Abstract
Background
Chronic myeloid leukaemia (CML) is a type of blood cancer that begins in the hematopoietic stem cells. It is primarily characterized by a specific chromosomal aberration, the Philadelphia chromosome. While the fusion gene is a major contributor to CML, several other genes including ADGRE2, that are reported as highly expressed in hematopoietic stem cells and could be utilized as a therapeutic marker in leukemic patients are implicated in the disease’s progression. Until recently, little research had been conducted to identify single nucleotide polymorphisms (SNPs) associated with CML. Therefore, this study aims to investigate the influence of non-synonymous variants on the structure and function of the gene encoding adhesion G protein-coupled receptor E2, ADGRE2, and to evaluate their association with CML and its clinical and pathological characteristics.
Methods
Non-synonymous SNPs of ADGRE2 were retrieved from the ENSEMBL, COSMIC, and gnomAD genome browsers, and the pathogenicity of deleterious variants was assessed using several established computational tools, including SIFT, CADD, REVEL, PolyPhen, and MetaLR.
Results
Various in silico analyses explored the impact of damaging SNP on the function, stability, and structure of EGF-like modules containing mucin-like hormone receptor-like2 (EMR2) protein encoded by the ADGRE2 gene. Genotype analysis was performed on collected blood samples, revealing that altered genotype TT of variant rs765071211 (C/T) was associated significantly with CML patients compared to the control. Further in vitro and in vivo analyses suggest that this SNP holds potential for clinical translation.
Keywords: Non-synonymous, CML, Bioinformatics tools, ARMS PCR, In silico analysis
Introduction
CML is a type of myeloproliferative malignancy, described by the unregulated growth and proliferation of myeloid cells at various stages of their development. The characterization and diagnostic criteria of CML defined by European Leukemia Net (ELN) has described 3 phases of this disease: chronic phase (CP), acceleration phase (AP), and blast transformation phase (BP), which is described by elevated levels of blasts of myeloid and/or lymphoid lineages [1, 2]. According to International Consensus Classification (ICC), 10–19% of bone marrow or peripheral blood blasts and ≥ 20% peripheral blood basophils are considered AP, while if the bone marrow or peripheral blood myeloid blast count is more than 20% then it is considered as the criteria for BP [3]. Most of the patients of CML are categorized under CP, however, if left untreated, this stage can progress to AP and a small fraction of patients may develop BP. The symptoms of CML are generally unspecific including fever, fatigue and weight loss, due to anemia and splenomegaly. Patients who have developed the blast phase, their symptoms may get severe which may include bleeding and bone pain. However, 50% patients of with CML in the chronic phase are asymptomatic and can only be diagnosed after routine blood tests. Reciprocal translocation between chromosomes 9 and 22 resulting in chimeric BCR-ABL is the major cause of CML in more than 90% of cases of this disease. BCR-ABL fusion gene encodes for an aberrant tyrosine kinase enzyme, which leads towards the increased proliferation of immature granulocytes, increased production of reactive oxygen species and genetic instability [4]. Genetic alterations in various other genes have been reported to correlate with CML, indicating the involvement of various other genetic factors in the development and progression of this disease [5]. The incidence rate of CML is reported to be 1-1.5 cases per 100,000 individuals, annually, without any geographic or racial bias. However, the prevalence of leukaemia is expected to increase and it is estimated that by the year 2040, the prevalence of CML will reach 0.18 million cases per year [6].
Single nucleotide polymorphism (SNP) is the most common type of genetic alteration occurring in DNA. SNPs occur when a single nucleotide in DNA is replaced with another nucleotide, resulting in a change in the genetic sequence. SNPs present in the non-coding or intronic regions can impact the regulation of the expression of the gene [1]. It was shown that intronic SNPs in P53 were associated with poor response and disease progression in CML patients [2]. SNPs located in the coding region of DNA can alter the structure, functions, and interactions of the subsequent protein, which may lead to the development of diseases such as cancers [7]. In the past decade, studies have shown various SNPs to be associated with various patient features such as response to therapy, survival, and prognosis in AML patients. Likewise, in CML, SNPs have been shown to impact the patient’s response to treatment and the overall prognosis of the disease. SNPs in BCR-ABL have been shown to induce resistance in patients against TKIs. A genetic association study has shown that SNPs in genes PSMB10, TNFRSF10D, PSMB2, PPARD and CYP26B1 were associated with CML predisposition [8, 9]. Therefore, it is essential to investigate SNPs in various genes that can result in increased cancer susceptibility to understand the underlying pathogenesis of various cancers.
Adhesion G protein-coupled receptors (GPCRs), commonly referred to as the ADGRE gene family, are a class of cell surface receptors that are vital to many physiological functions [10]. Numerous physiological processes, including cell adhesion, migration, immune response control, and tissue homeostasis are mediated by ADGRE receptors [11]. Their interactions with immune cells and extracellular matrix constituents impact various processes, including inflammation, tissue healing, and leukocyte trafficking [10, 12]. ADGRE2 is located on human chromosome 19 at position 19q13.31. ADGRE2, a member of the ADGRE family, is expressed in various tissues, including immune cells and the nervous system [13]. While its precise functions are still under investigation, the dysregulation of ADGRE2 can cause several diseases, including cancer, autoimmune disorders, and inflammatory ailments [14]. A recent study showed that the ADGRE2 gene has aberrant expression in AML which is associated with poor patient outcomes [15]. Targeting of the ADGRE2 gene with siRNA resulted in anti-leukemic effects in both in vitro and in vivo settings [10]. Furthermore, ADGRE2 has been shown to activate various signalling cascades such as PI3K/AKT, and PKC/MEK/ERK pathways to enhance the maintenance of proteostasis in leukaemia [15]. Furthermore, another study showed that various members of the ADGRE family including ADGRE2 were associated with poor prognosis and short survival in AML patients [16]. ADGRE2 gene has also been used to target AML via the CAR-T therapy approach [17]. Furthermore, ADGRE2 has been shown to activate various signalling cascades such as PI3K/AKT, and PKC/MEK/ERK pathways to enhance the maintenance of proteostasis in leukaemia [15]. This emerging evidence suggests that the ADGRE2 gene might have a role in CML. Moreover, since non-synonymous SNPs can alter the resultant protein’s structural and functional aspects, the presence of these variants in the ADGRE2 gene can have a damaging effect.
A previous study indicated that a missense SNP in the ADGRE2 gene resulted in amino acid substitution at residue 492 (p. C492Y) and was associated with vibratory urticaria [18]. The presence of this SNP led to an extension of the degranulation, an increase in the number of responsive mast cells, and a lowering of the activation threshold [19], suggesting that SNPs in ADGRE2 might be associated with diseases. However, most of the genetic variants in ADGRE2 are still uncharacterized in terms of their association with CML. Therefore, the current study aimed to explore the impact of the ADGRE2 non-synonymous variants by employing various computational tools that identified damaging SNPs in the gene and then analyzing the effects of those variants on protein function and structural stability. Furthermore, experimental analysis was performed to investigate the association of the variant exhibiting the most damaging potential with CML through genotyping analysis.
Methods
Data retrieval and mapping
Data on total genetic variants of ADGRE2 was retrieved from Ensembl [20], COSMIC [21], and gnomAD (https://gnomad.broadinstitute.org) [22]. The non-synonymous SNPs from all three databases were filtered out and merged while removing any redundant data. Information regarding exons and their corresponding amino acid residues was obtained from Ensembl and NCBI. The ADGRE2 protein sequence data was obtained from the UniProt database (https://www.uniprot.org/) [23], which is believed to be the most reliable database for protein sequences.
Prediction of pathogenicity
In the present study, various computational tools were utilized to analyze the pathogenic impact of non-synonymous SNPs. These tools evaluated ADGRE2 non-synonymous variants and assigned each variant a specific score based on its potential pathogenicity as determined by the algorithms. PolyPhen tool (http://genetics.bwh.harvard.edu/pph2/) predicts variant damaging effects based on the structural and evolutionary characteristics of the amino acids. This tool provides scores ranging from 0 to 1, with scores from 0.4 to 0.8 and 0.9-1 being possibly damaging and probably damaging, respectively. Variants with PolyPhen scores ≥ 0.999 were chosen [24]. SIFT tool (https://sift.bii.a-star.edu.sg) scores range from 0 to 1, where 0 signifies deleterious and 1 signifies tolerated, and for this study, variants with scores ≤ 0.05 were chosen. This tool predicts the damaging potential of the variant through the physical properties of the amino acid and the sequence homology [25]. REVEL categorizes variants as benign (< 0.5) or likely disease-causing (> 0.5), with variants scoring ≥ 0.5 chosen. REVEL predicts the pathogenicity of a missense variant by combining the scores for a particular variant from 13 individual bioinformatics tools [26]. MetaLR categorizes SNPs as damaging (0.5–0.9) or tolerated (0-0.4), and the variants scoring ≥ 0.5 were selected for this study. MetaLR employs logistic regression analysis to integrate the allele frequency and independent variant damaging score to estimate the pathogenicity of the non-synonymous variants [27]. Lastly, CADD (https://cadd.gs.washington.edu) utilizes sequence conservation and functional information to predict if variants are benign or damaging by scoring them from 0 to 30 and 31 to 35, respectively [28].
Effect of polymorphisms on RNA stability
In this study, the effect of selected non-synonymous SNP on mRNA stability was also determined. For this purpose, the RNAfold web server was used to predict the impact of the genetic variant by determining the minimum free energy and base pair probabilities of the mRNA secondary structure and comparing the results of the variant with wild-type mRNA (https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) [29].
Structure prediction and validation
The complete structure of the EMR2 protein encoded by ADGRE2 was unavailable in the protein data bank, so protein structure prediction was performed through I-TASSER (Iterative Threading ASSEmbly Refinement) [30], which employs the threading technique to predict the structure on the basis of the peptide sequence, assigning the confidence score ranging from − 5 to 2 based on the significance of template alignment. For the subsequent analysis, the model exhibiting the highest C-score was chosen, and the 3-D structure was visualized using the PyMOL v.3.0. (The PyMOL Molecular Graphics System, Version 3.0 Schrödinger, LLC) [31]. The predicted structure was validated through ERRAT [32], which determined the overall quality factor of the protein, while PROCHECK was used to determine the stereochemical quality of the protein structure by producing Ramachandran plots [33]. The information regarding the domains of protein and their subsequent amino acids was obtained from InterPro (https://www.ebi.ac.uk/interpro/search/sequence/). The structures were validated through InterPro and highlighted in PyMOL.
Stability analysis
The impact of the non-synonymous variant on protein stability and flexibility was also analyzed. The stability of the protein structure is determined through free energy. Evaluating the impact of a non-synonymous variant on the free energy of the protein can give a clear picture of its structural stability. For this purpose, various bioinformatic tools were applied to investigate any changes in the free energy of the protein induced by the pathogenic non-synonymous SNP in the ADGRE2 gene. I-Mutant 2.0 predicts the alteration in the stability of protein caused by variation in terms of DDG (Kcal/mol) (http://folding.uib.es/i-mutant/i-mutant2.0.html). A DDG value < 0 indicates a decrease, while > 0 indicates an increase in the stability of protein as a result of variation [34]. MUpro was also used to determine the structural stability of the variant protein (http://mupro.proteomics.ics.uci.edu/). This tool works on machine-learning algorithms trained on large mutational datasets. This tool also predicted the protein stability in terms of DDG values [35]. Lastly, the DynaMut2 tool was also used in this study(https://biosig.lab.uq.edu.au/dynamut2/). This tool predicted the impact of non-synonymous SNP on protein structural stability and changes in the intramolecular interactions due to the presence of the variant amino acid residue [36].
Variant effect on molecular characteristics
Genetic variations also affect the structures and functions of proteins. For this purpose, project HOPE was utilized (https://www3.cmbi.umcn.nl/hope/). This tool was designed to describe the role of variations in the human proteins in the molecular basis of disease-associated phenotype. This tool gathers information from various sources to determine protein sequence annotation, predict 3-dimensional coordinates of the protein, predict the impact of the amino acid variant on the protein’s functional and physio-chemical properties, and generate a heterogeneous report comprising of tables, figures, and texts [37].
Evolutionary conservation analysis of ADGRE2
The evolutionary conservation of amino acids in ADGRE2 encoded protein was predicted through the ConSurf tool, which is a web server that utilized the Bayesian method to highlight the protein regions that are important for its structure and functions (https://consurfdb.tau.ac.il). It gives the residue conservation score ranging from 1 to 9, where a score of 1 indicates the least conserved residue, while a score of 9 represents the most highly conserved residues [38].
Sub-cellular localization
The Subcellular localization of the ADGRE2 protein was predicted using Deeploc1.0 (https://services.healthtech.dtu.dk/services/DeepLoc-1.0/). This algorithm predicts the location of protein based on information regarding amino acid sequences by identifying those sequences that are important for the localization of a protein in a particular cellular compartment [39].
IRB and sample collection
Prior to the initiation of the experimental analysis, IRB approval (IRB number; 2024-IRB-A-24/24, approved on 6th may, 2024), was attained from the Ethical Review Committee, ASAB, NUST. Blood samples from the 105 patients suffering from CML were obtained from the Combined Military Hospital (CMH), Rawalpindi, Pakistan. CML patients with other morbidities were excluded from this study. Samples from 102 healthy individuals were taken as controls.
Primer designing
The Primers for ARMS PCR were designed using the tool Primer 1 (https://primer1.soton.ac.uk/primer1.html), and the primer sequences are shown in Table 1. Primers were designed to investigate the genotype of the ADGRE2 gene in the Control and Patient data. The primers were validated through UCSC in silico PCR (https://genome.ucsc.edu/cgi-bin/hgPcr).
Table 1.
ADGRE2 primers for ARMS PCR
| D67N (rs765071211) | Common Primer | CCCCATGGAGACTTGTGCCG |
| C primer (Wild type) | GCTGCCCTCAAGCCTCTGTCCT | |
| T Primer (variant) | GCCCAGGCTGGTCTTGAATTCC |
Genotype analysis
In this study, the genomic DNA from the blood samples was isolated using the phenol-chloroform extraction method. The genotyping primers designed in the previous step were used to detect the presence of the target genetic variant in the ADGRE2 gene. ARMS PCR was used to perform genotyping analysis using the Applied Biosystems™ Veriti™ 96-Well Thermal Cycler. For each sample, 12 µl PCR reaction mixture was made, which was composed of 6 µl Solis Biodyne FIREpol master mix, 1 µl common primer, 1 µl allele-specific primers,1 µl DNA sample, and 3 µl PCR water. Conditions used for PCR are given in Table 2.
Table 2.
PCR conditions
| Steps | Temperature | Time |
|---|---|---|
| Initial Denaturation | 95◦C | 10 min |
| Denaturation | 95◦C | 45 s |
| Annealing | 60◦C | 45 s |
| Extension | 72◦C | 45 s |
| Final extension | 72◦C | 7 min |
| Storage | 4◦C | ∞ |
Statistical analysis
The genotype data were statistically analyzed through Graph Pad Prism 9 (La Jolla, California, United States). Fisher’s exact test was performed on the variant data attained from healthy controls and CML patients. Additionally, odds ratio, relative risk, and 95% confidence intervals were also determined. Statistical significance was determined at a P-value < 0.05.
Results
Variant identification
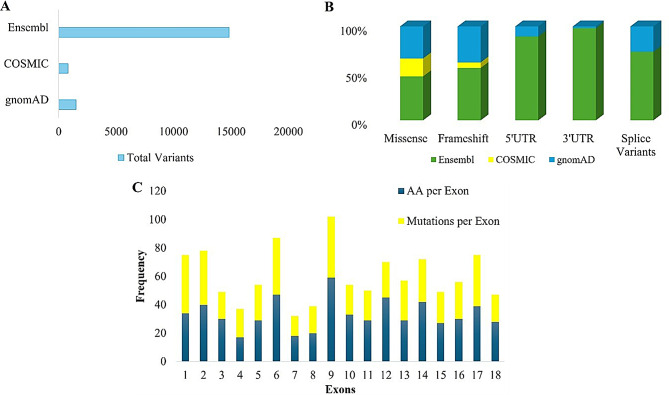
A total of 17,162 variants were retrieved from all three databases, of which 1,514 were from gnomAD, 802 from COSMIC, and 14,846 from Ensemble, as shown in Fig. 1A. Variants were then grouped into various categories based on their consequences, which include missense variants, frameshift variants, 5`UTR, 3`UTR, and splice variants (Fig. 1B), and from this data, 674 unique non-synonymous ADGRE2 gene variants were determined for further analyses. ADGRE2 gene is comprised of 21 exons that encode their respective amino acids. This study also found that exon 16 of the ADGRE2 gene encodes the highest number of amino acid residues, while in respect of non-synonymous variant frequency, exon number 9 contained the highest number of variants, as shown in Fig. 1C.
Fig. 1.
Identification of ADGRE 2 genetic variants (A) ADGRE2 variants retrieved from Ensembl, COSMIC, gnomAD (B) Total count of various ADGRE2 variants retrieved from the three databases (C) Frequency of amino acids per exon and variants per exon
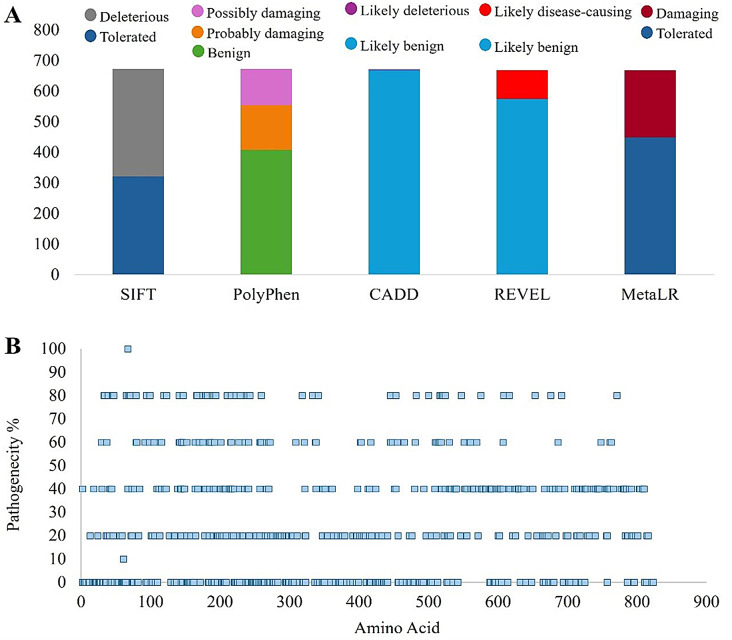
Variant pathogenicity analysis
The pathogenicity of the filtered non-synonymous variants was evaluated through various computational tools mentioned in the methodology. For that purpose, the pathogenicity scores given by the tools for each variant were determined, and the pathogenicity percentage for each variant was calculated (Fig. 2A). For this study, a pathogenicity percentage of > 80% was determined as the threshold. Out of 674 initial non-synonymous variants, only one variant was found to have a pathogenicity percentage of > 80% (Fig. 2B). A thorough analysis of the selected variants led to the selection of the non-synonymous variant rs765071211 for further in silico analysis due to its highest pathogenicity percentage, i.e., 100%. This variant caused amino acid alteration (D67N) at residue number 67 in the Egfca_6 domain of ADGRE2 encoded protein EMR2.
Fig. 2.
Analysis of pathogenicity of ADGRE2 non-synonymous variants (A) Filtration of SNPs through various algorithms (B) Pathogenicity of non-synonymous SNPs predicted through computational tools (SIFT, PolyPhen, CADD, REVEL, MetaLR)
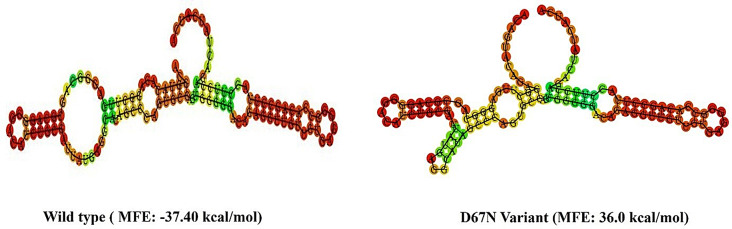
Effect of rs765071211 polymorphism on RNA secondary structure
The impact of selected SNP on the secondary structure of ADGRE2 mRNA was also predicted. For this purpose, minimum free energy (MFE) for the variant and wild-type mRNA were determined and compared. The secondary structure of variant mRNA exhibited a significant change compared to the wildtype mRNA, revealing a prominent impact of altered allele on the structure and stability of ADGRE2 mRNA. MFE value determined for the wildtype and rs765071211 variant demonstrated that reference allele C was stabilizing in nature with a low MFE value (-37Kcal/mol) while its corresponding variant allele T destabilized the mRNA secondary structure and had a high MFE value of 36Kcal/mol (Fig. 3). Decreased mRNA stability can alter the levels of the translated proteins [40].
Fig. 3.
Effect of ADGRE2 variant on its RNA secondary structure. The lower the value of MFE, the higher the stability of mRNA secondary structure and vice versa
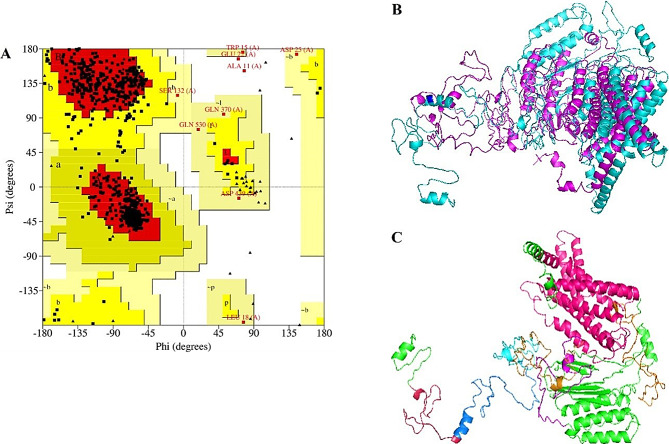
Structure prediction and validation
EMR2 is the protein encoded by the ADGRE2 gene. The structure of EMR2 was predicted through I-TASSER, which uses the threading technique to predict the structure of the protein. I-TASSER predicted five models of EMR2 protein, and the model with the highest C-score value was selected for further analysis. The 3-dimensional protein structure was visualized through PyMOL. The predicted structure was also validated through ERRAT. The overall quality factor of the model was determined to be 94.4809. According to the Ramachandran plot analysis conducted through PROCHECK, 88.4% of residues were in the highly favored region, followed by 10.4% of residues that were in the additional allowed region, and only 0.3% of the residues were found to be in the disallowed region (Fig. 4A). D67N variant 3D structure was attained by inducing amino acid modification in PyMOL through the mutagenesis plugin. The wildtype and variant proteins were superimposed, and it was determined that there was a high degree of structural deviation in the variant protein compared to the wildtype, as indicated by the RMSD value of 6.34Å (Fig. 4B). Higher values of RMSD indicate that there is a higher structural dissimilarity between the two protein structures [41].
Fig. 4.
Prediction and validation of EMR2 3D structure. (A) Ramachandran plot of EMR2 predicted structure showing 98.8% residues I allowed region. (B) Superimposition of native EMR2 (cyan) and D67N variant (magenta), RMSD was 6.34Å. (C) ADGRE2 Domains including egf_5 (raspberry color), egfca_6 (1)(marine blue), egfca_6 (2)(sand color), egfca_6 (3) (magenta color), EGF_3, GPS_3 (cyan color ) and 7tmB2 EMR domain (orange color)
EMR2 protein domains were also analyzed through InterPro, and their constituting amino acids were highlighted in PyMOL (Fig. 4C). It was revealed that EMR2 had seven domains. Domain Egf-like 1 contained amino acids from 28 to 66. Amino acid residues from 67 to 118, 119–162, 163–211, and 212–260 were part of Egfca-like 2, Egfca-like 3, Egfca-like 4, and Egfca-like 5 domains, respectively. GPS_3 domain was comprised of 52 amino acid residues from 478 to 529. Lastly, it was found that 7tmB2 EMR was the largest domain with 262 amino acids from 533 to 795.
Stability analysis
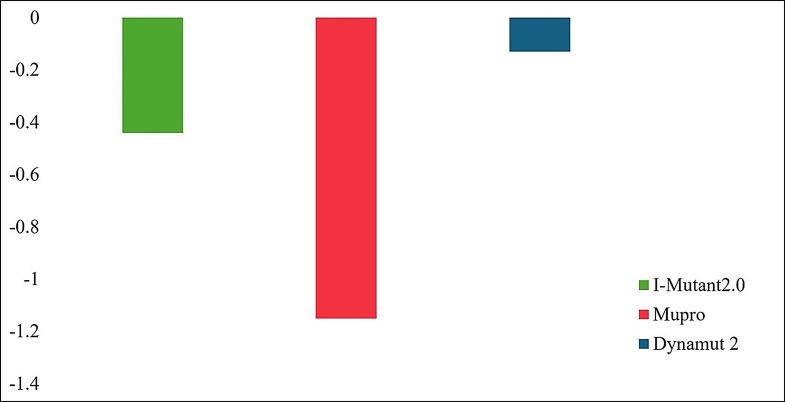
It is a general consensus that the majority of pathogenic non-synonymous SNPs affect protein structure stability. The effect of variant D67N on EMR2 stability was determined through various computational tools, including MUpro, I-Mutant, and Dynamut, and the predicted free energies for the variant were compared. I-Mutant2.0 indicated a DDG value of -0.44 kcal/mol for the D67N variant, indicating that the stability of the variant was decreased. Free energy analysis by MUpro also showed similar results, indicating that the stability of the variant was decreased with DDG value of -1.1507558 kcal/mol. Results from DynaMut 2 revealed that D67N decreased the DDG value of the protein (-0.04 kcal/mol), lowering its stability. All three computational tools predicted the D67N variant of EMR2 as destabilizing in nature (Fig. 5), which can potentially disrupt the normal structure and functioning of the protein.
Fig. 5.
Stability analysis of D67N through I-Mutant 2.0, MUpro, and Dynamut 2
Molecular characteristics analysis
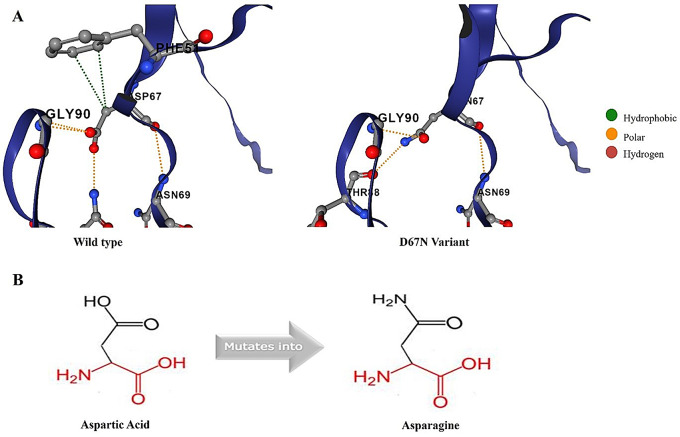
The molecular characteristics of non-synonymous SNP D67N were also investigated. Intramolecular interactions of the wild-type and variant amino acids were determined and compared through DynaMut 2. It was found that wildtype amino acid made nine intramolecular bonds (two hydrophobic bonds, four polar bonds, and three hydrogen bonds), while the variant amino acid made five intramolecular interactions, including two hydrogen bonds and three polar bonds (Fig. 6A). Project HOPE revealed that the D67N variation resulted in the change in protein net charge. The wildtype residue was a negatively charged amino acid, which was substituted with a neutral amino acid. Furthermore, the analysis also showed that this SNP can also result in the loss of interactions with other molecules and abolish the function of the protein (Fig. 6B).
Fig. 6.
Molecular characteristics of wildtype and D67N EMR2 proteins. (A) Analysis and comparison of intramolecular interactions of wildtype and variant amino acids with neighboring residues. (B) Structural analysis through Project HOPE indicating that negatively charged aspartic acid (D) mutates into electrochemically neutral amino acid asparagine (N)
Evolutionary conservation analysis
Amino acids located in the biologically active regions demonstrate high sequence conservation. Any variation within these residues can result in disruption of the normal biological activities of the protein. ConSurf server was used to evaluate the evolutionary conservation of the EMR2 protein at individual amino acid residues. This tool gave a complete analysis of the EMR2 protein. However, only that amino acid residue was focused, which was selected as the most highly pathogenic non-synonymous SNP. ConSurf analysis revealed that Aspartic acid at residue number 67 was highly conserved with a conservation score of 9. Furthermore, ConSurf also predicted D67N as a conserved and exposed residue with high functional significance (Fig. 7). The presence of highly conserved amino acids on the surface of the protein assists in showing their structural or functional significance.
Fig. 7.
Illustration of evolutionary conservation of EMR2 amino acid residues
Sub-cellular localization
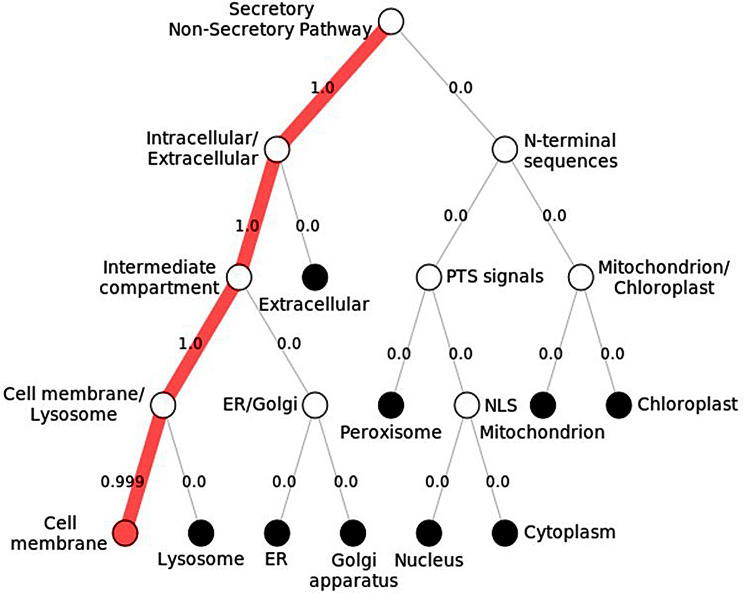
Subcellular localization of ADGRE2 encoded protein was also predicted, and Deeploc 1.0 gave the potential localization sites as well as the likelihood scores. It was revealed that EMR2 is a membrane-associated protein, and it is primarily allocated in the plasma membrane of cells, with a likelihood score of 0.9995, which is in line with the literature, as ADGRE2 is famous for its significant role in cell-cell interaction. The inner workings of the cell are also shown in Fig. 8.
Fig. 8.
Subcellular localization of EMR2 protein
Genotype analysis
Genotype analysis was performed on extracted DNA samples from both controls and CML patients to determine the presence of rs765071211 (C/T) in the ADGRE2 gene. For this purpose, ARMS PCR was used. The distribution frequency of both alleles of ADGRE2 genetic variant rs765071211 for CML positive samples and control are given in Table 3. This study showed that wild-type genotype CC showed no statistical significance with either control or disease group. In contrast, variant genotype TT showed statistical significance (P = < 0.005), and it was associated with an elevated risk of CML with an Odds ratio (OR) of 7.278 and a relative risk (RR) value of 2.381. In contrast, the heterozygous genotype CT was also found to be statistically significant (P < 0.005), but it was found to have a protective effect in this regard (RR = 0.3000; OR = 0.1250).
Table 3.
Genotype analysis of rs765071211 (C/T) through ARMS PCR
| Genotype | Patient (%) |
Control (%) |
Odds Ratio |
95% CI Odds Ratio |
Relative risk | 95% CI Relative risk |
P value |
|---|---|---|---|---|---|---|---|
| CC | 22.41 | 30.00 | 0.6741 | 0.2967 to 1.626 | 0.8254 | 0.5078 to 1.226 | < 0.005 |
| TT | 67.24 | 22.00 | 7.278 | 2.991 to 17.27 | 2.381 | 1.638 to 3.610 | |
| CT | 10.34 | 48.00 | 0.1250 | 0.04608 to 0.3473 | 0.3000 | 0.1406 to 0.5739 | |
| Alleles | |||||||
| C-Allele | 27.59 | 54.00 | 0.3245 | 0.1473 to 0.7241 | 0.5759 | 0.3658 to 0.8566 | < 0.005 |
| T-Allele | 72.41 | 46.00 | 3.082 | 1.381 to 6.789 | 1.737 | 1.167 to 2.734 | |
Allele frequencies for ADGRE2 variant rs765071211 were also determined (Table 3). It was revealed that the frequency of reference allele C was higher in the healthy controls compared to the CML group and had a protective effect (P < 0.005, OR = 0.3245, RR = 0.5759). On the other hand, allele T was more abundantly present in the CML group (72.41%) compared to the control group and was found to be significantly associated with the disease (OR = 3.082, RR = 1.737, P < 0.005).
ADGRE2 polymorphic variant was also compared in CML patients and control with respect to gender (Table 4). Variant genotype TT was found to be statistically significant in both males and females and showed an association with the disease. Meanwhile, heterozygous genotype CT was found to have a significant protective effect in both sexes (Table 4) Comparison of ADGRE2 polymorphismrs765071211 (C/T) in CML patients and control with respect to gender.
Table 4.
Genotype analysis on the basis of gender
| Genotype | Gender | Patient (%) |
Control (%) |
Odds Ratio | 95% CI Odds Ratio |
Relative risk | 95% CI Relative risk |
P value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | Female | 21.74 | 25.14 | 0.5128 | 0.1713 to 1.682 | 0.6481 | 0.2747 to 1.348 | < 0.005 | ||
| TT | 62.50 | 24.32 | 5.185 | 1.787 to 14.76 | 2.569 | 1.372 to 4.961 | ||||
| CT | 13.04 | 50.54 | 0.2200 | 0.06191 to 0.8694 | 0.3500 | 0.1181 to 0.8912 | ||||
| CC | Male | 22.86 | 15.38 | 1.630 | 0.3591 to 8.516 | 1.126 | 0.6708 to 1.552 | < 0.005 | ||
| TT | 68.57 | 15.38 | 12.00 | 2.375 to 58.11 | 1.846 | 1.279 to 3.032 | ||||
| CT | 8.57 | 69.23 | 0.04167 | 0.01010 to 0.2478 | 0.2813 | 0.09956 to 0.6083 |
Discussion
CML is a myeloproliferative neoplasm that is primarily caused by genetic alteration. However, the role of SNPs in the development and progression of CML is not completely established for several reasons, including a smaller number of candidate genes that are associated with CML [42, 43]. A previous study has described the association of genetic loci (17p11.1) with the risk of CML in the Korean population [44]. Non-synonymous SNPs can cause an alteration in protein structure and function and can lead towards a disease phenotype [45]; however, there is limited information regarding the impact of ADGRE2 polymorphism on its protein structure and activity. Hence, it is crucial to determine the pathogenic non-synonymous SNPs in the ADGRE2 gene, as these variants can have a damaging role and alter the structure and function of the protein.
The current study analyzed a total of 17,162 variants gathered from three databases, and only non-synonymous variants were filtered and selected for further analysis. These variants were then evaluated for pathogenicity using the computational tools Polyphen2, SIFT, REVEL, MeraLR, and CADD [46]. Based on their results, one pathogenic variant, D67N, was selected. This variant involves the substitution of aspartic acid with asparagine at position 67 of the EMR2 protein and is identified by the rsID rs765071211 (C/T). Following these pathogenicity analyses, this SNP was chosen and further investigated via insilico and experimental analysis. I-Tasser was utilized to predict the 3D structure of the EMR2 protein. The tool generated five different models, and the model with the highest confidence score (-1.12) was selected for further analysis. The impact of this variant on mRNA secondary structure was also predicted, and the variant was found to decrease the mRNA stability. Previously, studies have demonstrated that altered mRNA stability can lead to aberrant protein levels that can cause disease [47].
It was also shown that the selected D67N variant of ADGRE2 was located on the Egfca_2 domain of EMR2. This domain has been shown to be involved in interactions with other proteins. Tools such as I-Mutant and Project HOPE were used to study the structural and functional effects of the D67N missense variant on ADGRE2, which resulted in decreased protein stability. A decrease in the protein stability can affect the functions of the protein and can lead to damaging effects [48]. Destabilization of cell surface receptors can disrupt their interactions with ligand molecules that can alter their signaling cascade [49]. The Dynamut 2 showed that the molecular flexibility of the variant structure was decreased. Structural flexibility is an essential attribute of protein, and without it, proteins cannot perform their normal biological functions. It has been reported that altered protein flexibility can disrupt protein-protein interactions and can cause disorders [50]. Aspartic acid at residue 67 in EMR2 is a highly conserved and exposed amino acid, and its substitution with asparagine can alter protein functionality, which can have a damaging impact and cause disease. Alteration in the evolutionarily conserved residues can affect the protein activity and may cause diseases [51].
This study did not determine how the allelic frequencies of the identified SNP may differ across CML risk groups and their impact on prognosis, treatment response to first- and second-generation TKIs, and the attainment of deep molecular response (DMR). Additionally, while our focus was on genetic analysis, further exploration of the clinical characteristics of patients with CML carrying this SNP could enhance underlying molecular impact of this SNP on the pathogenesis of CML.
To investigate the association of selected variant rs765071211 with CML, ARMS PCR was performed. The data showed that genotype GG had no statistical significance, while variant genotype TT was associated with CML, and heterozygous genotype CT was found to have a protective effect in both sexes. A past study has also reported that the missense variant of ADGRE2 (C492Y) resulted in altered protein activity and was found to be associated with vibratory urticaria disease [18]. The association of genetic variants with diseases can give an insight into disease susceptibility, and they can also be used as markers for early diagnosis [52]. Hence, this non-synonymous SNP rs765071211 could also be a potential maker for CML.
Conclusion
Previous studies on CML have not provided insights into the role of SNPs in disease development and progression, emphasizing the need to identify new pathogenic variants that could potentially serve as novel treatments. This study focused on the pathogenic non-synonymous SNPs of the ADGRE2 gene and variant rs765071211 (D67N), identified as the most pathogenic with 100% pathogenicity prediction. This SNP also altered the protein’s structural stability, function, and flexibility, as determined through in silico analysis. This study further showed that the damaging SNP rs765071211 was associated with CML. Additional studies, including larger cohort analyses and functional investigations, are needed to validate the clinical significance of rs765071211 as a genetic marker for CML. Furthermore, the expression profile of ADGRE2 should also be explored. Further in vitro and in vivo studies are required to establish the role of this variant at molecular levels.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting project number (RSPD2024R729), King Saud University, Riyadh, Saudi Arabia, for funding this project.
Author contributions
Conceptualization, AF, MS, and YS; methodology, MS; experimentation validation AF, MS, TA, SR, YB, FMH, HJ, SZ MK, formal analysis, AF, HJ; investigation, MK, FMH, SZ; resources, MS, JHT, TA; data curation; writing—original draft preparation SR; writing—review and editing, AF; visualization, MS, supervision, YB; project administration, MS; All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Researchers Supporting project number (RSPD2024R729), King Saud University, Riyadh, Saudi Arabia, for funding this project. The funding body has no role in study design.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yasmin Badshah, Email: yasmeenb_1982@yahoo.com.
Suhail Razak, Email: smarazi@ksu.edu.sa.
References
- 1.Baccarani M, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osman AEG, Deininger MW. Chronic Myeloid Leukemia: Modern therapies, current challenges and future directions. Blood Rev. 2021;49:100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gianelli U, et al. International Consensus Classification of myeloid and lymphoid neoplasms: myeloproliferative neoplasms. Virchows Arch. 2023;482(1):53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minciacchi VR, Kumar R, Krause DS. Chronic myeloid leukemia: a model disease of the past, present and future. Cells. 2021;10(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdulmawjood B, et al. Genetic biomarkers in chronic myeloid leukemia: what have we learned so far? Int J Mol Sci. 2021;22(22):12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am J Hematol. 2020;95(6):691–709. [DOI] [PubMed] [Google Scholar]
- 7.Deng N, et al. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8(66):110635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soltani I, et al. Comprehensive in-silico analysis of damage associated SNPs in hOCT1 affecting Imatinib response in chronic myeloid leukemia. Genomics. 2021;113(1, Part 2):755–66. [DOI] [PubMed] [Google Scholar]
- 9.Bruzzoni-Giovanelli H, et al. Genetic polymorphisms associated with increased risk of developing chronic myelogenous leukemia. Oncotarget. 2015;6(34):36269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.I KY, et al. Activation of Adhesion GPCR EMR2/ADGRE2 Induces Macrophage Differentiation and Inflammatory Responses via Gα(16)/Akt/MAPK/NF-κB Signaling Pathways. Front Immunol. 2017;8:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tseng WY, Stacey M, Lin HH. Role of Adhesion G Protein-Coupled Receptors in Immune Dysfunction and Disorder. 2023. 24(6). [DOI] [PMC free article] [PubMed]
- 12.Waddell LA, et al. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front Immunol. 2018;9:2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CH, et al. Increased EMR2 expression on neutrophils correlates with disease severity and predicts overall mortality in cirrhotic patients. Sci Rep. 2016;6:38250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhudia N, et al. G Protein-Coupling of Adhesion GPCRs ADGRE2/EMR2 and ADGRE5/CD97, and Activation of G Protein Signalling by an Anti-EMR2 Antibody. Sci Rep. 2020;10(1):1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang D, et al. Adhesion GPCR ADGRE2 Maintains Proteostasis to Promote Progression in Acute Myeloid Leukemia. Cancer Res. 2024;84(13):2090–108. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Wu S, Alachkar H. Characterization of upregulated adhesion GPCRs in acute myeloid leukemia. Transl Res. 2019;212:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atilla E, Benabdellah K. The black hole: CAR T cell therapy in AML. Cancers. 2023;15(10):2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyden SE et al. Vibratory Urticaria Associated with a Missense Variant in ADGRE2. New England Journal of Medicine, 2016. 374(7): pp. 656–663. [DOI] [PMC free article] [PubMed]
- 19.I K-Y et al. Stimulation of Vibratory Urticaria-Associated Adhesion-GPCR, EMR2/ADGRE2, Triggers the NLRP3 Inflammasome Activation Signal in Human Monocytes. Front Immunol, 2021. 11. [DOI] [PMC free article] [PubMed]
- 20.Martin FJ, et al. Ensembl 2023. Nucleic Acids Res. 2023;51(D1):D933–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate JG, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis NM, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99(4):877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rentzsch P, et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber AR, et al. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):pW70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40(1):82–92. [Google Scholar]
- 32.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, et al. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–91. [Google Scholar]
- 34.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33(suppl2):pW306–W310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct Funct Bioinform. 2006;62(4):1125–32. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigues CH, Pires DE, Ascher DB. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2021;30(1):60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venselaar H, et al. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burley SK, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almagro Armenteros JJ, et al. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33(21):3387–95. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q et al. Translation affects mRNA stability in a codon-dependent manner in human cells. 2019. 8. [DOI] [PMC free article] [PubMed]
- 41.Amajala K et al. Homology Modeling and Structural Analysis of DNA Binding Response Regulator of Bacillus anthracis. Int J Sci Res, 2013. 2.
- 42.Hehlmann R. Chronic myeloid leukemia in 2020. Hemasphere. 2020;4(5):e468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, et al. Management of chronic myeloid leukemia and pregnancy: A bibliometric analysis (2000–2020). Front Oncol. 2022;12:826703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DH, et al. A genome-wide association study identifies novel loci associated with susceptibility to chronic myeloid leukemia. Blood J Am Soc Hematol. 2011;117(25):6906–11. [DOI] [PubMed] [Google Scholar]
- 45.Dakal TC, et al. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms in IL8 gene. Sci Rep. 2017;7(1):6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan Y-Y, et al. MBOAT1 homozygous missense variant causes nonobstructive azoospermia. Asian J Androl. 2022;24(2):186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Q, Bazzini AA. Translation and mRNA Stability Control. Annu Rev Biochem. 2023;92:227–45. [DOI] [PubMed] [Google Scholar]
- 48.Teilum K, Olsen JG, Kragelund BB. Protein stability, flexibility and function. Biochim et Biophys Acta (BBA)-Proteins Proteom. 2011;1814(8):969–76. [DOI] [PubMed] [Google Scholar]
- 49.Froning K et al. Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics. 2020. 11(1): p. 2330. [DOI] [PMC free article] [PubMed]
- 50.Teilum K, Olsen J, Kragelund BB. Functional aspects of protein flexibility. Cell Mol Life Sci. 2009;66:2231–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashenberg O, Gong LI, Bloom JD. Mutational effects on stability are largely conserved during protein evolution. Proceedings of the National Academy of Sciences, 2013. 110(52): pp. 21071–21076. [DOI] [PMC free article] [PubMed]
- 52.Khan N, et al. Investigating pathogenic SNP of PKCι in HCV-induced hepatocellular carcinoma. Sci Rep. 2023;13(1):12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.