ABSTRACT
In Gram-negative bacteria, the outer membrane (OM) is asymmetric, with lipopolysaccharides (LPS) in the outer leaflet and glycerophospholipids (GPLs) in the inner leaflet. The asymmetry is maintained by the Mla system (MlaA-MlaBCDEF), which contributes to lipid homeostasis by removing mislocalized GPLs from the outer leaflet of the OM. Here, we ascribed how Pseudomonas aeruginosa ATCC 27853 coordinately regulates pathways to provide defense against the threats posed by the deletion of mlaA. Especially, we explored (i) the effects on membrane lipid composition including LPS, GPLs, and lysophospholipids, (ii) the biophysical properties of the OM such as stiffness and fluidity, and (iii) the impact of these changes on permeability, antibiotic susceptibility, and membrane vesicles (MVs) generation. Deletion of mlaA induced an increase in total GPLs and a decrease in LPS level while also triggering alterations in lipid A structures (arabinosylation and palmitoylation), likely to be induced by a two-component system (PhoPQ-PmrAB). Altered lipid composition may serve a physiological purpose in regulating the mechanobiological and functional properties of P. aeruginosa. We demonstrated an increase in cell stiffness without alteration of turgor pressure and inner membrane (IM) fluidity in ∆mlaA. In addition, membrane vesiculation increased without any change in OM/IM permeability. An amphiphilic aminoglycoside derivative (3’,6-dinonyl neamine) that targets P. aeruginosa membranes induced an opposite effect on ∆mlaA strain with a trend toward a return to the situation observed for the WT strain. Efforts dedicated to understanding the crosstalk between the OM lipid composition, and the mechanical behavior of bacterial envelope, is one needed step for designing new targets or new drugs to fight P. aeruginosa infections.
IMPORTANCE
Pseudomonas aeruginosa is a Gram-negative bacterium responsible for severe hospital-acquired infections. The outer membrane (OM) of Gram-negative bacteria acts as an effective barrier against toxic compounds, and therefore, compromising this structure could increase sensitivity to antibiotics. The OM is asymmetric with the highly packed lipopolysaccharide monolayer at the outer leaflet and glycerophospholipids at the inner leaflet. OM asymmetry is maintained by the Mla pathway resulting in the retrograde transport of glycerophospholipids from the OM to the inner membrane. In this study, we show that deleting mlaA, the membrane component of Mla system located at the OM, affects the mechanical and functional properties of P. aeruginosa cell envelope. Our results provide insights into the role of MlaA, involved in the Mla transport pathway in P. aeruginosa.
KEYWORDS: P. aeruginosa, outer membrane, membrane biophysics, lipids, cell envelope
INTRODUCTION
In Gram-negative bacteria, like Pseudomonas aeruginosa, the cell envelope is made up of an inner phospholipid bilayer (IM), a periplasm that contains a thin peptidoglycan cell wall and an outer membrane (OM) presenting an asymmetric distribution of lipids on both leaflets. The outer leaflet is almost exclusively composed of lipopolysaccharides (LPS), whereas the inner leaflet mainly contains glycerophospholipids (GPLs). LPS is a negatively-charged amphiphilic molecule composed of three covalently linked moieties: lipid A, a core oligosaccharide, and a polysaccharide called O-antigen. GPLs are mainly phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), which are also found in the inner membrane (1).
The asymmetric distribution of LPS and GPLs in the OM is critical for maintaining membrane mechanical properties required for cell fitness and protection against environmental stress and antibiotics. This is elucidated by the fact that a loss of OM asymmetry is responsible for a decrease in permeability barrier function (2, 3), a reduction in virulence (4–7), and the dwindling of intrinsic resistance to some antimicrobials (8–10).
The maintenance of lipid asymmetry is therefore crucial and is in part controlled by the Mla (MlaA/VacJ-MlaC-MlaFEDB) system. The Mla system, composed of 6 proteins, facilitates retrograde phospholipid transport from the OM back to the IM (11). The MlaA (also annotated as VacJ) component is highly conserved among Gram-negative bacteria (Table 1). It was first identified in Shigella flexeneri as virulence-associated protein chromosome locus J (6). It is an integral alpha-helical OM protein (12) that transfers phospholipids from the outer leaflet of OM to the soluble periplasmic MlaC with no preference for specific phospholipids (13). MlaC (14) transfers the phospholipids to the multicomponent system MlaFEDB (13, 15). This ATP-binding-cassette (ABC) transport system in the IM comprises the IM permease MlaE, the IM ABC-type ATPase MlaF, the periplasmic subunit of MlaFEDB MlaD, which is anchored in the IM by a transmembrane helix, and a cytoplasmic accessory protein MlaB (13, 16).
TABLE 1.
MlaA amino acids sequence conservation in P. aeruginosa and other Gram-negative bacteria
| Microorganisms | Accession number | MlaA/VacJ | Amino acids |
|---|---|---|---|
| P. aeruginosa | AVK06939.1 | 100% | 234 |
| Pseudomonas putida | AFO47095.1 | 69.36% | 235 |
| Pseudomonas syringae | QWB08392.1 | 66.67% | 233 |
| Acinetobacter baumannii | BCR39766.1 | 41.75% | 299 |
| Neisseria gonorrhoeae | WP_010951410.1 | 38.39% | 277 |
| Campylobacter jejuni | EDP6655880.1 | 36.65% | 232 |
| Caulobacter crescentus | QXZ52028.1 | 36.70% | 286 |
| Vibrio cholerae | ACP06275.1 | 35.84% | 254 |
| Haemophilus influenzae | AIT66950.1 | 33.82% | 250 |
| Shigella flexneri 2 a | NP_708228.1 | 33.04% | 251 |
| Escherichia coli | QKU50363.1 | 33.04% | 251 |
| Salmonella enterica typhimurium | WP_000776787.1 | 32.17% | 251 |
In addition to the Mla system, lipid asymmetry in Gram-negative bacteria is maintained by the OM-phospholipase PldA (17, 18) and the palmitoyl transferase PagP (19). PldA cleaves GPLs at the sn-1 or sn-2 position, generating fatty acids and lysophospholipids (20), which can flip to the inner leaflet of the OM, where they are trafficked back to the IM by poorly understood mechanisms. Phospholipids at the inner leaflet of the OM are typically inaccessible to the active site of PldA (17, 21, 22). Disturbance of the OM integrity and asymmetry leads to the activation of PldA via a Ca2+-dependent dimerization mechanism (22, 23). PagP regulates lipid asymmetry of the OM by transferring sn-1 palmitate (C:16) from GPLs at the inner leaflet to lipid A at the outer leaflet, producing palmitoylated lipid A (11). Palmitoylated lipid A increases membrane hydrophobicity (24) and activates the periplasmic σE stress response (25). PagP activity is an established marker for monitoring ectopic GPL exposure in the OM of E. coli (26, 27) and accumulation of GPLs in the outer leaflet of the OM in cells lacking MlaA (4, 11). On Klebsiella pneumoniae (24) and Salmonella typhimurium (28), PagP showed a bifunctional activity with additional capacity to transfer a palmitoyl moiety to PG, which also increases membrane hydrophobicity (24) and promotes the generation of membrane vesicles (MVs) (28).
Regulating cell wall properties, including lipid asymmetry, plays a key role in antibiotic resistance and bacterial survival and thus might provide a source for antibiotic targets. Since the OM is the first place where antibiotics interact with the bacteria, we selected to focus on the protein from the Mla system, located at the OM, MlaA. On P. aeruginosa WT and ∆mlaA, we characterized the lipidic composition, the mechanical and biophysical properties of the cell envelope, and the significance of changes induced by mlaA deletion. Especially, we explored the effects of mlaA deletion on (i) the lipid composition of membranes including LPS, GPLs, and lysophospholipids, (ii) the biophysical properties of the OM (stiffness, fluidity, etc.), and (iii) the impact of these changes on permeability, antibiotic susceptibility, and MVs generation. All studies were conducted in the presence and absence of the amphiphilic aminoglycoside derivative, 3',6-dinonyl neamine (3’,6-diNn), known to bind to the main components of the OM, LPS (29), and GPLs (30, 31).
RESULTS
Characterization of P. aeruginosa ∆mlaA strain
Gene-specific PCR followed by electrophoresis was performed on DNA extracted from P. aeruginosa WT and ∆mlaA. We confirmed the deletion of mlaA (Fig. S1C). The mRNA expression of other mla genes (mlaB, mlaC, mlaD, mlaE, and mlaF) in ∆mla over WT strain showed an increase from 1.3-fold to 2.6-fold (Fig. S1D). P. aeruginosa strains, WT, ∆mlaA, and ∆mla att7::mlaA did not show any differences in growth curves, intracellular survival, and morphology (5).
Deletion of mlaA induced an increase in glycerophospholipids (GPLs) at the OM of P. aeruginosa. 3’,6-dinonyl neamine increased GPLs in the WT strain but not in P. aeruginosa ∆mlaA
The Mla system maintains OM-lipid asymmetry and traffics GPLs between the OM and IM bilayers (11, 32, 33). We hypothesized that mlaA deletion induced altered OM lipid composition. Therefore, we determined the total amounts of GPLs in the OM of P. aeruginosa WT and ∆mlaA. We then investigated the changes in individual GPLs, including phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL). To assess the potential role of PldA and/or PagP, we monitored the amounts of lyso-derivatives (lysophosphatidylethanolamine, LPE; lysophosphatidylglycerol, LPG). The degree of unsaturation was determined as well.
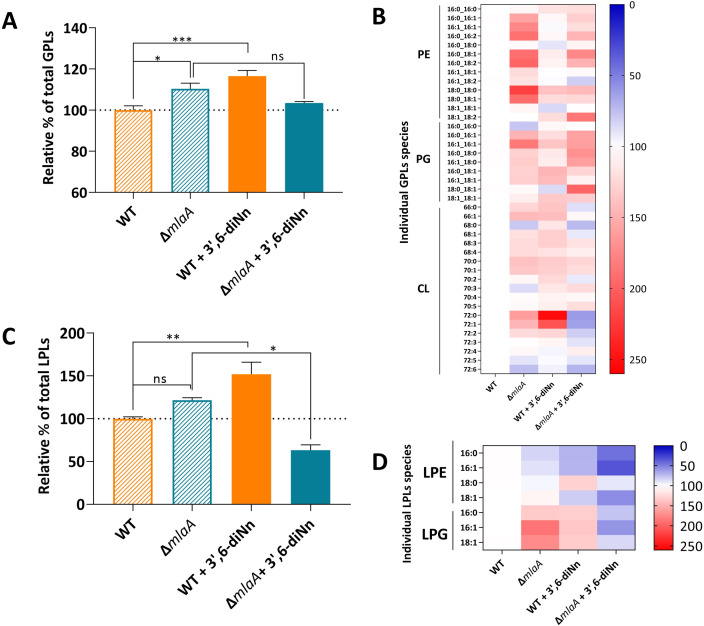
Focusing on total GPLs, we showed that deletion of mlaA (∆mlaA) increased total GPLs in P. aeruginosa OM (110% ± 6%) in comparison with P. aeruginosa WT (Fig. 1A). A deeper analysis of individual lipid headgroups (PE, PG, and CL) (Fig. 1B) revealed a significant increase in PE in ∆mlaA strain compared with WT bacteria (Fig. S3A). The increase was significant for the PE species (16:0_16:1, 16:0_16:2, 16:0_18:1, and 18:1_18:2) (Fig. S2A).
Fig 1.
Glycerophospholipids and lysophospholipids profile P. aeruginosa WT and ∆mlaA in the presence or absence of 3’,6-dinonyl neamine (1× MIC, 2 µg/mL, 1 h). (A and C) Total glycerophospholipids. (B and D) Heat maps of individual glycerophospholipids and lysophosphoglycerides species. Red-filled, blue-filled, and white-filled lines referred to increased, decreased, and unchanged levels of lipid structures, respectively. Total lipids were determined by using a fluorescent probe FM4-64, and individual lipids PE, PG, CL, LPE, and LPG were analyzed by LC-MS. The results are expressed in % where WT is considered 100%. Statistical analysis was performed using one-way ANOVA with Tukey’s multiple-comparison test. (A and C) ***P < 0.001, **P < 0.01, *P < 0.05; ns P > 0.05.
The addition of 3’,6-dinonyl neamine on P. aeruginosa WT induced a significant increase in GPLs (117% ± 7%), whereas no significant change was observed when adding the compound to P. aeruginosa ∆mlaA (Fig. 1A). In WT strain, an analysis on the individual GPL species showed a significant increase in one CL species (72:0) (Fig. S2C) without any significant global change for PE, PG, and CL (Fig. S3A). In P. aeruginosa ∆mlaA, incubation with 3’,6-dinonyl neamine did not alter the glycerophospholipids species (Fig. S2A through C and S3A).
The presence of higher amounts of GPLs at the OM of P. aeruginosa ∆mlaA might activate enzymes like PldA and/or PagP, resulting in the production of lyso-derivatives (34). Lysophospholipids are the products of GPLs hydrolysis by endogenous PldA or by-products of PagP. Interestingly, the profile of total lyso-derivatives was similar to that observed for GPLs with a tendency that lyso-derivatives in OM of P. aeruginosa ∆mlaA strain increased (122% ± 4%) when compared with WT (100% ± 4%) (Fig. 1C). At the headgroup level (Fig. 1D), the increase mostly resulted from LPG compared with LPE (Fig. S2D and S3B). As for GPLs, the addition of 3’,6-dinonyl neamine resulted in an increase in total lyso-derivatives in WT (152% ± 20%) (Fig. 1C) resulting from the increase in LPG (Fig. S3B). In P. aeruginosa ∆mlaA, a huge decrease (66% ± 8%) in lyso-derivatives was observed upon the addition of the compound (Fig. 1C), which affected both LPE and LPG (Fig. S3B).
Comparing lipid acyl chains, we observed that monounsaturated and polyunsaturated acyl chains increased in P. aeruginosa ∆mlaA compared with WT (Fig. 1B; Fig. S3C). Addition of 3’,6-dinonyl neamine on P. aeruginosa WT increased the three subtypes of acylated chains with a significant effect observed for saturated chains (Fig. 1B and D; Fig. S2A through D and S3C), whereas no significant change was observed for ∆mlaA strain (Fig. 1B; Fig. S2A through D and S3C).
In conclusion, the deletion of mlaA induced an increase in the amounts of GPLs at the OM of P. aeruginosa with a major effect on PE. 3’,6-dinonyl neamine increased GPLs in the WT strain without any significant changes in P. aeruginosa ∆mlaA.
Deletion of mlaA in P. aeruginosa induced a decrease in LPS and a increase in lipid A modifications by increasing palmitoylation and arabinosylation. Higher expression of two two-component systems, PhoPQ and PmrAB, is observed as well
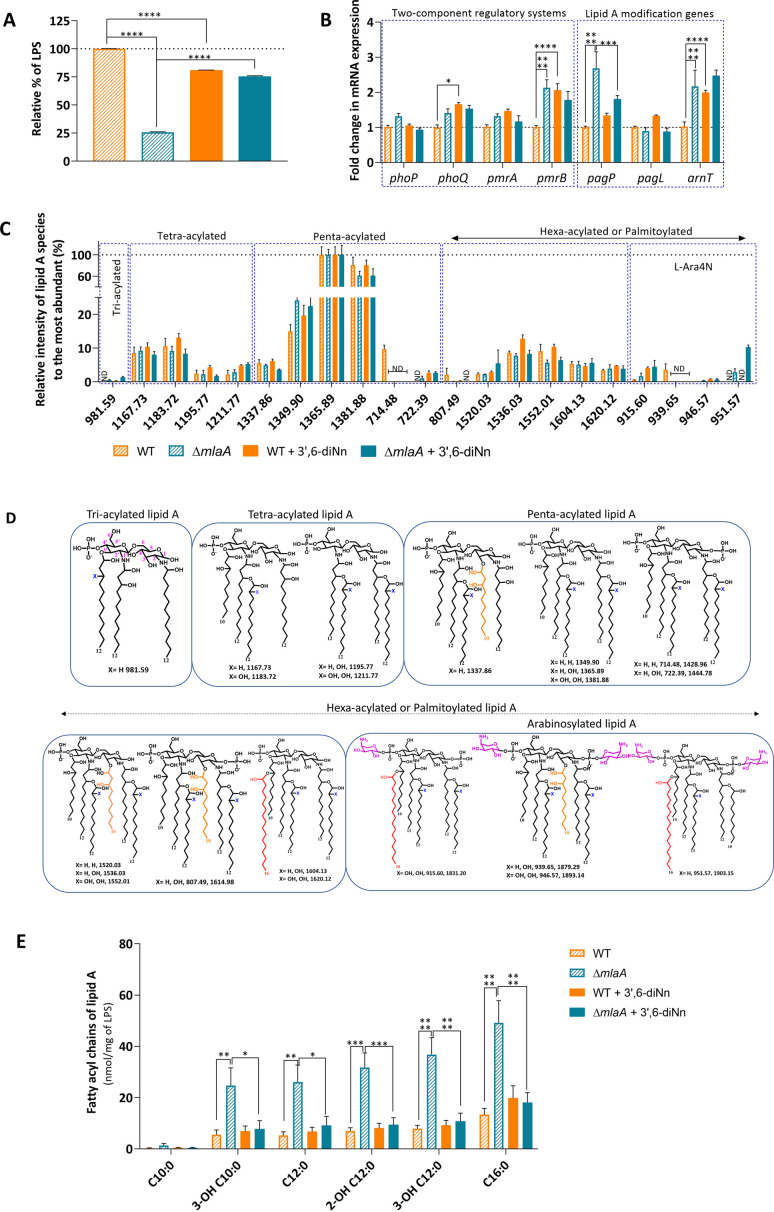
Perturbing OM asymmetry results in signal transduction across the periplasm and a lipid-mediated regulatory mechanism that controls LPS biosynthesis (17, 35). Accordingly, we hypothesized that mlaA deletion might influence the LPS levels in P. aeruginosa. Relative to the WT strain, the LPS levels of the ΔmlaA strain were significantly decreased (by 75%) (Fig. 2), similar to what was reported for ∆mlaF mutant of A. baumannii (36). The effect of 3’,6-dinonyl neamine is dependent upon the presence of MlaA, with a decrease in WT (by 19%) and a huge increase in ∆mlaA (Fig. 2A) strain (293%).
Fig 2.
Lipopolysaccharides (LPS) and lipid A modifications in P. aeruginosa WT and ∆mlaA non-treated and treated with 3’,6-dinonyl neamine (1× MIC, 2 µg/mL, 1 h). (A) Total LPS was measured by purpald assay. The results are expressed in relative % over P. aeruginosa WT, considered 100% (13,444 ± 16 µg/mL). (B) mRNA expression of two-component regulatory systems (phoP, phoQ, pmrA, and pmrB,) and lipid A modifications genes (pagP, pagL, and arnT). The results are expressed in fold change over P. aeruginosa WT. (C) MS peaks (m/z) obtained by ESI-MS (negative ion mode) shown in different boxes as tri-acylated, tetra-acylated, penta-acylated, hexa-acylated, palmitoylated, and arabinosylated with their relative intensity compared with the major peak for each condition (m/z 1365.89). (D) Predicted structures of lipid A structures from m/z were obtained. The positions of the acyl chain are hypothetical; 4-aminoarabinose added by ArnT is shown in purple, and palmitate chain added by PagP is shown in red on lipid A structures. (E) Fatty acids were isolated from lipid A structures and analyzed as FAMEs using gas chromatography - mass spectrometry (GC-MS). Statistical analysis was performed by two-way ANOVA with multiple comparisons (B, C, E) and one-way ANOVA with Tukey’s multiple-comparison test (A) ****P < 0.0001, ***P < 0.001, *P < 0.05, ND undetectable. All values are given in mean ± SEM, and the experiment was repeated thrice.
To know whether the decrease in LPS content in P. aeruginosa ∆mlaA is accompanied by structural modifications of the LPS moiety embedded in lipids (lipid A), we explored the activation of two two-component systems (TCSs), PhoP-PhoQ and PmrA-PmrB (37). In these systems, a sensory kinase, that is, PhoQ or PmrB, is located at the IM and is activated by an environmental signal, which in turn phosphorylates a cytoplasmic transcriptional regulator, PhoP or PmrA, respectively. PhoPQ and PmrAB are involved in OM remodeling by lipid A modifications in LPS through palmitoylation via PagP, which transfers a palmitoyl group from a GPL to the β-OH of the 3′-O-acyl fatty acyl group, deacylation via PagL, which removes 3-OH C:10, arabinosylation via ArnT, which adds 4-amino-4-deoxy-L-arabinose to one or both of the 1 and 4’-phosphate groups, and/or hydroxylation of secondary acyl chains via LpxO1 and LpxO2.
Deletion of mlaA (∆mlaA) led to the increased expression of the pmrB (2.1-fold), as well as the TCSs-induced lipid A modification genes (pagP 2.7-fold and arnT 2.2-fold) without any effect on pmrA, phoP, phoQ, and pagL (Fig. 2B). Upregulation of pagP, and arnT also reported in Salmonella enterica subsp. enterica serovar Typhimurium, helps to stabilize the OM, with an ultimate decrease in membrane fluidity and an increase in the surface charge (38, 39). Exposure of P. aeruginosa WT to 3’,6-dinonyl neamine induced an increase in the expression of pmrB (2.1-fold), phoQ (1.7-fold), and arnT (2.2-fold) (Fig. 2B). In the ∆mlaA strain, 3’,6-dinonyl neamine exposure decreased the expression of pagP (by 33%) (Fig. 2B).
To further assess changes in lipid A, we used high-resolution mass spectrometry. Overall, the mass spectrum of lipid A characterized 14 single-charged [M-H]- and seven double-charged [M-2H]2- lipid A structures that were tri-, tetra-, penta-, or hexa-acylated. Among these, some were palmitoylated and/or arabinosylated (Fig. 2C and D; Tables S2 and S3). In P. aeruginosa WT and ∆mlaA, before as well as after the treatment with 3’,6-dinonyl neamine, the most intense ion was m/z of 1365.89 (PLA1), which represents a mono-phosphate penta-acylated lipid A form (see detailed results in the supplementary part). Furthermore, we determined the levels of lipid A acyl chains after normalization to total LPS via derivatization/gas chromatography (Fatty Acid Methyl Esters; FAMEs) (Fig. 2A). We observed an increase in 2-OH C12:0, 3-OH C12:0 in P. aeruginosa ∆mlaA compared with WT strain. How these changes are related to the activity of LpxO1/2 known to α-hydroxylate the 2’- and 2- secondary acyl chains of lipid A (40) is unclear. C16:0 also increased in the P. aeruginosa ∆mlaA but decreased upon treatment with 3’,6-dinonyl neamine (Fig. 2E), consistent with the expression of pagP (Fig. 2B).
In summary, the absence of mlaA induced lipid A modifications through potential activation of PhoPQ and PmrAB TCSs, which in turn induced lipid A palmitoylation and arabinosylation. 3’,6-dinonyl neamine also induced lipid A modifications through activation of PhoPQ and PmrAB, increasing mRNA expression of pagP, pagL, and arnT in P. aeruginosa WT, whereas a decrease in mRNA expression of pagP was observed in ∆mlaA strain. The increase of arabinosylation is in line with the increase in zeta potential (−31.8 mV in ∆mlaA strain) compared with the WT strain (−37.6 mV) (Fig. S5). If and how these changes in OM lipid composition affect the mechanobiological properties of OM was the next question we addressed by using atomic force microscopy (AFM).
Deletion of mlaA induced an increase in the stiffness of the cell envelope without altering the turgor pressure. 3’,6-dinonyl neamine induced a higher softening of the cell envelope of the P. aeruginosa ΔmlaA
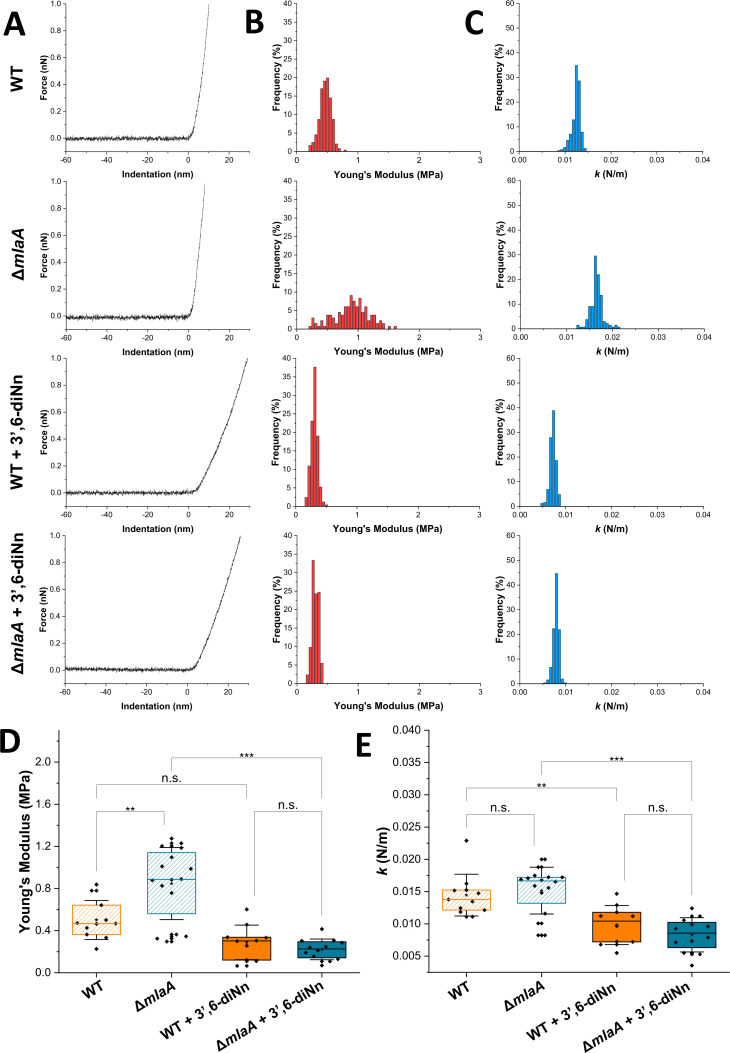
AFM has emerged as a valuable multifunctional technique to investigate the structure and mechanical properties of microbial cells. In mechanobiology, AFM-based indentation experiments have enabled to quantify the mechanical properties of whole bacterial cells and isolated cell walls (41). Spatially resolved force-distance curves were collected on top of the bacteria (Fig. 3A), and respective elasticity (Fig. 3B) and stiffness (Fig. 3C) information was extracted from them (41, 42). Representative force-indentation curves are shown in Fig. 3A, for both P. aeruginosa WT and ΔmlaA strains before and after treatment with 3’,6-dinonyl neamine at the minimum inhibitory concentration (MIC) for 1 h. As expected for Gram-negative bacteria, the curves featured two regions, that is, a nonlinear regime at low forces reflecting cell wall elasticity, followed by a linear regime at high forces associated with turgor pressure. From these two regions, we calculated the bacterial Young’s modulus, E, and the bacterial spring constant, k (43, 44). Representative distributions of E and k values are shown in Fig. 3B and C, respectively.
Fig 3.
Characterization of the cell envelope mechanics by AFM. (A) Representative force-indentation curves and distributions of (B) Young’s modulus and (C) spring constant of one representative P. aeruginosa cell of WT and ΔmlaA strains, before and after treatment with 3’,6-dinonyl neamine (1× MIC, 2 µg/L, 1 h). (D) Box plot shows the Young’s modulus, and (E) spring constant values were calculated from the force-indentation curves for both strains, before and after antibiotic treatment. (D and E) Statistical analyses were carried out using a statistical Tukey’s multiple-comparison test. Stars indicate the mean values, lines the medians, boxes indicate the 25%–75% quartiles, and whiskers indicate the standard deviation obtained from at least 10 independent cells over at least three independent experiments ***P ≤ 0.001, **P ≤ 0.01, and ns P > 0.01, determined by Tukey’s multiple-comparison test.
A Young’s modulus of 0.50 ± 0.18 MPa (mean ± SD, n = 11 cells) was measured for P. aeruginosa WT (Fig. 3D), a value higher than that of 0.24 MPa ± 0.04 published by Formosa et al (45), a difference that we attribute to intercellular variability and differences in experimental conditions.
Interestingly, the Young’s modulus value here calculated for P. aeruginosa WT was slightly lower than those published for E. coli (ranging from 1 to a few MPa) (46–48). This difference can be explained by (i) the thinner peptidoglycan layer of P. aeruginosa compared with E. coli (49), (ii) a reduced interaction between peptidoglycan and the OM (50) and, (iii) differences in lipid composition as lower amounts of very long chain fatty acids covalently bonded to hopanoids (51) or the shorter fatty acyl chains of lipid A (C12) from P. aeruginosa compared with other Gram-negative bacteria including A. baumannii (C12-C14) and Burkholderia mallei (C14-C16) (52). Deletion of mlaA significantly increased Young’s modulus (Fig. 3D) as it reached 0.84 ± 0.34 MPa (n = 16 cells), indicating a stiffening of the cell envelope.
A decrease in Young’s modulus (Fig. 3D) to 0.29 ± 0.164 MPa (n = 10 cells) and 0.22 ± 0.10 MPa (n = 12 cells) was observed upon incubation with 3’,6-dinonyl neamine, for the WT and ∆mlaA strains, respectively.
The k values (Fig. 3E) were found to be similar for WT and mlaA deleted strains (0.015 ± 0.003 and 0.015 ± 0.004 N/m, n = 11, 16 cells), indicating similar levels of turgor pressure. These values are in the range of those reported by Yao et al for P. aeruginosa (0.02 N/m) (49) as well as for Shewanella putrefaciens (0.02–0.05 N/m), another Gram-negative bacterium (53). The spring constants here calculated for P. aeruginosa were lower compared with E. coli (0.04 N/m) (54, 55). Treatment with 3’,6-dinonyl neamine resulted in a decrease of k to 0.010 ± 0.003 N/m (WT, n = 10 cells) and 0.008 ± 0.003 N/m (ΔmlaA, n = 12 cells), indicating a turgor pressure decrease as a result of the antibiotic action.
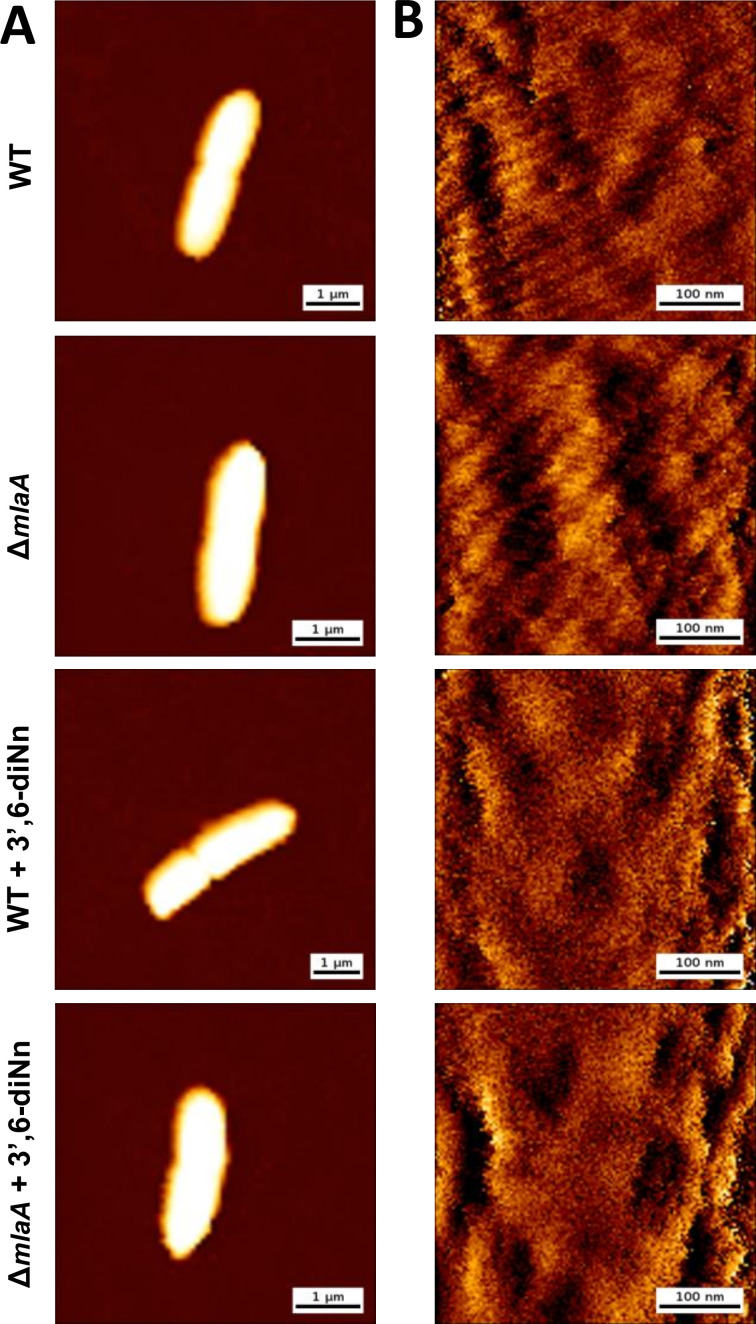
In parallel, AFM topographical characterization of the cell morphology was performed in quantitative imaging mode (Fig. 4). As can be seen in Fig. 4A, both WT and ΔmlaA strains showed the characteristic rod shape of P. aeruginosa cells. No morphological differences were observed between the two strains, as confirmed by the high-resolution images recorded on top of the cells (Fig. 4B). Similar morphological features were observed upon treatment with 3’,6-dinonyl neamine, with no significative changes detected on the surface ultrastructure as evaluated by AFM. Identical height values, extracted from the AFM images, were observed for P. aeruginosa cells from both before and after antibiotic treatment (Fig. S5). Importantly, this observation validates the use of AFM in the present study, excluding the role of possible AFM artifacts/interferences to the conclusions described above.
Fig 4.
AFM topographical characterization of P. aeruginosa WT and ∆mlaA non-treated and treated with 3’,6-dinonyl neamine (1× MIC, 2 mg/L, 1 h). (A) Representative AFM height images of P. aeruginosa WT and ΔmlaA strains, before and after treatment with antibiotic 3’,6-dinonyl neamine (1× MIC, 2 µg/mL, 1 h). Color scale: 650 nm. (B) High-resolution height images were recorded on top of the cells. Color scale: 20 nm. The images were recorded in phosphate-buffered saline (PBS), and the cells were immobilized on glass-bottom dishes previously functionalized with poly-L-lysine.
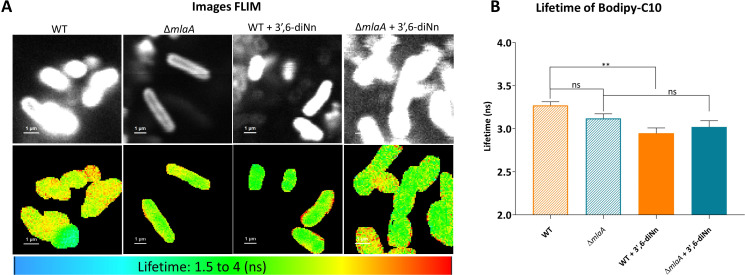
Deletion of mlaA did not change IM fluidity in P. aeruginosa. 3’,6-dinonyl neamine increased fluidity of IM in WT without any effect on ∆mlaA strain
Since the mechanical parameters of the cell envelope were changed, we were wondering if alterations of membrane biophysical parameters such as IM fluidity would also be affected. By using fluorescence-lifetime imaging microscopy (FLIM), we monitored the fluorescence lifetime of BODIPY-C10, which correlates with the viscosity of the environment in which the probe is embedded (56). In our analysis, a longer fluorescence lifetime of the probe indicates a higher viscosity or a lower fluidity of the membrane (57). The FLIM analysis showed that membrane fluidity of ∆mlaA is similar to that of WT (Fig. 5A and B). 3’,6-dinonyl increased fluidity in P. aeruginosa WT without any significant effect on P. aeruginosa ∆mlaA (Fig. 5B).
Fig 5.
FLIM images with their lifetime. P. aeruginosa WT and ∆mlaA in the presence and absence of 3’,6 dinonyl neamine (1× MIC, 2 mg/L, 1 h). (A) Top panels are intensity images, and the below panels are pseudo-colored FLIM images. (B) Lifetime measurements of the bacteria stained with BODIPY-C10. The data were obtained from two individual experiments, and in each experiment, at least 10 bacteria or three slides were considered to measure the lifetimes. The data are presented as means ± SD from three independent replicates. Statistical analysis was performed by one-way ANOVA with Tukey’s multiple-comparison test. **P < 0.01 and ns P >0.05.
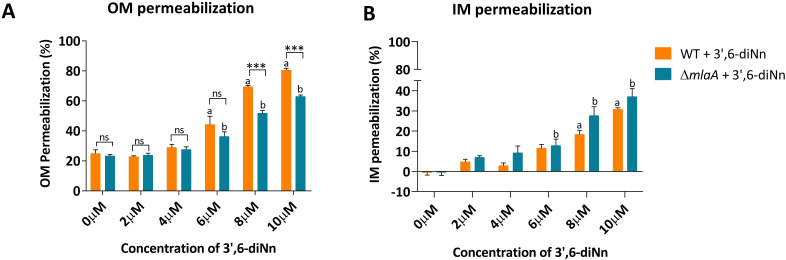
Deletion of mlaA did not change OM and IM permeability. P. aeruginosa ∆mlaA was less sensitive to OM permeabilization induced by 3’,6 dinonyl neamine compared with WT, whereas no difference was observed in IM
The amphipathic nature of LPS contributes to the permeability barrier of the OM (3). Changes in LPS contents and structural variations in lipid A may increase or decrease the permeation of hydrophobic compounds (58). We first determined OM and IM permeability in P. aeruginosa WT and ∆mlaA. On both strains, we also evaluated the effect of 3’,6-dinonyl neamine known to interact with LPS in the OM of P. aeruginosa (29). The OM permeability (Fig. 6A) was determined by using nitrocefin, a chromogenic β-lactam compound that undergoes a color change from yellow to red upon hydrolysis. In whole-cell Gram-negative bacteria, β-lactamases are primarily located in the periplasm; therefore, the nitrocefin hydrolysis rate is primarily limited by its diffusion rate across the OM (59). In the absence of antibiotics, both strains did not show any difference in OM permeability (10), suggesting no impact of mlaA deletion on OM permeability. Upon exposure to increasing concentrations of 3’,6-dinonyl neamine (0–10 µM), an increase in OM permeability was seen for both strains with a significant effect observed at 6 µM. At 8 µM and 10 µM, the effect was significantly higher for WT compared with ∆mlaA strain (26% and 22%, respectively) (Fig. 6A). The decrease in LPS and the larger Young’s modulus we observed for P. aeruginosa ∆mlaA could explain the lower susceptibility of the OM to be permeabilized by 3’,6-dinonyl neamine.
Fig 6.
Membrane permeabilization. (A) Outer membrane permeabilization was assessed by nitrocefin assay (B). Inner membrane permeabilization was assessed by propidium iodide assay where WT (orange) and ∆mlaA (blue) were incubated with different concentrations (0–10 µM) of 3’,6-dinonyl neamine. Statistical analysis was performed using two-way ANOVA with multiple comparisons. ***P < 0.001; ns P >0.05; (A and B) significant increase in membrane permeabilization compared with untreated P. aeruginosa WT (A) and ∆mlaA (B).
The IM permeability (Fig. 6B) was studied by propidium iodide, which fluoresces when it binds DNA. In the absence of an exogenous compound, no difference was observed for WT and P. aeruginosa ∆mlaA. 3’,6-dinonyl neamine induced a dose-dependent (0–10 µM) increase in IM permeability in both strains. At 6 µM and 8 µM, a significant effect was observed for ∆mlaA and WT strains, respectively. In contrast with data for OM, no difference in IM permeabilization was observed between WT and ∆mlaA, although the effect seemed slightly higher for P. aeruginosa ∆mlaA (Fig. 6B).
Deletion of mlaA induced a decrease (2-fold to 8-fold) in MIC of fluoroquinolones
To tentatively link membrane permeability to the antimicrobial activity of antibiotics, we determined the MICs in P. aeruginosa WT and ∆mlaA for several antibiotics (abbreviations defined in Table 2). These included glycopeptides (VAN), lipoglycopeptides (TEL, DAP), oxazolidinone (LZD), cephalosporins (FEP, CAZ), penicillin (TMO), fluoroquinolones (CIP, LVX, MOX), polymyxin (COL), aminoglycoside (AMK, GEN, TOB, NEO), an amphiphilic aminoglycoside (3’,6-dinonyl neamine), carbapenems (IMI, MEM), rifamycin (RIF), macrolide (ERY), and aminocoumarin (NOV) (Table 2). Interestingly, fluoroquinolones have a MIC value lower (2-fold to 8-fold) against P. aeruginosa ∆mlaA compared with WT, a decrease that was reversed in complemented strain. In contrast, no major change was observed for cephalosporins, polymyxin E, aminoglycoside (conventional or amphiphilic), and carbapenem. Also, antibiotics that were inefficient against P. aeruginosa WT, such as penicillin, glycopeptide, lipoglycopeptides, oxazolidinone, rifamycin, macrolide, and aminocoumarin, showed no decrease in MIC values in ∆mlaA strain, suggesting that the sensitivity to antimicrobial compounds cannot be attributed to a global impairment of the OM/IM permeability. Changes in physicochemical parameters (i.e., hydrophobicity) could optimize the diffusion of some antibiotics, like fluoroquinolones, resulting in appropriate intracellular drug concentrations for antibacterial activity. A change in membrane fluidity might also impact the function of fluoroquinolone-specific efflux pumps or porins.
TABLE 2.
Antibiotic susceptibility: MIC of different antibiotics determined for P. aeruginosa WT and ∆mlaA (N = 3) , ND not determined
| Antibiotics | Generic abbreviation | Target/Process | MICs (µg/mL) | ||
|---|---|---|---|---|---|
| Wild-type | ∆mlaA | ||||
| Aminoglycosides | Amikacin | AMK | Binding to the aminoacyl site of 16S ribosomal RNA within the 30S ribosomal subunit | 8 | 8 |
| Gentamicin | GEN | 4 | 4 | ||
| Tobramycin | TOB | 1 | 2 | ||
| Neomycin | NEO | 128 | >128 | ||
| Neamine | NEA | >512 | >512 | ||
| Amphiphilic aminoglycoside derivative | 3’,6-dinonyl neamine | 3’,6-diNn | Binding to the LPS & GPL domains | 4 | 4 |
| Polymyxin | Colistin | COL | Binding to the LPS | 4 | 2 |
| Fluoroquinolones | Ciprofloxacin | CIP | Inhibiting DNA gyrase or topoisomerase IV | 0.25 | 0.125 |
| Levofloxacin | LVX | ||||
| 1 | 0.25 | ||||
| Moxifloxacin | MXF | 2 | 0.5 | ||
| Carbapenems | Imipenem | IPM | Binding to the penicillin-binding proteins | 4 | 4 |
| Meropenem | MEM | 0.5 | 1 | ||
| Cephalosporins | Cefepime | FEP | Inhibiting penicillin-sensitive enzymes (transpeptidases, carboxypeptidases) | 2 | 2 |
| Ceftazidime | CAZ | 1 | 2 | ||
| Penicillin | Temocillin | TMO | Inhibiting the transpeptidase that catalyzes the final step in cell wall biosynthesis | >64 | >64 |
| Glycopeptide | Vancomycin | VAN | Binding to the acyl-d-alanyl-d-alanine terminus of the growing peptidoglycan | >512 | >512 |
| Lipoglycopeptides | Telavancin | TEL | Inserting the lipophilic tail into the bacterial cell membrane causes rapid membrane depolarization and a potassium ion efflux | >512 | >512 |
| Daptomycin | DAP | >512 | >512 | ||
| Rifamycin | Rifampicin | RIF | Inhibiting bacterial DNA-dependent RNA polymerase | 16 | 16 |
| Macrolide | Erythromycin | ERY | Binding to the 23S ribosomal RNA molecule in the 50S subunit of ribosomes | >64 | >64 |
| Aminocoumarin | Novobiocin | NVB | Inhibiting GyrB subunit of the bacterial DNA gyrase | >64 | >64 |
| Oxazolidinone | Linezolid | LZD | Binding a site on the bacterial 23S ribosomal RNA of the 50S subunit | 0.5 | 1 |
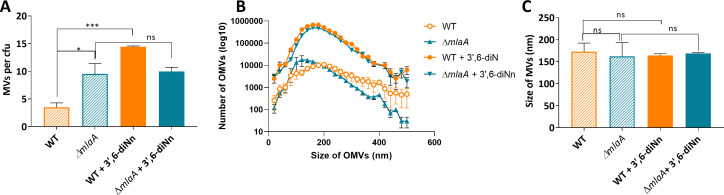
Deletion of mlaA increased P. aeruginosa membrane vesiculation. 3’,6-dinonyl neamine increased MVs number in WT and to a lesser extent in ∆mlaA strain
The differences in P. aeruginosa WT and ∆mlaA concerning their membrane lipid composition (Fig. 1 and 2) as well as differences in Young’s modulus (Fig. 3D) motivated us to investigate the formation of membrane-derived vesicles. The results showed that ∆mlaA produced more MVs (2.7-fold) compared with WT strain (Fig. 7A). This increase was largely reversed in complemented strain (Fig. S7B). The incubation of P. aeruginosa WT with 3’,6-dinonyl neamine markedly increased the MVs production with an increase of 4.1-fold (Fig. 7A) with no effect on ∆mlaA.
Fig 7.
MVs quantification. MVs P. aeruginosa WT and ∆mlaA in the presence and absence of 3’,6-dinonyl neamine (1× MIC, 2 mg/L, 1 h) were analyzed using a nanoparticle tracking analyzer. (A) Number of MVs produced per colony forming unit (CFU). (B) MVs size distribution in nanometers (nm). Figure 6B was used to calculate the mean size of the membrane vesicles, which is shown in (C). Statistical analysis was performed by one-way ANOVA with Tukey’s multiple-comparison test. ****P < 0.0001, **P < 0.01, *P < 0.05; nsp >0.05.
Regarding the MVs size distribution (Fig. 7B), no difference in the average mean size was observed when comparing MVs generated from WT (172.1 ± 19.5 nm) and ∆mlaA (162.3 ± 30.9 nm) strains (Fig. 7C). The addition of 3’,6-dinonyl neamine to WT (164.4 ± 3.8 nm) and ∆mlaA (168.5 ± 1.7 nm) did not change mean size of MVs.
DISCUSSION
The maintenance of OM lipid asymmetry is critical for resisting antibiotics and host stresses, as reported for numerous Gram-negative bacteria including E. coli (60), H. influenzae (61), Burkholderia cepacia complex structures (8), A. baumannii (62), and P. aeruginosa (10, 63). LPS levels and localization are involved in preserving the stiffness and permeability barrier of Gram-negative bacteria (64). As such, the Mla (maintenance of the OM lipid asymmetry) transport, composed of MlaA/VacJ-MlaCBDEF and facilitating the retrograde phospholipid transport from the outer back to the inner membrane (11), has been proposed as a target for drug development (14).
Here, we focused on MlaA, contributing to removing phospholipids from the outer leaflet of OM and transferring them to MlaC located in the periplasm. We explored the consequences of mlaA deletion on the mechanical (cell stiffness, turgor pressure), biophysical (surface charge, fluidity), and functional (MVs generation, antibiotic susceptibility, permeability) properties of the OM in relation to changes in GPLs and LPS. All experiments were conducted in the absence and presence of one amphiphilic aminoglycoside, 3’,6-dinonyl neamine, known to interact with P. aeruginosa OM, and especially with LPS and GPLs (29–31, 65).
First, the work shows the changes in membrane lipid composition induced by P. aeruginosa to defend against the deletion of mlaA. Total GPLs increased with a prominent effect on PE. In parallel, bacterial LPS levels decreased, and changes in lipid A structures (arabinosylation, palmitoylation) were observed. These changes were associated with increased cell stiffness without alteration of turgor pressure and height profile. Remodeling of OM induced by mlaA deletion induced an increase in MVs generation without any significant effect on either IM fluidity or the IM/OM membrane permeability.
In the presence of 3',6-dinonyl neamine, P. aeruginosa WT also showed an increase in GPLs and a decrease in LPS with lipid A modifications (arabinosylation and palmitoylation), but differences of P. aeruginosa WT and ∆mlaA were observed with no significant effect on GPLs, a huge increase in LPS (associated with a decrease in lipid A hydroxylation and palmitoylation), decrease in cell stiffness and turgor pressure, and no effect on IM fluidity observed in ∆mlaA strain. This might result in a slight and non-significant increase in MVs generation and less permeabilization of the OM in ∆mlaA as observed in WT strain. These differences of P. aeruginosa WT and ∆mlaA in response to 3',6-dinonyl neamine might result not only from the extent of changes in GPLs and LPS contents but also from changes in lipid A structures, which may result in differences in drug lipid insertion.
At a glance, the study first highlighted how P. aeruginosa copes with mlaA deletion and modifies its membrane lipid composition. The data confirm a link between maintaining OM lipid asymmetry and regulating the level of LPS molecules at the surface of Gram-negative bacteria (11, 17, 60). Intriguingly, there is a decrease in LPS contents in ∆mlaA compared with P. aeruginosa WT. The first hypothesis was the impact of mlaA deletion on LPS synthesis. We excluded a decrease in the stability of LpxC, one of the nine critical enzymes involved in lipid A synthesis, related to a partial or complete loss of YejM activity (64, 66, 67), since YejM was overexpressed in P. aeruginosa ∆mlaA (Fig. S4A), in agreement with Guest and co-authors (64). Whether a decrease in lipid A synthesis could be afforded by a reduced LpxK activity induced by changes in the ratio of unsaturated and saturated fatty acids (68) or a competition for a common substrate for GPLs and LPS synthesis, that is, β-hydroxymyristoyl–ACP (69, 70), is unknown. We also ruled out a restriction of LPS transport to OM related to a decrease in CL synthesis (69, 71, 72) since we did not observe any major change in cardiolipin content in P. aeruginosa ∆mlaA compared with WT (Fig. S3A). Finally, an increased release of LPS from the MVs in P. aeruginosa ∆mlaA compared with WT must be also excluded (Fig. S8B).
Interestingly, LPS and GPLs varied in the opposite way. Since MVs are enriched in GPLs and not in LPS (Fig. S8A), they are unlikely involved in maintaining the ratio between LPS and GPLs. One other explanation could be changes in the directionality of phospholipid transport (from IM to OM-anterograde transport and from OM to IM-retrograde transport), even anterograde transport described for A. baumannii (62, 73) and E. coli (33), is largely disputed. For P. aeruginosa, to our knowledge, nothing has yet been published.
Beyond the effect induced by mlaA deletion on LPS levels and the ratio between LPS and GPLs, an effect on lysophospholipids was also observed with a higher LPG content compared with LPE. This could result (i) from a lack of activity of PldA and PagP toward PE, (ii) a higher stability of OM when PE flipped due to its zwitterionic character and small headgroup size (74), and (iii) a higher release of LPE in MVs derived from ∆mlaA strain. In P. aeruginosa ATCC27853 WT and ∆mlaA, PCR products of pagP and pldA were present (Fig. 2B; Fig. S4C). However, a significant increase in mRNA expression was observed only in the case of pagP (2.7-fold) (Fig. 2B; Fig. S4C).
Second, the consequences of mlaA deletion on the mechanical and biophysical properties of P. aeruginosa membranes were evidenced. P. aeruginosa ∆mlaA showed an increase in cell surface stiffness likely linked with changes in lipid composition, including (i) longer acyl chains increasing the interactions between neighboring LPS molecules (3) and (ii) increase of phospholipids containing saturated acyl chains. However, the relationship between OM lipid composition and OM stiffness might be more complex, implying the presence of peptidoglycan and/or the link between peptidoglycan and OM (75, 76). Drugs interacting with LPS and/or GPLs like 3’,6-dinonyl neamine also played a critical role in the mechanoproperties of bacterial membranes by decreasing cell stiffness and turgor pressure independently of the mlaA deletion or not. This could result from competition with Ca2+ and interaction with negatively-charged LPS, which could resolve the destabilization of the OM. No change was observed for the IM fluidity by comparing P. aeruginosa ∆mlaA with WT, but a decrease of the surface charge was noticed, in agreement with lipid A modifications, including arabinosylation.
Third, focusing on the functional impact of mlaA deletion, a huge increase in the generation of MVs was observed in P. aeruginosa ∆mlaA. This is in agreement with literature (77–80) and complements data reported on Haemophilus influenzae, Vibrio cholera (80), Neisseria gonorrhoae (77), Campylobacter jejuni (79), and E. coli mlaA knockouts (81). Increased vesiculation might provide a transient advantage during the adaptation process. MlaA, by regulating vesiculation, could allow bacteria to spare the consumption of energy requested by the continuous liberation of compounds of the OM and periplasm (80). The mechanism responsible for hypervesiculation in the ∆mlaA strain is likely complex. Curvature inducing lipid molecules (negative and positive) at particular sites might favor MVs release from bacteria (34). Since we observed an increase in PE (Fig. S3A), which displays positive and negative intrinsic molecular curvatures, respectively (82–84), we could imagine the formation of highly curved structures such as membrane vesicles. However, other hypotheses might be raised like (i) weaker interactions between LPS molecules resulting from the insertion of GPLs into the outer leaflet of OM, favoring thus OM bulging and formation of vesicles (85), (ii) changes in thermotropic behavior and modifications of fluidity resulting from changes in unsaturated/saturated lipid acyl chain ratio (86, 87) or changes in the polar head may facilitate membrane vesiculation. The PE, PG, and CL ratio might be critical in this respect as the phase transition temperature (Tm) of PE (DPPE, 41°C) is lower than that of PG (DPPG, 63°C). Also, CL is a lipid-containing four associated fatty acyl chains characterized by a high degree of unsaturation (88), which can form very rigid, gel-like membrane domains in the otherwise more fluid membrane promoting the initiation of vesicle formation (89). In addition, lipid A modifications resulting from environmental changes and PhoPQ-PmrAB activation (Fig. 2B), potentially interleaflet coupling with modified acylated lipid A and the pre-existing covalent linkages to peptidoglycans, could induce an increase of MVs production as reported for Salmonella (90). A last hypothesis is the accumulation of the PQS signaling molecule (2-heptyl-3-hydroxy-4-quinolone) in the outer leaflet of the OM. An increase in PQS was reported in P. aeruginosa ∆mlaA compared with the WT (5), and this might promote OM curvature and vesiculation (91, 92). Besides these effects, MVs generation could also be influenced by changes in the covalent OM-peptidoglycan crosslinks resulting from integral proteins (i.e., Lpp) (93).
In the presence of 3',6-dinonyl neamine, a huge enhancement in the generation of MVs was noticed, especially for P. aeruginosa WT. Modulating the LPS biosynthesis, changes in lipid A structure might directly facilitate the formation of MVs or favor the insertion of the drug, promoting MVs generation (38, 94). The reduced effect on ∆mlaA strains could be related to the lower content in LPS in comparison to WT or changes in lipid A modifications. This could be responsible for changes in the insertion of 3’,6-dinonyl neamine into the outer leaflet of the OM with an increase in the surface area of the outer leaflet, leading to enhanced interleaflet tension that is relieved through induction of membrane curvature and subsequent MVs release. This agrees with mechanisms behind the MVs biogenesis that were previously described (82, 91).
In the end, the question of the potential impact on antibiotic susceptibility was raised. The reduced LPS levels in the ΔmlaA strain were not sufficient to modify sensitivity to antibiotics like colistin or amphiphilic aminoglycoside, in agreement with the absence of OM permeabilization induced by these compounds at their MIC values. Two hypotheses could be raised (i) the decreased LPS may contribute to raise bacterial surface hydrophobicity, therefore, jeopardizing the outcome of P. aeruginosa interaction with hydrophobic and lipophilic molecules, and (ii) LPS modifications such as the lipid A palmitoylation and amino-arabinosylation or aminoacylation of phospholipids as reported for P. aeruginosa (95, 96) may contribute to antimicrobial peptide resistance. These effects on LPS might explain why OM is less prone to be permeabilized in P. aeruginosa ∆mlaA compared with WT strain impairing the activity of antibiotics like colistin for which the OM permeabilization is required to reach IM and induce cell lysis (97). More generally, if sensitivity to antimicrobial compounds is unlikely to be due to a global impairment of OM/IM permeability, changes in physicochemical parameters (i.e., hydrophobicity) could optimize the diffusion of some antibiotics such as fluoroquinolones. This could lead to appropriate intracellular drug concentrations for antibacterial activity. A change in membrane fluidity could also affect the function of fluoroquinolone-specific efflux pumps or porins.
Moving to the potential interest of MlaA in clinics, since this study does not consider the complexity of biological, clinically relevant conditions (98–100), it is difficult to extrapolate our conclusions to the in vivo situation directly. Other Pseudomonas strains (PAO1, PA14) or clinical strains (from patients suffering from cystic fibrosis) should be selected. Especially, it would have been interesting to compare the P. aeruginosa clinical isolates from the different sites of infection and geographical areas to understand the role of MlaA in bacterial physiology and pathology in a healthcare setting. Interestingly, during lower respiratory tract infection in non-typeable H. influenzae (NTHi) modulation of outer leaflet of OM through increased expression of mlaA (vacJ) genes could minimize recognition by bactericidal anti-oligosaccharide antibodies (61).
In conclusion, this work highlights some aspects of the membrane dynamics of P. aeruginosa: (i) the cross-talk between LPS and glycerophospholipids mislocated at the outer leaflet of the OM of P. aeruginosa (34) and (ii) the role of the individual glycerophospholipids and lipid A structures for changes in mechanical properties of OM, including cell stiffness, turgor pressure, and IM fluidity. The impact of the changes in membrane lipid composition on biophysical and functional membrane properties, including vesiculation and permeability, was also assessed. Although it is risky to define mechanical hypotheses linking lipid composition and membrane biophysical properties, especially since the system maintaining OM asymmetry is complex and involves several proteins probably acting in a coordinated manner, a better understanding of membrane biophysics as the maintenance of OM lipid asymmetry in link with lipid membrane composition is a pre-requisite for opening new research avenues in the design of innovative antibacterial drugs.
MATERIALS AND METHODS
3’,6-dinonyl neamine (3’,6-diNn) was synthesized by Decout and colleagues (101). P. aeruginosa strain ATCC27853 (WT) was obtained from the Pasteur Institute (Brussels, Belgium; Prof. R. Vanhoof). Muller-Hinton broth cation-adjusted (MHB-Ca) and Luria-Bertani broth (LB) were purchased from RothTM. LPE-17:1, LPG-17:1, and CL-14:0, used as internal standards, were purchased from Avanti Polar Lipids. FAME standards were ordered from Larodan (Solna, Sweden). CH3CN, CHCl3, CH3OH, and hexane from Fisher Scientific (Leicestershire, UK) were of high-performance liquid chromatography (HPLC) grade and were used without further purification. All other reagents and chemicals used were of the highest grade of purity commercially available. Milli-Q H2O was filtered through a 0.22 µm filter and obtained using a Milli-Q Millipore system (Milli-Q plus 185).
Bacterial strains and culture conditions
Strains were grown on Tryptic Soy Agar (TSA), Luria-Bertani (LB) agar plates, or in LB media. P. aeruginosa ∆mlaA (PA_2800) mutant was created using two-step allelic exchange (102–104) as described earlier (5). Complementation was carried out to ensure that the deletion of the mlaA gene did not have a polar effect on adjacent genes or mutations elsewhere in the genome during the construction process. Wild-type mlaA under the control of its native promoter (PmlaA) was cloned into the mini-Tn7 vector, which was then used to integrate a single copy of PmlaA-mlaA at the attTn7 site into the chromosome of P. aeruginosa ATCC27853 ΔmlaA by transposition as previously described in Choi and Schweizer (105).
P. aeruginosa ATCC27853 and P. aeruginosa ∆mlaA strains grew until the mid-exponential phase. For some experiments, strains were then incubated with 3’,6-dinonyl neamine at its MIC (2 µg/mL) against P. aeruginosa for 1 h.
Some of the phenotypes with complemented strains (∆mlaA att7::mlaA) were characterized and found to be similar to those from WT for growth rate, PQS levels, motility including swarming, swimming and twitching (5), or MVs generation and MICs (this study).
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
RT-qPCR was done to quantify the mRNA transcripts from the WT, the ∆mlaA, and the ∆mla att7::mlaA. Total RNA was isolated using InviTrap Spin Universal RNA Minit Kit (INVITEK). DNA contamination was removed by TURBO DNA-freeTM kit from Invitrogen, and RNA was quantified using nanodrop (OD260). One microgram of total RNA was heated at 65°C for 15 min and then cooled down at 0°C, mixed with buffer, MgCl2, random hexamers, dNTP mix, RNase inhibitor, and reverse transcriptase to synthesize complementary DNA (cDNA) using first-strand cDNA synthesis Kit (ref 11483188001, Roche). Reverse transcription was performed on iCycleriQ (BIO-RAD ) with thermal cycler programming as follows: 25°C for 10 min; 42°C for 60 min; 99°C for 5 min, and 4°C for 5 min. Real-time PCR was performed with SYBR green (SsoAdvanced Universal SYBR Green Supermix, BIO- RAD) and using the primers specified in Table S1 using a CF96 Real-Time System (C1000 Touch Thermal cycler, BIO-RAD).
Lipid analysis
The lipid content of the different samples was analyzed by LC-MS, as previously reported (56). Briefly, lipids from the membranes were analyzed after liquid/liquid extraction (CH2Cl2-CH3OH-H2O, 4:2:1, vol/vol/vol) under an acidic condition in the presence of internal standards (LPE-17:1, LPG-17:1, and CL-14:0). Phospholipid analysis was performed using a Xevo TQ-S (Waters, USA), and cardiolipin analysis was carried out on an LTQ-orbitrap (from Thermo Fisher Scientific).
For the phospholipids, an HSS LC-18 column 100 × 2.1 mm, 1.8 µm (Waters, USA) was used at a temperature of 40°C. The mobile phase consisted of a gradient between A: CH3OH-CH3CN (9:1, vol/vol) 75% H2O 25%; B: CH3OH-CH3CN (9:1, vol/vol) and C: CH3(CH2)2-2-OH, all containing ammonium acetate (5 mM). An ESI probe, operated in negative mode, was used for sample ionization. The mass spectrometer parameters were as follows: capillary voltage: 2.9kV; cone voltage: 70V; desolvation temperature: 400°C; desolvation gas flow: 1,000 L/h; cone gas flow: 150 L/h; and nebulizer: six bars.
For the analysis of cardiolipins, a Nucleosil C8 column 150 × 4 mm, 5 µm (Macherey-Nagel, GmbH & Co. KG) was used. The same mobile phase as used for the other phospholipids was selected. An ESI probe operated in negative mode was used for cardiolipin ionization. The mass spectrometer parameters were as follows: capillary voltage: −40V; capillary temperature: 275°C; tube lens voltage: −173V; sheath gas flow: 20AU; auxiliary gas flow: 10AU, and sweep gas flow: 8AU.
The relative quantification of the lipids was based on the ratio between the area under the curve (AUC) of the lipid structures and the AUC of the respective internal standard. The obtained data were normalized to the total lipid content determined via labeling the lipids with FM4-64.
Lipopolysaccharides (LPS) extraction and quantification
To quantify LPS levels, LPS molecules were extracted from bacterial suspensions at mid-log growth phases and quantified by purpald assay, with data normalized to colony-forming units.
LPS was extracted from bacteria using an LPS extraction kit (iNtRON Biotechnology). Briefly, P. aeruginosa cells were harvested at 2978 g for 5 min. One milliliter of lysis buffer was added to the pellet and vortexed vigorously. Two hundred microliters of CHCl3 were added to the suspension, vortexed, and incubated at room temperature for 5 min. The suspension was centrifuged to pipette out the upper layer (400 µL). Eight hundred microliters of purification buffer were mixed, and LPS was precipitated by incubation at −20°C for 20 min. After centrifugation, the LPS pellet was washed with 70% ethanol and dissolved in 10 mM Tris-HCl. Samples were then lyophilized to get powder until further use.
LPS was quantified using purpald assay based on periodate oxidation of formaldehyde generated from 2-keto-3-deoxyoctonate (KDO) (106, 107). Briefly, 50 µL of LPS samples or standards (LPS from P. aeruginosa serotype 10, 0–1,200 µg/mL) was added to 50 µL of 32 mM NaIO4 in a 96-well plate and incubated for 25 min. Fifty microliters of 136 mM purpald reagent in 2 N NaOH were added to each well, followed by incubation for 20 min, and the addition of 50 µL of 64 mM NaIO4. After 20 min of incubation, absorbance was measured at 550 nm with a SpectraMax M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
By using standard curve for quantification (determined for each independent assay), and for sake of comparison, the results are expressed in %. We considered as 100%, the value obtained for P. aeruginosa WT and untreated and the values obtained for ∆mlaA strain and for WT/∆mlaA P. aeruginosa treated with 3’,6-dinonyl neamine or colistin were compared to this value (100%).
Lipid A sample preparation and analysis
From LPS extracts, lipid A samples were prepared by mild hydrolysis with 200 µL 1% acetic acid at 100°C for 2 h (108). The solution was left at room temperature to cool down, and 400 µL of CHCl3 and 200 µL of H2O were added. The solution was then centrifuged to obtain a bottom layer that was dried under N2 stream. Lipid A samples were resuspended in CHCl3:CH3OH (4:1, vol/vol) and analyzed by direct injection with an LTQ-orbitrap mass spectrometer (Thermo Fisher Scientific, Bleiswijk, The Netherlands) in negative mode with electrospray ionization. The samples were directly infused at a flow rate of 10 µL/min for 3 min. The MS conditions were: capillary temperature: 275°C, capillary voltage: –10 V, and tube lens: –125 V. Sheath gas, auxiliary gas, and sweep gas flow rates were set at 5 AU.
Fatty acid extraction from lipid A and analysis
The lipid A extract was treated according to a procedure adapted from Myron Sasser (109). Briefly, after adding the internal standard C20:0 (Larodan, Solna, Sweden), the extract was saponified by adding NaOH, CH3OH, and water. The methylation of fatty acids was done by adding HCl, water, and CH3OH to the solution. FAMEs were extracted using a mixture of hexane and methyl tert-butyl ether (1/1; vol/vol). Before injection in gas chromatography, the extract (hexane, methyl tert-butyl ether) was purified by adding water and sodium hydroxide.
FAMEs were injected and separated by gas chromatography (GC) (Trace 2000; Thermo Finnigan, Milan, Italy) equipped with an autosampler (GC PAL, CTC Analytics, Zwingen, Switzerland) and a column Ultra 2 (25 m × 0.32 mm ID, film thickness 0.52 µm; Agilent Technologies, Santa Clara, CA, USA), continuously flowed with H2 as a carrier gas at a constant pressure of 60 kPa. The GC temperature program was the following: an initial temperature of 80°C, which increased to 170°C with a temperature slope of 25°C/min, then from 170°C to 300°C with a slope of 5°C/min. The temperature of 300°C was maintained for 2 min to clean the column. FAMEs were detected with flame ionization detector (Thermo Finnigan, Milan, Italy) kept at a constant temperature of 255°C and flowed by air (350 mL/min) and H2 (35 mL/min). An external standard composed of FAME C10:0, 3OH-C10:0, C12:0, 2OH-C12:0, 3OH-C12:0, C16:0, and C20:0 (Larodan, Solna, Sweden) was used to identify the unknown peaks with the retention time and quantify the peaks through the known concentrations. Chromatographs were processed by using ChromQuest 5.0 software (Thermo Finnigan, Milan, Italy). The results were normalized with the total LPS amount and expressed in µg.
MVs isolation and characterization
P. aeruginosa WT and ∆mlaA strains were grown in one-fifth of the volume of Erlenmayer’s flasks until mid-logarithmic phase and treated with 3’, 6-dinonyl neamine (1× MIC, 2 µg/mL) for 1 h. The bacterial cells were pelleted by centrifugation at 2,978 g at 4°C (Eppendorf 5810 R centrifuge: A-4–62 rotor). The supernatant was filtered via a 0.45 µm polyvinyl difluorine (PVDF) filter (Whatman) and then ultracentrifuged at 150,000 g for 3 h at 4°C (BeckmanTM; 80 Ti rotor). The supernatant was discarded, and the pellet was resuspended in 10 mM HEPES-0.85% NaCl buffer, pH7.2 (MVs buffer). The MVs were purified using gradient density of OptiPrep-iodixanol (Sigma-Aldrich) in MV buffer, as described previously (110). ZetaVIEW S/N 18–400 (x30 series, Inning am Ammersee, Germany) was used to determine the size distribution and the amount of the MVs.
Outer membrane isolation
Outer membrane from P. aeruginosa WT and ∆mlaA, non-treated and treated with 3’,6-dinonyl neamine, was extracted using Mizuno and Kageyama’s protocol (111). Briefly, bacteria were grown in LB media at 37°C, until OD620: 0.8. Cell pellet (1.5 g) was suspended in 9 mL of 2 M sucrose, 10 mL of 0.1 M Tris-HCl (pH 7.8 at 25°C), 0.8 mL of 1% Na-EDTA ( ethylenediaminetetraacetic acid disodium salt) (pH 7.0), and 1.8 mL of 0.5% lysozyme and incubated at 37°C for 60 min. After 30 min of incubation, DNase (3 µg/mL) was added, and incubation was continued for a further 30 min. Samples were centrifuged at 15,000 g, for 15 min, at 37°C to remove the spheroplasts. The supernatant was ultracentrifuged at 100,000 g, 60 min, 4°C to recover the outer membranes. Total lipids and proteins were quantified with a fluorescent probe FM4-64 and BCA protein assay kit (Pierce, Thermoscientific), respectively.
The absence of IM as a contaminant was confirmed with NADH oxidase assay, an IM-specific enzyme. Upon time, OM extracted from P. aeruginosa WT strain showed no change in absorbance, whereas IM extracts from P. aeruginosa and E. coli decreased in absorbance reflecting NADH consumption (Fig. S1A). To further confirm the absence of IM, western blot against LepB, an IM protein, was performed using IM extracted from E. coli as a positive control. The WB did not show any band in OM of P. aeruginosa, whereas IM showed the presence of LepB (Fig. S1B).
Atomic force microscopy (AFM)
Bacterial growth conditions
P. aeruginosa WT and ΔmlaA cells were cultured overnight in 10 mL of LB medium, at 37°C, under shaking. Stationary phase cultures were then diluted to an OD600 = 0.1 in the same culture medium and allowed to grow until the exponential phase (OD600 = 0.8) under shaking at 37°C. P. aeruginosa WT and ΔmlaA strains were treated with 1× MIC (2 µg/mL) 3’,6-dinonyl neamine for 1 h. The cells were then centrifuged (2,000 g, 5 min), washed and resuspended in 1× PBS. These washing and resuspension steps were repeated twice, and at the end, the cellular suspension was diluted 1:100 in PBS.
Sample preparation
AFM studies were conducted with bacteria immobilized on glass-bottom Petri dishes previously coated with Poly-L-Lysine (PLL). PLL coating was achieved by letting the dish in contact with PLL solution for 20 min at room temperature and after rinsing twice with PBS to remove non-attached PLL molecules. One milliliter of bacterial suspension was subsequently added to the PLL-coated dish and incubated for 20 min at room temperature. After that, at least three rinsing steps with PBS were performed, and the dish was filled with 2 mL of this buffer.
AFM imaging and mechanical studies
Both imaging and mechanical studies were carried out with MSCT-C cantilevers (Bruker), in PBS, at room temperature, using a JPK NanoWizard 4 NanoScience AFM. Before the experiments, the cantilevers were thoroughly rinsed with water and ethanol, dried with N2 flow, and further cleaned in a UV ozone chamber for 15 min. Freshly cleaned cantilevers were calibrated by the thermal noise method (112), and spring constant values were found to be always approximately 0.02 N m−1. Bacteria were first imaged in quantitative imaging (QI) mode at 128 × 128 pixels resolution, using an applied force of 0.5 nN, a constant approach/retract speed of 30 µm/s, and a ramp size of 500 nm. High-resolution (256 × 256 pixels) QI images were then recorded by imaging a 400 × 400 nm2 area on top of the bacteria, keeping the same imaging parameters. Mechanical measurements were conducted in force volume (FV) mode. For each cell, 256 force-distance curves (16 × 16 pixels) were recorded on 250 × 250 nm2 areas using an applied force of 1 nN, a constant approach/retract speed of 1 µm/s, and a ramp size of 1 µm. Data were analyzed with the JPK Data Processing software (version 6.1.172). The approach segment of the force-distance curves was fitted with the Hertz/Sneddon model over a distance of 20 nm, using a Poisson ratio of 0.5 and considering a conical tip of 15° half-cone angle (113). Although the Young’s Modulus (E) was extracted from the nonlinear region of the curves, the spring constant (k) was calculated through the slope of their linear region.
Fluorescence-lifetime imaging microscopy (FLIM)
Sample preparation
BODIPY-C10 was obtained from the Molecular Design and Synthesis lab at KU Leuven. The bacterial slide preparation protocol was adapted from previous studies with some modifications (56, 114). Briefly, the cells (107 CFU/mL) were resuspended in the PBS containing BODIPY-C10 (0.5 µM) supplemented with 0.1% wt/vol of glucose. The cells were labeled by mixing and incubating at 37°C with shaking. Subsequently, 200 µL of the cell suspensions was immobilized on the 8-well Ibidi microscopy chamber, precoated with 0.01% poly-L-Lysine. BODIPY-C10 concentrates mostly in the inner membrane of P. aeruginosa (56).
FLIM imaging
The chambers containing the bacteria were mounted on a confocal & multiphoton microscope (LSM 980, ZEISS) equipped with a time-correlated single photon counting (TCSPC) system FLIM module (PicoQuant, Germany). BODIPY-C10 was excited with a coherent (Chameleon Discovery) pulsed laser (80 MHz) at 800 nm, and emission was recorded over a spectrum of 505–545 nm. The FLIM images were obtained at the resolution of 512 × 512 pixels.
The analysis of the FLIM images was performed with SymPhoTime 64 software, which allows fitting the fluorescence decay of the images’ pixels to mathematical decay models. An average lifetime is hence calculated for each pixel, which is color-coded in the FLIM images. Each decay curve and its corresponding image were acquired after collecting 1,000 photons.
Membrane permeabilization by 3’, 6-dinonyl neamine
OM permeabilization
The OM permeabilization was assessed by the method used by Angus et al (115). Briefly, the overnight culture of P. aeruginosa was grown. 0.25 µg/mL of imipenem was added to the bacterial culture to induce a higher level of β-lactamases and incubated for an hour. Bacteria were centrifuged at 3,000 g for 7 min and then, resuspended in 1× PBS (pH 7.2) to get an OD620 0.5. Thereafter, 50 µL of different concentrations (0–10 µM) of 3’, 6-dinonyl neamine was added to 100 µL bacterial suspension. Fifty microliters of nitrocefin (50 µg/mL) were added in the 96-well plates. Absorbance at 490 nm was monitored with a SpectraMax M3 microplate reader at 25°C (Molecular Devices, Sunnyvale, CA, USA). Ten micrometers of colistin was used as a positive control (100%).
IM permeability
The permeabilization of bacterial membranes was determined by using a membrane-impermeable probe, propidium iodide (PI), which fluoresces only when it binds to DNA (31). Briefly, an overnight culture of P. aeruginosa was grown and diluted to (OD620 0.5) and added to PI (dissolved in water) and different concentrations of 3’,6-dinonyl neamine (0–10 µM). Suspension was incubated in dark for 15 min, and fluorescence intensity for excitation, 540 nm, and emission, 610 nm, was measured with a SpectraMax M3 microplate reader at 25°C (Molecular Devices, Sunnyvale, CA, USA). Ten micrometers of colistin were used as a positive control (100%).
Antibiotic susceptibility assay
MICs of antibiotics were assessed by microdilution in cation-adjusted-Mueller Hinton Broth according to Clinical and Laboratory Standard Institute (2018).
Zeta potential
Dynamic light scattering (DLS) technology was performed using a Zetasizer Nano SZ equipment from Malvern Instruments (Grovewood Road, UK) with patented NIBS (non-invasive back scatter) technology, and the recommended software was used for zeta potential determination.
ACKNOWLEDGMENTS
The authors wish to thank Prof. Wim Dehaen and Dr. Tomas Opsomer for providing us with the BODIPY-C10 probe, Virginie Mohymont and Romain Tricot for providing dedicated technical assistance.
This work was supported by the F.S.R-FNRS, T.1003.14, J.0205.16, T.0175.20, and by UCL (ARC 17.22.085; Study Grants Covid [2022] and Patrimoine [2023]).
Contributor Information
M.-P. Mingeot-Leclercq, Email: marie-paule.mingeot@uclouvain.be.
Eric Cascales, Centre National de la Recherche Scientifique, Marseille, France.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.01484-24.
(A) NADH oxidase activity upon time; (B)Western Blot; (C)PCR products of mlaA, mlaB, mlaC, mlaD, mlaE, and mlaF; (D)Relative mRNA expression in the ∆mlaA strain.
Individual glycerophospholipids (PE, PG, CL) and lysophospholipids (LPE, LPG) species analysis.
(A) Individual glycerophospholipids; (B) lysophosphoglycerides; (C) and saturated, monounsaturated, and polyunsaturated fatty acyl chains.
mRNA expression.
AFM height characterization of P. aeruginosa WT and ∆mlaA non-treated and treated.
Zeta potential of the bacterial envelope of P. aeruginosa WT and ∆mlaA strains.
Membrane vesicles (MVs) quantification.
Composition of Membrane vesicles (MVs) (n=3) with (A) GPLs and (B) LPS.
Table S1: Primers used. Table S2: Predicted lipid A acyl chains. Table S3: Relative intensity of lipid A structures.
Supplemental captions of figures and tables, and additional experimental details.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Kondakova T, D’Heygère F, Feuilloley MJ, Orange N, Heipieper HJ, Duclairoir Poc C. 2015. Glycerophospholipid synthesis and functions in Pseudomonas. Chem Phys Lipids 190:27–42. doi: 10.1016/j.chemphyslip.2015.06.006 [DOI] [PubMed] [Google Scholar]
- 2. Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS. 2016. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278. doi: 10.1146/annurev-micro-102215-095308 [DOI] [PubMed] [Google Scholar]
- 3. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM. 2014. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun 82:660–669. doi: 10.1128/IAI.01057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaur M, Buyck JM, Goormaghtigh F, Decout J-L, Mozaheb N, Mingeot-Leclercq M-P. 2023. Deficient Pseudomonas aeruginosa in MlaA/VacJ outer membrane lipoprotein shows decrease in rhamnolipids secretion, motility, and biofilm formation, and increase in fluoroquinolones susceptibility and innate immune response. Res Microbiol 174:104132. doi: 10.1016/j.resmic.2023.104132 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x [DOI] [PubMed] [Google Scholar]
- 7. Zhao L, Gao X, Liu C, Lv X, Jiang N, Zheng S. 2017. Deletion of the vacJ gene affects the biology and virulence in Haemophilus parasuis serovar 5. Gene 603:42–53. doi: 10.1016/j.gene.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 8. Bernier SP, Son S, Surette MG. 2018. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200:e00156-18. doi: 10.1128/JB.00156-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDaniel C, Su S, Panmanee W, Lau GW, Browne T, Cox K, Paul AT, Ko SHB, Mortensen JE, Lam JS, Muruve DA, Hassett DJ. 2016. A putative ABC transporter permease is necessary for resistance to acidified nitrite and EDTA in Pseudomonas aeruginosa under aerobic and anaerobic planktonic and biofilm conditions. Front Microbiol 7. doi: 10.3389/fmicb.2016.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, Nizet V. 2017. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J Mol Med 95:1127–1136. doi: 10.1007/s00109-017-1579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. doi: 10.1038/s41564-017-0046-x [DOI] [PubMed] [Google Scholar]
- 13. Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285. doi: 10.1016/j.cell.2017.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y-MM, Miao Y, Munguia J, Lin L, Nizet V, McCammon JA. 2016. Molecular dynamic study of MlaC protein in Gram-negative bacteria: conformational flexibility, solvent effect and protein-phospholipid binding. Protein Sci 25:1430–1437. doi: 10.1002/pro.2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang X, Chang S, Qiao W, Luo Q, Chen Y, Jia Z, Coleman J, Zhang K, Wang T, Zhang Z, Zhang C, Zhu X, Wei X, Dong C, Zhang X, Dong H. 2021. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat Struct Mol Biol 28:81–91. doi: 10.1038/s41594-020-00532-y [DOI] [PubMed] [Google Scholar]
- 16. Thong S, Ercan B, Torta F, Fong ZY, Wong HYA, Wenk MR, Chng S-S. 2016. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife 5:1–19. doi: 10.7554/eLife.19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. May KL, Silhavy TJ. 2018. The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9:1–14. doi: 10.1128/mBio.00379-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Powers MJ, Trent MS. 2018. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol Microbiol 107:47–56. doi: 10.1111/mmi.13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Privé GG. 2004. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J 23:2931–2941. doi: 10.1038/sj.emboj.7600320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filkin SY, Lipkin AV, Fedorov AN. 2020. Phospholipase superfamily: structure, functions, and biotechnological applications. Biochemistry (Mosc) 85:S177–S195. doi: 10.1134/S0006297920140096 [DOI] [PubMed] [Google Scholar]
- 21. Bishop RE. 2008. Structural biology of membrane-intrinsic β-barrel enzymes: sentinels of the bacterial outer membrane. Biochim et Biophys Acta (BBA) - Biomembranes 1778:1881–1896. doi: 10.1016/j.bbamem.2007.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dekker N. 2000. Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol 35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x [DOI] [PubMed] [Google Scholar]
- 23. Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature New Biol 401:717–721. doi: 10.1038/44890 [DOI] [PubMed] [Google Scholar]
- 24. Sun L, Zhang Y, Cai T, Li X, Li N, Xie Z, Yang F, You X. 2022. CrrAB regulates PagP-mediated glycerophosphoglycerol palmitoylation in the outer membrane of Klebsiella pneumoniae. J Lipid Res 63:100251. doi: 10.1016/j.jlr.2022.100251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tam C, Missiakas D. 2005. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli. Mol Microbiol 55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x [DOI] [PubMed] [Google Scholar]
- 26. Jia W, El Zoeiby A, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. 2004. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem 279:44966–44975. doi: 10.1074/jbc.M404963200 [DOI] [PubMed] [Google Scholar]
- 27. Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella typhimurium outer membrane . Proc Natl Acad Sci USA 111:1963–1968. doi: 10.1073/pnas.1316901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sautrey G, Zimmermann L, Deleu M, Delbar A, Souza Machado L, Jeannot K, Van Bambeke F, Buyck JM, Decout J-L, Mingeot-Leclercq M-P. 2014. New amphiphilic neamine derivatives active against resistant Pseudomonas aeruginosa and their interactions with lipopolysaccharides. Antimicrob Agents Chemother 58:4420–4430. doi: 10.1128/AAC.02536-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Khoury M, Swain J, Sautrey G, Zimmermann L, Van Der Smissen P, Décout J-L, Mingeot-Leclercq M-P. 2017. Targeting bacterial cardiolipin enriched microdomains: an antimicrobial strategy used by amphiphilic aminoglycoside antibiotics. Sci Rep 7:10697. doi: 10.1038/s41598-017-10543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sautrey G, El Khoury M, Dos Santos AG, Zimmermann L, Deleu M, Lins L, Décout J-L, Mingeot-Leclercq M-P. 2016. Negatively charged lipids as a potential target for new amphiphilic aminoglycoside antibiotics: a biophysical study. J Biol Chem 291:13864–13874. doi: 10.1074/jbc.M115.665364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Powers MJ, Trent MS. 2019. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc Natl Acad Sci U S A 116:17147–17155. doi: 10.1073/pnas.1902026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shrivastava R, Jiang X, Chng S-S. 2017. Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli: a physiological function for the Tol-Pal complex. Mol Microbiol 106:395–408. [DOI] [PubMed] [Google Scholar]
- 34. Giordano NP, Cian MB, Dalebroux ZD. 2020. Outer membrane lipid secretion and the innate immune response to Gram-negative bacteria. Infect Immun 88:1–21. doi: 10.1128/IAI.00920-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cian MB, Giordano NP, Mettlach JA, Minor KE, Dalebroux ZD. 2020. Separation of the cell envelope for Gram-negative bacteria into inner and outer membrane fractions with technical adjustments for Acinetobacter baumannii. J Vis Exp 60517. doi: 10.3791/60517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer LD, Minor KE, Mettlach JA, Rivera ES, Boyd KL, Caprioli RM, Spraggins JM, Dalebroux ZD, Skaar EP. 2020. Modulating isoprenoid biosynthesis increases lipooligosaccharides and restores Acinetobacter baumannii resistance to host and antibiotic stress. Cell Rep 32:108129. doi: 10.1016/j.celrep.2020.108129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. doi: 10.3389/fmicb.2011.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF. 2016. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. MBio 7:1–12. doi: 10.1128/mBio.00940-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. King JD, Kocíncová D, Westman EL, Lam JS. 2009. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun 15:261–312. doi: 10.1177/1753425909106436 [DOI] [PubMed] [Google Scholar]
- 41. Krieg M, Fläschner G, Alsteens D, Gaub BM, Roos WH, Wuite GJL, Gaub HE, Gerber C, Dufrêne YF, Müller DJ. 2019. Atomic force microscopy-based mechanobiology. Nat Rev Phys 1:41–57. doi: 10.1038/s42254-018-0001-7 [DOI] [Google Scholar]
- 42. Mathelié-Guinlet M, Asmar AT, Collet JF, Dufrêne YF. 2020. Lipoprotein lpp regulates the mechanical properties of the E. coli cell envelope. Nat Commun 11:1789. doi: 10.1038/s41467-020-15489-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arnoldi M, Fritz M, Bäuerlein E, Radmacher M, Sackmann E, Boulbitch A. 2000. Bacterial turgor pressure can be measured by atomic force microscopy. Phys Rev E 62:1034–1044. doi: 10.1103/PhysRevE.62.1034 [DOI] [PubMed] [Google Scholar]
- 44. Touhami A, Nysten B, Dufrêne YF. 2003. Nanoscale mapping of the elasticity of microbial cells by atomic force microscopy. Langmuir 19:4539–4543. doi: 10.1021/la034136x [DOI] [Google Scholar]
- 45. Formosa C, Grare M, Jauvert E, Coutable A, Regnouf-de-Vains JB, Mourer M, Duval RE, Dague E. 2012. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain. Sci Rep 2:575. doi: 10.1038/srep00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cerf A, Cau JC, Vieu C, Dague E. 2009. Nanomechanical properties of dead or alive single-patterned bacteria. Langmuir 25:5731–5736. doi: 10.1021/la9004642 [DOI] [PubMed] [Google Scholar]
- 47. Longo G, Rio LM, Roduit C, Trampuz A, Bizzini A, Dietler G, Kasas S. 2012. Force volume and stiffness tomography investigation on the dynamics of stiff material under bacterial membranes. J Mol Recognit 25:278–284. doi: 10.1002/jmr.2171 [DOI] [PubMed] [Google Scholar]
- 48. Mathelié-Guinlet M, Grauby-Heywang C, Martin A, Février H, Moroté F, Vilquin A, Béven L, Delville MH, Cohen-Bouhacina T. 2018. Detrimental impact of silica nanoparticles on the nanomechanical properties of Escherichia coli, studied by AFM. J Colloid Interface Sci 529:53–64. doi: 10.1016/j.jcis.2018.05.098 [DOI] [PubMed] [Google Scholar]
- 49. Yao X, Jericho M, Pink D, Beveridge T. 1999. Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol 181:6865–6875. doi: 10.1128/JB.181.22.6865-6875.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mortensen NP, Fowlkes JD, Sullivan CJ, Allison DP, Larsen NB, Molin S, Doktycz MJ. 2009. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25:3728–3733. doi: 10.1021/la803898g [DOI] [PubMed] [Google Scholar]
- 51. Vitiello G, Oliva R, Petraccone L, Vecchio PD, Heenan RK, Molinaro A, Silipo A, D’Errico G, Paduano L. 2021. Covalently bonded hopanoid-lipid A from Bradyrhizobium: the role of unusual molecular structure and calcium ions in regulating the lipid bilayers organization. J Colloid Interface Sci 594:891–901. doi: 10.1016/j.jcis.2021.03.072 [DOI] [PubMed] [Google Scholar]
- 52. Korneev KV, Arbatsky NP, Molinaro A, Palmigiano A, Shaikhutdinova RZ, Shneider MM, Pier GB, Kondakova AN, Sviriaeva EN, Sturiale L, Garozzo D, Kruglov AA, Nedospasov SA, Drutskaya MS, Knirel YA, Kuprash DV. 2015. Structural relationship of the lipid A acyl groups to activation of murine toll-like receptor 4 by lipopolysaccharides from pathogenic strains of Burkholderia mallei, Acinetobacter baumannii, and Pseudomonas aeruginosa.. Front Immunol 6:595. doi: 10.3389/fimmu.2015.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaboriaud F, Bailet S, Dague E, Jorand F. 2005. Surface structure and nanomechanical properties of Shewanella putrefaciens bacteria at two pH values (4 and 10) determined by atomic force microscopy. J Bacteriol 187:3864–3868. doi: 10.1128/JB.187.11.3864-3868.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beckmann MA, Venkataraman S, Doktycz MJ, Nataro JP, Sullivan CJ, Morrell-Falvey JL, Allison DP. 2006. Measuring cell surface elasticity on enteroaggregative Escherichia coli wild type and dispersin mutant by AFM. Ultramicroscopy 106:695–702. doi: 10.1016/j.ultramic.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 55. Velegol SB, Logan BE. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir 18:5256–5262. doi: 10.1021/la011818g [DOI] [Google Scholar]
- 56. Mozaheb N, Van Der Smissen P, Opsomer T, Mignolet E, Terrasi R, Paquot A, Larondelle Y, Dehaen W, Muccioli GG, Mingeot-Leclercq M-P. 2022. Contribution of membrane vesicle to reprogramming of bacterial membrane fluidity in Pseudomonas aeruginosa. mSphere 7:e0018722. doi: 10.1128/msphere.00187-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuimova MK, Yahioglu G, Levitt JA, Suhling K. 2008. Molecular rotor measures viscosity of live cells via fluorescence lifetime imaging. J Am Chem Soc 130:6672–6673. doi: 10.1021/ja800570d [DOI] [PubMed] [Google Scholar]
- 58. Vaara M, Plachy WZ, Nikaido H. 1990. Partitioning of hydrophobic probes into lipopolysaccharide bilayers. Biochim et Biophys Acta (BBA) - Biomembranes 1024:152–158. doi: 10.1016/0005-2736(90)90218-D [DOI] [PubMed] [Google Scholar]
- 59. Hancock RE, Wong PG. 1984. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob Agents Chemother 26:48–52. doi: 10.1128/AAC.26.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. doi: 10.1073/pnas.1601375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. 2011. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog 7:e1001247. doi: 10.1371/journal.ppat.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, Miller SI. 2019. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 8:1–25. doi: 10.7554/eLife.40171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen L, Gao X, Wei J, Chen L, Zhao X, Li B, Duan K. 2012. PA2800 plays an important role in both antibiotic susceptibility and virulence in Pseudomonas aeruginosa. Curr Microbiol 65:601–609. doi: 10.1007/s00284-012-0196-2 [DOI] [PubMed] [Google Scholar]
- 64. Guest RL, Rutherford ST, Silhavy TJ. 2021. Border control: regulating LPS biogenesis. Trends Microbiol 29:334–345. doi: 10.1016/j.tim.2020.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ouberai M, El Garch F, Bussiere A, Riou M, Alsteens D, Lins L, Baussanne I, Dufrêne YF, Brasseur R, Decout JL, Mingeot-Leclercq MP. 2011. The Pseudomonas aeruginosa membranes: a target for a new amphiphilic aminoglycoside derivative? Biochim et Biophys Acta (BBA) - Biomembranes 1808:1716–1727. doi: 10.1016/j.bbamem.2011.01.014 [DOI] [PubMed] [Google Scholar]
- 66. Fivenson EM, Bernhardt TG. 2020. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. MBio 11:1–12. doi: 10.1128/mBio.00939-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nguyen M, Gautier T, Reocreux G, Pallot G, Maquart G, Bahr P-A, Tavernier A, Grober J, Masson D, Bouhemad B, Guinot P-G. 2021. Increased phospholipid transfer protein activity is associated with markers of enhanced lipopolysaccharide clearance in human during cardiopulmonary bypass. Front Cardiovasc Med 8:756269. doi: 10.3389/fcvm.2021.756269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wahl A, My L, Dumoulin R, Sturgis JN, Bouveret E. 2011. Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol Microbiol 80:1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x [DOI] [PubMed] [Google Scholar]
- 69. Emiola A, Andrews SS, Heller C, George J. 2016. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 113:3108–3113. doi: 10.1073/pnas.1521168113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thomanek N, Arends J, Lindemann C, Barkovits K, Meyer HE, Marcus K, Narberhaus F. 2018. Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front Microbiol 9:3285. doi: 10.3389/fmicb.2018.03285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anderson MS, Raetz CR. 1987. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem 262:5159–5169. doi: 10.1016/S0021-9258(18)61169-X [DOI] [PubMed] [Google Scholar]
- 72. Douglass MV, Cléon F, Trent MS. 2021. Cardiolipin AIDS in lipopolysaccharide transport to the gram-negative outer membrane. Proc Natl Acad Sci U S A 118:e2018329118. doi: 10.1073/pnas.2018329118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Powers MJ, Simpson BW, Trent MS. 2020. The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport. Elife 9:1–21. doi: 10.7554/eLife.56571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Son M, London E. 2013. The dependence of lipid asymmetry upon polar headgroup structure. J Lipid Res 54:3385–3393. doi: 10.1194/jlr.M041749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. de Jonge EF, van Boxtel R, Balhuizen MD, Haagsman HP, Tommassen J. 2022. Pal depletion results in hypervesiculation and affects cell morphology and outer-membrane lipid asymmetry in bordetellae. Res Microbiol 173:103937. doi: 10.1016/j.resmic.2022.103937 [DOI] [PubMed] [Google Scholar]
- 76. Paulsson M, Kragh KN, Su YC, Sandblad L, Singh B, Bjarnsholt T, Riesbeck K. 2021. Peptidoglycan-binding anchor is a Pseudomonas aeruginosa OmpA family lipoprotein with importance for outer membrane vesicles, biofilms, and the periplasmic shape. Front Microbiol 12:639582. doi: 10.3389/fmicb.2021.639582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baarda BI, Zielke RA, Le Van A, Jerse AE, Sikora AE. 2019. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog 15:e1007385. doi: 10.1371/journal.ppat.1007385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Batista JH, Leal FC, Fukuda TTH, Alcoforado Diniz J, Almeida F, Pupo MT, da Silva Neto JF. 2020. Interplay between two quorum sensing-regulated pathways, violacein biosynthesis and VacJ/Yrb, dictates outer membrane vesicle biogenesis in Chromobacterium violaceum. Environ Microbiol 22:2432–2442. doi: 10.1111/1462-2920.15033 [DOI] [PubMed] [Google Scholar]
- 79. Davies C, Taylor AJ, Elmi A, Winter J, Liaw J, Grabowska AD, Gundogdu O, Wren BW, Kelly DJ, Dorrell N. 2019. Sodium taurocholate stimulates Campylobacter jejuni outer membrane vesicle production via down-regulation of the maintenance of lipid asymmetry pathway. Front Cell Infect Microbiol 9:177. doi: 10.3389/fcimb.2019.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. doi: 10.1038/ncomms10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nasu H, Shirakawa R, Furuta K, Kaito C. 2022. Knockout of mlaA increases Escherichia coli virulence in a silkworm infection model. PLoS One 17:e0270166. doi: 10.1371/journal.pone.0270166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cooke IR, Deserno M. 2006. Coupling between lipid shape and membrane curvature. Biophys J 91:487–495. doi: 10.1529/biophysj.105.078683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dowhan W. 1997. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66:199–232. doi: 10.1146/annurev.biochem.66.1.199 [DOI] [PubMed] [Google Scholar]
- 84. Graham TR, Kozlov MM. 2010. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol 22:430–436. doi: 10.1016/j.ceb.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Werneburg M, Zerbe K, Juhas M, Bigler L, Stalder U, Kaech A, Ziegler U, Obrecht D, Eberl L, Robinson JA. 2012. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13:1767–1775. doi: 10.1002/cbic.201200276 [DOI] [PubMed] [Google Scholar]
- 86. Chowdhury C, Jagannadham MV. 2013. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim et Biophys Acta (BBA) - Proteins Proteomics 1834:231–239. doi: 10.1016/j.bbapap.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 87. Kulkarni HM, Jagannadham MV. 2014. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology (Reading, Engl) 160:2109–2121. doi: 10.1099/mic.0.079400-0 [DOI] [PubMed] [Google Scholar]
- 88. Schlame M, Rua D, Greenberg ML. 2000. The biosynthesis and functional role of cardiolipin. Prog Lipid Res 39:257–288. doi: 10.1016/s0163-7827(00)00005-9 [DOI] [PubMed] [Google Scholar]
- 89. Kudryakova IV, Suzina NE, Vinokurova NG, Shishkova NA, Vasilyeva NV. 2017. Studying factors involved in biogenesis of Lysobacter sp. XL1 outer membrane vesicles. Biochemistry Moscow 82:501–509. doi: 10.1134/S0006297917040125 [DOI] [PubMed] [Google Scholar]
- 90. Bonnington KE, Kuehn MJ. 2016. Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. MBio 7:e01532-16. doi: 10.1128/mBio.01532-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. MBio 8:1–13. doi: 10.1128/mBio.01034-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature New Biol 437:422–425. doi: 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- 93. Schwechheimer C, Kulp A, Kuehn MJ. 2014. Modulation of bacterial outer membrane vesicle production by envelope structure and content. BMC Microbiol 14:324. doi: 10.1186/s12866-014-0324-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kawasaki K, Manabe T. 2010. Latency of the lipid A deacylase PagL is involved in producing a robust permeation barrier in the outer membrane of Salmonella enterica. J Bacteriol 192:5837–5840. doi: 10.1128/JB.00758-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gunn JS, Belden WJ, Miller SI. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb Pathog 25:77–90. doi: 10.1006/mpat.1998.0217 [DOI] [PubMed] [Google Scholar]
- 96. Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J. 2009. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol 71:551–565. doi: 10.1111/j.1365-2958.2008.06562.x [DOI] [PubMed] [Google Scholar]
- 97. Sabnis A, Hagart KL, Klöckner A, Becce M, Evans LE, Furniss RCD, Mavridou DA, Murphy R, Stevens MM, Davies JC, Larrouy-Maumus GJ, Clarke TB, Edwards AM. 2021. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 10:e65836. doi: 10.7554/eLife.65836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Diaz Iglesias Y, Van Bambeke F. 2020. Activity of antibiotics against Pseudomonas aeruginosa in an in vitro model of biofilms in the context of cystic fibrosis: influence of the culture medium. Antimicrob Agents Chemother 64:e02204-19. doi: 10.1128/AAC.02204-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pompilio A, Crocetta V, Pomponio S, Fiscarelli E, Di Bonaventura G. 2015. In vitro activity of colistin against biofilm by Pseudomonas aeruginosa is significantly improved under “cystic fibrosis-like” physicochemical conditions. Diagn Microbiol Infect Dis 82:318–325. doi: 10.1016/j.diagmicrobio.2015.01.006 [DOI] [PubMed] [Google Scholar]
- 100. Quinn RA, Comstock W, Zhang T, Morton JT, da Silva R, Tran A, Aksenov A, Nothias L-F, Wangpraseurt D, Melnik AV, Ackermann G, Conrad D, Klapper I, Knight R, Dorrestein PC. 2018. Niche partitioning of a pathogenic microbiome driven by chemical gradients. Sci Adv 4:eaau1908. doi: 10.1126/sciadv.aau1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Baussanne I, Bussière A, Halder S, Ganem-Elbaz C, Ouberai M, Riou M, Paris JM, Ennifar E, Mingeot-Leclercq MP, Décout JL. 2010. Synthesis and antimicrobial evaluation of amphiphilic neamine derivatives. J Med Chem 53:119–127. doi: 10.1021/jm900615h [DOI] [PubMed] [Google Scholar]
- 102. Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 102:8006–8011. doi: 10.1073/pnas.0503005102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ude J, Tripathi V, Buyck JM, Söderholm S, Cunrath O, Fanous J, Claudi B, Egli A, Schleberger C, Hiller S, Bumann D. 2021. Outer membrane permeability: antimicrobials and diverse nutrients bypass porins in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 118:e2107644118. doi: 10.1073/pnas.2107644118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Choi K-H, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- 106. Lee CH, Tsai CM. 1999. Quantification of bacterial lipopolysaccharides by the purpald assay: measuring formaldehyde generated from 2-keto-3-deoxyoctonate and heptose at the inner core by periodate oxidation. Anal Biochem 267:161–168. doi: 10.1006/abio.1998.2961 [DOI] [PubMed] [Google Scholar]
- 107. Quesenberry MS, Lee YC. 1996. A rapid formaldehyde assay using purpald reagent: application under periodation conditions. Anal Biochem 234:50–55. doi: 10.1006/abio.1996.0048 [DOI] [PubMed] [Google Scholar]
- 108. Park HG, Sathiyanarayanan G, Hwang CH, Ann DH, Kim JH, Bang G, Jang KS, Ryu HW, Lee YK, Yang YH, Kim YG. 2017. Chemical structure of the lipid A component of Pseudomonas sp. strain PAMC 28618 from thawing permafrost in relation to pathogenicity. Sci Rep 7:2168. doi: 10.1038/s41598-017-02145-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sasser M. 2006. Technical note # 101 Microbial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). T N:1–6. [Google Scholar]
- 110. Cooke AC, Nello AV, Ernst RK, Schertzer JW. 2019. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS ONE 14:e0212275. doi: 10.1371/journal.pone.0212275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mizuno T, Kageyama M. 1979. Isolation and characterization of a major outer membrane protein of Pseudomonas aeruginosa: evidence for the occurrence of a lipoprotein. J Biochem 85:115–122. doi: 10.1093/oxfordjournals.jbchem.a132300 [DOI] [PubMed] [Google Scholar]
- 112. Sader JE, Larson I, Mulvaney P, White LR. 1995. Method for the calibration of atomic force microscope cantilevers. Rev Sci Instrum 66:3789–3798. doi: 10.1063/1.1145439 [DOI] [Google Scholar]
- 113. Thomas G, Burnham NA, Camesano TA, Wen Q. 2013. Measuring the mechanical properties of living cells using atomic force microscopy. J Vis Exp:50497. doi: 10.3791/50497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mika JT, Thompson AJ, Dent MR, Brooks NJ, Michiels J, Hofkens J, Kuimova MK. 2016. Measuring the viscosity of the Escherichia coli plasma membrane using molecular rotors. Biophys J 111:1528–1540. doi: 10.1016/j.bpj.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother 21:299–309. doi: 10.1128/AAC.21.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) NADH oxidase activity upon time; (B)Western Blot; (C)PCR products of mlaA, mlaB, mlaC, mlaD, mlaE, and mlaF; (D)Relative mRNA expression in the ∆mlaA strain.
Individual glycerophospholipids (PE, PG, CL) and lysophospholipids (LPE, LPG) species analysis.
(A) Individual glycerophospholipids; (B) lysophosphoglycerides; (C) and saturated, monounsaturated, and polyunsaturated fatty acyl chains.
mRNA expression.
AFM height characterization of P. aeruginosa WT and ∆mlaA non-treated and treated.
Zeta potential of the bacterial envelope of P. aeruginosa WT and ∆mlaA strains.
Membrane vesicles (MVs) quantification.
Composition of Membrane vesicles (MVs) (n=3) with (A) GPLs and (B) LPS.
Table S1: Primers used. Table S2: Predicted lipid A acyl chains. Table S3: Relative intensity of lipid A structures.
Supplemental captions of figures and tables, and additional experimental details.