Abstract
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionic acid receptors (AMPARs) are cation-selective ion channels that mediate most fast excitatory neurotransmission in the brain. Although their gating mechanism has been studied extensively, understanding how cations traverse the pore has remained elusive. Here, we investigated putative ion and water densities in the open pore of Ca2+-permeable AMPARs (rat GRIA2 flip-Q isoform) at 2.3-2.6Å resolution. We show that the ion-permeation pathway attains an extracellular Ca2+ binding site (Site-G) when the channel gate moves into the open configuration. Site-G is highly selective for Ca2+ over Na+ favoring the movement of Ca2+ into the selectivity filter of the pore. Seizure related N619K mutation, adjacent to site-G, promotes channel opening, but attenuates Ca2+ binding and thus diminishes Ca2+ permeability. Our work identifies the importance of site-G which coordinates with the Q/R-site of the selectivity filter to ensure the preferential transport of Ca2+ through the channel pore.
Introduction
AMPARs mediate most of the fast excitatory synaptic transmission in the central nervous system1. The neurotransmitter, L-glutamate, activates postsynaptic AMPARs, which triggers channel gating and ion permeation, causing dendritic membrane depolarization. AMPARs are non-selective cation channels that are able to permeate various cations, however, Na+ and Ca2+ are the main permeating ions in the context of synaptic transmission. Ca2+-permeable and - impermeable AMPARs (CP- and CI-AMPARs) are present in the brain, typically associated with auxiliary subunits that modulate their function2. CP-AMPARs regulate neuronal excitability but also transport the second messenger Ca2+ to augment signal transduction3. Although they are less abundant than CI-AMPARs, CP-AMPARs are critical for synaptic plasticity underlying learning and addiction, excitatory synaptic transmission at the afferent synapses of inhibitory neurons, and synaptic maintenance mediated by glial cells, while their dysfunctions are related to developmental epilepsy, glioma, amyotrophic lateral sclerosis, and excitotoxicity3. Despite the importance in elucidating brain physiology and designing therapeutics that could control synaptic activity, the structural basis for ion permeation in AMPARs remains elusive.
The AMPAR ion channel assembles as homo- or hetero-tetramers of homologous subunits GluA1-42; each subunit consists of an amino terminal domain (NTD), a ligand-binding domain (LBD), a transmembrane domain (TMD), and a cytoplasmic domain (CTD) (Fig. 1A). The NTD and LBD build the pseudo-2-fold symmetric extracellular architecture, while the TMDs form a pseudo-4-fold symmetric ion channel4–6. The TMD contains three membrane spanning helices (M1, M3, and M4), a re-entrant helix (M2), and a selectivity filter (SF). The pore is made exclusively by the M2, M3, and the SF (Fig. 1B–D). At the extracellular entrance of the pore, the M3 helices converge and form the channel gate. Binding of L-glutamate to the LBD induces a domain closure that transduces to the TMD7–11, which dilates the gate into a 2-fold symmetric architecture9–11 (Fig. 1C and D). According to the symmetry, the B/D (or A/C) subunits are geometrically equivalent4 (Fig. 1B). The LBDs in the B/D subunits are pivotal for dilation, as they pull and kink the M3, whereas the M3 remains straight in the A/C subunits (Fig1. C and D)9–11. The ion permeation path below the gate is subdivided into the upper vestibule and the SF (Fig. 1C). The SF is surrounded by the lateral vestibule (Fig. 1C).
Figure 1. The ion permeation path of A2iQ/γ2(KKEE).
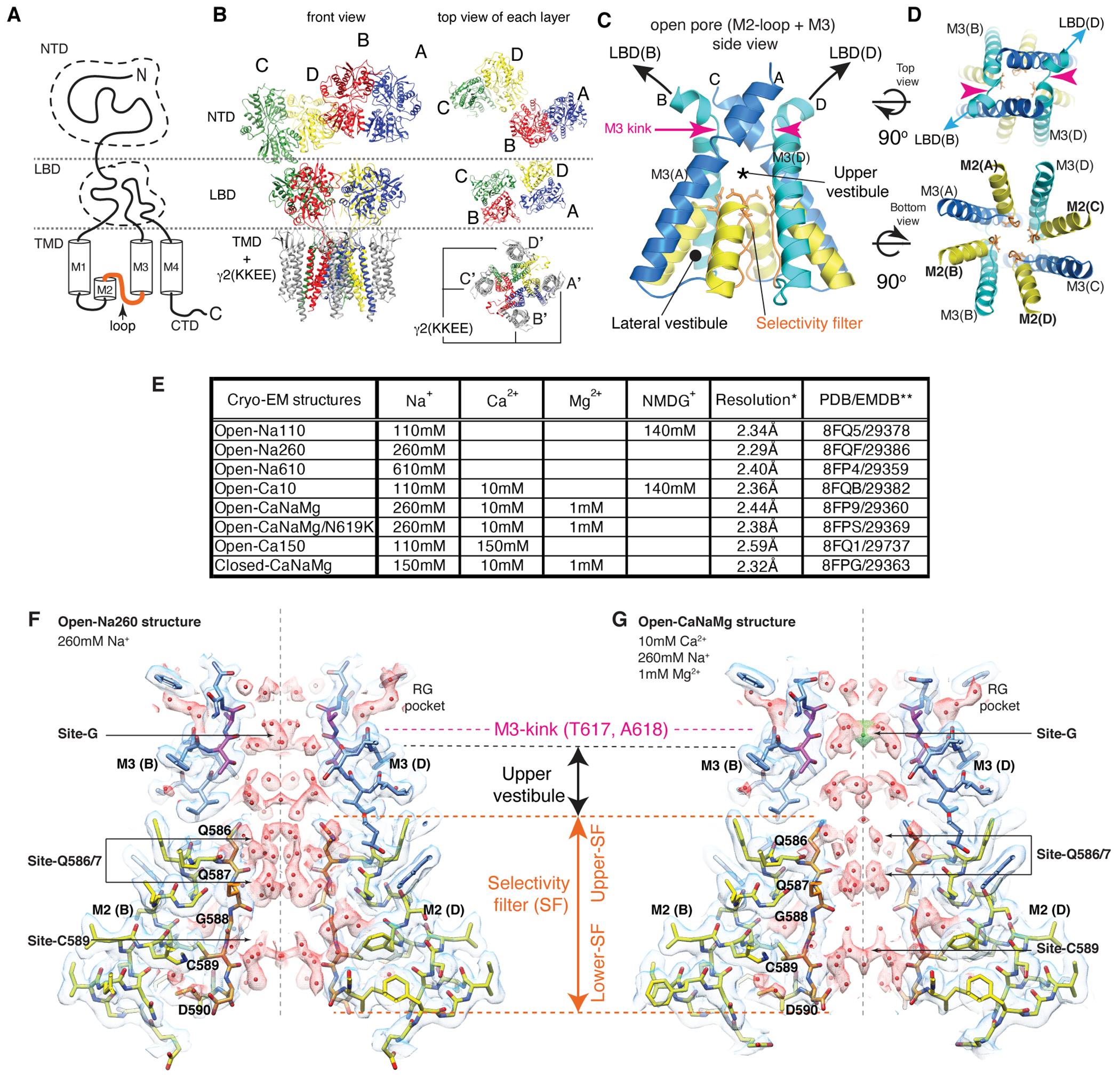
A. Domain organization of AMPAR subunit. The orange loop contains the selectivity filter. B. Tetrameric assembly of A2iQ/γ2(KKEE), whose global structure is indistinguishable from the WT complex. GluA2 subunits; A=blue, B=red, C=green, and D=yellow. Four γ2(KKEE)s are in gray and their positions are labeled A’, B’, C’ and D’. C. Architecture of the open pore. Blue=M3 of A/C subunits, cyan=M3 of B/D subunits, yellow=M2, and orange=selectivity filter. D. Top: The open pore viewed from the extracellular space. Bottom: The selectivity filter viewed from the cytoplasmic side. E. Summary of ion conditions used in each cryo-EM structure. (*) C2 map resolutions of the transmembrane region are listed. See Table 1 and Supplementary Table 1 for details of refinement statistics. (** ) PDB/EMDB codes are for the transmembrane region. F and G. The central slices of the pore of Open-Na260 (E, map threshold=6.0σ) and Open-CaNaMg (F, map threshold=3.3σ). The cryo-EM density map (mesh) and the atomic model are superimposed. Putative water and Ca2+ are in red and green spheres, respectively. Regions of intense density accumulation are indicated as sites-G, -Q586/587, and -C589. Dashed gray lines are pore symmetry axes. The RG pocket indicates a cavity filled with water that is on the rear side of site-G and commonly found in the open structures.
CI-AMPARs arise from RNA editing that converts the encoding of amino acid at the Q/R-site in the SF of GluA2 (residue 586 in rat) from neutral glutamine (Q) to a positively charged arginine (R)3,12. The unedited homotetrameric GluA2 flip(Q) isoform (A2iQ), a frequently used model for the CP-AMPAR, has a linear current-voltage relationship in the absence of polyamines13. When the permeating cations on both sides of the membrane are the same, the AMPAR conductance is near identical at a wide range of voltages with a reversal potential of 0mV. Therefore, in the absence of voltage, the distribution of permeating cations and water in the open pore is predicted to provide critical insights into the mechanism of conductance and ion permeation14,15.
Cryo-EM structures of the open pore
To facilitate, and potentially stabilize, the energetically unstable open state, the wild type (WT) A2iQ was co-expressed with the K52E/K53E (KKEE) mutant of auxiliary subunit TARPγ2 (γ2) which substantially slows the rate of AMPAR desensitization16. The KKEE mutations are in the flexible extracellular β1-β2 loop, unresolved in all TARP structures, but predicted to make transient contacts with the LBD9,11,17,18. To capture the open pore conformation, the agonist (100mM L-glutamate) and an inhibitor of desensitization (330μM cyclothiazide (CTZ)) were co-applied to the purified receptor prior to vitrification. The cryo-EM structures, at overall resolution in the range of 2.28-2.59Å, were obtained in various concentrations of Na+ and Ca2+ to identify ion specific features (Table 1, Extended Data Fig. 1–4, and Supplementary Table 1). It was critical to use large number of particles, approximately 600,000 to 1,000,000 particles, in the final refined map to reveal the signals of non-protein features in the open pore. When necessary, NMDG was supplemented to maintain ionic strength (Fig. 1E). Large cation NMDG+ permeates AMPAR substantially less (~2%) compared to small cations19, and thus the probability of finding NMDG+ in the pore is expected to drop accordingly and be negligeable.
Table1.
Cryo-EM data collection, refinement, and validation statistics
| Open-Na260, TMD-STG (EMDB-29386) (PDB 8FQF) | Open-CaNaMg, TMD-STG (EMDB-29360) (PDB 8FP9) | Open-Ca150, TMD-STG (EMBD-29737) (PDB 8FQ1) | Open-Na610, TMD-STG (EMDB-29359) (PDB 8FP4) | Open-Ca10, TMD-STG (EMDB-29382) (PDB 8FQB) | Open-Na110, TMD-STG (EMBD-29378) (PDB 8FQ5) | Open-CaNaMg/N619K, TMD-STG (EMDB-29369) (PDB 8FPS) | Closed-CaNaMg, TMD-STG (EMDB-29363) (PDB 8FPG) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Data collection and processing | ||||||||||||||||
| Magnification | 105,000x | 130,000x | 130,000x | 105,000x | 130,000x | 130,000x | 130,000x | 130,000x | ||||||||
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | ||||||||
| Electron exposure (e−/Å2) | 51 | 50 | 50 | 50 | 53 | 52 | 54.6 | 56 | ||||||||
| Defocus range (μm) | −0.7 to −2.1 | −0.5 to −1.5 | −0.8 to −2.0 | −0.8 to −2.0 | −0.8 to −2.0 | −0.6 to −2.0 | −0.8 to −2.0 | −0.8 to −1.8 | ||||||||
| Pixel size (Å) | 0.820 | 0.8195# | 0.8195# | 0.820 | 0.8195# | 0.8195# | 0.8195# | 0.8195# | ||||||||
| Energy filter band width (eV) | 20 | 15 | 20 | 20 | 20 | 20 | 20 | 15 | ||||||||
| Number of micrographs | 16,096 | 32,662 | 28,472 | 23,493 | 28,505 | 30,384 | 34,729 | 34,553 | ||||||||
| Initial particle images (no.) | 5,409,247 | 5,075,710 | 5,573,868 | 5,140,390 | 7,059,177 | 4,655,266 | 6,655,184 | 3,905,978 | ||||||||
| Final particle images (no.) | 1,093,405 | 822,038 | 540,796 | 901,154 | 986,075 | 692,680 | 693,935 | 847,436 | ||||||||
| Symmetry imposed | C1 | C2 | C1 | C2 | C1 | C2 | C1 | C2 | C1 | C2 | C1 | C2 | C1 | C2 | C1 | C2 |
| Map resolution (Å) | 2.38 | 2.29 | 2.59 | 2.44 | 2.78 | 2.59 | 2.50 | 2.40 | 2.48 | 2.36 | 2.42 | 2.34 | 2.50 | 2.38 | 2.42 | 2.32 |
| Map resolution with LBD* (Å) | 2.50 | n.d. | 2.63 | 2.57 | 2.84 | 2.73 | 2.57 | 2.46 | 2.52 | 2.44 | 2.46 | 2.38 | 2.59 | 2.48 | 2.52 | n.d. |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.3-3.7 | 2.2-3.5 | 2.5-4.1 | 2.4-3.8 | 2.6-4.3 | 2.5-4.1 | 2.4-3.9 | 2.3-3.9 | 2.4-4.1 | 2.3-3.9 | 2.3-4.0 | 2.2-3.7 | 2.4-4.2 | 2.3-3.9 | 2.3-3.9 | 2.2-3.7 |
| Refinement (C2) ^ | ||||||||||||||||
| Initial model used (PDB code) | 5WEO | 5WEO | 5WEO | 5WEO | 5WEO | 5WEO | 5WEO | 5WEO | ||||||||
| Model resolution (Å) | 2.0/2.3 (masked) | 2.2/2.5 (masked) | 2.2/2.7 (masked) | 2.2/2.4 (masked) | 2.1/2.5 (masked) | 2.1/2.4 (masked) | 1.9/2.6 (masked) | 2.0/2.4 (masked) | ||||||||
| FSC threshold | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | 0.143/0.5 | ||||||||
| Map sharpening B factor (Å2) | −56.0 | −69.5 | −72.1 | −70.8 | −64.9 | −58.3 | −62.8 | −62.0 | ||||||||
| Model composition | ||||||||||||||||
| Non-hydrogen atoms | 10,895 | 10,851 | 10,876 | 10,876 | 10,823 | 10,876 | 10,602 | 10,674 | ||||||||
| Protein residues | 1,366 | 1,366 | 1,366 | 1,366 | 1,366 | 1,366 | 1,338 | 1,348 | ||||||||
| Ligands | CL:2 | CA:1, CL:4 | CA:2 | CL:2 | CA:1, CL:2 | CL:4 | CL:4 | CL:4 | ||||||||
| Water | 231 | 190 | 170 | 218 | 176 | 220 | 152 | 187 | ||||||||
| B factors (Å2) | ||||||||||||||||
| Protein | 38.03 | 57.87 | 55.01 | 40.64 | 41.01 | 39.99 | 35.33 | 42.86 | ||||||||
| Ligand | 59.77 | 76.98 | 87.40 | 73.02 | 75.39 | 63.98 | 66.30 | 71.43 | ||||||||
| Water | 25.84 | 44.19 | 45.02 | 36.14 | 33.39 | 37.15 | 27.41 | 38.77 | ||||||||
| R.m.s. deviations | ||||||||||||||||
| Bond lengths (Å) | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.003 | ||||||||
| Bond angles (°) | 0.428 | 0.478 | 0.403 | 0.421 | 0.451 | 0.441 | 0.370 | 0.489 | ||||||||
| Validation | ||||||||||||||||
| MolProbity score | 1.30 | 1.13 | 1.21 | 1.36 | 1.45 | 1.23 | 1.12 | 1.73 | ||||||||
| Clashscore | 4.62 | 3.35 | 3.70 | 4.15 | 4.96 | 3.44 | 3.32 | 4.90 | ||||||||
| Poor rotamers (%) | 0.54 | 0.98 | 1.17 | 1.25 | 1.79 | 1.16 | 1.01 | 1.84 | ||||||||
| Ramachandran plot | ||||||||||||||||
| Favored (%) | 97.72 | 98.03 | 98.01 | 97.57 | 98.10 | 97.80 | 98.76 | 96.00 | ||||||||
| Allowed (%) | 2.28 | 1.97 | 1.99 | 2.43 | 1.90 | 2.20 | 1.24 | 4.00 | ||||||||
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||
The pixel size (Å) was 0.647 at the detector level prior to binning
Resolution of the map generated by focused refinement of the LBD-TMD-STG
Model refinement was conducted against the C2 map of TMD-STG
n.d. Not determined
The specific cation conditions examined (followed by the name of obtained structure) are; 110mM Na+/140mM NMDG+ (Open-Na110), 260mM Na+ (Open-Na260), 610mM Na+ (Open-Na610), 10mM Ca2+/110mM Na+/140mM NMDG+ (Open-Ca10), 10mM Ca2+/260mM Na+/1mM Mg2+ (Open-CaNaMg), and 150mM Ca2+/110mM Na+ (Open-Ca150) (see Fig. 1E). The concentrations listed above include the extra 110mM Na+ from the agonist solution, because the L-glutamate stock solution is titrated at pH7.4 using NaOH (see Methods). For example, the particles in Open-Na110 were purified in 140mM NMDG+ and adding the L-glutamate stock solution that contains Na+ made the final ionic condition of 110mM Na+/140mM NMDG+ (see Methods for other conditions). Some conditions contain 1mM Mg2+ but its effect was not systematically explored. Additionally, a reference structure of the closed state (Closed-CaNaMg) was obtained in the presence of antagonist (100μM CNQX) and 330μM CTZ in 10mM Ca2+/150mM Na+/1mM Mg2+.
The architectures of the complexes in both the open and closed states were at the stoichiometry of A2iQ:γ2(KKEE)=4:4 and agree with the WT complex10 (Fig. 1B and Extended Data Fig. 5F–H). The extracellular region spanning the KKEE mutations is flexible and unresolved, like the WT (Extended Data Fig. 1G–H). Gating is accompanied with global motions of the γ2(KKEE) equivalent to those observed in TARPγ811 (Extended Data Fig. 5I–N). The core gating machinery of the A2iQ/γ2(KKEE) complex is virtually identical to the WT complexes9,10. In all open structures, the LBDs were closed, fully occupied with CTZ and L-glutamate. The dimer-of-dimers organization of the tetrameric LBD (LBD gating ring) adopts alternative architectures characterized by small displacements between the two LBD dimers (Extended Data Fig. 5A–E and Supplementary Table 1), consistent with the mobile nature of the LBDs. The cytoplasmic half of the SF, at residues G588 and C589, was more flexible than the surroundings based on the lower local resolution (Extended Data Fig. 3I–L). Consistently, alternative side chain orientation was observed at C589 under a subset of ion conditions.
Identification of a novel Ca2+ binding site
Numerous non-protein densities occupy the gate, vestibules, and SF (Fig. 1F–G and Extended Data Fig. 6–7), with the majority being interpreted as putative water molecules, although, we cannot fully exclude the possibility that some are ions or even noise. The large densities at site-G (i.e., at the gate residues T617/A618), as well as at site-Q586/7 and site-C590 within the SF (Fig. 1F–G), if present, were consistent in the half-maps and regardless of applying C2 symmetry (Extended Data Fig. 3–4). In fact, the density accumulation at the upper SF, around site-Q586/7, has been observed in previously reported open structures that were resolved at around 3-3.5Å resolution10,20. With improved resolution, we extend the previous observations and find that the pore-occupying densities in the upper SF often form parallel vertical columns (Fig. 1F and Extended Data Fig. 6–7), indicative of energetically equivalent ion conduction pathways, contrasting with the single file configuration observed in potassium channels14. The distribution of putative ion and water density within the SF was variable between ion condition and morphologically complex (Fig. 1F–G and Extended Data Fig. 6–7), making it difficult to identify features that are specific to any cations. In contrast, the positions of many putative waters in the lateral vestibule match between structures, highlighting their common accumulation points (Extended Data Fig. 8). The distribution of the putative water in the lateral vestibule may suggest their role in providing structural flexibility to the SF.
While it was challenging to interpret the non-protein densities in the lower ion permeation path, robust cation specific features were identifiable at the upper element of the pore. The central pore axis density at site-G was present only in ionic conditions that included Ca2+ (i.e., Open-Ca10, Open-CaNaMg, and Open-Ca150), regardless of imposing symmetry, while the surrounding polypeptides adopt virtually identical conformations, in all cases (Fig. 2A–F, H, and Extended Data Fig. 4C–E). Therefore, the correlation between the density and ion composition suggests that the central density contains Ca2+. The higher local resolution of the density in 150mM Ca2+ (Open-Ca150) compared to 10mM Ca2+ (Open-Ca10 and Open-CaNaMg) is consistent with more stable Ca2+ binding at site-G at higher Ca2+ concentrations (Extended Data Fig. 3I–K). Although the central density should be regarded as an ensemble of Ca2+ and water due to their dynamic nature and the limited resolution, it is possible to place a Ca2+ surrounded by four waters at 2.4Å spacing, partially fulfilling the requirements for the hydration shell21 (Fig. 2H–I). Importantly, the central pore occupying density at site-G did not emerge even when the Na+ concentration was raised from 110mM (in Open-Na110) to 610mM (in Open-Na610) (compare Fig. 2C and E), indicative of high selectivity of site-G for Ca2+ over Na+. In the open-gate conformation, site-G is located immediately outside of the membrane, thus it may be viewed as an extracellular vestibule immediately outside of the membrane electric field. The vestibule is formed only in the open state and thus site-G is a transient state-dependent Ca2+ binding site (Extended Data Fig. 6A–C, and H). Furthermore, site-G is in the C-terminal portion of the M3 helix encoded by the evolutionary-conserved amino acid sequence, SYTANLAAF, making it likely to be a common structural element in almost all ionotropic glutamate receptors (iGluRs) (Fig. 2G).
Figure 2. Site-G as a Ca2+ binding site at the gate.
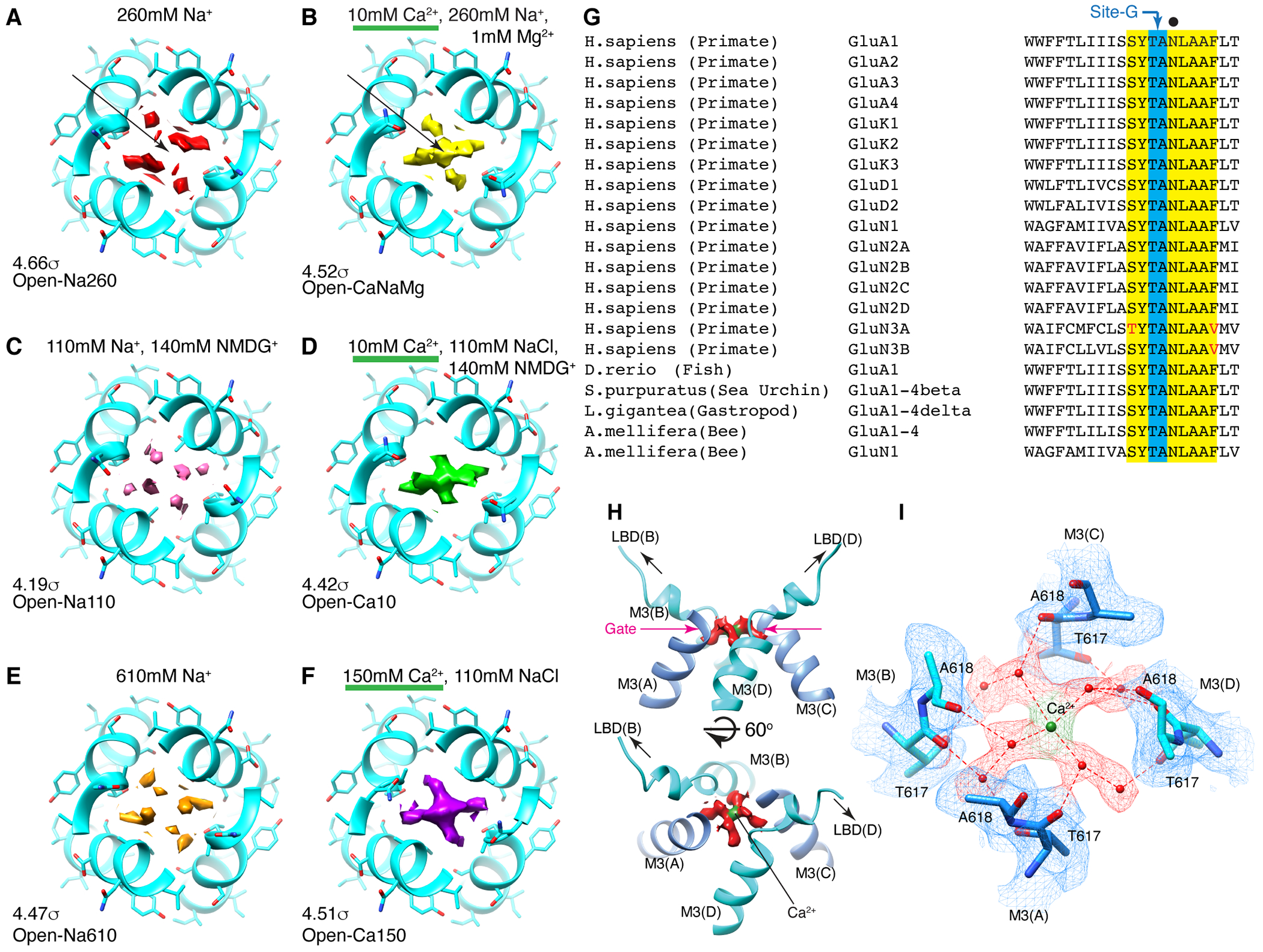
A-F. The top views of site-G in the open gates under different ionic conditions. The M3 helices in cyan. Pore occupying densities are in red (A. Open-Na260), yellow (B. Open-CaNaMg), pink (C. Open-Na110), green (D. Open-Ca10), orange (E. Open-Na610), and purple (F. Open-Ca150). The sigma values indicate display thresholds. Overall density occupying site-G is stronger when Ca2+ is present. For example, compare the arrows in A and B. See Extended Data Fig. 6-7 for sideview. G. Alignment spanning the SYTANLAAF motif. Atypical residue in red. The non-vertebrate subunit nomenclature follows reference49. H. Side (top) and diagonal (bottom) view of site-G in Open-CaNaMg. The M3 helices in cyan and blue. The EM density map of putative water and putative Ca2+ are in red and green, respectively. I. T617 and A618 make site-G. The atomic model and density map are superimposed. The dashed red lines are the putative hydrogen bonds and ion-water interactions.
External Ca2+ block predicted from the open structures
Given that 10mM Ca2+ can dominate the occupancy of site-G even in the presence of excess Na+ (Fig. 2A and B), we postulated that Ca2+ binding to site-G would cause block by excluding Na+ from entering the pore. Since Site-G lies outside the membrane electric field, the mechanism of block would be expected to be voltage-independent. To test this, we recorded steady-state membrane currents evoked by 10 mM L-glutamate with 100 μM CTZ on outside-out patches expressing A2iQ/γ2 receptors using external solution of 150 mM NaCl with varying concentrations of added Ca2+ (0.1-108mM). L-Glutamate was applied in the presence of cyclothiazide (100 μM), to attenuate AMPAR desensitization, and the steady-state response observed at each concentration of external Ca2+ was ramped from −100 mV to +100 mV. As anticipated, we observed block of AMPAR-mediated membrane currents by external Ca2+ in a dose-dependent manner over a wide range of membrane potentials (Fig.3A ). At −100mV, the apparent affinity for inhibition (IC50) and steady-state response (Gmin) was estimated to be 5.21 ± 0.64 mM and 19.67 % (n=8), respectively (Fig. 3B and Supplementary Table 2). Interestingly, although site-G is in an extracellular location, external Ca2+-block was voltage-dependent whose data points were well fit by a single permeant ion blocker model22 with estimated Kd(0mV) of 3.2 mM at physiological Ca2+ levels (Fig. 3C and Supplementary Table 3). This latter point suggests that external Ca2+ not only impedes the flow of Na+ at site-G but also hinders Na+ permeation at another location within the membrane that is sensitive to voltage. In keeping with this there is a large density accumulation in the SF of Open-Ca150 (Extended Data Fig. 6C and 7C), implying that the SF also traps Ca2+ that interferes with the passage of Na+, accounting for the voltage-dependent component of Ca2+ block.
Figure3. External Ca2+ block of GluA2/γ2 and its voltage dependence.
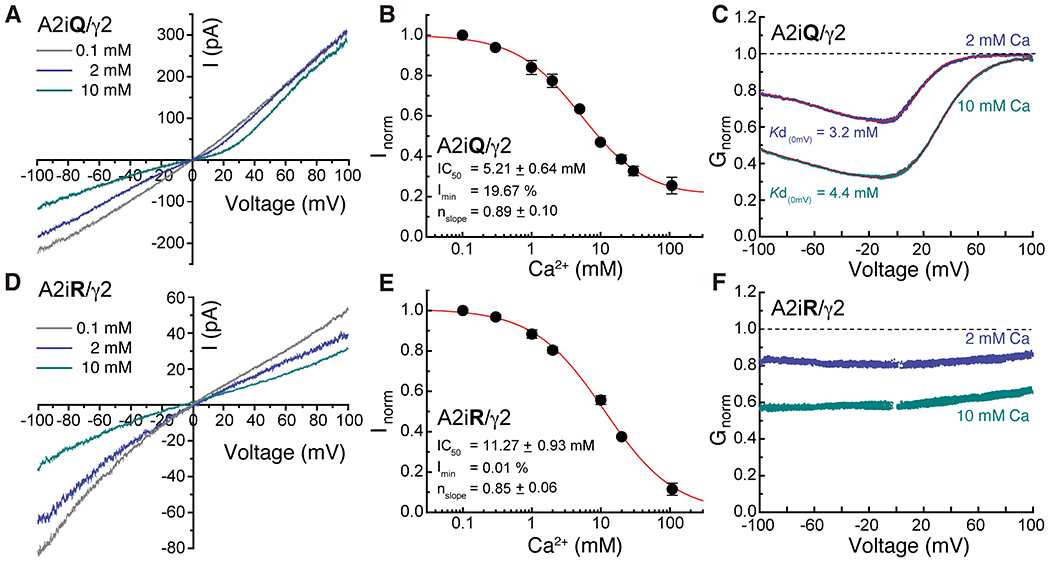
A and D. Current records observed in presence of 10mM glutamate and 100μM CTZ at a voltage range of −100 to +100 mV in presence of 0.1mM (gray), 2mM (light blue) and 10mM (dark blue) Ca2+ for A2iQ/γ2 (Patch # 220914p7) and A2iR/γ2 (Patch # 221025p2) receptors. B and E. Inhibition plots of block by external Ca2+ of A2iQ/γ2 (B, n=8) and A2iR/γ2 (E, n=6) receptors at −100 mV. Data are presented as mean values ± SEM. C and F. Conductance-voltage plots of block by 2 mM or 10 mM external Ca2+ of A2iQ/γ2 (C) and A2iR/γ2 (F) receptors. The voltage-dependence of block of A2iQ/γ2 receptors was well fit by a single permeant ion blocker model (red line) whereas block of A2iR/γ2 receptors was voltage-insensitive.
To explore this point further, we repeated experiments with Ca2+-impermeable GluA2flip(R)/TARPγ2 (A2iR/γ2) receptors, which do not permeate Ca2+ beyond the SF but may allow Ca2+ to access and bind to site-G. Here, we observed that external Ca2+ was still effective in blocking A2iR mediated Na+ currents (Fig. 3E), though the mechanism was now voltage-independent (Fig. 3F). The apparent affinity for inhibition (IC50, 11.27 ± 0.93 mM, n=6) was lower, suggesting there may be coupling between site-G and the SF in A2iQ/γ2 receptors, or alternatively, the R586 residue may weaken the Ca2+ affinity at site-G in A2iR/γ2 receptors. Conductance was negligeable at the highest Ca2+ concentration (108 mM), which would be expected since A2iR receptors poorly conduct Ca2+ (Fig. 3E and Supplementary Table 2). Collectively, both voltage-dependent and -independent mechanisms contribute to external Ca2+ block of A2iQ receptors, consistent with the involvement of an upper vestibular structure, site-G, that lies outside the electrostatic field generated by the membrane potential.
The N619K mutation reveals site-G function
To further test if Ca2+ block is mediated by site-G, the N619 residue (Fig. 2G, black dot) adjacent to site-G was interrogated. The N619K mutation in GluA2 corresponds to seizure related N650K mutation in GluN1 subunit of NMDA receptor (NMDAR)23. The cryo-EM structure of N619K mutation of A2iQ in complex with γ2(KKEE) (Open-CaNaMg/N619K), in the same ionic condition as the WT (Open-CaNaMg) (Fig. 1E and 4A), revealed that its open conformation lacks the M3 kink present in the WT receptor (Fig. 4B black arrowhead and Extended Data Fig. 2M–N, 6G, 7G, Table 1, and Supplementary Table 1), while the predicted pore radius along the permeation path was similar (Extended Data Fig. 9). In the B/D subunits of WT, a cavity with a density that can accommodate three water molecules is found at the rear side of site-G next to N619 (RG pocket), contributing to the M3 kink by disrupting the α-helix fold (Fig. 4B and Extended Data Fig. 7A–F). The mutated residue, K619, is space occupying and prevents the RG pocket from forming, which may keep the M3 intact (Fig. 4A and Extended Data Fig. 7G).
Figure 4. N619K mutation reduces Ca2+ binding at site-G, external Ca2+ block, and Ca2+ permeability.
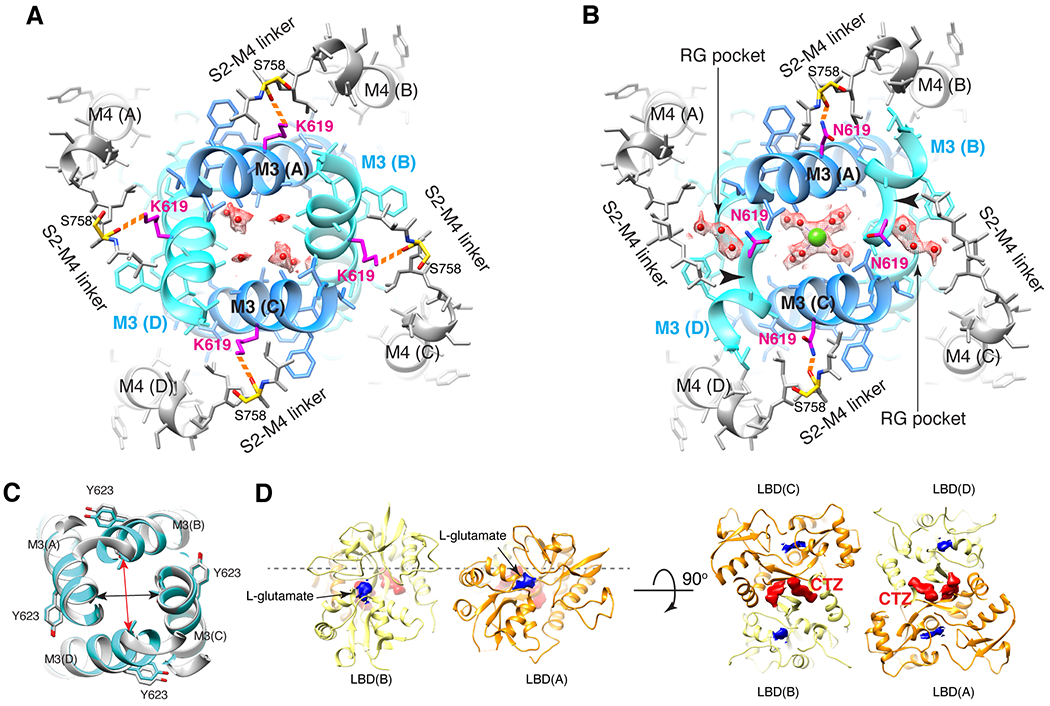
A-B. The site-G of the mutant A2iQ(N619K)/γ2(KKEE) (A) and WT A2iQ/γ2(KKEE) (B) viewed from the extracellular side. The M3 kink (arrowhead in B) is only present in WT. The central pore density is absent in the mutant, while the Ca2+ (green sphere) is found in the WT. Water in red sphere. Density map in mesh. The side chains of mutated K619 and WT counterpart N619 are in magenta. The S758 in yellow. The K619 interacts with the carbonyl oxygens of S758 (orange dashed lines). In WT the RG pocket filled with water occupies the space between N619 and S758 in the B/D subunits. In A/C subunits, the N619 interacts with the carbonyl oxygens of S758 (orange dashed lines). C. Overlay of site-G of WT (light gray) and N619K (cyan) receptors, aligned using the M3 segment residues 610-615. The pore is narrower in the B/D subunit axis (red arrow: distances of T617 Cα of B/D; 12.38Å in N619K vs. 13.57Å in WT) and wider in the A/C axis (black arrow: distances of T617 Cα of A/C; 11.84 in N619K vs. 11.56Å in WT). D. The ribbon model of LBD gating ring of LBDconf1 in A2iQ(N619K)/γ2(KKEE) complex (Supplementary Table 1) displayed at two orthogonal views. LBD monomers of A/C and B/D subunits in orange and light yellow, respectively. The densities for glutamate and CTZ are in blue and red, respectively. The conf2 is also fully liganded as in conf1 (not shown).
Focused classification and refinement of the LBD gating ring (Fig. 4D) revealed that the LBDs are fully liganded like the WT, but the mutant gate adopts an alternative open conformation (Fig. 4A). Substituting the intrinsically dynamic water in the RG pocket with the well-structured side chain of K619, that is greater in size than the N619 in WT (Fig. 4A and B), may contribute to stabilizing the alternative open conformation in the N619K mutant complex. In fact, the N619K mutant A2iQ bound to γ2 (N619K/γ2) spontaneously opens to about 30% of the peak agonist response (Fig. 5A, D, Extended Data Fig. 10A–F, and Supplementary Table 4–6). Interestingly, the leak current was weakly sensitive to AMPAR antagonists, GYKI-52466 and NBQX (Extended Data Fig. 10G–L, and Supplementary Table 4–6), as well as being sensitive to block by cytoplasmic spermine, which is a hallmark of the open pore of the AMPAR (Extended Data Fig. 10A–F). Unlike γ2-bound WT A2iQ (WT/γ2), the N619K/γ2 receptors fail to desensitize in the continued presence of the agonist and exhibit slow agonist off-kinetics suggesting that they have a much higher affinity for L-glutamate (Fig. 5A–F). Interestingly, the leak and agonist evoked responses of the N619K/γ2 receptor have differing affinities and voltage-sensitivity to cytoplasmic polyamine block suggesting that the N619K mutation profoundly affects ion transport through the pore (Extended Data Fig. 10B, C, E, F and Supplementary Table 7).
Figure 5. N619K mutation promotes opening and alters kinetic properties of the ion channel.
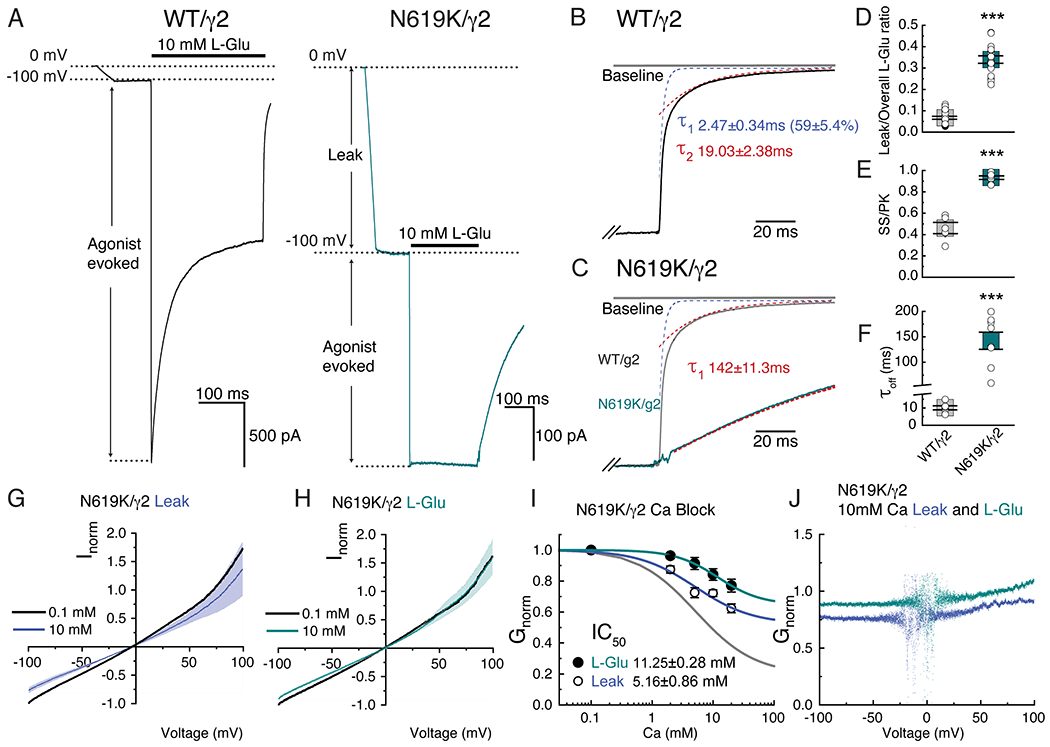
A. Example traces of leak and agonist-evoked responses of WT (in black, Patch # 200121p2) and mutant N619K (in cyan, Patch # 221201p5) receptors at −100 mV. Note the substantial leak current in the mutant. B. Example of deactivation kinetics of WT (black trace) fit by bi-exponential function (blue and red dashed lines are the τ1 and τ2 component, respectively). C. Example of deactivation kinetics of N619K superimposed with the WT example from panel B. N619K deactivation kinetics was fit by mono-exponential function. Trace response (cyan) and model fit (red). D-F. Scatter plots showing all data points from each group. Average represented with grey and cyan squares with SEM. D. Ratio of leak over overall evoked currents (ratio leak current of overall current WT: 0.07 ± 0.01, n=6; N619K: 0.34 ±0.02, n=8; two-sided Mann-Whitney test, p-value <0.001). E. Steady state current over peak current ratio (WT: 0.46 ± 0.05, n=8; N619K=0.93±0.02, n=8; unpaired two-sided t-test with Welch correction, p-value <0.001). F. Deactivation kinetics measured using the offset kinetics. Off kinetics in WT and N619K were fit to bi-exponential (τ1 and τ2) and mono-exponential functions, respectively (WT: 10.10 ± 1.18, n=6;N619K: 142.00 ±11.30, n=8; unpaired two-sided t-test with Welch correction, p-value <0.001). Also see in Supplementary Table 4. G and H. Normalized I-V currents recorded in N619K/γ2 mutant at holding potential ganging from −100 mV to +100mV. Black; average response from all recordings in 0.1 mM Ca solution. Blue; average responses in 10 mM Ca2+ for leak current (n=6). Cyan; 10 mM glutamate and 100μM CTZ evoked responses (n=6). SEM of leak and evoked currents in dim colors. I. Inhibition plots of external Ca2+ block for leak (open circles, n=6) and agonist-evoked (filled circles, n=6) responses in N619K receptors at −100 mV. Data are presented as mean values ± SEM. (Also see in supplementary table 3) J. Conductance-voltage plots of block by 10 mM external Ca2+ for the agonist-evoked curve (cyan) and the leak curve (blue).
A closer examination of site-G in the N619K mutant reveals that it is narrower in the B/D subunit axis (distances of T617 Cα of B/D; 12.38Å in N619K vs. 13.57Å in WT) and wider in the A/C axis (distances of T617 Cα of A/C; 11.84 in N619K vs. 11.56Å in WT) compared to the WT (Fig. 4C). These structural changes were correlated with the absence of central Ca2+ density with the mutant lacking the structural signature for Ca2+ binding to site-G (Fig. 4A and B). In keeping with this, external Ca2+ exhibited a substantially reduced ability to block both the leak or agonist-evoked responses of N619K/γ2 receptors with Gmin values of 52.18 % (leak) and 66.07 % (Agonist) (n=6), respectively (Fig. 5G–J and Supplementary Table 2). The residual external Ca2+ block in N619K/γ2 may be caused by the interaction between the Ca2+ with elements in the ion permeation pathway other than site-G. Considering that the external Ca2+ block is only weakly voltage-dependent in the N619K/γ2 (Fig. 5J), and that the polyamine sensitivity is substantially altered in N619K/γ2 compared to WT/γ2 (Extended Data Fig. 10), as described above, there may be a weak voltage-dependent interaction between Ca2+ and the SF of N619K/γ2 contributing to the residual external Ca2+ block. Long-range allosteric coupling between the architectures surrounding the gate and the M211,20, which is adjacent to the SF, could potentially explain the mechanism by which the cytoplasmic polyamine block affinity is affected in the N619K mutant.
Importantly, the mutant N619K/γ2 receptors exhibited markedly reduced Ca2+ permeability compared to WT/γ2 receptors (Fig. 6 and Supplementary Table 8), with an estimated PCa2+/PNa+ ratio of 0.32 ± 0.04 (n=10) for N619K/γ2 receptors compared to 5.28 ± 0.21 (n=8) for WT/γ2 receptors (unpaired t-test, p-value < 0.001, Fig. 6D and Supplementary Table 8), which is consistent with the observation in GluA2 receptors expressed in the absence of auxiliary subunits24. The K619 side chains point away from the pore, their negatively charged ε-amino group (pKa=9.5) engaged by the carbonyl oxygens in the S785 residue within 3Å distance, and therefore unlikely to produce strong charge repulsion to the permeating cations (Fig. 4A). Positive charge at position 619 is unlikely to be the determinant because mutating N619 to a neutral cysteine also attenuates Ca2+permeability24. Collectively, we suggest that site-G is a Ca2+ binding site that is a key determinant of Ca2+ permeability in AMPARs.
Figure 6. Calcium block and permeation are affected by N619K mutation.
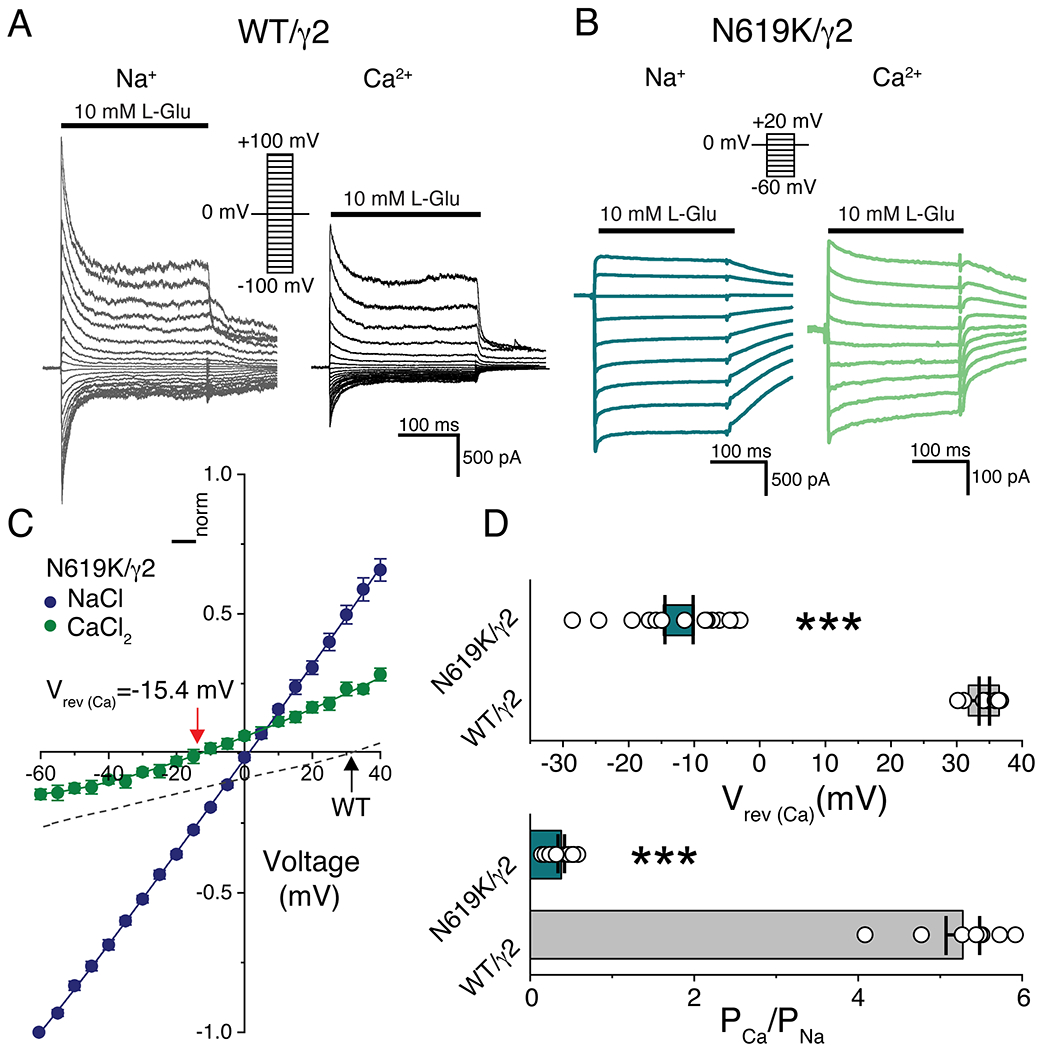
A and B. Example traces of responses from WT/;2 (grey in Na+, black in Ca2+) and N619K/γ2 (dark cyan in Na+, green in Ca2+) at various membrane potentials (in 10 mV increments) in absence of spermine. Receptors were activated with 10mM L-glutamate. The external solutions; Na+ = 150mM Na+ recording solution (patch # 190117p5 for WT/γ2, patch # 221124p2 for N619K/γ2), and Ca2+ = 108 mM Ca2+ recording solution (patch # 190117p5 for WT/γ2 and # 221124p2 for N619K/γ2). C. Current-voltage (I-V) plots of mutant N619K/γ2 receptors (n=10) in external 150 mM Na+ (blue circles) or 108 mM Ca2+ (green circles). In dashed black line it is shown IV plot of WT/γ2 receptors (n=8) in 108 mM Ca2+ (dotted line). Data are presented as mean values ± SEM. D. (Top) Scatter plot comparing the reversal potential in 108 mM Ca2+ from WT/γ2 and N619K/γ2. Grey (WT/γ2) and cyan (N619K/γ2) squares indicate average with SEM (Vrev = 34.1 ± 0.8 mV for WT/γ2, n=8; Vrev = −15.4 ± 2.3 mV N619K/γ2, n=10; unpaired two-sided t-test with Welch correction, p-value < 0.001). D. (Bottom) Comparison of the relative divalent permeabilities (PCa2+/PNa+) of WT/γ2 and mutant N619K/γ2 receptors (PCa2+/PNa+ =5.28 ± 0.21 for WT/γ2, n=8; PCa2+/PNa+ = 0.32 ± 0.04 for N619K/γ2, n=10) unpaired two-sided t-test with Welch correction, p-value < 0.001). (Also see in Supplementary Table 8)
Discussion
State dependent Ca2+ binding site critical for ion transport
In this work, we have presented high resolution cryo-EM structures of the open pore of the AMPAR in complex with the gain-of-function mutant of TARPγ2(KKEE). The difference observed in pore occupying densities with and without Ca2+ revealed a novel Ca2+ binding site (i.e., site-G) at the open gate that lies outside the membrane electric field (Fig. 7). Importantly, site-G is formed only when the channel enters the open state, making it a transient state-dependent Ca2+ binding site. Occupancy of Ca2+ at site G offers a mechanism whereby Ca2+ preferentially accumulates at the open gate to ensure Ca2+ entry to the pore and SF, even though the physiological extracellular concentration and diffusion coefficient of Ca2+ are 70-fold and 2-fold lower than those of Na+, respectively (Fig. 7). Increase in external Ca2+ concentration promotes Ca2+ occupancy at site-G that, in turn, blocks the passage of Na+ (Fig. 7). Furthermore, the function of site-G was disrupted by introducing the N619K mutation immediately next to site-G (Fig. 7). Collectively, site-G offers a structural explanation for the: (1) facilitation of Ca2+ permeation in CP-AMPAR, and (2) dose-dependent external Ca2+ block of AMPARs which is a new pharmacological observation.
Figure 7. Schematic model for the function of site-G.
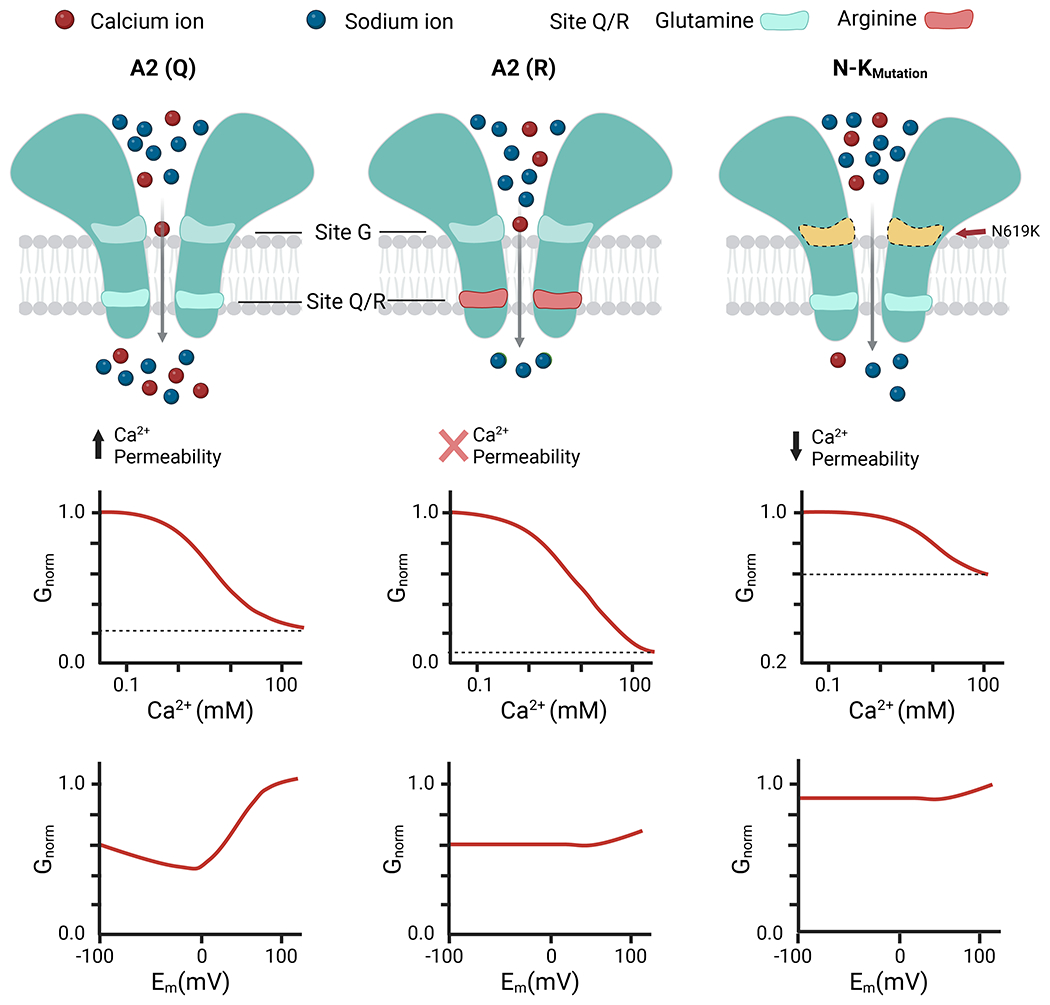
Transport of Ca2+ through the AMPAR pore requires both Site-G and the Q/R site. Site-G is at the channel gate outside the membrane electric field, while the Q/R site of the selectivity filter sits within the membrane electric field. (left) When Ca2+ binds to site-G it partially prevents the passage of Na+, causing external Ca2+ block of currents carried by Na+. Due to this arrangement, Ca2+ entry to the Q/R site is maintained even though physiological external Ca2+ concentration is about 50-fold lower than Na+. This distinction accounts for the efficient divalent permeability in GluA2-iQ (A2(Q)) channels. (middle) In GluA2-iR (A2(R)), Ca2+ can still access site-G but cannot permeate beyond the Q/R site. External Ca2+ block is present, and voltage-independent, consistent with site-G being located outside the membrane electric field. (right) For GluA2-iQ N619K mutant channels (N-K mutant), site-G can no longer bind Ca2+, and thus external Ca2+ block is weak. Attenuated site-G reduces the ability of Ca2+ to reach the Q/R site, ultimately leading to lower Ca2+ permeability.
Significance of extracellular vestibules in Ca2+ permeation
In AMPARs, gate-opening removes the physical obstruction for ion permeation9–11,20. This simple view is revised by our identification of site-G in CP-AMPARs. The gate-opening and ion binding is in fact coupled because the pore dilation not only creates a sieve for ions and water to pass through, but also reveals a surface that preferentially capture Ca2+ over Na+, which facilitates Ca2+ permeation. More generally, negatively charged surfaces exist at the extracellular vestibules of Ca2+-selective ion channels to efficiently capture extracellular Ca2+, such as seen in the L5-6 loops of calcium channel Cav1.125, extracellular vestibule of engineered Ca2+-selective bacterial ion channel CavAb26, and divalent cation recruitment site (D517, E518, and D547) in the Ca2+-selective transient receptor potential channel V6 (TRPV6)27. We suggest that site-G in the AMPAR/γ2 complex performs an analogous role as the Ca2+-accumulating extracellular vestibules found in other Ca2+-selective channels. Since the gate is highly conserved amongst all iGluR families (Fig. 2G), site-G may be critical in providing sufficient influx of Ca2+ through Ca2+ permeable iGluRs, such as CP-AMPARs and NMDA receptors (NMDAR), to trigger intracellular signaling28. In the case of NMDARs, dilation of the SF and additional Ca2+ binding sites are proposed to account for their greater Ca2+ permeability compared to AMPARs29–31.
Site-G and Q/R site are key determinants to Ca2+ permeation
Previous work in AMPARs emphasized the sole role of the Q/R site that sits at the top of the SF in determining Ca2+ permeability28. Accordingly, recent structural analysis and molecular dynamic simulation has focused on the role of the Q/R-site and SF in their modelling and theoretical framework32–34. However, site-G was not among the ion binding sites predicted by molecular dynamic simulations in the iGluR pore32,35, which highlights the importance of taking experimental approach to identifying pore ion binding sites (Note: Ca2+ binding Site 4 in reference32 differs from Site-G). In fact, previous experimental observations have implicated that factors outside of the Q/R-site and SF may be critical for Ca2+ permeation. First, N619K and N619C mutations in the extracellular vestibule of GluA2, which is adjacent to site-G, substantially attenuate Ca2+ permeability24, indicative of a structural element outside the Q/R-site crucial for Ca2+ permeability. Second, a hypothetical Ca2+ binding site in the upper pore has been proposed to explain for the external Ca2+ block in NMDARs36,37. The postulate was based on the observation that the block was insensitive to applied voltage and, therefore, the hypothetical Ca2+ binding site must be outside the membrane electric field. Subsequently, it was shown that the DRPEER motif, present only in the GluN1 subunit and located in the extracellular vestibule, plays a substantial role to facilitating Ca2+ permeability and captures putative Ca2+ before entering the pore31,38. However, disruption of DRPEER motif could not fully eliminate Ca2+ permeation, indicating the involvement of another Ca2+ binding site31. Our work offers an explanation to these finding by providing the structural basis for a novel Ca2+ binding site (i.e., site-G) at the vestibule of the AMPAR pore, which is located in the evolutionary-conserved SYTANLAAF motif and upstream of the well-established pore Q/R-site in the SF that determines Ca2+ permeability28 (Fig. 7).
Whether Ca2+ block of AMPARs fulfills a physiological role in regulating glutamatergic synapses is not immediately clear. Assuming an extracellular calcium concentration ([Ca2+]ex) of 2 mM, which is typical of brain slice experiments, AMPARs would be blocked by about 20%. Measurements have shown that in human brain the physiological [Ca2+]ex is ~1.2mM, lower than what is used in slice physiology experiments 39. The [Ca2+]ex decreases by 0.45mM in seizures or following high activity 40. These reported variations in [Ca2+]ex may impact the amplitudes of the AMPAR mediated response by less than ~10% through the external calcium block reported in this work, which does not offer a sizeable range for regulation. Perhaps a more plausible explanation is that the modest block exerted by physiological levels of Ca2+ is a necessary tradeoff to allow the selective transport of Ca2+ through the open pore of unedited AMPARs over the more abundant Na+ ions. This line of reasoning is consistent with our current understanding of NMDARs which are similarly blocked in a voltage-dependent manner by extracellular Ca2+ (see Figs 7 and 8 of 41) with the added distinction that they are also blocked by Mg2+ 42,43 which permits them to act as coincident detectors at glutamatergic synapses. By extension, we predict that the more abundantly expressed edited AMPARs, which do not transport Ca2+, also exhibit Ca2+ block, not because it plays a significant role in the selective transport of divalent ions, but rather by virtue of possessing an extracellular vestibule (i.e. site-G) which is common to all AMPARs.
Ca2+ recognition site outside the SF
The SF plays a central role in controlling ion transport in the cation permeating pore-loop ion channel family with four-fold (pseudo)symmetry44, which includes, potassium channels, calcium channels, sodium channels, cyclic nucleotide gated (CNG) channels, iGluRs, and TRP channels. In potassium channels, to achieve high selectivity to K+, the SF maintains an optimal architecture for dehydrated K+ but deforms into a non-permeating conformation with other cations14. In contrast, selectivity for Ca2+ in calcium channels is achieved by the negatively charged glutamic acid side chain in the SF that forms a high affinity Ca2+ binding site26. The presence of Ca2+ in the pore prevents Na+ from entering, while Na+ permeates efficiently in the absence of Ca2+, exhibiting anomalous mole-fraction effect15. The Ca2+ in the SF must be knocked-off by a charge repulsion from another Ca2+ entering the pore26. A similar mechanism is proposed for the Ca2+-selective TRPV6 channel27. The CNG channel is non-selective, permeable to both Ca2+ and Na+, but exhibit Ca2+ mediated channel block45,46, whose characteristic ion conduction properties is also supported by the mechanism within the SF47. In contrast, our observation in CP-AMPAR suggests that efficient Ca2+ transport requires the preferential capture of Ca2+ at the open gate in the upper pore prior to Ca2+ reaching the SF in the lower pore. We suggest that site-G is a weak Ca2+ binding site compared to those found in the SF of Ca2+-selective channels, because the surrounding side chains are neutral in site-G whereas in highly Ca2+-selective channels they are acidic. The Ca2+ remains hydrated at site-G but with only four water molecules resolved at the distance (2.4Å) of a primary hydration shell (Fig. 2I), which is consistent with a weak binding site. Indeed, the hydration of Ca2+ at site-G contrasts from the octameric oxygen coordination offered by four carboxylic acidic side chains in the high affinity Ca2+ binding site in the SF of Cav1.1, Cav3.1, and TRPV625,27,48. In the case of CavAb, it is proposed that the high affinity binding of Ca2+ to the SF is mediated indirectly by the hydration shell that consists of eight water molecules26, which agrees with the idea that coordination of Ca2+ by only four water molecule at site-G would form a weaker binding site.
Does Site-G have a shared function in the iGluR family?
Given the prevalence of the SYTANLAAF motif in all mammalian iGluR subfamilies, it may be envisaged that Site-G fulfills a common role in Ca2 permeation. However, site-G may not convert to a Ca2+ binding site in all cases, based on the precedence of de-coupling between gating and Ca2+ binding observed in the N619K mutant (Fig. 7). We predict Ca2+ binding to site-G, and thus the coupling, is less efficient in a subset of invertebrate iGluRs that encode lysine (K) at the position equivalent to N619 in rat GluA249. The efficacy of open gate converting to an efficient Ca2+ binding site may depend on how close the open conformation approaches those that are optimal for Ca2+ binding. We suggest that the exact open gate architecture, and by implication the composition of subunits and dynamics associated with gating, determines whether site-G converts to an efficient Ca2+ binding site. In this context, gate-opening and Ca2+ binding may not be coupled at site-G in kainate-type iGluRs (KARs), given its low divalent permeability. The NMDARs have greater Ca2+ permeability than CP-AMPARs and the gate of both receptors is encoded by the identical amino acid sequence (SYTANLAAF). Thus, given that NMDARs are subject to external Ca2+ block that may be mediated by a Ca2+ binding site in the upper pore, as discussed above, it seems natural to presume that the gate of NMDARs would adopt a conformation similar to site-G with efficient Ca2+ binding capacity. Taken together, future work is necessary to solve the open state structure of KARs and examine whether alternative conformations exist for the open gate of NMDARs other than those that are currently known50,51.
In conclusion, we have identified that the gate of the AMPAR contains a transient Ca2+ binding site, site-G, that is revealed only in the open state. The evidence for ion recognition and regulation of ion permeation mediated by site-G, outside of the selectivity filter, advances our understanding on the nature of divalent cation permeation in AMPARs as well as non-selective cation channels, in general.
Methods
Expression and purification
The cDNA construct of rat GluA2flipQ isoform was tagged with a FLAG epitope near its C-terminal, as described52,53. Rat TARPγ2 (stargazin) KKEE mutant construct was generated as described16 and a 1D4 tag was fused at the very C-terminal. The two cDNAs were cloned into the DualTetON plasmid54 to generate a plasmid named DualTetON-A2iQFLAG-KKEE1D4, which DOX dependently expresses both proteins simultaneously. The two proteins were co-expressed without using any tether (Extended Data Fig. 1A). A stable TetON HEK cell line was generated by co-transfecting DualTetON-A2iQFLAG-KKEE1D4 and a plasmid that confers hygromycin (hyg) resistance, using established method52–54. Clone was isolated in the presence of 30μM NBQX and 120μg/ml hyg. Clone#53 was chosen, based on its growth rate and expression level of the complex, and adapted to FreeStyle293 media (Gibco/ThermoFisher) in suspension.
The N619K mutation was introduced using site directed mutagenesis to the wild type GluA2flipQ construct. DualTetON-A2iQ(N619K)FLAG-KKEE1D4 plasmid was used to generate a stable cell line (clone#8) that co-expresses GluA2flipQ(N619K)-FLAG and TARPγ-2 (stargazin) KKEE-1D4, DOX dependently. The expression level of the mutant complex was comparable to the WT complex. The biochemical property was indistinguishable from WT, and thus identical purification protocol was used for both complexes.
Approximately 1.2L to 2.4L of near saturating suspension culture of clone#53 (or clone#8 in the case of N619K) in FreeStyle293 media supplemented with 30μM NBQX and 1:500 diluted anti-clumping agent (Gibco, ThermoFisher, Cat#0010057DG) was used as a starting material for each experiment. Cells were induced with 7.5γg/ml DOX, 1mM sodium butylate, and 1%FCS for 28hrs as described53. Following procedures were conducted on ice or at 4ΰC. Cells were centrifuged at 931xg for 10 min, washed with D-PBS once, centrifuged again, and the pellet flash frozen in liquid nitrogen for storage. Approximately 10-12ml of frozen pellets was resuspended in Resuspend buffer (25mM TrisHCl pH8.0, 150mM NaCl, 2mM TCEP, 15μM NBQX, and protease inhibitors: 1mM PMSF, 10μg/ml aprotinin, 0.5mM benzamidine, 1μg/ml pepstatin A, 5μg/ml leupeptin), making the final volume 90ml. 10ml of 10xdigi (25mM TrisHCl pH8.0, 150mM NaCl, 7.5% digitonin) was added and the mixture was nutated at 4ΰC for 2.5hr to dissolve the membrane. The large debris was removed by low-speed centrifuge (3000rpm, 10min, at 4ΰC) and its supernatant was ultracentrifuged at 235,400xg in 45Ti rotor (Beckman) for 1hr. The resulting supernatant was incubated in batch with 1ml of FLAG M2 agarose beads (Sigma) for 2 hr. The beads were collected by centrifugation at 58xg for 5min and transferred into an empty column. The beads were washed for 6 column volumes with wash buffer (0.03% GDN, 20mM TrisHCl pH8.0, 150mM NaCl). The proteins were eluted using 6ml of wash buffer containing 0.5mg/ml FLAG peptide. The eluate was concentrated down to 0.55ml using Ultrafree 100KDa MWKO ultrafiltration (Millipore). The concentrated sample was ultracentrifuged at 75,325xg for 15 min and applied to Superdex200 Increase column (GE Healthcare) equilibrated with GF buffer (For Open-Na260; 0.03% GDN, 20mM TrisHCl pH8.0, 150 mM NaCl. For Open-Na610; 0.03% GDN, 20mM TrisHCl pH8.0, 500 mM NaCl. For Open-CaNaMg, Closed-CaNaMg, and Open-CaNaMg(N619K); 0.03% GDN, 20mM TrisHCl pH8.0, 150 mM NaCl, 10mM CaCl2, 1mM MgCl2. For Open-Ca150; 0.03% GDN, 20mM TrisHCl pH8.0, 150 mM CaCl2. For Open-Na110 and Open-Ca10; 0.03% GDN, 20mM TrisHCl pH8.0, 140 mM NMDG). 1M NMDG was buffered with HCl to pH8.0 and used as a stock solution to prepare other buffers. The isocratic elution (0.5ml per fraction) was conducted at flow rate 0.5 ml/min (Extended Data Fig. 1C). The peak fractions were combined and concentrated down to 30μl using Ultrafree 100KDa MWKO ultrafiltration. Purity was checked by SDS-PAGE (Extended Data Fig. 1B). To prepare Open-Ca10, complex prepared in NMDG was mixed with 1M CaCl2 at 9:1 ratio. The final protein concentration ranged from 7mg/ml to 14mg/ml.
Grid preparation
To prepare samples in the open states, one volume of proteins was mixed with 0.1 volume of 3.3mM CTZ for 30min and, before freezing, 0.1 volume of 1M L-glutamate (pH7.4) was added. The 1M L-glutamate solution contains 1.1M NaOH, thus the final concentration of Na+ increases by 110mM after adding the agonist to the purified receptors. To prepare samples in the closed state, one volume of proteins was mixed with 0.1 volume of 3.3mM CTZ and 0.1 volume of 100mM CNQX disodium salt (Tocris) and incubated for 15min prior to freezing.
2μl of protein solution was applied to Quantifoil1.2/1.3 (300 mesh, Cu grid, holy carbon membrane) and plunged into liquid ethane using Vitrobot Mark4 (ThermoFisher). The standard parameters were, blot force=12, blot time=4.5 sec, temp=4 ΰC, humidity=100%, wait time=30sec, and drain time=none. The samples containing 140mM NMDG or greater than 500mM NaCl were more viscous than the others, so the filter papers were doubled, and blot time was extended to 6~7 sec. Optimal freezing condition were determined by inspecting the grids using TF20 (FEI/ThermoFisher) or Glacios (ThermoFisher).
Cryo-EM imaging
All data was collected using TitanKriosG4i (ThermoFisher) equipped with BioQuantumK3 detector at Vanderbilt University cryo-EM facility. Images were collected at 50 frames/movie. Aberration free image shift function was used in EPU (ThermoFisher) semiautomated data collection software. The microscope is equipped with fringe free optics which enables smaller beam diameter for imaging. Objective aperture was not used. Data collection was completed from one grid in a single imaging session that lasted for 3.5 to 5 days. Detector dose rate was at 15.3-15.7e−/pix/sec (measured over ice). Total dose was at 50-56 e−/Å2 (measured over vacuum). Each movie contained 50 frames. At 105kx, 2 shots per hole were taken. At 130kx, 3-4 shots per hole were taken. A representative raw motion corrected micrograph of Open-Na260 is shown in Extended Data Fig. 1D. Detailed parameters used for data collection in each sample are summarized in Table 1 and Supplementary Table 1.
Common image processing pipeline
The image processing of each dataset follows a common pipeline. Minor modifications were made when necessary. All image processing in the current work was done using Relion355. Each raw movie stack (50 frames) was motion corrected (at 4x4 patches) and dose weighted using Motioncorr256. CTFFIND4 was used to estimate CTF from non-dose weighted images using 1024x1024 pixel tiles57. All dataset produced excellent statistics in rlnCtfMaxResolution and rlnCtfFigureOfMerit. C2 symmetry was imposed only at the last step of 3D refinement. The NTD, which is flexible relative to the rest of the architecture5, was included in the 2D and initial 3D classification, but was outside the mask used for the focused classification and refinement in 3D. EMD-2680 was used as initial model. The LBD adopted multiple conformations in all datasets except for the Open-Ca150, which was noticeable as ill-defined LBDs when the particles were aligned using the mask containing LBD-TMD and TARPγ-2(KKEE). The alignment was guided towards improving the resolution of the membrane embedded region at the cost of degrading the alignment of the LBD, because the former contains many bundles of alpha helices that generate strong signals. The heterogeneity of LBD conformations was resolved by further focused classification and refinement imposing C2 symmetry and using the LBD mask, which contains the LBD gating ring (the tetrameric assembly of the LBD). The particles contained in the final refinement of the TMD-STG were used as input to the LBD classification. The differences between the LBD conformations were characterized as translation and rotation between the two LBD dimers (Extended Data Fig. 5), which were small conformational differences in the organization of the LBDs in the gating ring9. When necessary, the composite maps made of LBD map plus TMD-STG map were generated using EMDA58. The local resolution of the map was estimated using Relion3 and ResMap59. The overall resolutions of the maps were estimated by FSC=0.143 cutoff in Relion60 (Extended Data Fig. 2). The image processing and model statistics are summarized in Table 1 and Supplementary Table 1. Angular distributions of assigned angles were inspected to ensure the coverage of the Fourier space. Visual inspection of the map showed no signs of artifacts.
Image processing of Open-Na260
The workflow is summarized graphically in the Extended Data Fig. 1F, which follows the general procedure described above. 5,409,247 particles were selected using Autopick with templates obtained from 2D class averages generated from manually picked ~3000 particles. Particles were extracted at box size 360pixel and reduced to 64pixel. The extracted particles were divided into four subsets. Each subset was subject to 2D classification, specifying 160 classes (Extended Data Fig. 1E). After two rounds of 2D classification, 2,110,000 particles contributed to generating well-defined class averages that contain features of AMPARs and were subject to further 3D classification. For 3D classification, particles were re-extracted at box size 360pixel and reduced to 180pixel, resulting in a binning factor of 2. Particles were classified into 6 classes without mask and without imposing symmetry (C1) for 40 iterations. Additional 20 iteration was conducted by applying a mask covering the LBD and TMD (LBD-TMD) with regularization parameter at T=15. Three well-defined classes (class3, 4, and 6) were selected, totaling 1,240,685 particles (Extended Data Fig. 1F1).
The particles combined from the 3 classes were re-extracted at box size 360pixel without binning. After CTF refinement, Refine3D and PostProcess were conducted again followed by Bayesian polishing. Shiny particles were subject to a second round of CTF refinement. Duplicate particles were removed, a total of 1,093,405 particles proceeding to final Refne3D. Refinement and PostProcess were first conducted with C1 (no symmetry) using the TMD-STG mask, that covers the TMD of GluA2 and TARPγ-2(KKEE), generating a map with overall resolution of 2.38Å. LBD-TMD mask was also used to generate a 2.50Å overall resolution map, whose LBDs were ill-resolved due to conformational heterogeneity. The final refinement, Refine3D and PostProcess, were done, imposing C2, using the TMD-STG mask resulting in a map with overall resolution of 2.29Å. The angular distributions of the assigned angles show that the side views were the most abundant (Extended Data Fig. 1F2).
To resolve the heterogeneity of the LBDs, the particles were classified into 4 classes (T=50) using a mask that covers the LBD gating ring (LBD mask) (Extended Data Fig. 1F3). Three classes (Class 1, 2, and 4, which will be referred to as conf1, 2, and 3, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, 2, and 3 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 3.01Å, 3.43Å, and 3.04Å, respectively (Extended Data Fig. 1F4). The angular distributions of the assigned angles were very similar in L1-3 and consistent with the distribution of the consensus reconstruction described above, indicating that the classification of the LBD is based on conformation rather than angular distribution.
The 3D classification and refinement from the binned images (boxsize=180pixels) resulted in a 4.4Å map containing the NTD. The tetrameric NTD, organized in dimer-of-dimers, was positioned in the asymmetric (AS) orientation relative to the rest of the particle as described previously53 and no classes adopted pseudo symmetric (PS) orientation. Atomic models of the LBD, TMD, TARPγ-2(KKEE) generated below and the GluA2-NTD (PDB:6U6I) were rigid body fit into the 4.4Å map to generate Fig. 1B.
Image processing of Open-CaNaMg
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. ~5,000,000 particles were selected using Autopick with templates obtained from 2D class averages generated from manually picked ~3000 particles. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were divided into four subsets. Each subset was subject to 2D classification, specifying 160 classes. After two rounds of 2D classification, ~1,200,000 particles contributed to generating well-defined class averages and were subject to further 3D classification. For 3D classification, particles were re-extracted at box size 456pixel and reduced to 180pixel. Particles were classified into 6 classes without mask and without imposing symmetry for 37 iterations with regularization parameter at T=6. Four well-defined classes (class2=235,679particles, class3=217,442particles, class5=130,488particles, and class4=238,429particles) were selected, totaling 822,038 particles. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the TMD-STG mask resulted in a reconstruction of an overall resolution of 2.61Å. After CTF refinement, Refine3D and PostProcess were conducted again, producing a 2.63Å map (the resolution was reduced but map quality improved). The particles were subject to Bayesian polishing, training followed by polishing. Refine3D and PostProcess produced a map at overall resolution of 2.59Å. LBD-TMD mask was also used to generate a 2.63Å overall resolution map, whose LBDs were ill-resolved due to conformational heterogeneity. The final refinement, Refine3D and PostProcess, were done, imposing C2, using the TMD-STG mask, producing a map with overall resolution of 2.44Å.
To resolve the heterogeneity of the LBDs, the particles were classified into 4 classes (T=50) using the LBD mask for 40 iterations. Two classes (class2=204,515particles, and class3=210,811particles which will be referred to as conf1 and 2, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, and 2 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 2.78Å and 3.14Å, respectively.
Image processing of Open-Na610
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. About 5,100,000 particles were selected using Autopick with templates used for Open-Na150. Particles were extracted at box size 360pixel and reduced to 64pixel. The extracted particles were divided into four subsets, and each subset was subject to 2D classification, specifying 160 classes. Well-defined classes selected during the first round was re-extracted and re-scaled to 180pixel box size. After the second round of 2D classification, ~2,285,000 particles contributed to generating well-defined class averages and were subject to further 3D classification. For 3D classification, particles were re-extracted at box size 456pixel and reduced to 180pixel. Particles were classified into 6 classes without mask and without imposing symmetry for 36 iterations with regularization parameter at T=4. Then, the classification was continued up to 60 iterations using the LBD-TMD mask with T=15. Three well-defined classes (class1=244,459particles, class3=300,808particles, and class4=355,887particles) were selected, totaling 901,154 particles. The particles were re-extracted at box size 360pixel without rescale. Refine3D and PostProcess using the LBD-TMD mask was conducted (C1) for 21 iterations, generating a reconstruction of an overall resolution of 2.79Å. The focused refinement was then continued from iteration 19 using the TMD-STG mask, generating a reconstruction of an overall resolution of 2.59Å. After CTF refinement, Refine3D and PostProcess were conducted (C1) again in two steps using both masks, producing maps of overall resolution of 2.66Å and 2.61Å with the LBD-TMD mask and TMD-STG mask, respectively. The resolution was reduced but map quality improved by the CtfRefine. The particles were subject to Bayesian polishing, training followed by polishing. The Refine3D and PostProcess produced C1 maps of overall resolution of 2.57Å and 2.50Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask, the LBDs were ill-resolved due to conformational heterogeneity. The final focused refinement, Refine3D and PostProcess, were done, imposing C2, in two steps using both the LBD-TMD mask and TMD-STG mask, resulting in maps with overall resolution of 2.46Å and 2.40Å, respectively.
To resolve the heterogeneity of the LBDs, the particles were classified, imposing C2, into 4 classes (T=50) using the LBD mask for 40 iterations. Three classes (class1=207,914particles, class2=220,191 particles, and class4=299,853particles, which will be referred to as conf1, 2 and 3, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, 2, and 3 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 3.08Å, 2.98Å, and 3.01Å, respectively.
Image processing of Open-Ca150
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. ~5,573,000 particles were selected using Autopick with templates used in Open-CaNaMg. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were divided into four subsets. Each subset was subject to 2D classification, specifying 160 classes. After 2D classification, ~1,640,000 particles contributed to generating well-defined class averages and were subject to further 3D classification. For 3D classification, particles were re-extracted at box size 456pixel and reduced to 180pixel. Particles were classified into 3 classes without mask and without imposing symmetry for 40 iterations with regularization parameter at T=4. Then the classification was continued up to 60 iterations using the LBD-TMD mask with T=15. The well-defined class1, containing 540,796particles, was selected. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the LBD-TMD mask was conducted (C1) and converged in 24 iterations, generating a reconstruction of an overall resolution of 3.04Å. The focused refinement was then continued from iteration 13 using the TMD-STG mask, generating a reconstruction of an overall resolution of 2.86A. After CTF refinement, Refine3D and PostProcess were conducted (C1) again in two steps using both masks, producing maps of overall resolution of 2.95Å and 2.84Å with the LBD-TMD mask and TMD-STG mask, respectively. The particles were subject to Bayesian polishing, training followed by polishing. The Refine3D and PostProcess produced C1 maps of overall resolution of 2.84Å and 2.78Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask the LBDs were ill-defined. The final focused refinement, Refine3D and PostProcess, were done imposing C2 in two steps using both the LBD-TMD mask and TMD-STG mask, resulting in maps with overall resolution of 2.73Å and 2.58Å, respectively. The overall yield of particle and resolution was lower than the other dataset, possibly due to stronger beam induced motion which may be caused by using the 150mM CaCl2.
To resolve the heterogeneity of the LBDs, the particles were classified, imposing C2, into 3 classes (T=50) using the LBD mask for 40 iterations. One classes (class3=192,490particles) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. Refine3D and PostProcess, produced maps with overall resolutions of 3.14Å.
Image processing of Open-Na110
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. ~8,603,000 particles were selected using Autopick with templates used in Open-CaNaMg. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were divided into six subsets. Each subset was subject to 2D classification, specifying 160 classes. After two rounds of 2D classification, ~2,892,000 particles contributed to generating well-defined class averages and were subject to further 3D classification. For 3D classification, particles were extracted at box size 456pixel and reduced to 180pixel, and classified into 3 classes without mask and without imposing symmetry (C1) for 18 iterations with regularization parameter at T=4. The classification continued with the LBD-TMD mask up to 40 iterations with T=20. One well-defined class was selected, totaling 692,680 particles. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the LBD-TMD mask and TMD-STG mask resulted in C1 maps of overall resolution of 2.59Å and 2.46Å, respectively. After CTF refinement, Refine3D and PostProcess was conducted (C1) again in two steps using both masks, producing maps of overall resolution of 2.59Å and 2.52Å with the LBD-TMD mask and TMD-STG mask, respectively. The particles were subject to Bayesian polishing, training followed by polishing.
The Refine3D and PostProcess produced C1 maps of overall resolution of 2.46Å and 2.42Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask the LBDs were ill-defined. The final focused refinement, Refine3D and PostProcess, were done, imposing C2, in two steps using both the LBD-TMD mask and TMD-STG mask, resulting in maps with overall resolution of 2.38Å and 2.34Å, respectively.
To resolve the heterogeneity of the LBDs, the particles were classified into 3 classes (T=50) using the LBD mask for 40 iterations. Two classes (class2=189,181particles, and class3=218,818particles which will be referred to as conf1 and 2, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, and 2 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 3.11Å and 3.35Å, respectively.
Image processing of Open-Ca10
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. ~7,059,000 particles were selected using Autopick with templates used in Open-CaNaMg. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were divided into four subsets. Each subset was subject to 2D classification, specifying 160 classes. After two rounds of 2D classification, ~1,952,000 particles contributed to generating well-defined class averages that contain features of AMPARs and were subject to further 3D classification. For 3D classification, particles were extracted at box size 456pixel and reduced to 180pixel, and classified into 3 classes without mask and without imposing symmetry (C1) for 31 iterations with regularization parameter at T=4. The classification continued with the LBD-TMD mask up to 50 iterations with T=20. One well-defined class was selected, totaling 986,075 particles. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the LBD-TMD mask and TMD-STG mask resulted in C1 maps of overall resolution of 2.56Å and 2.46Å, respectively. After CTF refinement, Refine3D and PostProcess were conducted (C1) again in two steps using both masks, producing maps of overall resolution of 2.59Å and 2.56Å with the LBD-TMD mask and TMD-STG mask, respectively. The particles were subject to Bayesian polishing, training followed by polishing. The Refine3D and PostProcess produced C1 maps of overall resolution of 2.52Å and 2.48Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask the LBDs were ill-defined. The final focused refinement, Refine3D and PostProcess, were done, imposing C2, in two steps using both the LBD-TMD mask and TMD-STG mask, resulting in a map with overall resolution of 2.44Å and 2.36Å, respectively.
To resolve the heterogeneity of the LBDs, the particles were classified into 4 classes (T=50) using the LBD mask for 40 iterations. Two classes (class 1=215,389particles, and class 2=278,786particles which will be referred to as conf1 and 2, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, and 2 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 3.01Å and 3.14Å, respectively.
Image processing of Open-CaNaMg/N619K
The common pipeline was followed with minor modification during the first 3D classification. Motion correction and CTF estimation was done as described. 2D class averages were generated from 109,101 particles that were selected using Autopick with the template used in Open-CaNaMg. A subset of well-defined class averages was further used as templates to select ~7,006,000 particles using Autopick from all micrographs. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were divided into five subsets. Each subset was subject to 2D classification, specifying 160 classes. After two rounds of 2D classification, ~1,957,000 particles contributed to generating well-defined class averages and were subject to further 3D classification. For 3D classification, particles were re-extracted at box size 456pixel and reduced to 180pixel. Particles were classified into 6 classes without mask and without imposing symmetry (C1) for 40 iterations with regularization parameter at T=4. The classification continued with the LBD-TMD mask up to 60 iterations with T=15. Two well-defined classes (class1=357,587particles and class5=336,348particles) were selected, totaling 693,935 particles. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the LBD-TMD mask and TMD-STG mask resulted in C1 maps of overall resolution of 2.61Å and 2.44Å, respectively. After CTF refinement, Refine3D and PostProcess were conducted (C1) using TMD-STG mask, producing a map of overall resolution of 2.61Å. The particles were subject to Bayesian polishing, first the training and then the actual polishing. The Refine3D and PostProcess produced C1 maps of overall resolution of 2.59Å and 2.50Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask the LBDs were ill-defined. The final focused refinement, Refine3D and PostProcess, were done, imposing C2, in two steps using both the LBD-TMD mask and TMD-STG mask, resulting in maps with overall resolution of 2.48Å and 2.38Å, respectively.
To resolve the heterogeneity of the LBDs, the particles were classified into 4 classes (T=50) using a mask that covers the LBD gating ring (LBD mask) for 40 iterations. Two classes (class 1=203,784particles, and class 3=210,604particles which will be referred to as conf1 and 2, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, and 2 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 3.14Å and 3.17Å, respectively.
Image processing of Closed-CaNaMg
The common pipeline was followed with minor modification during the first 2D and 3D classification. The data processing was initiated for the first 14,999 micrographs when it became available during the session after the first two days of data collection. The remaining 19,554 micrographs were processed upon completion of the entire session. As described above, the entire dataset derives from single grid, and thus splitting the data into two batches is for the purpose of initiating data processing sooner, and to evaluate if prolonged imaging of the identical grid would cause any deterioration to the image quality, which turned out to be no issues as described in the following. Motion correction and CTF estimation was done as described for Open-Na260. ~3,900,000 particles (batch1) and ~4,600,00 particles (batch2) were selected using Autopick. Particles were extracted at box size 456pixel and reduced to 64pixel. The extracted particles were subject to 2D classification, resulting in ~1,020,000 particles (batch1) and ~1,007,00 particles (batch2) contributing to generating well-defined class averages that contain features of AMPARs. Each batch was subject to identical 3D classification, refinement, and PostProcess separately and produced maps of the TMD-STG with similar overall resolution of 2.66Å (Batch1) and 2.56Å (Batch2).
Particles of two batches after 2D classification were combined (~2,027,000 particles) and re-extracted at box size 456pixel and re-scaled to 180pixel. Particles were classified into 6 classes without mask and without imposing symmetry (C1) for 40 iterations with regularization parameter at T=6. Two well-defined classes (class3=484,918particles and class4=362,460particles) were selected, totaling 847,378 particles. The particles were re-extracted at 456pixel and rescaled to 360pixel, which results in apix=0.819Å/pix equivalent to the apix in Open-Na260. Refine3D and PostProcess using the STG-TMD mask resulted in C1 maps of overall resolution of 2.57Å. After CTF refinement, Refine3D and PostProcess were conducted (C1) using TMD-STG mask, producing a map of overall resolution of 2.59Å. The particles were subject to Bayesian polishing, training followed by polishing. The Refine3D and PostProcess produced C1 maps of overall resolution of 2.52Å and 2.42Å, using the LBD-TMD mask and TMD-STG mask, respectively. In the map generated using the LBD-TMD mask the LBDs were ill-defined. The final focused refinement, Refine3D and PostProcess, were done, imposing C2, using the TMD-STG mask, resulting in a map with overall resolution of 2.32Å.
The heterogeneity of the LBDs was resolved by classifying the particles into 4 classes (T=50) using LBD mask for 40 iterations. Two classes (class 1=215,404particles, and class 3=262,250particles which will be referred to as conf1 and 2, respectively) produced well-defined map beyond 3.5Å resolution. The remaining class was ill-defined, indicating substantial heterogeneity of conformations. The LBDs of conf1, and 2 were subject to Refine3D and PostProcess, producing maps with overall resolutions of 2.76Å and 2.81Å, respectively.
Modeling
The reference model, PDB 5weo, was divided into domains and rigid body fit into the EM density map using Chimera61. The fit was further adjusted using the jiggle fit function in Coot62. Further manual adjustment with real space refine zone function in Coot was used to generate an atomic model. H2O was modeled manually to densities that are in hydrogen bond distance to potential acceptors and doners. The pore occupying densities in the Open-Na110 (i.e., containing 140mM NMDG+ and 110mM Na+) were interpreted to be all water. Although some may be Na+ ions, we did not have sufficient experimental basis to assign them as ions. In addition, none of the densities were large enough to accommodate NMDG. The Open-Na110 provided the reference to find waters in other structures. In fact, many waters were found within 1.2Å distance from where they were found in the Open-Na110 (for example see Extended Data Fig. 8). In addition, the knowledge on their average intermolecular distance (2.8Å) and average neighbors (4.4) were used when a network of H2O was observed 15. Validation was conducted using coot. The generated model was further refined using the real_space_refine tool in Phenix63. Real space refinement was conducted by imposing secondary structure restrains by annotating helices and sheets in the PBD file. To prevent overfitting of the models into the density, refinement was run for five cycles with strict geometric restraints of 0.005-0.01 for bond length and 0.5-1 for bond angle while non-crystallographic symmetry (NCS) consistent with C2 symmetry of the map was imposed when necessary. With the default restraints library provided by real_space_refine, the distances between Ca2+ and nearest water was in the range of 2.5-2.6Å. The refinement outcome was virtually identical when custom geometry restraints of target distance 2.39±0.1Å was imposed21, which resulted in 2.4Å spacing between Ca2+ and nearest water. Ca2+ was modeled and refined without conflict at site-G in Open-Ca10, Open-CaNaMg, and Open-Ca150. In addition, model refinement was successful when Ca2+ was placed at site-Q586/7 in Open-Ca150. FCS of model vs map was output by Mtriage. Cryo-EM validation tools MolProbity and Mtriage were used for validation (Table 1 and Supplementary Table 1). The maps containing both the LBD and the TMD-STG were interpreted by rigid body fitting the atomic models of individual fragments. To interpret the connection between the fragments, composite maps were used. Pymol (Schrödinger) and Chimera61 were used to further analyze the structure and generate figures. The pore radius accessible to solvent was estimated using CHAP software64.
Multiple sequence alignment
Clustal Omega65 was used to align the iGluR sequences from various species.
Cell culture and transfection for electrophysiology
HEK293T-TetON cells were maintained in DMEM high glucose containing glutamine and without sodium pyruvate. The media was supplemented with 10% fetal bovine serum (Gibco) and 1% Penicillin and Streptomycin stock solution (Gibco, 10.000 units/ml of penicillin and 10.000 μg/mL of streptomycin). Cells were plated at low density (1.6 × 104 cells/ml) on poly-D-lysine–coated, 35-mm, plastic dishes and were transiently transfected 48 h later using the calcium phosphate technique, as previously described66. To promote synthesis of GluA2(Q)-γ2, GluA2(R)- γ2, GluA2(Q)(N619K)-γ2, all flip isoforms, encoded in the plasmids, doxycycline was added approximately 24 hours before performing experiments at a final concentration of 2γg/ml.
Electrophysiology
Experiments were performed 24 hours after adding doxycycline to transfected cells. Agonist or antagonist solutions were rapidly applied to outside-out patches excised from transfected cells using a piezoelectric stack (Physik Instrumente). Solution exchange (10–90% rise time of 250–350 μs) was determined by measuring the liquid junction current at the end of an experiment. All recordings were performed using an Axopatch 200B (Molecular Devices) with thick-walled, borosilicate glass pipettes (3–6 MΩ) coated with dental wax to reduce electrical noise. Series resistance (3–12 MΩ) was compensated by 95%. To study calcium block in AMPA receptors voltage ramps were recorded at range of holding potentials from −100 to +100 mV. Data acquisition was performed using pClamp10 software (Molecular Devices) and was tabulated using Excel (Microsoft Corp.). All experiments were performed at room temperature.
All chemicals were purchased from Sigma-Aldrich, unless otherwise indicated. The external solution contained (in mM) 150 NaCl, 5 HEPES, and 0.1 CaCl2, at pH 7.3–7.4. The internal solution contained (in mM) 115 NaCl, 10 NaF, 5 HEPES, 5 Na4BAPTA (Life Technologies), 1 MgCl2, 0.5 CaCl2, and 10 Na2ATP, pH 7.3–7.4. The osmotic pressure of all solutions was adjusted to 295–300 mOsm with sucrose, if necessary. For polyamine block experiments, instead of 10 mM Na2ATP, 30 μM spermine was included in the internal solution. For calcium block experiments, external Ca2+ was added into 150 mM NaCl to give a final concentration of either 0.3, 1, 2, 5, 10, 20 or 30 mM, except for the highest (108 mM) Ca2+ solution which contained only 108 mM CaCl2 (+ HEPES) with no added NaCl. In all cases, cyclothiazide was added at a final concentration 100 μM to the control and agonist external solutions to reduce receptor desensitization. Currents recorded in control solution were used to leak-subtract experimental records. For calcium permeation experiments (Fig. 6), high concentration external Ca2+ contained (in mM) 108 CaCl2 and 5 HEPES (in mM) at pH 7.3–7.4 (pH adjusted with Ca (OH)2).
Concentrated (10×) agonist stock solutions were prepared by dissolving L-glutamate in the appropriate external solution and adjusting the pH to 7.3–7.4 and were stored frozen at −20°C. Stocks were thawed on the day of the experiment and used to prepare agonist-containing external solutions.
Single permeant blocker model for external Ca2+ and internal polyamines
Conductance-voltage (G-V) relationships of block by external Ca2+ or internal polyamines were fit using Origin Pro 2020 (OriginLab) with the following equation from 22:
| eq. 1 |
Kd is the dissociation constant at 0 mV and it is defined as:
| eq. 2 |
And redefined as shown:
| eq. 3 |
Where h and k represent the voltage dependence of g (koff/kon) and L (kperm/kon), respectively.
Calcium permeability
I-V relationships were used to calculate reversal potentials in 150mM external Na+ and in 108 mM external Ca2+ solutions. Fittings were calculated with a polynomial function with order of 4. The x-intercept of the polynomial fit was determined to be the reversal potential (Vrev). Ca2+ permeability relative to Na+ ratio (PCa/PNa) was calculated by the equation:
| eq. 4 |
Where PCa and PNa are the permeability coefficients for Ca2+ and Na+, respectively. F is Faraday’s constant, R is the gas constant, and T is the temperature in Kelvin.
Kinetics
Gating properties of AMPA receptors was assessed by applying a single voltage ramp (40 ms) from 0mV to -100 mV to assess the leak current and then followed by 250 ms application of L-glutamate (10 mM) to measure the steady-state equilibrium of agonist-induced receptor desensitization and the time constant of deactivation (indicated as off kinetics in Fig. 5). Off kinetics (τoff) were fit using a mono- or bi-exponential function as described previously67 and presented as weighted means.
Inhibition plots of external Ca2+ block
Dose-inhibition curves of external Ca2+ block were fit using a single binding site isotherm function as described previously68:
| eq. 5 |
Where IC50 is the concentration of external Ca2+ that elicits half-maximal block, N is the slope and Gmin is the steady-state response.
Statistics
Results are expressed as mean ± SEM. Values for n refer to the numbers of individual patches. Statistical analysis was performed using Origin Pro 2020 (Originlab). Two-tailed unpaired t-test with Welch correction was performed on the populations where normal distribution assumption could not be rejected (Shapiro-Wilk analyses, ps > 0.15, Fig 5E, F and Fig 6D). Mann-Whitney U-test was performed on the population that was not normally distributed (Fig 5D). ps < 0.05 were considered to be statistically significant. *** indicates ps<0.001.
All data were illustrated using Origin Pro 2020 and Adobe Illustrator CS5.
Extended Data
Extended Data Fig. 1. Expression, purification, and cryo-EM data processing.
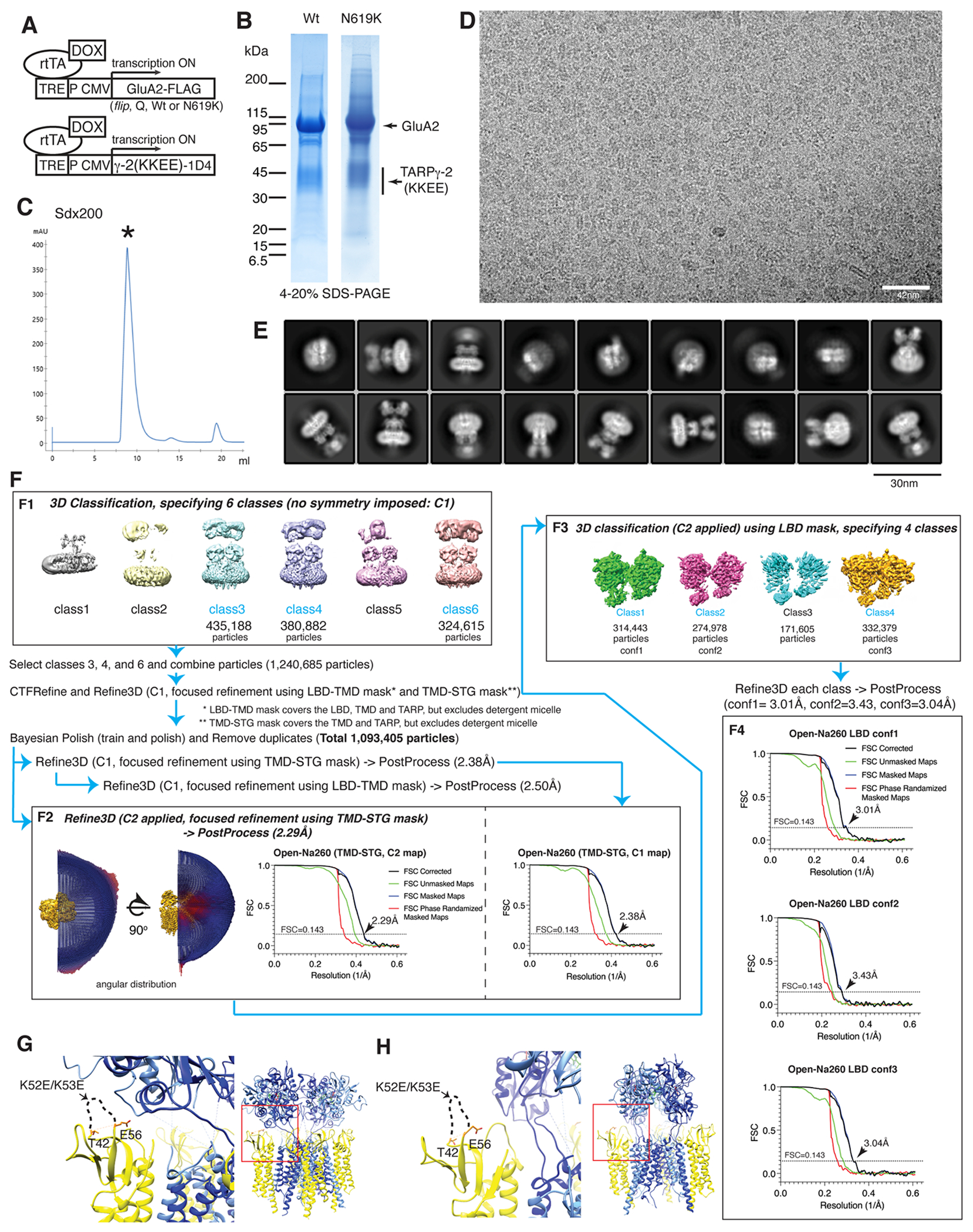
A. DOX inducible expression scheme used in stable HEK cell line. In clone#53, GluA2(flip-Q isoform) and TARPγ-2(KKEE) were stably co-expressed, independently without tether. Clone #8 expresses the GluA2(flip-Q isoform, N619K mutant) and TARPγ-2(KKEE). TARPγ-2 is abbreviated as γ-2. B. Purified complex resolved in SDS-PAGE. C. Superdex200 (Sdx200) size exclusion chromatograph in the final step of purification. The peak with asterisk contains the complex formed of GluA2(flip-Q) and TARPγ-2(KKEE). The N619K mutant complex exhibit identical chromatography profile and not shown. D. A representative motion corrected cryo-EM image of purified GluA2/TARPγ-2(KKEE) complex of Open-Na260. The displayed image is representative among a total of ~16,000 micrographs. E. A set of representative 2D class averages obtained during the image processing of Open-Na260. The class averages displayed are from 1/4 of the total dataset. F. Image processing of Open-Na260. Other structures were obtained using similar procedures and described in the methods. G-H. The atomic model of Open-Na260. GluA2=blue, TARPγ-2(KKEE)=yellow. TARPγ-2(KKEE) in the A’/C’ positions (G) and B’/D’ positions (H) are shown. The zoomed views (left panels) correspond to the red square in the right panels. In all the structures determined in this work (see Table1), the regions containing the KKEE mutations were unresolved due to structural flexibility of the extracellular loop.
Extended Data Fig. 2. Fourier Shell Correlation (FSC) curves.
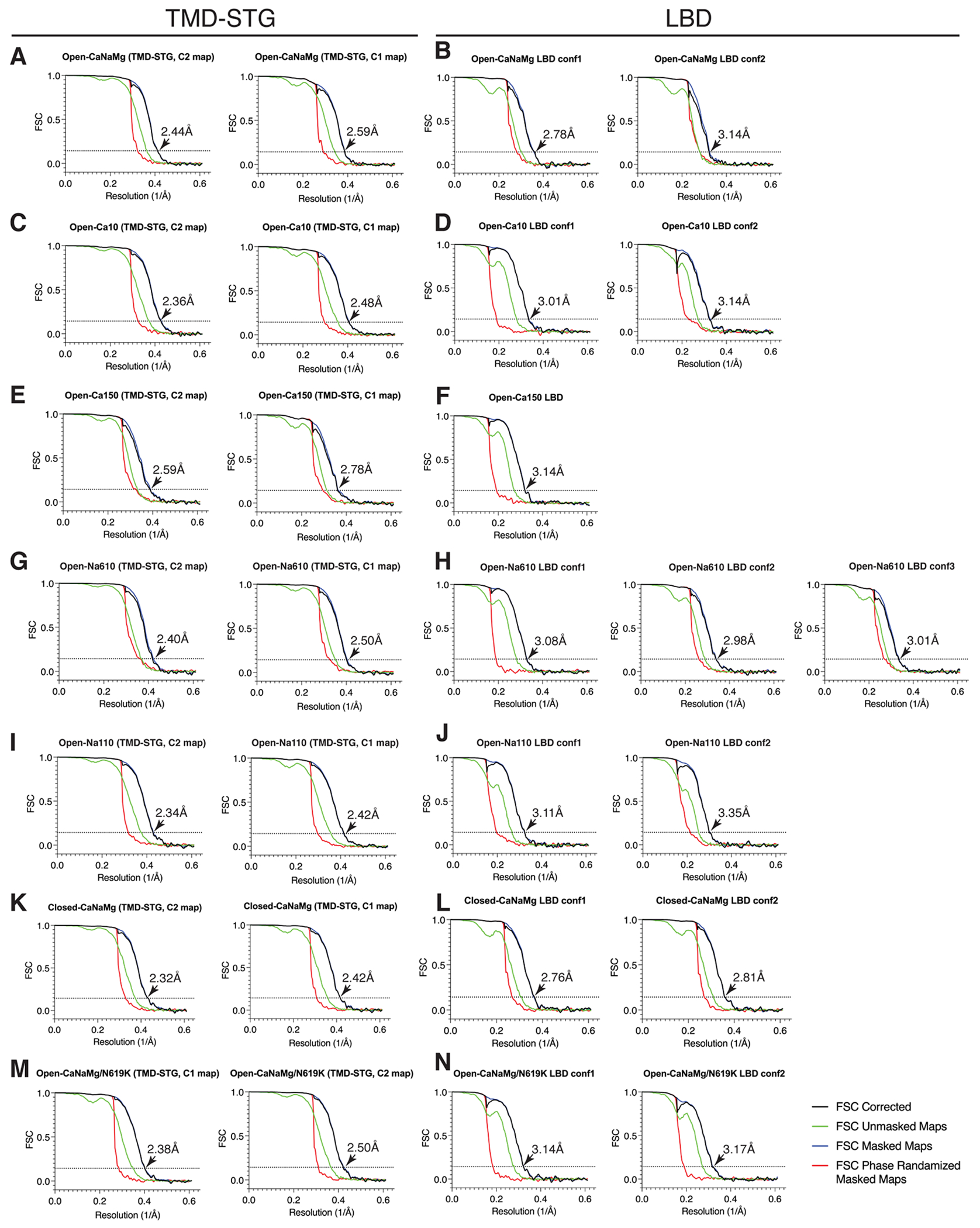
The FSC curves of the GluA2/TARP complexes presented in this work. The membrane embedded portion of the structures were refined using the TMD-STG mask, with (C2) and without (C1) imposing symmetry (A, C, E, G, I, K, and M). The LBDs were refined using the LBD mask with C2 symmetry imposed (B, D, F, H, J, L, and N). A-B. Open-CaNaMg. C-D. Open-Ca10. E-F. Open-Ca150. G-H. Open-Na610. I-J. Open-Na110. K-L. Open-CaNaMg/N619K. M-N. Closed-CaNaMg. The FSCs and final resolutions were calculated using Relion. FSC=0.143 is indicated by dashed line. Resolution is indicated by the arrows and numbers. Note: FSC curves of Open-Na260 are provided in Extended Data Fig. 1 F2 and F4.
Extended Data Fig. 3. Local resolution and representative fit between map and model.
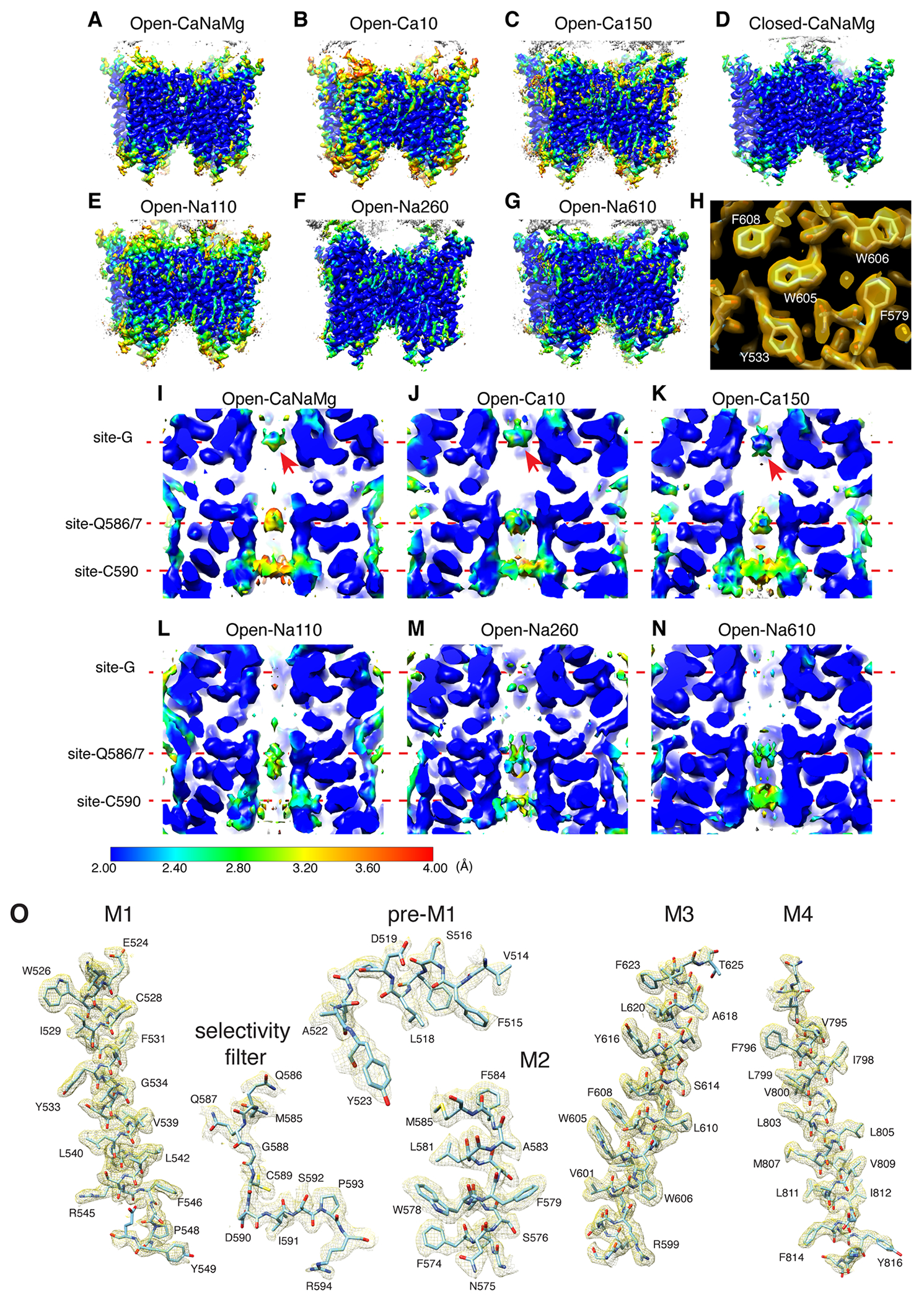
A-G. Local resolution was estimated by ResMap 59, using as input the half maps generated by 3D refinement in Relion. The heatmap for resolution is shown on the bottom right. Side view of each indicated structure. H. Overlay of map and model in a subregion of M1-3 helices. With an optimal threshold, the holes in the aromatic sidechains are discernable. I-N. Cross sections of local resolution heatmaps of the open pores. The arrows in I-K are the Ca2+ density at site-G. Note higher local resolution of the density in 150mM Ca2+ (Open-Ca150) compared to 10mM Ca2+ (Open-Ca10 and Open-CaNaMg), consistent with more binding at higher concentration. Note, the maps displayed are half-maps that are unfiltered/unsharpened, and thus the resolution appear lower than the full-maps displayed in other figures. O. Representative overlay of map and model in the TMD. Open-Na260 is shown. Others were at similar quality. Also see Extended Data Fig. 5-6.
Extended Data Fig. 4. Consistency of pore densities between the C1 and C2 maps.
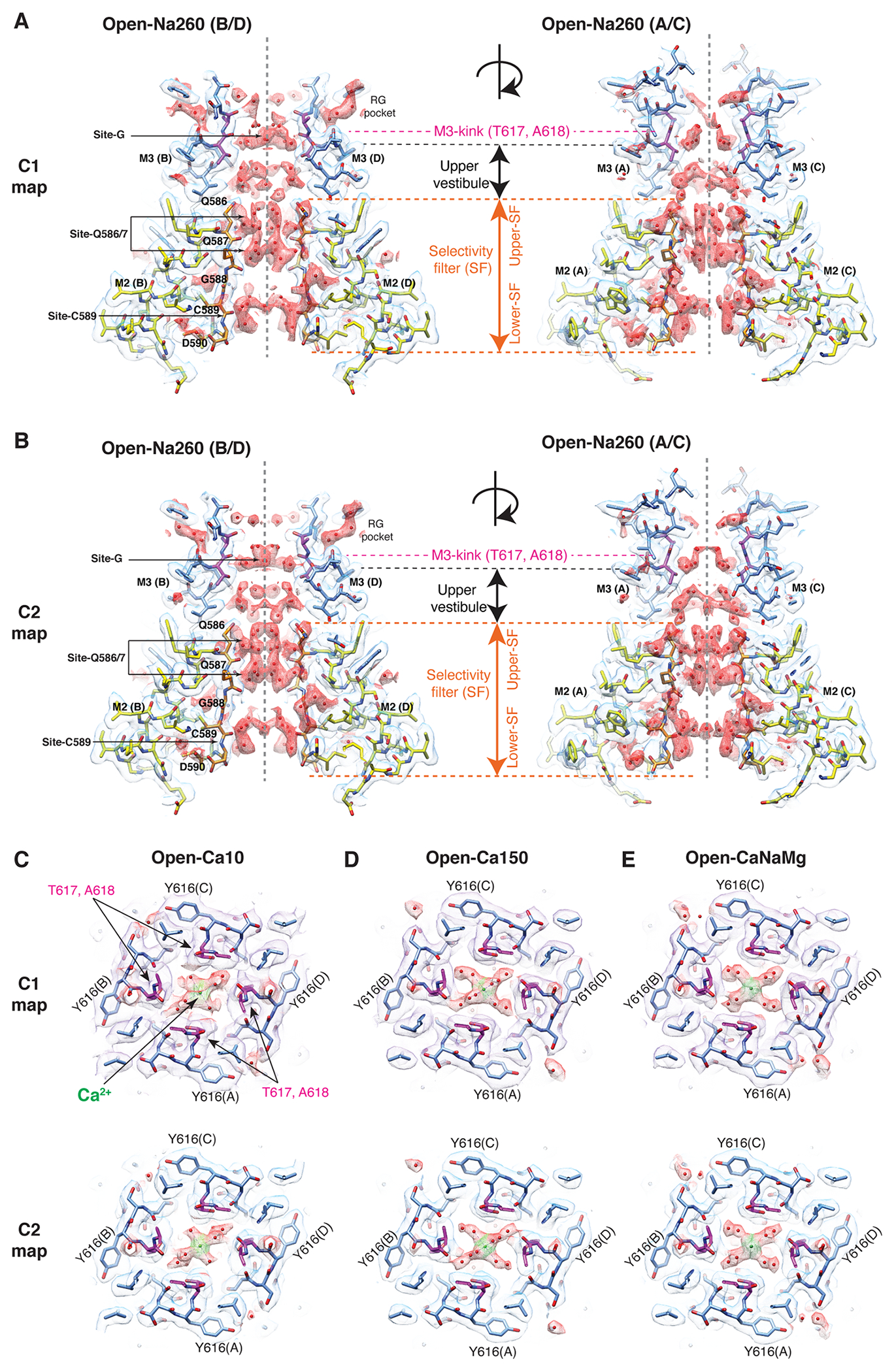
A-B. Cross sections of the pores of Open-Na260. (A) C1 maps. (B) C2 maps. All maps were filtered according to the local resolution calculated by Relion. The cryo-EM density map (mesh) and the atomic model are superimposed. The atomic models were built based on the C2 map and superimposed into the C1 map in A. Putative water and calcium ion are in red mesh. The atomic models of the polypeptides of M2, SF, and M3 are shown in yellow, orange, and sky blue, where elements are colored according to oxygen=red, nitrogen=blue, and sulfate=yellow. Resides that define the gate in the M3 of each structure are colored magenta. C-E. Overlay of the model and map at site-G are shown. C. Open-Ca10. D. Open-Ca150. E. Open-CaNaMg. Top: C1 map. Bottom: C2 map. The C1 map was filtered using the local resolution function in Relion. T617 and A618 are in magenta. The Ca2+ is green.
Extended Data Fig. 5. Global structures of the A2iQ/γ2(KKEE).
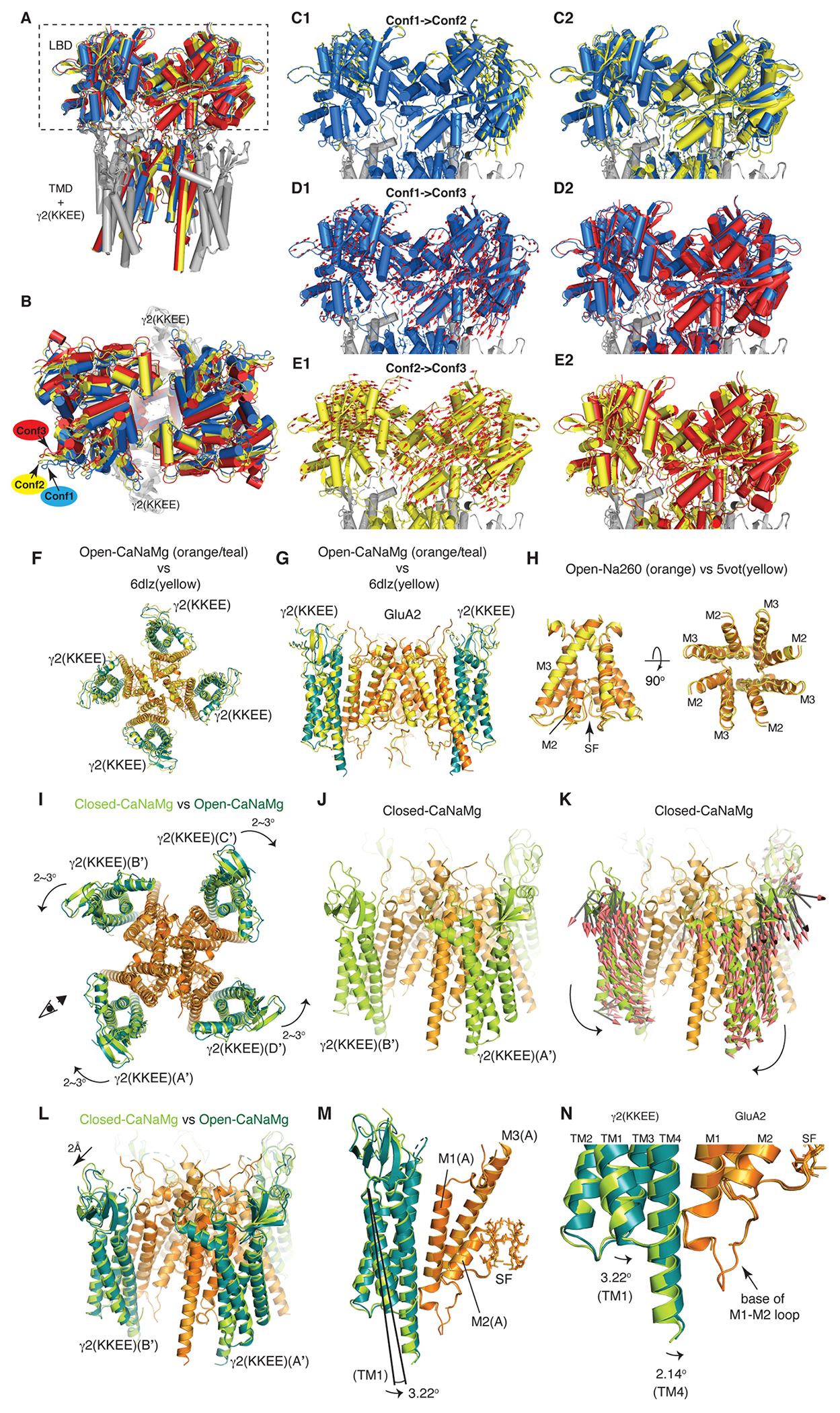
A-E. Conformational heterogeneity of the LBD gating ring in Open-Na260. A. LBD-TMD sectors were reconstructed from particles that produced LBD Conf1, Conf2, and Conf3. The LBD-TMD were aligned and superimposed using the cytoplasmic half of the SF. The TMD and γ2(KKEE) are nearly perfectly aligned as expected; there was no obvious global conformational heterogeneity of the TMD and γ2(KKEE). Blue=Conf1, Yellow=Conf2, and Red=Conf3. The rectangle indicates the regions that are highlighted in panels C-E. B. The LBD gating ring of the aligned and superimposed structure shown in panel A is viewed from above. The helices of the LBDs do not superimpose. C. Conformational transition from Conf1 to Conf2. The Conf1 conformation is shown with the mode vectors (yellow arrows) that schematically represent the displacements of the Cα during the conformational transition from Conf1 to Conf2 (panel C1). The lengths of the vectors are increased by 50% than actual displacements for clarity. The Conf1 and Conf2 conformations are superimposed in the same view as in panel C1. D and E. The transition from Conf1 to Conf3 (panels D1 and D2) and from Conf2 to Conf3 (panels E1 and E2) are shown as in panel C. F-N. Global architecture of A2iQ/γ-2(KKEE). F and G. The atomic model of Open-CaNaMg at 2.4Å resolution, where γ2(KKEE) in teal and GluA2 in orange, is superimposed to the A2iQ/γ2 wild-type complex in yellow (PDB:6dlz, 3.9Å resolution). The root mean square deviation (RMSD) of the Cα is 1.909Å. Top view (A) and cross section side view (B). H. The pore (i.e., M2, SF, and M3) of Open-Na260 at 2.3Å resolution is superimposed to the A2iQ/γ2 wild-type complex in yellow (PDB:5vot, 4.9Å resolution). The RMSD of Cα of the M3 in B/D subunit pairs is 0.642Å. I. Conformational difference between Closed-CaNaMg and Open-CaNaMg. The two structures were aligned at the cytoplasmic half of M3 (residue 597-610) at RMSD=0.3Å. GluA2 are colored in ogrange and light orange in Open-CaNaMg and Closed-CaNaMg, respectively. The four γ2(KKEE)s of Open-CaNaMg are colored in teal, while in Closed-CaNaMg they are in light green. The position of the γ2(KKEE)s relative to the GluA2 are indicated by the A’-D’ labels, as defined in Fig1B. The γ2(KKEE)s undergo counter rotations of 2-3° within the A’/B’ and C’/D’ pairs (curved arrows). The eye and arrow indicate the viewpoint of the structures in J-K. J. Closed-CaNaMg without the mode vector arrows. K. Closed-CaNaMg with the mode vector arrows that describe the directions and magnitudes of displacement of the Cα between Closed-CaNaMg and Open-CaNaMg. Arrows are shown for all motion greater than 0.5Å. The counter rotation of the γ2(KKEE)s in the A’/B’ pairs are shown. L. Superposition of the Open-CaNaMg and Closed-CaNaMg shows the ~2Å outward motion of the extracellular domain of the TARPs. M and N. The activation is also accompanied by a 2-3° tilt of the TARPs, which brings close the cytoplasmic extension of TM4 of γ2(KKEE) to the base of M1-M2 loop of GluA2. In the magnified view (I), the difference in the tilt angle in TM1 and TM4 of γ2(KKEE) is shown.
Extended Data Fig. 6. Comparison of cryo-EM maps and models in the pores in the cross-section containing the A/C subunits.
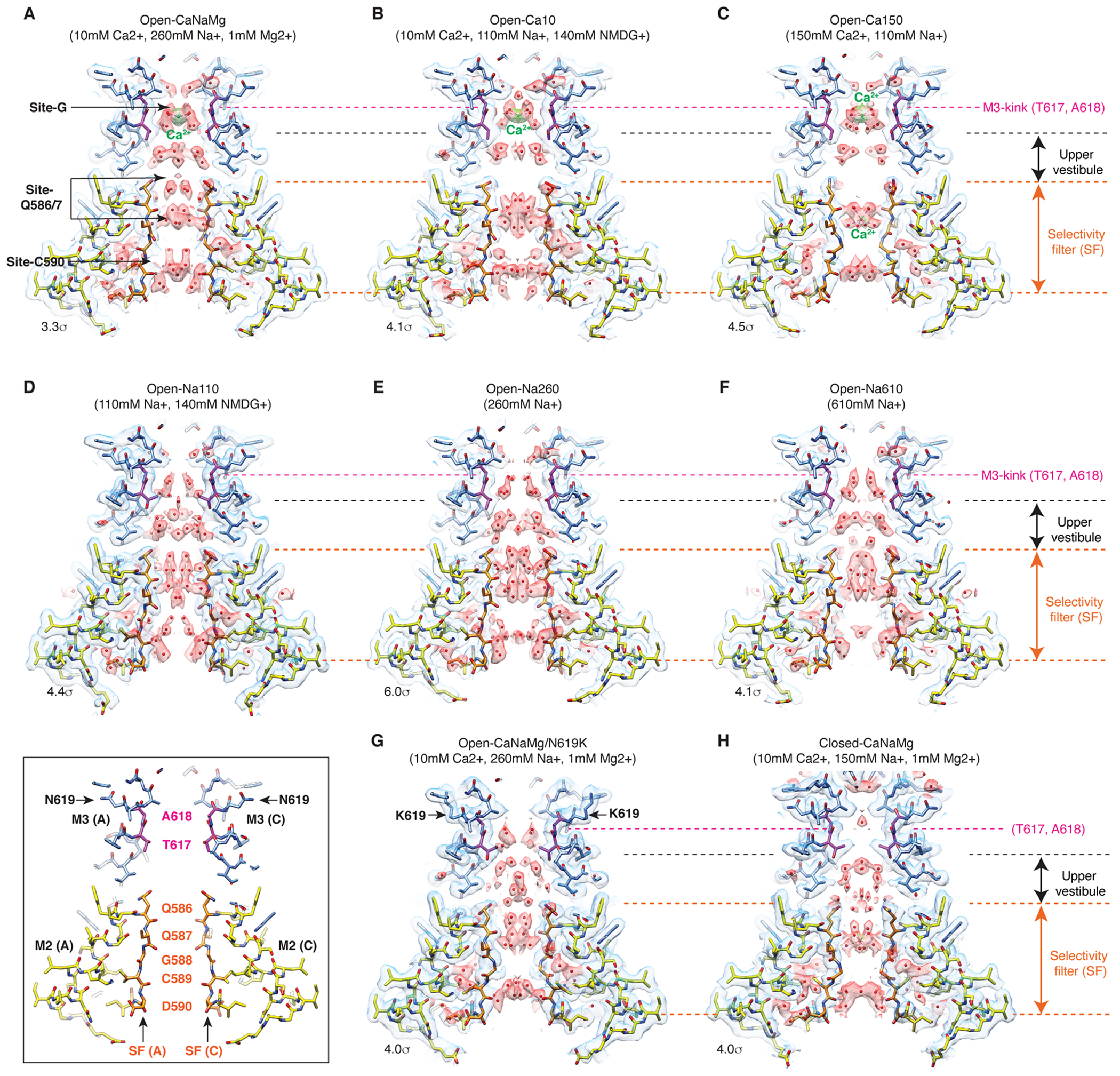
Cross sections containing A/C subunits of the pores of Open-CaNaMg (A), Open-Ca10 (B), Open-Ca150 (C), Open-Na110 (D), Open-Na260 (E), Open-Na610 (F), Open-CaNaMg/N619K (G), and Closed-CaNaMg (H) are displayed. The cryo-EM density map (mesh) and the atomic model are superimposed. The cryo-EM maps are displayed using the indicated sigma values. Putative water and calcium ion are in red and green spheres, respectively. The atomic models of the polypeptides of M2, SF, and M3 are shown in yellow, orange, and sky blue, where elements are colored according to oxygen=red, nitrogen=blue, and sulfate=yellow. Resides that define the gate in the M3 of each structure are colored magenta. The left bottom inset shows the locations of the key residues in the wild type open pore. C2 maps are displayed (also see Table1).
Extended Data Fig. 7. Comparison of cryo-EM maps and models in the pores in the cross-section containing the B/D subunits.
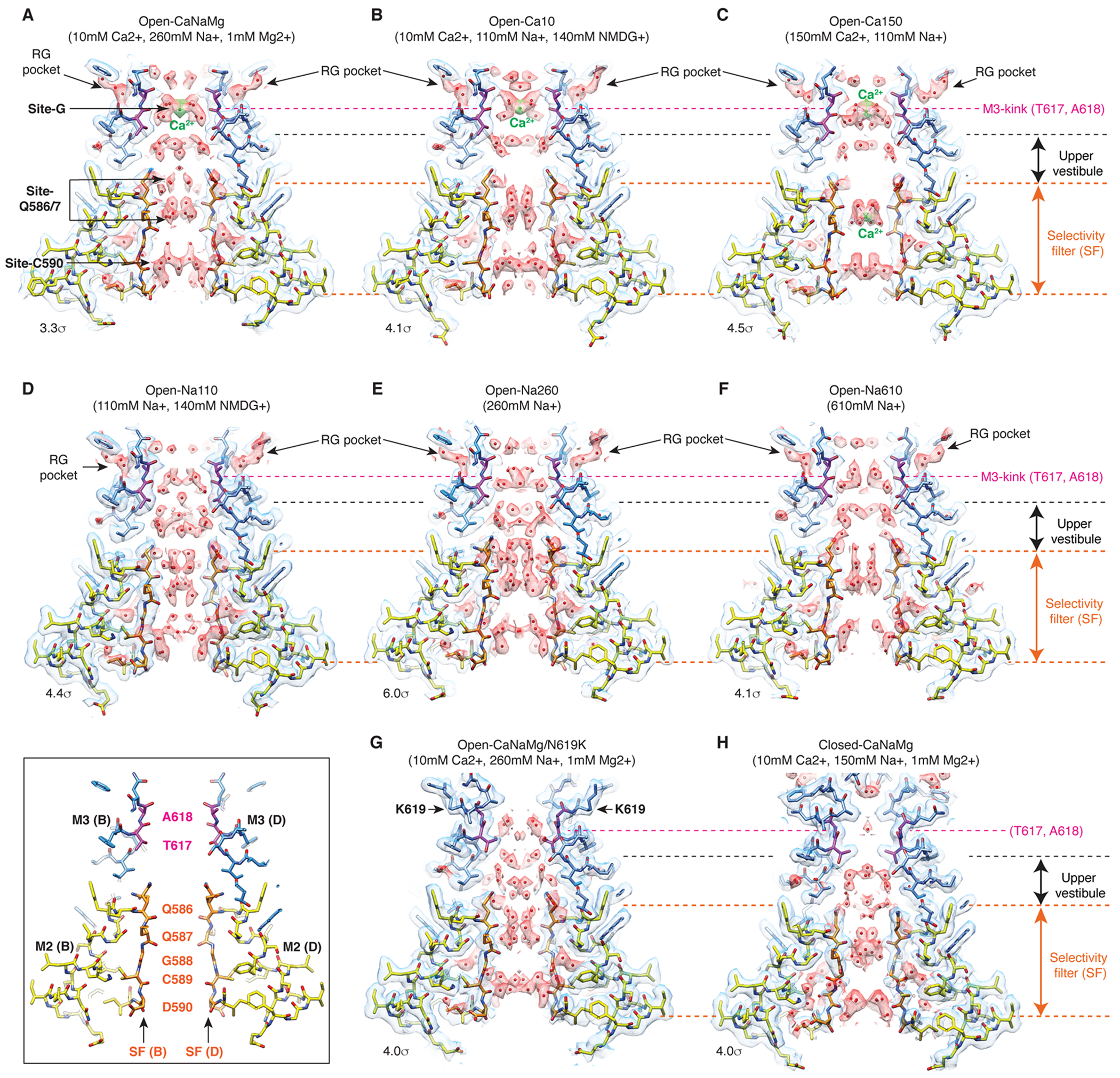
Cross sections containing B/D subunits of the pores of Open-CaNaMg (A), Open-Ca10 (B), Open-Ca150 (C), Open-Na110 (D), Open-Na260 (E), Open-Na610 (F), Open-CaNaMg/N619K (G), and Closed-CaNaMg (H) are displayed. The cryo-EM density map (mesh) and the atomic model are superimposed. The cryo-EM maps are displayed using the indicated sigma values. Putative water and calcium ion are in red and green spheres, respectively. The atomic models of the polypeptides of M2, SF, and M3 are shown in yellow, orange, and sky blue, where elements are colored according to oxygen=red, nitrogen=blue, and sulfate=yellow. Resides that define the gate in the M3 of each structure are colored magenta. The left bottom inset shows the locations of the key residues in the wild type open pore. Each view is the orthogonal to the corresponding structure shown in the previous figure. C2 maps are displayed (also see Table1).
Extended Data Fig. 8. Putative water in the vestibules that are common in Open-Na260 and Open-Na110.
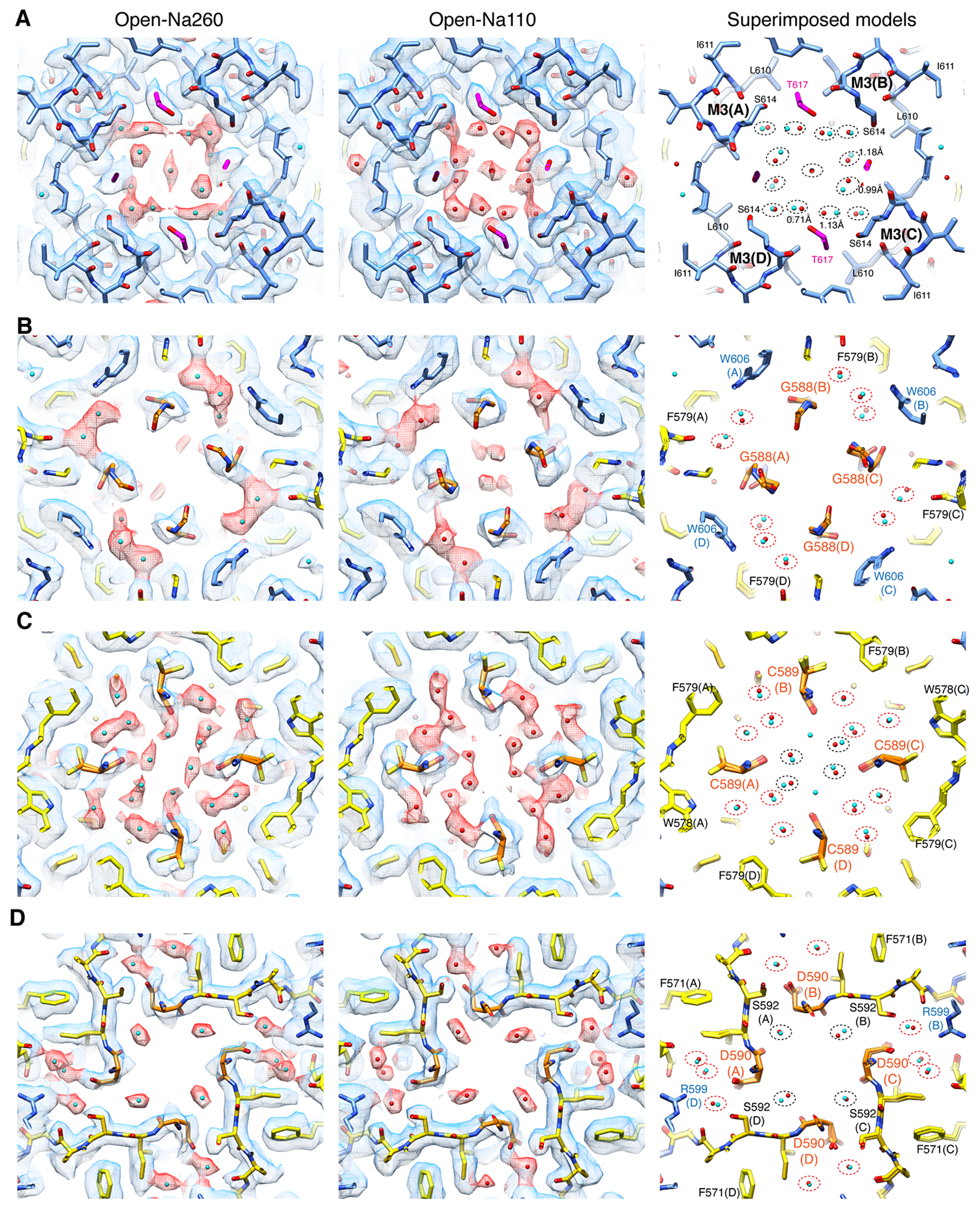
The horizontal sections through the upper (A) and lateral vestibule (B-D). The sections were made at different levels that contain residue S614 (B), G588 (B), C589 (C), and D590 (D) of the SF. In each row, Open-Na260 (left), Open-Na110 (middle), and superimposed models of two structures are displayed. Blue and red mesh are density map of polypeptides and putative water, respectively. The M3 (blue model), M2 with M2-M3 linker (yellow model), and SF (orange model) are shown. Cyan and red spheres are putative water in Open-Na260 and Open-Na110, respectively. The location of putative water is nearly identical in both structures, as indicated by dotted ovals surrounding the cyan and red water pairs in the right panels. The water surrounded by the red dashed oval are the ones in the lateral vestibules. Cyan-red sphere pair distances were below 1Å in most cases. In A (right) the actual distances are provided next to the oval for subset of cyan-red sphere pairs. The densities in the lateral vestibule interpreted as water, described above, are unlikely to be cation because these is always a nearby - NH group within 3Å distance.
Extended Data Fig. 9. Solvent accessible pore radius.
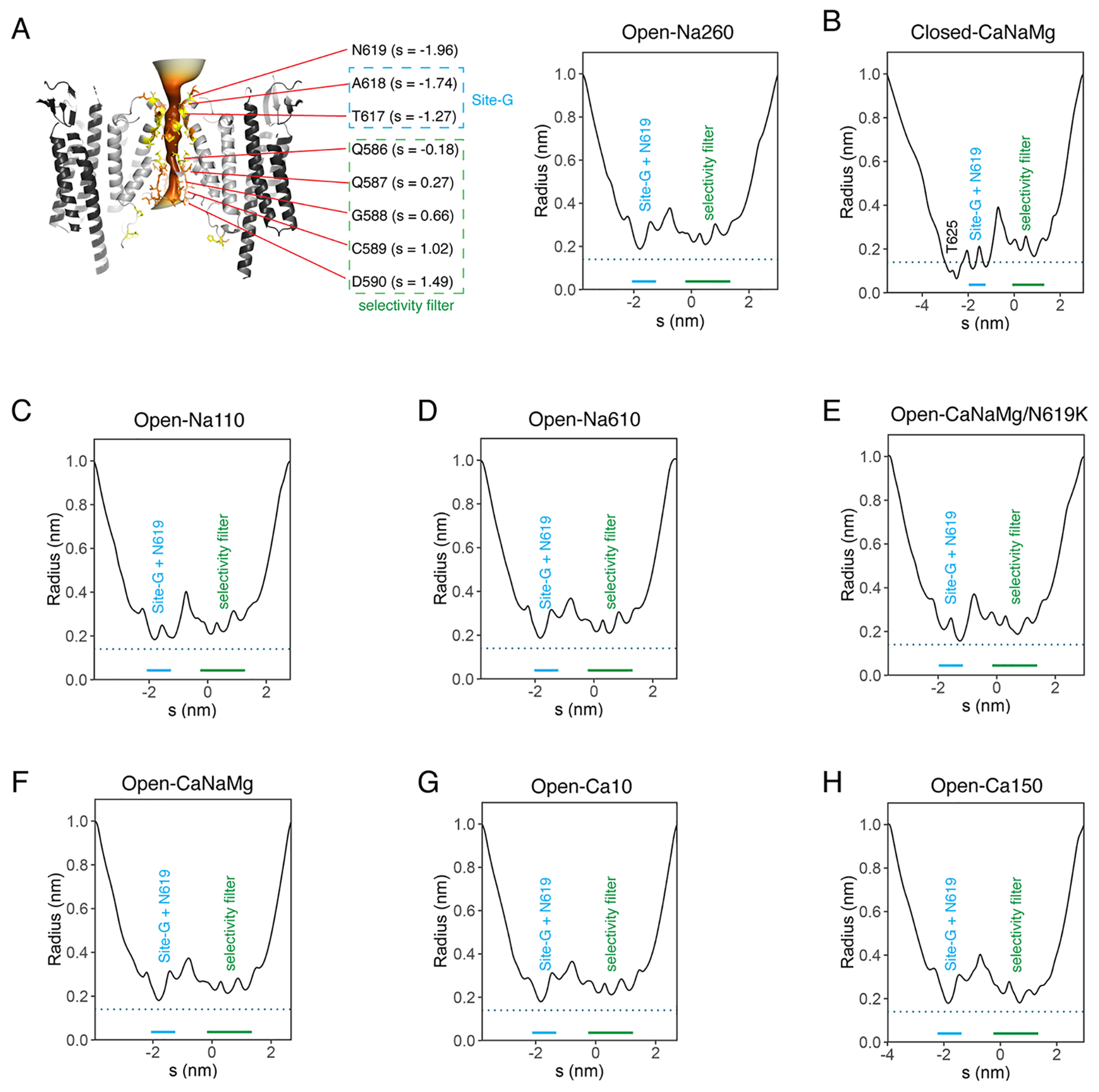
The pore radius accessible to solvent was estimated using CHAP software 64. A. (Left) The surface representation of the accessible pore is shown with the ribbon diagram of Open-Na260. GluA2: light gray. TARPγ-2: dark gray. Pore facing and pore lining residues are in yellow and orange, respectively. The darkness of orange surface corelates with narrower radius of the pore. The value of s is the coordinate along the pore pathway, whose origin is set at the center of mass of the pore forming residues. The relations between the key pore residues and the value of s (in the unit of nm) are shown. (right) Radius vs. s plot. The location of the site-G plus N619 and SF are in blue and green, respectively. The dashed line indicates 1.4Å. The radius of closed pore (Closed-CaNaMg) plotted in B crosses below the 1.4Å radius but the open pores don’t. Radius vs. s plots are shown for Open-Na110 (C), Open-Na610 (D), Open-CaNaMg/N619K (E), Open-CaNaMg (F), Open-Ca10 (G), and Open-Ca150 (H).
Extended Data Fig. 10. Characterization of A2iQ(N619K)/γ2 complex.
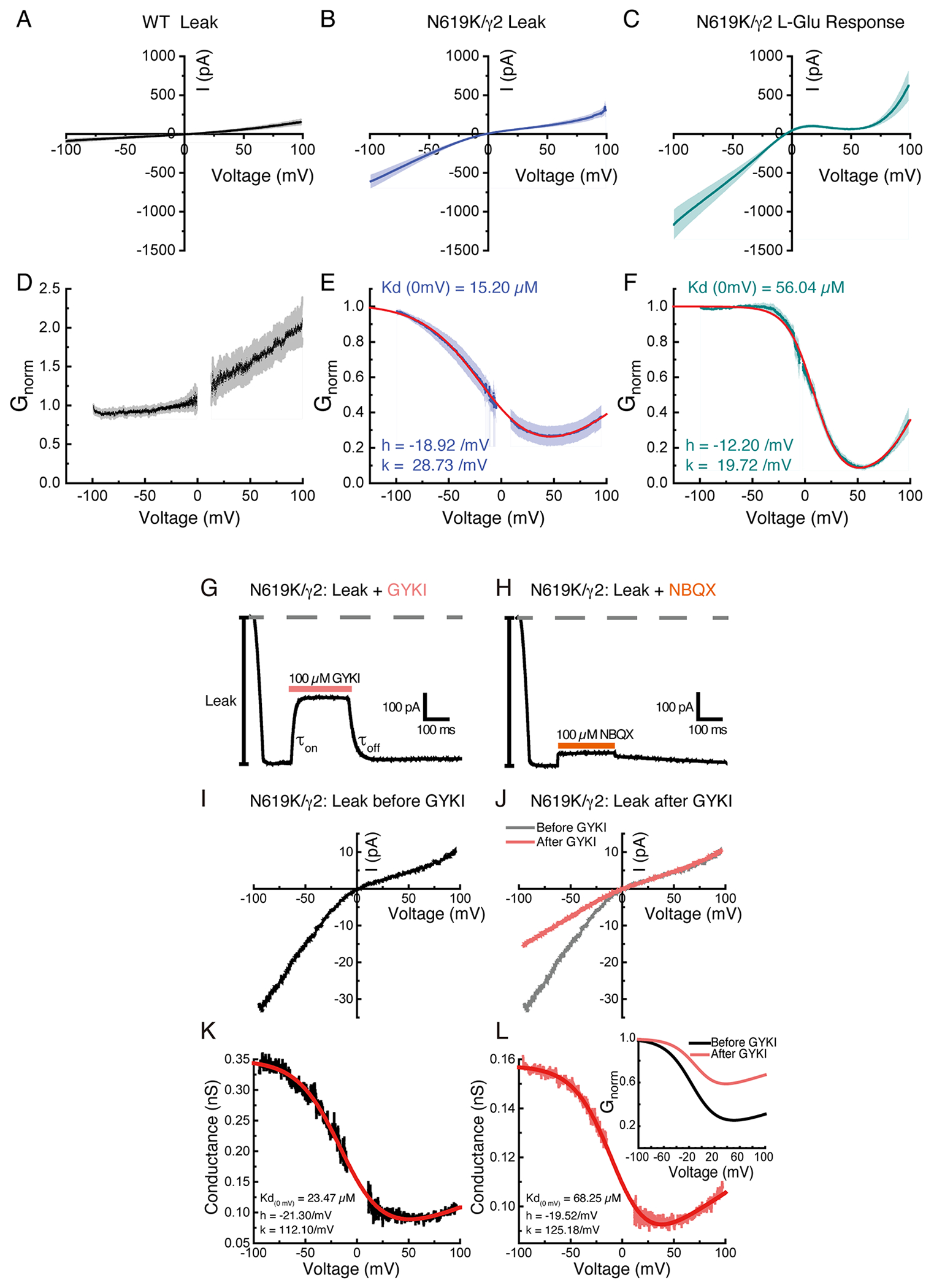
A-C. Averaged ramp currents (I-V) recorded in excised patches expressing wild-type A2iQ/γ2 prior to agonist stimulation (A, black), N619K/γ2 leak (B, blue) and agonist-evoked (C, dark cyan) responses using an internal patch solution containing 30 μM spermine. Dim colours show the SEM. The I-V plots both show bi-rectifying I-V relationships demonstrating that cytoplasmic spermine blocks the leak (B) and agonist-gated state (C) of the mutant A2iQ(N619K)/γ2 receptors. Data are presented as mean values ± SEM. (also see Supplementary Table 4). D-F. Conductance-voltage (G-V) plots converted from A-C for wild-type leak (D, black), N619K/γ2 leak (E, blue) and agonist-evoked (F, dark cyan) responses. In dim colours it is shown the SEM of the averaged conductance curves, respectively. Red lines in E and F are the fit for the G-V relationships for leak and agonist-evoked responses of N619K/γ2 receptors, respectively, using the single permeant blocker model (eq.1). Data are presented as mean values ± SEM. (Also see Supplementary Table 7) G and H. Fast jumps experiments comparing the ability of the AMPAR negative allosteric modulator, GYKI 52466 (100 μM) (G, Patch # 221129p3) or the competitive antagonist, NBQX (100 μM) (H, Patch # 221129p3) to block the leak currents mediated by N619K/γ2 receptors. The onset and off kinetics of GYKI (G) were estimated as τon = 12.7 ± 0.9 ms (n=10) and τoff = 37.1 ± 3.9 ms (n=10), respectively (also see Supplementary Table 5 and 6). I. I-V plot of the leak current of N619K/γ2 receptors with an internal patch solution containing 30 μM spermine in the absence of agonist. J. Typical I-V plots of leak currents of N619K/γ2 receptors before (grey) and after (pink) applying 100 μM GYKI (Patch # 221201p4), in the absence of agonist and containing 30 μM spermine in the internal solution. K and L. Conductance-voltage (G-V) plots of the leak N619K/γ2 receptors, converted from J (Patch # 221201p4), before (K, black) and after (L, pink) applying 100 μM GYKI. The G-V curves are fit (red in K and L) using a single permeant ion blocker model (eq. 1). L. (inset) Normalized G-V plots before (black) and after (pink) the application of GYKI are superimposed.
Supplementary Material
Acknowledgements:
We acknowledge the use of cryo-EM facility at the Center for Structural Biology and DORS Data Storage Core at Vanderbilt. Melissa Chambers, Scott Collier, and Mariam Haider maintained the cryo-EM facility and facilitated data collection. We thank Kwangho Kim and Plamen Christov at Vanderbilt Chemical Synthesis Core for synthesizing chemicals. Software was provided by SBGrid. The work was supported by funding from NIH grant R56/R01MH123474 (to T.N.), S10OD030292-01 (to T.N), Stanley Cohen Innovation Fund (to T.N.) and the Canadian Institutes of Health Research (FRN 163317, D.B.). X.W. was funded by a Max Binz fellowship from McGill’s Faculty of Medicine.
Footnotes
Competing interests: The authors have no competing interests.
Data and materials availability:
The structural data in this work is deposited to PDB (and EMDB) under the accession numbers: 8FQF(EMD-29386), 8FP9(EMD-29360), 8FQ1(EMD-29737), 8FP4(EMD-29359), 8FQB(EMD-29382), 8FQ5(EMD-29378), 8FPS(EMD-29369), 8FPG(EMD-29363), 8FQG(EMD-29387), 8FQH(EMD-29388), 8FQ0(EMD-29394), 8FPC(EMD-29361), 8FPH(EMD-29364), 8FQ6(EMD-29379), 8FPV(EMD-29370), 8FPY(EMD-29371), 8FPZ(EMD-29372), 8FQD(EMD-29384), 8FQE(EMD-29385), 8FQ8(EMD-29380), 8FQA(EMD-29381), 8FQ2(EMD-29375), 8FQ3(EMD-29376), 8FPK(EMD-29367), 8FPL(EMD-29368). The C1 maps are associated with each entry. Request for materials (plasmids and cell lines) will be fulfilled for reasonable inquiries and should be addressed to Terunaga Nakagawa.
References
- 1.Hansen KB et al. Structure, Function, and Pharmacology of Glutamate Receptor Ion Channels. Pharmacol Rev 73, 298–487, doi: 10.1124/pharmrev.120.000131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greger IH, Watson JF & Cull-Candy SG Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins. Neuron 94, 713–730, doi: 10.1016/j.neuron.2017.04.009 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Cull-Candy SG & Farrant M Ca(2+) -permeable AMPA receptors and their auxiliary subunits in synaptic plasticity and disease. J Physiol 599, 2655–2671, doi: 10.1113/JP279029 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobolevsky AI, Rosconi MP & Gouaux E X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756, doi:nature08624 [pii] 10.1038/nature08624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa T, Cheng Y, Ramm E, Sheng M & Walz T Structure and different conformational states of native AMPA receptor complexes. Nature 433, 545–549 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Meyerson JR et al. Structural mechanism of glutamate receptor activation and desensitization. Nature, doi: 10.1038/nature13603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong N & Gouaux E Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181, doi:S0896-6273(00)00094-5 [pii] (2000). [DOI] [PubMed] [Google Scholar]
- 8.Mayer ML The Challenge of Interpreting Glutamate-Receptor Ion-Channel Structures. Biophys J, doi: 10.1016/j.bpj.2017.07.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S et al. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246 e1214, doi: 10.1016/j.cell.2017.07.045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature, doi: 10.1038/nature23479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Watson JF, Matthews PM, Cais O & Greger IH Gating and modulation of a hetero-octameric AMPA glutamate receptor. Nature 594, 454–458, doi: 10.1038/s41586-021-03613-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnashev N et al. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257, 1415–1419 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Brown P, McGuire H & Bowie D Stargazin and cornichon-3 relieve polyamine block of AMPA receptors by enhancing blocker permeation. J Gen Physiol 150, 67–82, doi: 10.1085/jgp.201711895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Morais-Cabral JH, Kaufman A & MacKinnon R Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414, 43–48 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Hille B Ion channels of excitable membranes. 3rd edn, (Sinauer, 2001). [Google Scholar]
- 16.Hawken NM, Zaika EI & Nakagawa T Engineering defined membrane-embedded elements of AMPA receptor induces opposing gating modulation by cornichon 3 and stargazin. J Physiol, doi: 10.1113/JP274897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94, 569–580 e565, doi: 10.1016/j.neuron.2017.04.025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawe GB et al. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron 89, 1264–1276, doi: 10.1016/j.neuron.2016.01.038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soto D, Coombs ID, Gratacos-Batlle E, Farrant M & Cull-Candy SG Molecular Mechanisms Contributing to TARP Regulation of Channel Conductance and Polyamine Block of Calcium-Permeable AMPA Receptors. J Neurosci 34, 11673–11683, doi: 10.1523/JNEUROSCI.0383-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herguedas B et al. Mechanisms underlying TARP modulation of the GluA1/2-γ8 AMPA receptor. Nat Commun 13, 734, doi: 10.1038/s41467-022-28404-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding MM Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr 62, 678–682, doi: 10.1107/S0907444906014594 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Bowie D, Lange GD & Mayer ML Activity-dependent modulation of glutamate receptors by polyamines. J Neurosci 18, 8175–8185, doi: 10.1523/JNEUROSCI.18-20-08175.1998 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolcheva M et al. The pathogenic N650K variant in the GluN1 subunit regulates the trafficking, conductance, and pharmacological properties of NMDA receptors. Neuropharmacology 222, 109297, doi: 10.1016/j.neuropharm.2022.109297 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Jatzke C, Hernandez M & Wollmuth LP Extracellular vestibule determinants of Ca2+ influx in Ca2+-permeable AMPA receptor channels. J Physiol 549, 439–452, doi: 10.1113/jphysiol.2002.034413 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J et al. Structure of the voltage-gated calcium channel Ca(v)1.1 at 3.6 A resolution. Nature 537, 191–196, doi: 10.1038/nature19321 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Tang L et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505, 56–61, doi: 10.1038/nature12775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saotome K, Singh AK, Yelshanskaya MV & Sobolevsky AI Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511, doi: 10.1038/nature17975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonas P & Burnashev N Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron 15, 987–990, doi: 10.1016/0896-6273(95)90087-x (1995). [DOI] [PubMed] [Google Scholar]
- 29.Amin JB et al. Two gates mediate NMDA receptor activity and are under subunit-specific regulation. Nat Commun 14, 1623, doi: 10.1038/s41467-023-37260-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amin JB, Leng X, Gochman A, Zhou HX & Wollmuth LP A conserved glycine harboring disease-associated mutations permits NMDA receptor slow deactivation and high Ca(2+) permeability. Nat Commun 9, 3748, doi: 10.1038/s41467-018-06145-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe J, Beck C, Kuner T, Premkumar LS & Wollmuth LP DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci 22, 10209–10216 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schackert FK et al. Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel. J Chem Inf Model, doi: 10.1021/acs.jcim.2c01494 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biedermann J, Braunbeck S, Plested AJR & Sun H Nonselective cation permeation in an AMPA-type glutamate receptor. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2012843118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minniberger S, Abdolvand S, Braunbeck S, Sun H & Plested AJR Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors. J Mol Biol 435, 167970, doi: 10.1016/j.jmb.2023.167970 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Mesbahi-Vasey S, Veras L, Yonkunas M, Johnson JW & Kurnikova MG All atom NMDA receptor transmembrane domain model development and simulations in lipid bilayers and water. PLoS One 12, e0177686, doi: 10.1371/journal.pone.0177686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma G & Stevens CF Interactions between two divalent ion binding sites in N-methyl-D-aspartate receptor channels. Proc Natl Acad Sci U S A 93, 14170–14175, doi: 10.1073/pnas.93.24.14170 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Premkumar LS & Auerbach A Identification of a high affinity divalent cation binding site near the entrance of the NMDA receptor channel. Neuron 16, 869–880, doi: 10.1016/s0896-6273(00)80107-5 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Karakas E & Furukawa H Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997, doi: 10.1126/science.1251915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsberg M et al. Ionized calcium in human cerebrospinal fluid and its influence on intrinsic and synaptic excitability of hippocampal pyramidal neurons in the rat. J Neurochem 149, 452–470, doi: 10.1111/jnc.14693 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Heinemann U & Louvel J Changes in [Ca2+]o and [K+]o during repetitive electrical stimulation and during pentetrazol induced seizure activity in the sensorimotor cortex of cats. Pflugers Arch 398, 310–317, doi: 10.1007/BF00657240 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Ascher P & Nowak L The role of divalent cations in the N-methyl-D-aspartate responses of mouse central neurones in culture. J Physiol 399, 247–266, doi: 10.1113/jphysiol.1988.sp017078 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak L, Bregestovski P, Ascher P, Herbet A & Prochiantz A Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307, 462–465, doi: 10.1038/307462a0 (1984). [DOI] [PubMed] [Google Scholar]
- 43.Mayer ML, Westbrook GL & Guthrie PB Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263, doi: 10.1038/309261a0 (1984). [DOI] [PubMed] [Google Scholar]
- 44.Lee CH & MacKinnon R Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell 168, 111–120 e111, doi: 10.1016/j.cell.2016.12.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue J, Han Y, Zeng W & Jiang Y Structural mechanisms of assembly, permeation, gating, and pharmacology of native human rod CNG channel. Neuron 110, 86–95 e85, doi: 10.1016/j.neuron.2021.10.006 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derebe MG, Zeng W, Li Y, Alam A & Jiang Y Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore. Proc Natl Acad Sci U S A 108, 592–597, doi: 10.1073/pnas.1013643108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X et al. Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel. Nat Struct Mol Biol 27, 625–634, doi: 10.1038/s41594-020-0433-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y et al. Cryo-EM structures of apo and antagonist-bound human Ca(v)3.1. Nature 576, 492–497, doi: 10.1038/s41586-019-1801-3 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Ramos-Vicente D. et al. Metazoan evolution of glutamate receptors reveals unreported phylogenetic groups and divergent lineage-specific events. Elife 7, doi: 10.7554/eLife.35774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou TH, Tajima N, Romero-Hernandez A & Furukawa H Structural Basis of Functional Transitions in Mammalian NMDA Receptors. Cell 182, 357–371 e313, doi: 10.1016/j.cell.2020.05.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H. et al. Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors. Neuron, doi: 10.1016/j.neuron.2021.05.031 (2021). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 52.Shanks NF, Maruo T, Farina AN, Ellisman MH & Nakagawa T Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci 30, 2728–2740, doi:30/7/2728 [pii] 10.1523/JNEUROSCI.5146-09.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa T. Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366, 1259–1263, doi: 10.1126/science.aay2783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farina AN et al. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors Journal of Neuroscience (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warshamanage R, Yamashita K & Murshudov GN EMDA: A Python package for Electron Microscopy Data Analysis. bioRxiv, 2021.2007.2026.453750, doi: 10.1101/2021.07.26.453750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65, doi: 10.1038/nmeth.2727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenthal PB & Henderson R Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF et al. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544, doi: 10.1107/S2059798318006551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klesse G, Rao S, Sansom MSP & Tucker SJ CHAP: A Versatile Tool for the Structural and Functional Annotation of Ion Channel Pores. J Mol Biol 431, 3353–3365, doi: 10.1016/j.jmb.2019.06.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sievers F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539, doi: 10.1038/msb.2011.75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown PM, Aurousseau MR, Musgaard M, Biggin PC & Bowie D Kainate receptor pore-forming and auxiliary subunits regulate channel block by a novel mechanism. J Physiol 594, 1821–1840, doi: 10.1113/JP271690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dawe GB et al. Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors. Neuron 102, 976–992 e975, doi: 10.1016/j.neuron.2019.03.046 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Wong AY, Fay AM & Bowie D External ions are coactivators of kainate receptors. J Neurosci 26, 5750–5755, doi: 10.1523/JNEUR0SCI.0301-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structural data in this work is deposited to PDB (and EMDB) under the accession numbers: 8FQF(EMD-29386), 8FP9(EMD-29360), 8FQ1(EMD-29737), 8FP4(EMD-29359), 8FQB(EMD-29382), 8FQ5(EMD-29378), 8FPS(EMD-29369), 8FPG(EMD-29363), 8FQG(EMD-29387), 8FQH(EMD-29388), 8FQ0(EMD-29394), 8FPC(EMD-29361), 8FPH(EMD-29364), 8FQ6(EMD-29379), 8FPV(EMD-29370), 8FPY(EMD-29371), 8FPZ(EMD-29372), 8FQD(EMD-29384), 8FQE(EMD-29385), 8FQ8(EMD-29380), 8FQA(EMD-29381), 8FQ2(EMD-29375), 8FQ3(EMD-29376), 8FPK(EMD-29367), 8FPL(EMD-29368). The C1 maps are associated with each entry. Request for materials (plasmids and cell lines) will be fulfilled for reasonable inquiries and should be addressed to Terunaga Nakagawa.


