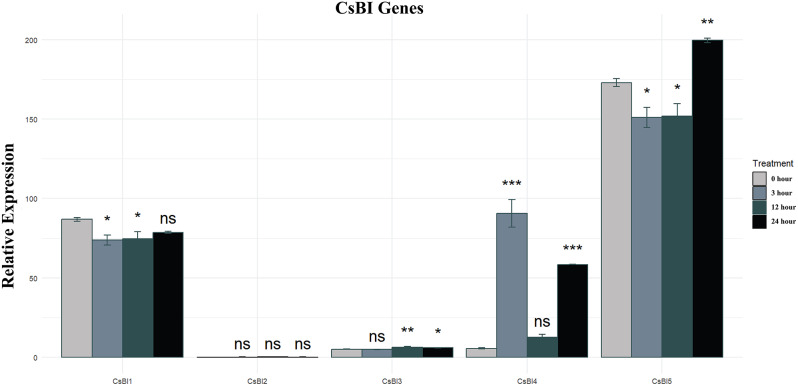
Chitosan oligosaccharide induces cold tolerance
To investigate the beneficial effects of chitosan oligosaccharide against cold stress, cucumber seedlings were exposed to temperatures of 12 °C during the day and 6 °C at night. Seedlings were pretreated with 50 mg L − 1 chitosan oligosaccharide, while distilled water served as the control. Samples were collected at 0, 3, 12, and 24 h after cold stress initiation for transcriptome analysis. During the progression of cold stress, the expression levels of CsBI3, CsBI4, and CsBI5 genes were significantly elevated compared to those in the control group (Fig. 11). Conversely, CsBI1 was significantly downregulated with increasing cold stress, while CsBI2 was also downregulated but not significantly.
Fig. 11.
Graphs representing the expression levels of C. sativus BI-1 genes under cold tolerance induced by exogenous chitosan oligosaccharide (Less than 0.05=*, Less than 0.01=**, Less than 0.001=*** ns = non significant). Graph was constructed with R programming language
Discussion
In the post-genomics era, the availability of genomic resources has expanded the possibilities for crop breeding to address the challenges posed by biotic and abiotic stresses, [65, 77] which significantly impact crop growth, development, and the ultimate quality and yield of agricultural products [78, 79]. As Pakistan, is an agrarian nation, it takes advantage of a suitable climate that allows year-round cultivation of vegetables Current study was aimed for a better understanding of genome-wide analysis and involvement of BI-1 gene for resistance against abiotic and abiotic stress. BI-1 inhibited cell death and was found in both animals and plants [80]. The mRNA expression of BI-1 has been studied in different tissues of plants which boost up during aging, biotic and abiotic stresses [81].
In this study, identification and analysis of BI-1 genes offers crucial insights into their function and evolutionary history [41]. It revealed five distinct genes (CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5) in cucumber, each encoding proteins of varying lengths and sizes. Comparative phylogenetic analysis, involving BI-1 genes from cucumber and other species like A. thaliana, C. melo, O. sativa, T. aestivum, B. hispida, S. lycopersicum, and C. pepo, highlighted four main clades (1, 2, 3, and 4). These clades underscore different gene lineages and evolutionary divergence among BI-1 family members: Clades (3and 4) were largest with 23 members of each, clade 1 of 21 members, and clade 2 of 14 members. Each clade member may have similar function with each other. Sub-cellular localization analysis indicated the localization of these five BI-1 genes in distinct cellular compartments such as plasmid, Golgi apparatus, vacuole and ER, thus suggesting the potential of BI-1 proteins to play part in various cellular processes [82]. Sub-cellular localization of this gene had previously been studied by various other researchers indicating its presence in distinct cellular compartments particularly in ER and BI-1 was found conserved across eukaryotes [83]. The maximum localization of cucumber and tobacco BI-1 proteins in the plasma membrane and vacuoles suggests that the BI-1 gene plays a role in regulating processes within these cellular structures. Specifically, this means that the BI-1 gene may be involved in functions related to the plasma membrane, such as signal transduction, ion transport, and interaction with the external environment, as well as in vacuoles, which are important for storage, waste disposal, and maintaining cellular homeostasis. The high localization of BI-1 proteins in these areas indicates their potential importance in maintaining cellular stability and responding to stress conditions in cucumber and tobacco [42].
The non-redundant BI-1 protein sequences from C. sativus reveal highly conserved domains. Each gene in C. sativus maintains its conserved domain. Notably, Motif 1 is present in all genes, suggesting its potential functional role in encoding the BI-1 protein. While CsBI1, CsBI2, and CsBI3 genes share certain conserved motifs, CsBI4 and CsBI5 possess their unique set of motifs. However, despite these variations, all five BI-1 genes maintain the BI-1 domain, underscoring its conservation across them [82]. Gene structure analysis of cucumber’s BI-1 genes indicates variability in the number of introns, ranging from 3 to 5. By aligning exons and introns, researchers gain insights into the evolutionary history and interrelationships among these genes in C. sativus [84]. This alignment not only reveals the fundamental gene structures but also provides valuable information on the evolutionary associations between genes in different organisms [85].
Protein-protein interaction network analysis of cucumber BI-1 proteins revealed that CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5 demonstrated interactions with themselves within the C. sativus genome (Fig. 4). This indicated that CsBI1 proteins were linked with CsBI4 and CsBI5, while CsBI2 proteins also exhibited linkages with CsBI4 and CsBI5. Similarly, CsBI3 proteins were connected with CsBI4 and CsBI5. These interactions suggested that these proteins might have had similar functions in cucumber [84]. These interactions reflect crucial functions and processes in plants, including signal transduction pathways, plant development, physiology, and responses to pathogens [86]. Analysis of chromosomal location indicates that CsBI1 is situated on chromosome 6, while CsBI2 and CsBI3 are both located on chromosome 2. Similarly, CsBI4 and CsBI5 are positioned on chromosome 3. This data offers insights into the genomic organization and distribution of BI-1 genes within the cucumber genome. Furthermore, studying the chromosomal location of genes facilitates the investigation of gene duplications and their evolutionary implications [87].
Genes located on the same chromosome are likely the result of tandem duplication, indicating the presence of two or more copies of the gene [88]. Conversely, when genes are found on different chromosomes, they may have arisen from segmental duplication, which involves the duplication of genetic segments across chromosomes [89].
Synteny analysis revealed that BI-1 genes in cucumber have undergone tandem duplications. Specifically, CsBI2 is genetically linked with CsBI3, and CsBI4 is linked with CsBI5. The CsBI2 and CsBI4 genes are located on chromosome 2, whereas CsBI3 and CsBI5 are located on chromosome 3. Synteny acts as a structural framework that helps for the determination of preserved homologous genes and their particular order across genomes of various species [90]. The analysis of dual synteny indicates that segmented duplication is responsible for the presence of syntenic relationships among all the BI-1 genes in Cucumis sativus, Cucumis melo, and Cucumis moschata. Specifically, CsBI1, CsBI2, CsBI3, CsBI4, and CsBI5 in C. sativus exhibit syntenic connections with genes belonging to their respective groups located on different chromosomes in C. melo and C. moschata.
All 7 paralogous pairs of BI-1 had Ka/Ks ratio less than 1 indicating that these genes had passed through strong purifying selection pressure and positive selection might have acted on only few sites during the process of evolution among these paralogs [91]. Gene duplication plays a pivotal role in the emergence of new gene subfamilies within genomes and genetic systems during evolutionary events. The emergence of novel gene families primarily occurs through mechanisms such as tandem duplication, polyploidy, and segmental duplications [92]. These duplication events lead to the creation of additional copies of genes, providing raw material for evolutionary innovation and adaptation [93]. Tandem duplication involves the consecutive duplication of genes on the same chromosome, resulting in gene clusters. Polyploidy involves whole-genome duplication events, leading to an increase in the number of copies of all genes within an organism [94]. Segmental duplications involve the duplication of chromosomal segments, contributing to the expansion of specific gene families [95]. Together, these mechanisms drive the diversification and evolution of gene families, enabling organisms to adapt to changing environments and ecological niches over time [96].
The cis-regulatory elements analysis of BI-1 genes unveils insights into their spatial and temporal expression patterns [97]. Various cis-regulatory elements (CREs) present within the promoter regions of CsBI genes are associated with specific responses and signaling pathways. Cis-elements were grouped into phytohormone responses, stress responses, and growth and development. Among the elements associated with growth and development, numerous motifs are prevalent in promoter regions, including the Skn-1_motif, GCN4_motif, MRE, Box-4, CAT-box, O2-site, and circadian elements. Regarding phytohormone responses, ABRE, P-box, TGACG motif, TCA-element, and CGTCA motif are identified, associated respectively with salicylic acid (SA), abscisic acid (ABA), ethylene, and MeJA responses. Furthermore, various stress-responsive elements including ARE, LTR, MBS, and W box correlate with light stress, cold stress, heavy metal, drought and biotic stress responses, providing insights into the regulatory mechanisms enhancing the CsBI gene family’s responses to abiotic and biotic stress. For instance, the presence of Myb-binding sites suggests involvement in fungal infection responses, while AP-1 sites indicate participation in pathogen response mechanisms. TGA elements are linked to salicylic acid signaling and pathogen defense, whereas AE-boxes are associated with anaerobic induction [92, 93]. Overall, the arrangement and presence of these CREs within the promoter region influence gene expression patterns. In silico analysis of CREs aids in estimating the potential functions of genes based on their regulatory elements [98]. These cis-elements collectively contribute to the intricate regulatory network governing plant responses to both abiotic and biotic stresses.
Analysis of putative miRNA targets of BI-1 genes in cucumber suggests that these genes may undergo post-transcriptional regulation by miRNAs. This implies that miRNAs may play a role in the expression levels of BI-1 genes by targeting their mRNA transcripts for degradation or translational repression. Such post-transcriptional control mechanisms mediated by miRNAs are important for regulating gene expression and maintaining cellular homeostasis in response to various developmental and environmental cues [99]. The interaction between miRNAs and BI-1 can restrict expression levels and potentially alter the function of BI-1 proteins in cucumber. miRNA, the fundamental regulators in plants, play a key part in different biological processes, growth, development, defense and regulation of internal equilibrium [100]. These small RNA molecules display an exceptional conservation across distinct species showing that each microRNA executes its specified task disregarding of the organism it belongs to [100]. Both types of miRNAs have been identified, such as csa-novel-mir97 and csa-novel-mir109, which play roles in regulating plant responses to salt and drought stresses through the abscisic acid pathway [101]. In addition, csa-miR2950 is involved in floral development in the context of biotic stress [102], csa-miR58 plays a major role in enhancing the production of antifungal compounds to bolster the plant’s resistance to pathogen infections [103], and csa-miR68 is crucial for the development of the plant’s immune system [104].
Virus-stressed plants exhibited high levels of antioxidant activity. Our findings align with previous data, showing increased levels of antioxidants such as superoxide dismutase, peroxidase, and catalase [105]. Catalase reduces the harmful effects of toxic peroxides, while superoxide dismutase decomposes superoxide radicals [60]. The increase in antioxidant enzymes likely results from the activation of plant defense mechanisms under stress [106].
Begomovirus-infected cucumber plants were found to have lower concentrations of chlorophyll a, chlorophyll b, and total chlorophyll compared to the control group. Virus-infected plants showed a significant decrease in photosynthetic pigment concentration [107], which corresponds to decreased photosynthetic activity due to the down-regulation of specific genes during the viral attack [108, 109]. The RT-qPCR study revealed that the expression pattern of BI-1 genes was up regulated in ToLCNDV-infected cucumber, indicating their involvement in defense responses against both biotic and abiotic stresses. This finding underscores the potential role of BI-1 genes in mediating plant defense mechanisms in response to pathogen invasion. Furthermore, the role of BI-1 gene expression in defense responses has been extensively studied in other plant species such as A. thaliana, barley, and rice, suggesting its evolutionary conservation across different plant taxa. This suggests that BI-1 genes may play a crucial role in modulating plant immunity and stress tolerance across a wide range of plant species [80]. In A. thaliana, the AtBI-1 expression is quickly boosted up when subjected to injuries or pathogens [110]. Additionally, the expression study revealed that the BI-1 genes control the growth of cucumber stem, leaf, and fruit, demonstrating a direct connection between the BI-1 gene family and the development of cucumber.
During our transcriptomic analysis, comparing the control group with the treatment group revealed distinct trends in gene expression levels under cold stress and chitosan oligosaccharide (CsBI) treatment in cucumber seedlings. Notably, CsBI3, CsBI4, and CsBI5 consistently showed significant increases in expression levels in the treatment group, suggesting CsBI’s potential to potentiate gene upregulation under cold stress. Conversely, CsBI1 and CsBI2 displayed less pronounced downregulation in the treatment group. The gene expression analysis of BI-1 genes under cold tolerance caused by exogenous chitosan oligosaccharide gives understanding about how these genes respond to abiotic stress thus suggesting that BI-1 genes may have a functional role against cold stress in cucumber. Additional studies are required to reveal the function of BI-1 genes in mechanisms that are related to cold stress. The collective findings of the whole study indicate that the CsBI gene family plays a highly significant role in both biotic and abiotic stress responses. This study paves the way for enhancing cucumber crop performance under stressful conditions.
Conclusion
The study provides a comprehensive understanding of the BI-1 gene family in C. sativus, including gene structures, phylogenetic relationships, chromosomal locations, and expression patterns. Five BI-1 genes were identified and categorized, with conserved gene structures observed across evolution. CsBI3, CsBI4 and CsBI5 showed high expression levels during cold stress, indicating their importance in abiotic stress defense. Up-regulation of BI-1 genes in ToLCNDV-infected cucumber suggests their involvement in defense against both biotic and abiotic stresses. Overall, this research enhances our understanding of the molecular mechanisms regulating stress responses in C. sativus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors extend their appreciation to the Deanship of Biosciences, University of Birmingham, Edgbaston, Birmingham, B15 2TT, United Kingdom and Department of Plant Breeding and Genetics, Faculty of Agricultural Sciences, University of the Punjab, P.O BOX. 54590, Lahore, Pakistan for funding this work.
Author contributions
The contributions of the authors to this article are as follows: SA played a significant role in data curation, formal analysis, and drafting the original manuscript. MZH and AS were responsible for conceptualization, data curation, methodology development, and co-authored the original draft. MS and RS were actively involved in data curation, formal analysis, software development, supervision, and contributing to the original draft. SA, MAJ, and JT participated in data curation, investigation, methodology development, and played crucial roles in reviewing and editing the manuscript. HA led the efforts in securing funding, contributed to methodology development, provided essential resources for the study, and co-authored the original draft. Finally, JA, BJ, RSL and BUN focused on reviewing and editing the manuscript, enhancing the methodology, and providing additional resources to support the research.
Funding
Not applicable.
Data availability
I affirm that all necessary data and permissions have been provided for this study. Any interested researchers can access the required data to support the findings and conclusions of this article. For publicly archived datasets, hyperlinks are provided in this manuscript in appropriate place for convenience. Rest assured, the plants were collected from Departments of Horticulture Punjab University Lahore with voucher HDpu7598 and submitted to Horticultural bank Punjab University Lahore. I have ensured that all data, materials, software applications, and custom code supporting the claims made in this article are in full compliance with field standards. It’s important to note that I have taken into account the possibility of individual journal policies regarding research data sharing, considering the norms and expectations of our discipline.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data transparency
Rest assured, I have ensured that all data, materials, software applications, and custom code supporting the claims made in this article are in full compliance with field standards. It’s important to note that I have taken into account the possibility of individual journal policies regarding research data sharing, considering the norms and expectations of our discipline.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Muhammad Shafiq, Email: shafiq.iags@pu.edu.pk.
Javaria Tabassum, Email: javariatabassum.pbg@pu.edu.pk.
Muhammad Arshad Javed, Email: chairman.pbg@pu.edu.pk.
References
- 1.Navathe S, Singh S, Singh VK, Chand R, Mishra VK, Joshi AK. Genome-wide mining of respiratory burst homologs and its expression in response to biotic and abiotic stresses in Triticum aestivum. Genes Genomics. 2019;41(9):1027–43. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa T, Aki T, Yanagisawa S, Uchimiya H, Kawai-Yamada M. Overexpression of BAX INHIBITOR-1 links plasma membrane microdomain proteins to stress. Plant Physiol. 2015;169(2):1333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramiro DA, Melotto-Passarin DM, Barbosa MA, Fd S, Gomez SGP, Massola Junior NS, Lam E, Carrer H. Expression of Arabidopsis Bax Inhibitor‐1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol J. 2016;14(9):1826–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharif R, Su L, Chen X, Qi X. Involvement of auxin in growth and stress response of cucumber. Vegetable Res. 2022;2(1):1–9. [Google Scholar]
- 5.Lu P-P, Yu T-F, Zheng W-J, Chen M, Zhou Y-B, Chen J, Ma Y-Z, Xi Y-J, Xu Z-S. The wheat bax Inhibitor-1 protein interacts with an aquaporin TaPIP1 and enhances disease resistance in Arabidopsis. Front Plant Sci. 2018;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unnikrishnan BV, Pradeepkumar T, Sidharthan PP, Mohan M. Microbe mediated abiotic stress tolerance in cucurbitaceous vegetables. Biologia. 2023;78(10):2863–73. [Google Scholar]
- 7.Ningombam B, Devi AS, Darvhankar M. Effects of abiotic stresses on crop yield: a review. Pharm Innovations. 2021;10(5):418–22. [Google Scholar]
- 8.Seitaj B, Maull F, Zhang L, Wüllner V, Wolf C, Schippers P, La Rovere R, Distler U, Tenzer S, Parys JB. Transmembrane BAX inhibitor-1 motif containing protein 5 (TMBIM5) sustains mitochondrial structure, shape, and function by impacting the mitochondrial protein synthesis machinery. Cells. 2020;9(10):2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich A. Role of the Coxiella burnetii type IV effector protein CaeB in cell death suppression and ER stress signaling. Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU); 2021.
- 10.Park W, Wei S, Kim B-S, Kim B, Bae S-J, Chae YC, Ryu D, Ha K-T. Diversity and complexity of cell death: a historical review. Exp Mol Med 2023:1–22. [DOI] [PMC free article] [PubMed]
- 11.Geng Y, Faber KN, de Meijer VE, Blokzijl H, Moshage H. How does hepatic lipid accumulation lead to lipotoxicity in non-alcoholic fatty liver disease? Hep Intl. 2021;15:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe N, Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006;45(6):884–94. [DOI] [PubMed] [Google Scholar]
- 13.Sami A, Haider M, Meeran M, Ali M, Abbas A, Ali Q, Umar M. Exploring morphological traits variation in chenopodium murale: a comprehensive multivariate analysis. Bull Biol Allied Sci Res. 2023;2023(1):43–43. [Google Scholar]
- 14.Chae H-J, Ke N, Kim H-R, Chen S, Godzik A, Dickman M, Reed JC. Evolutionarily conserved cytoprotection provided by bax Inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101–13. [DOI] [PubMed] [Google Scholar]
- 15.Sami A, Haider M, Iqbal M, Bhatti M, Ahmad S, Khalid M. Deterrence effect of colored diversion sheets on the population density of melon fruit flies bactrocera cucurbitae (coquillett) and yield parameters of bitter gourd (momordica charantia L). Biol Agricultural Sci Res J. 2023;2023(1):17–17. [Google Scholar]
- 16.Irfan U, Haider M, Shafiq M, Sami A, Ali Q, Genome editing for early and late flowering in plants. Bull Biol Allied Sci Res. 2023;2023(1):45–45. [Google Scholar]
- 17.Ahmad S, Khan K, Saleh IA, Okla MK, Alaraidh IA, AbdElgawad H, Naeem M, Ahmad N, Fahad S. TALE gene family: identification, evolutionary and expression analysis under various exogenous hormones and waterlogging stress in Cucumis sativus L. BMC Plant Biol. 2024;24(1):564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herath V, Gayral M, Adhikari N, Miller R, Verchot J. Genome-wide identification and characterization of Solanum tuberosum BiP genes reveal the role of the promoter architecture in BiP gene diversity. Sci Rep. 2020;10(1):11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Li X, Shi L, Jing Y, Song Q, Chen Y, He L, Wang F, Gao J, Bi Y. Genome-wide identification and analysis of the cytochrome B5 protein family in chinese cabbage (Brassica rapa L. ssp. Pekinensis). International Journal of Genomics, 2019. [DOI] [PMC free article] [PubMed]
- 20.Hückelhoven R, Dechert C, Kogel K-H. Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria Graminis. Proc Natl Acad Sci. 2003;100(9):5555–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaguancela OA, Zúñiga LP, Arias AV, Halterman D, Flores FJ, Johansen IE, Wang A, Yamaji Y, Verchot J. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol Plant Microbe Interact. 2016;29(10):750–66. [DOI] [PubMed] [Google Scholar]
- 22.Nagano M, Kakuta C, Fukao Y, Fujiwara M, Uchimiya H, Kawai-Yamada M. Arabidopsis Bax inhibitor-1 interacts with enzymes related to very-long-chain fatty acid synthesis. J Plant Res. 2019;132(1):131–43. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R. Overexpression of Bax inhibitor suppresses the fungal elicitor‐induced cell death in rice (Oryza sativa L.) cells. Plant J. 2003;33(3):425–34. [DOI] [PubMed] [Google Scholar]
- 24.Eichmann R, Schultheiss H, Kogel K-H, Hückelhoven R. The barley apoptosis suppressor homologue BAX inhibitor-1 compromises nonhost penetration resistance of barley to the inappropriate pathogen blumeria graminis f. sp. tritici. Mol Plant Microbe Interact. 2004;17(5):484–90. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Tang C, Huang X, Li F, Chen X, Zhang G, Sun Y, Han D, Kang Z. Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia Striiformis. J Exp Bot. 2012;63(12):4571–84. [DOI] [PubMed] [Google Scholar]
- 26.Almas MH, Shah RA, Tahir SMH, Manzoor M, Shafiq M, Shah MH, Hashmi MM, Ali M, Bhatti MHT, Sami A. The effect of substrate, Growth Condition and Nutrient Application methods in Morphological and Commercial attributes of Hybrid Rose (Rosa indica L.) Cv. Kardinal. J Appl Res Plant Sci. 2023;4(01):356–62. [Google Scholar]
- 27.Mushtaq R, Khan M, Manzoor M, Shafiq M, Bilal M, Manzoor T, Ali M, Mazhar HS-U-D, Hashmi M, Anees M. Genome-wide analysis of the Ethylene-Insensitive3-Like Gene Family in Cucumber (Cucumis sativus). J Appl Res Plant Sci. 2023;4(02):702–10. [Google Scholar]
- 28.Gelaye Y. Cucumber (Cucumis sativus) production in Ethiopia: Trends, prospects and challenges: a review. Cogent Food Agric. 2023;9(1):2221103. [Google Scholar]
- 29.Park G, Choi Y, Jung J-K, Shim E-J, Kang M-y, Sim S-C, Chung S-M, Lee GP, Park Y. Genetic diversity assessment and cultivar identification of cucumber (Cucumis sativus L.) using the fluidigm single nucleotide polymorphism assay. Plants. 2021;10(2):395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JX, Liu SS, Gu QS. Transmission efficiency of Cucumber green mottle mosaic virus via seeds, soil, pruning and irrigation water. J Phytopathol. 2016;164(5):300–9. [Google Scholar]
- 31.Shafiq M, Ahmad M, Nisar A, Manzoor MT, Abid A, Mushtaq S, Riaz A, Ilyas M, Sarwar W, Nawaz-ul-Rehman MS. Molecular characterization and phylogenetic analysis of tomato leaf curl Palampur virus, a bipartite begomovirus, associated with Cucumis sativus L. in Pakistan. 3 Biotech. 2019;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cathrin PB, Ghanim M. Recent advances on interactions between the whitefly Bemisia tabaci and begomoviruses, with emphasis on Tomato yellow leaf curl virus. Plant Virus–Host Interact 2014:79–103.
- 33.Islam MAU, Nupur JA, Shafiq M, Ali Q, Sami A, Shahid MA. In silico and computational analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes in Chilli (Capsicum annuum L). BMC Genomics. 2023;24(1):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sami A, Haider MZ, Shafiq M. Microbial nanoenzymes: Features and applications. In: Fungal Secondary Metabolites. Elsevier; 2024: 353–367.
- 35.Sami A, Haider MZ, Shafiq M, Sadiq S, Ahmad F. Genome-wide identification and in-silico expression analysis of CCO gene family in sunflower (Helianthus Annnus) against abiotic stress. Plant Mol Biol. 2024;114(2):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilodeau SM. Ecological process in pattern generation in tropical coral-seagrass reefscapes. Wake Forest University; 2019.
- 37.Taha N, Abdalla N, Bayoumi Y, El-Ramady H. Management of greenhouse cucumber production under arid environments: a review. Environ Biodivers Soil Secur. 2020;4(2020):123–36. [Google Scholar]
- 38.Giordano M, Petropoulos SA, Rouphael Y. Response and defence mechanisms of vegetable crops against drought, heat and salinity stress. Agriculture. 2021;11(5):463. [Google Scholar]
- 39.Ali MM, Bachik NA, Muhadi NA, Yusof TNT, Gomes C. Non-destructive techniques of detecting plant diseases: a review. Physiol Mol Plant Pathol. 2019;108:101426. [Google Scholar]
- 40.Veyer DL, Maluquer de Motes C, Sumner RP, Ludwig L, Johnson BF, Smith GL. Analysis of the anti-apoptotic activity of four Vaccinia virus proteins demonstrates that B13 is the most potent inhibitor in isolation and during viral infection. J Gen Virol. 2014;95(12):2757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haider MZ, Sami A, Shafiq M, Anwar W, Ali S, Ali Q, Muhammad S, Manzoor I, Shahid MA, Ali D. Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress. Front Plant Sci. 2023;14:1269995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hückelhoven R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. [DOI] [PubMed] [Google Scholar]
- 43.Sami A, Haider M, Imran M, Abbas A, Javed M, Synergizing, food safety, quality and genetic improvement: the intersection of food microbiology and processing. Bull Biol Allied Sci Res. 2023;2023(1):44–44. [Google Scholar]
- 44.Watanabe N, Lam E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J Biol Chem. 2008;283(6):3200–10. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad B, Mahmood A, Sami A, Haider M, Impact of climate change on fruits and crops production in South Punjab. Farmer‘S Perspective. Biol Agricultural Sci Res J. 2023;2023(1):22–22. [Google Scholar]
- 46.Sáez C, Ambrosio LG, Miguel SM, Valcárcel JV, Díez MJ, Picó B, López C. Resistant sources and genetic control of resistance to ToLCNDV in cucumber. Microorganisms. 2021;9(5):913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haider M, Sami A, Mazhar H, Akram J, NISA B, Umar M, Meeran M. Exploring morphological traits variation in Gomphrena globosa: a multivariate analysis. Biol Agricultural Sci Res J. 2023;2023(1):21–21. [Google Scholar]
- 48.Haider MZ, Sami A, Shafiq M, Anwar W, Ali S, Ali Q, Muhammad S, Manzoor I, Shahid MA, Ali D. Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress. Front Plant Sci 2023, 14. [DOI] [PMC free article] [PubMed]
- 49.Zeshan Haider M, Sami A, Shafiq M, Anwar W, Ali S, Ali Q, Shahid MA, Ali D, Alarifi S. Genome-wide identification and In-Silico expression analysis of Carotenoid cleavage oxygenases (CCO) Gene Family CCOCCO Gene Family in Oryza sativaOryza sativa (rice) in response to abiotic stress. Front Plant Sci, 14:1269995. [DOI] [PMC free article] [PubMed]
- 50.Dekomah SD, Wang Y, Qin T, Xu D, Sun C, Yao P, Liu Y, Bi Z, Bai J. Identification and expression analysis of calcium-dependent protein kinases gene family in potato under drought stress. Front Genet. 2022;13:874397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shafiq M, Manzoor M, Bilal M, Manzoor T, Anees MM, Rizwan M, Haider MZ, Sami A, Haider MS. Genome-Wide Analysis of Plant Specific YABBY Transcription Factor Gene Family in Watermelon (Citrullus lanatus) and Arabidopsis. J Appl Res Plant Sci. 2024;5(01):63–78. [Google Scholar]
- 52.HU L-p, Zhang F, SONG S-h TANG, X-w, Hui X, LIU G-m, Yaqin W. HE H-j: genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J Integr Agric. 2017;16(7):1486–501. [Google Scholar]
- 53.Zhang H, Han F, Yan X, Liu L, Shu X, Hu H. Prevalence and phylogenetic analysis of spike gene of porcine epidemic diarrhea virus in Henan province, China in 2015–2019. Infect Genet Evol. 2021;88:104709. [DOI] [PubMed] [Google Scholar]
- 54.Lei Y, Cui Y, Cui R, Chen X, Wang J, Lu X, Wang D, Wang S, Guo L, Zhang Y. Characterization and gene expression patterns analysis implies BSK family genes respond to salinity stress in cotton. Front Genet. 2023;14:1169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sami A, Haider MZ, Shafiq M, Sadiq S, Ahmad F. Genome-Wide Identification and In-silico Expression Analysis of CCO Gene Family in Sunflower (Helianthus annnus). 2023. [DOI] [PMC free article] [PubMed]
- 56.Sami A, Saeed M, Shafiq M, Abbas SM, Anum A, Haider H, Bhatti MHT, Raza MA, Khan N, Shahid NA. Role of horticulture in Disaster Risk Management. Disaster risk reduction in Agriculture. Springer; 2023. pp. 393–406.
- 57.Hussain M, Javed MM, Sami A, Shafiq M, Ali Q, Mazhar HS-U-D, Tabassum J, Javed MA, Haider MZ, Hussain M, et al. Genome-wide analysis of plant specific YABBY transcription factor gene family in carrot (Dacus carota) and its comparison with Arabidopsis. BMC Genomic Data. 2024;25(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan C, Li N, Wang Y, Yu X, Yang L, Cao R, Ye X. Integrated physiological and transcriptomic analyses revealed improved cold tolerance in cucumber (Cucumis sativus L.) by exogenous chitosan oligosaccharide. Int J Mol Sci. 2023;24(7):6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang L, Yang X, Gao Y, Yang S. Genome-wide identification and characterization of TALE superfamily genes in soybean (Glycine max L). Int J Mol Sci. 2021;22(8):4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali Q, Sami A, Haider MZ, Ashfaq M, Javed MA. Antioxidant production promotes defense mechanism and different gene expression level in Zea mays under abiotic stress. Sci Rep. 2024;14(1):7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shang J, Tian J, Cheng H, Yan Q, Li L, Jamal A, Xu Z, Xiang L, Saski CA, Jin S. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020;21(1):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali M, Shafiq M, Haider MZ, Sami A, Alam P, Albalawi T, Kamran Z, Sadiq S, Hussain M, Shahid MA et al. Genome-wide analysis of NPR1-like genes in citrus species and expression analysis in response to citrus canker (Xanthomonas axonopodis Pv. Citri). Front Plant Sci 2024, 15. [DOI] [PMC free article] [PubMed]
- 63.Bhatti MHT, Sami A, Haider MZ, Shafiq M, Naeem S, Tariq MR, Ahmad S, Irfan U. Genetic Diversity of Vegetable Crops and Utilization in Food and Nutritional Security. In: Sustainable Utilization and Conservation of Plant Genetic Diversity. Edited by Al-Khayri JM, Jain SM, Penna S. Singapore: Springer Nature Singapore; 2024: 171–197.
- 64.Sami A, Han S, Haider MZ, Khizar R, Ali Q, Shafiq M, Tabassum J, Khalid MN, Javed MA, Sajid M, et al. Genetics aspect of vitamin C (ascorbic acid) biosynthesis and signaling pathways in fruits and vegetables crops. Funct Integr Genom. 2024;24(2):73. [DOI] [PubMed] [Google Scholar]
- 65.Jadoon S, Ali Q, Sami A, Haider MZ, Ashfaq M, Javed MA, Khan MA. DNA damage in inhabitants exposed to heavy metals near Hudiara drain, Lahore, Pakistan. Sci Rep. 2024;14(1):8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naz H, Akram NA, Ashraf M. Impact of ascorbic acid on growth and some physiological attributes of cucumber (Cucumis sativus) plants under water-deficit conditions. Pak J Bot. 2016;48(3):877–83. [Google Scholar]
- 67.Naz S, Perveen S. Response of wheat (Triticum aestivum L. var. galaxy-2013) to pre-sowing seed treatment with thiourea under drought stress. Pak J Bot. 2021;53(4):1209–17. [Google Scholar]
- 68.Ulfat M, Fazal S, Javed MM, Khan ZUD. Evaluation of peroxidase activity in Beta vulgaris L., Cucumis sativus L. and Raphanus sativus L. Pak J Bot. 2012;44:91–6. [Google Scholar]
- 69.Ravelo-Pérez LM, Hernández-Borges J, Rodríguez-Delgado M, Borges-Miquel T. Spectrophotometric analysis of lycopene in tomatoes and watermelons: a practical class. Chem Educ. 2008;13(1):11–3. [Google Scholar]
- 70.Alici EH, Arabaci G. Determination of SOD, POD, PPO and cat enzyme activities in Rumex obtusifolius L. Annual Res Rev Biology 2016:1–7.
- 71.Zhou B, Wang J, Guo Z, Tan H, Zhu X. A simple colorimetric method for determination of hydrogen peroxide in plant tissues. Plant Growth Regul. 2006;49:113–8. [Google Scholar]
- 72.Venkataravanappa V, Reddy CL, Shankarappa K, Jayappa J, Pandey S, Krishna Reddy M. Characterization of Tomato leaf curl New Delhi virus and DNA-satellites association with mosaic disease of cucumber. Int J Biotech Bioeng. 2019;5(6):93–109. [Google Scholar]
- 73.Shafiq M, Iqbal Z, Ali I, Abbas Q, Mansoor S, Briddon RW, Amin I. Real-time quantitative PCR assay for the quantification of virus and satellites causing leaf curl disease in cotton in Pakistan. J Virol Methods. 2017;248:54–60. [DOI] [PubMed] [Google Scholar]
- 74.Raes J, Van de Peer Y. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl Bioinf. 2003;2(2):91–101. [PubMed] [Google Scholar]
- 75.Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant–environment interactions. Annu Rev Plant Biol. 2019;70:489–525. [DOI] [PubMed] [Google Scholar]
- 76.Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399(2):257–61. [DOI] [PubMed] [Google Scholar]
- 77.Das A, Singh S, Islam Z, Munshi A, Behera T, Dutta S, Weng Y, Dey S. Current progress in genetic and genomics-aided breeding for stress resistance in cucumber (Cucumis sativus L). Sci Hort. 2022;300:111059. [Google Scholar]
- 78.Sahoo T, Singh D. Estimation of genetic variability, heritability and genetic advance in Cucumber (Cucumis sativus L.) for yield and its components under protected structure. Int J Curr Microbiol Appl Sci. 2020;9(4):2756–64. [Google Scholar]
- 79.Sami A, Han S, Haider MZ, Khizar R, Ali Q, Shafiq M, Tabassum J, Khalid MN, Javed MA, Sajid M. Genetics aspect of vitamin C (ascorbic acid) biosynthesis and signaling pathways in fruits and vegetables crops. Funct Integr Genom. 2024;24(2):1–15. [DOI] [PubMed] [Google Scholar]
- 80.Zhang L, Wang K, Han Y, Yan L, Zheng Y, Bi Z, Zhang X, Zhang X, Min D. Genome-wide analysis of the VQ motif-containing gene family and expression profiles during phytohormones and abiotic stresses in wheat (Triticum aestivum L). BMC Genomics. 2022;23(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jalili S, Ehsanpour AA, Javadirad SM. The role of melatonin on caspase-3-like activity and expression of the genes involved in programmed cell death (PCD) induced by in vitro salt stress in alfalfa (Medicago sativa L.) roots. Bot Stud. 2022;63(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Wang C, Lin Q, Gao F, Ma Y, Zhang M, Lin Y, Ma Q, Hua X. Genome-wide analysis of phylogeny, expression profile and sub-cellular localization of SKP1-Like genes in wild tomato. Plant Sci. 2015;238:105–14. [DOI] [PubMed] [Google Scholar]
- 83.Mamode Cassim A, Grison M, Ito Y, Simon-Plas F, Mongrand S, Boutté Y. Sphingolipids in plants: a guidebook on their function in membrane architecture, cellular processes, and environmental or developmental responses. FEBS Lett. 2020;594(22):3719–38. [DOI] [PubMed] [Google Scholar]
- 84.Zhu Y-X, Yang L, Liu N, Yang J, Zhou X-K, Xia Y-C, He Y, He Y-Q, Gong H-J, Ma D-F. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019;19:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larue GE, Roy SW. Where the minor things are: a pan-eukaryotic survey suggests neutral processes may explain much of minor intron evolution. Nucleic Acids Res 2023:gkad797. [DOI] [PMC free article] [PubMed]
- 86.Wei C, Li M, Li X, Lyu J, Zhu X. Phase separation:the Master Key to deciphering the physiological and pathological functions of cells. Adv Biology. 2022;6(7):2200006. [DOI] [PubMed] [Google Scholar]
- 87.Zhang G, Zhang Z, Luo S, Li X, Lyu J, Liu Z, Wan Z, Yu J. Genome-wide identification and expression analysis of the cucumber PP2C gene family. BMC Genomics. 2022;23(1):563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, Pan C, Wang Y, Ye L, Wu J, Chen L, Zou T, Lu G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics. 2015;16:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo W, Comai L, Henry IM. Chromoanagenesis from radiation-induced genome damage in Populus. PLoS Genet. 2021;17(8):e1009735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lallemand T, Leduc M, Landès C, Rizzon C, Lerat E. An overview of duplicated gene detection methods: why the duplication mechanism has to be accounted for in their choice. Genes. 2020;11(9):1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C, Chen X, Han J, Lu W, Ren Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He Y, Liu X, Zou T, Pan C, Qin L, Chen L, Lu G. Genome-wide identification of two-component system genes in cucurbitaceae crops and expression profiling analyses in cucumber. Front Plant Sci. 2016;7:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen X, Wang Z, Tang R, Wang L, Chen C, Ren Z. Genome-wide identification and expression analysis of Hsf and Hsp gene families in cucumber (Cucumis sativus L). Plant Growth Regul. 2021;95(2):223–39. [Google Scholar]
- 94.Liu P, Wang S, Wang X, Yang X, Li Q, Wang C, Chen C, Shi Q, Ren Z, Wang L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L). Plant Physiol Biochem. 2020;151:197–213. [DOI] [PubMed] [Google Scholar]
- 95.Xu X, Liu M, Lu L, He M, Qu W, Xu Q, Qi X, Chen X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol Genet Genomics. 2015;290:1403–14. [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Zhang J, Li H, Zhang G, Hu D, Zhang D, Xu X, Yang Y, Huang Z. Genome-wide identification of the phytocyanin Gene Family and its potential function in salt stress in soybean (Glycine max (L.) Merr). Agronomy. 2023;13(10):2484. [Google Scholar]
- 97.Zhang Z, Luo S, Liu Z, Wan Z, Gao X, Qiao Y, Yu J, Zhang G. Genome-wide identification and expression analysis of the cucumber PYL gene family. PeerJ. 2022;10:e12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gudi S, Saini DK, Halladakeri P, Singh G, Singh S, Kaur S, Goyal P, Srivastava P, Mavi G, Sharma A. Genome-wide association study unravels genomic regions associated with chlorophyll fluorescence parameters in wheat (Triticum aestivum L.) under different sowing conditions. Plant Cell Rep 2023:1–20. [DOI] [PubMed]
- 99.Ling J, Luo Z, Liu F, Mao Z, Yang Y, Xie B. Genome-wide analysis of microRNA targeting impacted by SNPs in cucumber genome. BMC Genomics. 2017;18:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumar K, Mandal SN, Neelam K, de Los Reyes BG. MicroRNA-mediated host defense mechanisms against pathogens and herbivores in rice: balancing gains from genetic resistance with trade-offs to productivity potential. BMC Plant Biol. 2022;22(1):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharif R, Zhu Y, Huang Y, Sohail H, Li S, Chen X, Qi X. microRNA regulates cytokinin induced parthenocarpy in cucumber (Cucumis sativus L). Plant Physiol Biochem. 2024;212:108681. [DOI] [PubMed] [Google Scholar]
- 102.Li C, Li Y, Bai L, Zhang T, He C, Yan Y, Yu X. Grafting-responsive miRNAs in cucumber and pumpkin seedlings identified by high‐throughput sequencing at whole genome level. Physiol Plant. 2014;151(4):406–22. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y, Zhang W, Dong M. The miR-58 microRNA family is regulated by insulin signaling and contributes to lifespan regulation in Caenorhabditis elegans. Sci China Life Sci. 2018;61:1060–70. [DOI] [PubMed] [Google Scholar]
- 104.He Y, Yang K, Zhang X. Viral microRNAs targeting virus genes promote virus infection in shrimp in vivo. J Virol. 2014;88(2):1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ali M, Shafiq M, Haider MZ, Sami A, Alam P, Albalawi T, Kamran Z, Sadiq S, Hussain M, Shahid MA. Genome-wide analysis of NPR1-like genes in citrus species and expression analysis in response to citrus canker (Xanthomonas axonopodis Pv. Citri). Front Plant Sci. 2024;15:1333286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma P, Jha AB, Dubey RS, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. Journal of botany 2012.
- 107.Gonçalves MC, Vega J, Oliveira JG, Gomes M. Sugarcane yellow leaf virus infection leads to alterations in photosynthetic efficiency and carbohydrate accumulation in sugarcane leaves. Fitopatologia Brasileira. 2005;30:10–6. [Google Scholar]
- 108.Balachandran S, Hurry V, Kelley S, Osmond C, Robinson S, Rohozinski J, Seaton G, Sims D. Concepts of plant biotic stress. Some insights into the stress physiology of virus-infected plants, from the perspective of photosynthesis. Physiol Plant. 1997;100(2):203–13. [Google Scholar]
- 109.Setiyobudi RH, Subiastuti AS, Daryono BS. The effect of Begomovirus infection on phenotypic characters of Cucumis melo L.‘Melona’. In: AIP Conference Proceedings. AIP Publishing, 2020.
- 110.Kiani BH. Metabolomic profiling of different cereals during Biotic and Abiotic stresses. Omics Approach to manage abiotic stress in Cereals. Springer; 2022. pp. 119–50.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I affirm that all necessary data and permissions have been provided for this study. Any interested researchers can access the required data to support the findings and conclusions of this article. For publicly archived datasets, hyperlinks are provided in this manuscript in appropriate place for convenience. Rest assured, the plants were collected from Departments of Horticulture Punjab University Lahore with voucher HDpu7598 and submitted to Horticultural bank Punjab University Lahore. I have ensured that all data, materials, software applications, and custom code supporting the claims made in this article are in full compliance with field standards. It’s important to note that I have taken into account the possibility of individual journal policies regarding research data sharing, considering the norms and expectations of our discipline.