ABSTRACT
Given the challenges that SARS-CoV-2 variants have caused in terms of rapid spread and reduced vaccine efficacy, a rapid and cost-effective assay that can detect new and emerging variants is greatly needed worldwide. We have successfully applied the xenonucleic acid-based molecular-clamping technology to develop a multiplex reverse-transcription quantitative real-time PCR assay for SARS-CoV-2 multivariant detection. The assay was used to test 649 nasopharyngeal swab samples that were collected for clinical diagnosis or surveillance. The assay was able to correctly identify all 36 Delta variant samples as it accurately detected the D614G, T478K, and L452R mutations. In addition, the assay was able to correctly identify all 34 Omicron samples by detecting the K417N, T478K, N501Y, and D614G mutations. This technique reliably detects a variety of variants and has an analytical sensitivity of 100 copies/mL. In conclusion, this novel assay can serve as a rapid and cost-effective tool to facilitate large-scale detection of SARS-CoV-2 variants.
IMPORTANCE
We have developed a multiplex reverse-transcription quantitative real-time PCR (RT-qPCR) testing platform for the rapid detection of SARS-CoV-2 variants using the xenonucleic acid (XNA)-based molecular-clamping technology. The XNA-based RT-qPCR assay can achieve high sensitivity with a limit of detection of about 100 copies/mL for variant detection which is much better than the next-generation sequencing (NGS) assay. Its turnaround time is about 4 hours with lower cost and a lot of Clinical Laboratory Improvement Amendments (CLIA) labs own the instrument and meet skillset requirements. This assay provides a rapid, reliable, and cost-effective testing platform for rapid detection and monitoring of known and emerging SARS-CoV-2 variants. This testing platform can be adopted by laboratories that perform routine SARS-CoV-2 PCR testing, providing a rapid and cost-effective method in lieu of NGS-based assays, for detecting, differentiating, and monitoring SARS-CoV-2 variants. This assay is easily scalable to any new variant(s) should it emerge.
KEYWORDS: SARS-CoV-2 variant detection, molecular-clamping technology, XNA
INTRODUCTION
More than 3 years after its initial emergence, the SARS-CoV-2 fueled COVID-19 pandemic and continues to spread globally with 755 million infections and 6.8 million deaths to date (13 February 2023) (1). In the United States alone, there have been 102 million documented cases, and the death toll has passed 1.1 millions (13 February 2023) (2). Despite increasing vaccination levels and a rising number of COVID-19-recovered people who have acquired some degree of immunity, the spread has continued. This is largely due to the appearance of more transmissible and partially vaccine-resistant novel variants of the virus. Since the inception of the pandemic, five variants of concern (VOC) and seven variants of interest (VOI) have emerged worldwide (3–8). These include the Alpha B.1.1.7 (501Y V1) (3), Beta B.1.351 (501Y.V2) (4), Gamma P.1 (501Y.V3) (5), Epsilon CAL.20C (20 C/S:452R, or B.1.429) (6–8), Delta B.1.617.2, and Omicron (B.1.1.529) (9). As of mid-2023, the infection rates have declined, leading to the removal of certain VOC from the World Health Organization, Centers for Disease Control and Prevention (CDC), and European Center for Disease Prevention and Control websites (10–12). As of June 2024, only VOI and variants under monitoring (VUM) lists are maintained. The prevalent VOIs currently include EG.5, BA.2.86, and JN.1, while the VUMs consist of JN.1.7, KP.2, and KP.3.
The Alpha B.1.1.7 strain, also known as 20I/501Y.V1 and VOC 20DEC-01 (VOC-20DEC-01, previously written as VOC-202012/01), was found initially in the southeast of England in early October 2020 and became prevalent in both Europe and the United States shortly thereafter (13–16). It is estimated to be 40%–80% more transmissible than the original SARS-CoV-2 strain (17, 18).
While the main variant from April to June 2021 in the United States was the Alpha variant, its decline from 70% of cases (8 May to 19 June 2021) to 9.0% of cases (17 July 2021) corresponded with an increase in the prevalence of the Delta variant, which rose from 0.6% (24 April 2021) to 82.8% (17 July 2021) (19–23). The Delta B.1.617.2 variant was first identified in India (24, 25) and is characterized by 13 mutations (26). In addition, a Delta plus variant, which has the Delta mutations plus an additional K417N mutation, has also been detected. Compared to Alpha, these variants are more contagious and are associated with a high degree of mortality. The Delta variants grow more rapidly in the human respiratory tract and have 1,000 times higher viral loads than the original strain (27). In addition, large numbers of breakthrough cases associated with the Delta variants were documented in previously vaccinated individuals. Since November 2021, the emerging Omicron variant (B.1.1.529) was found and reported in Botswana and South Africa. A big family of Omicon variants (BA.2, BA.4, BA.5, Botswana and South Africa, BF.7, BQ.1, etc.) has been reported and studied (28–30).
Each one of these main VOCs has a signature set of spike protein mutations that allow them to be uniquely identified. While each VOC has a unique set of spike protein mutations, the D614G mutation is shared by all five VOC. The D614G mutation involves an amino acid change from aspartic acid to glycine, which is caused by an A-to-G nucleotide transition at position 23403 of the viral genome. This change stabilizes the spike protein and enhances its fitness and infectivity (20) and is associated with increased viral infectivity and transmissibility (21, 22). Another key mutation, N501Y, is found in all the VOCs except for Delta. The N501Y mutation is located within the receptor-binding domain (RBD) of the spike protein and is associated with stronger binding to ACE2 receptor and higher viral infectivity. Of greater concern, though, is the fact that the efficacy of vaccine-mediated immunity against strains containing the N501Y mutation seems to be reduced (23). These two mutations were the two earliest mutations observed and remain the most prominent mutations detected in the SARS-CoV-2 virus.
To date, next-generation sequencing (NGS) has been the standard method used for SARS-CoV-2 variants detection (31). Although the NGS-based assays facilitate variant finding and confirmation, they are expensive and time consuming, require technical expertise, and are not readily available, particularly in low-income countries and regions. These factors limit their utility in large-scale testing and monitoring of the newly spreading and known SARS-CoV-2 variants. Certainly, whole-genome sequence (WGS) can discover the new variants for SARS-CoV-2 and cannot be replaced by qPCR. Thus PCR method and NGS method can well supplement each other in viral variant detection and tracking. Given their importance in fighting the pandemic, the MIT Technology Review magazine listed “COVID variant tracking” as one of the 10 breakthrough technologies in 2022 (32).
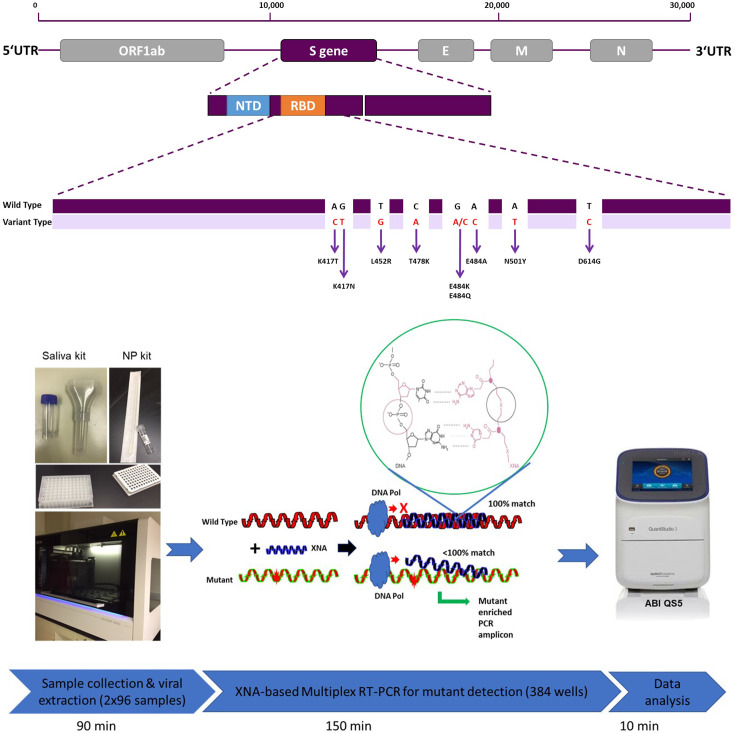
Considering the rapid evolution and emergence of various variants, testing platforms capable of rapid detection of specific variants in a cost-effective manner are urgently needed. Toward this end, target-specific RT-PCR screens for specific mutations within the spike protein had been developed (33, 34). In this study, we have developed a multiplex reverse-transcription quantitative real-time PCR (RT-qPCR) assay that can rapidly and reliably detect known and emerging SARS-CoV-2 variants based on their mutation profiles (Fig. 1). The assay is based on molecular-clamping technology and uses xenonucleic acids (XNA) as molecular clamping probes.
Fig 1.
Principle of SARS-COV-2 variant detection RT-qPCR. (A) SARS-CoV-2 genome structure and its spike gene target sites for our multivariant assay. (B) A high-throughput RT-qPCR workflow for SARS-CoV-2 variant detection, highlighting the mechanism of XNA-based molecular clamping technique capable of distinguishing a targeted mutation from its wild type. The circle indicates the primary structure of the XNA clamping molecule in a typical XNA/DNA duplex.
XNAs are artificial genetic polymers that retain the Watson-Crick base-pairing capability and exhibit high chemical and biological stability. In practical applications for disease diagnosis and treatment, XNAs have been used as a source of nuclease-resistant affinity reagents (aptamers) and catalysts (xenozymes). More notably, they can be employed as molecular clamps in RT-qPCR or as highly specific molecular probes for detecting nucleic acid target sequences due to the stronger hybridizing/binding capability seen in XNA/DNA duplexes than DNA/DNA duplexes (35). Furthermore, even a single base-pair mismatch near the center of the sequence of an XNA/DNA duplex can result in a big drop of 10°C–18°C in melting temperature (Tm) (36). This appealing feature allows for the highly specific clamping of the XNA molecule onto the targeted sequence (usually the wild type, WT) to block WT amplification, thus minimizing the WT background in RT-qPCR and selectively enhancing the signal of the mutant. Robust XNA-based assays have been extensively studied and used for in vitro molecular diagnostic assays for early detecting cancer-associated gene mutations (37), fusion gene detection (38) and companion diagnostic (39). XNA can also be used in NGS similarly against wild-type background to increase the specificity and sensitivity in mutant detections (40). Broad and diverse applications of XNAs in molecular diagnostics and other related fields including imaging and microscopy have been recently reviewed (35, 41). Based on their molecular properties and prior utility in cancer mutation detection, XNAs represent an ideal tool for developing highly sensitive RT-qPCR assays for detecting SARS-CoV-2 mutations. The XNA we used in this study is a class of chemically modified peptide nucleic acids (PNAs) with improved and more hydrophilic structure.
In this study, we demonstrated for the first time the feasibility, efficiency, reliability, and applicability of using an XNA-based RT-qPCR method to detect various SARS-CoV-2 variants. This assay provides a rapid, reliable, and cost-effective testing platform for the detection and monitoring of known and emerging SARS-CoV-2 variants.
MATERIALS AND METHODS
Study design
Deidentified patient nasopharyngeal swabs (NPS) collected and tested from early 2021 to early January 2022 at San Francisco VA Medical Center clinical laboratory and DiaCarta clinical laboratory for clinical diagnostic or screening purposes wwere used in this study. The second set of deidentified NPS samples collected and tested from June 2021 to early January 2022 at the COVID-19 screening lab at Franciscan University, Ohio, were also used.
Sample collection and RNA extraction
From early 2021 to early January 2022, patient NPS samples were collected in a virus transportation medium (VTM), tested for SARS-CoV-2 infection, and then stored at –80°C before being used for analysis in this study. An automatic RNA/DNA extraction instrument MGISP-960 (MGI Tech Co., Ltd.) and the MGI Easy Nucleic Acid Extraction Kit (Cat# 1000020261) were used to extract SARS-CoV-2 viral RNA according to the manufacturer’s instructions. The starting extraction volume for each sample is 200 µL of NPS sample in VTM. The final RNA extraction volume is 40 µL in RNase-free water, of which 5.5 µL was then used for a single RT-qPCR. The typical turnaround time from sample RNA extraction was about 90 min for 384 samples (Fig. 1) (42).
Multiplex primer and probe design
In order to design primers and probes to detect all the SARS-CoV-2 variants of concern, the conserved regions of ORF1ab gene (34) and areas adjacent to the N501Y, D614G, T478K, L452R, K417T, and K417N mutations in S protein RBD were targeted (Fig. 1A). Unique combinations of these mutants can be used to identify each specific VOC (Table S1).
Gene sequences were retrieved from GenBank and GISAID databases for primer and probe design to ensure coverage of all SARS-CoV-2 variant strains. Multiple alignments of the collected sequences were performed using Qiagen CLC Main Workbench 20.0.4., and conserved regions in each target gene were identified using BioEditor 7.2.5. Primers and probes were designed by using Primer3plus software. All primers were designed with a Tm of approximately 60℃, and the probes were designed with a Tm of about 65℃. The amplicon sizes were kept within the range of 70–150 bp for each primer pair in order to achieve high amplification efficiency and detection sensitivity (Table S2). All designed primers and probes were ordered from Integrated DNA Technologies, Inc. (Coralville, IA) and LGC Biosearch Technologies (Novato, CA), respectively.
XNA design, synthesis, analysis, and optimization
Five groups of XNAs, each specific to one of five SARS-CoV-2 mutations (D614G, N501Y, T478K, L452R, and K417T/K417N), were designed to be exact matches with the WT sequence in order to facilitate selective blocking of qPCR amplification of WT targets. For each mutation, a number of XNAs of different lengths were synthesized in our chemistry laboratory (Table S3a and b). Each XNA was also designed to partially overlap with and be of the same strand/sense as the corresponding qPCR probe. The best XNA probe sequences were selected based on iterative adjustment of multiple major physicochemical factors: probe sequence length, GC content, purine content and arrangement, self-complementarity, and melting temperature. Five groups of XNAs were synthesized, and each group was tested using qPCR to find the optimal size and concentration of XNA for each group. Fig. S1 showed matrix assisted laser desorption ionization (MALDI)-TOF mass spectra of XNAs.
Real-time reverse-transcription PCR
Two different detection kit assays were developed using this methodology. The QuantiVirus SARS-CoV-2 Variants Detection Kit assay consisted of three multiplex PCR tubes: tube A tested for the presence of D614G and L452R, tube B tested for the presence of K417 T, N501Y, and the human Rp gene as internal control, and tube C tested for the presence of T478K, K417 N, and ORF1ab as wild-type target. The QuantiVirus SARS-CoV-2 Delta Plus Detection Kit assay consisted of only one 4-plex PCR tube that tested for the presence of T478K, K417N, ORF1ab, and Rp.
XNA-based RT-qPCRs with a total volume of 10 µL were developed using the following reagents: 5.5 µL of viral RNA, 1.0 µL of primer and probe mixture (final concentration of 0.2 µM and 0.1 µM, respectively), 1.0 µL 10× XNAs mixes, and 2.5 µL of TaqPath 1-step Multiplex Master Mix (Catalog# A28526, Thermos Fisher, MA). Each test included a positive control (PC), a negative control (NC), and a no template control (NTC) to ensure there were no false positives, false negatives, or contamination in each qPCR run. The PC contained all mutation target genes, the NC included the wild-type gene for each target, and the NTC was water.
The qPCR was performed at 25℃ for 2 min with uracil-N-glycosylase (UNG) incubation to remove potential carryover, then 53℃ for 10 min for reverse transcription, followed by 95℃ for 2 min. Then 45 cycles of 95℃ for 3 s and 60℃ for 30 s were used for amplification. A Bio-Rad CFX384 (Bio-Rad, CA) was used for RT-qPCR amplification and detection (42–44). Assessment of the results for each individual assay should be based on the Ct values < 40 for each gene (Orf1 ab, D614G, N501Y , T478K, L452R, K417T, and K417N) as positive and Ct > 40 as negative. The results were interpreted according to the Table S1.
Analysis of assay sensitivity and cross-reactivity test
Twist SARS-CoV-2 RNA controls 16 (Beta), 17 (Gamma), 23 (Delta), and 48 (Omicron; Twist Bioscience, CA) were used as references to test the assay sensitivity. We used a twofold dilution series from 800 copies/mL down to 25 copies/mL of the templates in triplicates and identified the lowest concentration that was detectable with 95% confidence to determine the analytical sensitivity limit of detection (LoD).
As control standards, MERS-CoV and SARS-CoV coronavirus samples were ordered from ATCC (Manassas, Virginia), and the ZeptoMetrix NATtrol Respiratory Validation Panel was ordered from ZeptoMetrix (cat# NATRVP-3, Buffalo, NY). RNA/DNA was extracted from high-titer stocks of these potentially cross-reacting microorganisms (estimated 105 units/mL) using the MGI Easy Nucleic Acid Extraction Kit (Cat# 1000020261). The extracted sample RNA/DNA was eluted with sterile RNase-free water to give a 100 µL solution. A volume of 5.5 µL of each of the purified RNA/DNA samples was tested in triplicate with QuantiVirus SARS-CoV-2 Variants Detection Kit.
Clinical evaluation of samples
All the clinical samples were evaluated using the QuantiVirus SARS-CoV-2 Variants Detection Test kit. In addition, the presence/absence of SARS-CoV-2 was confirmed for all samples using the Food and Drug Administration (FDA) Emergency Use Authorization (EUA)-approved QuantiVirus SARS-CoV-2 Multiplex Test Kit at the DiaCarta CLIA-certified clinical laboratory or at the Franciscan University COVID screening lab.
Sanger sequencing and next-generation sequencing verification
All samples that were identified as variant positive by this assay were sent out for Sanger Sequencing to confirm their mutational status (SequeTech, CA), and all sequences were analyzed via the UCSC SARS-CoV-2 Genome Browser (45).
Sanger Sequencing was prepared according to the following procedure. The extracted RNA samples from the SARS-CoV2 virus were amplified in a 10 µL reaction of RT-PCR using the following reagents: 2.5 µL of 4× 1-step qRT-PCR Master Mix, 2.0 µL of primer mixture (final concentration of 0.2 µM), and 5.5 µL of viral RNA. The RT-PCR procedure consisted of the following steps, and a Bio-Rad CFX384 instrument (Bio-Rad, CA) was utilized: (i) incubation at 25℃ for 2 minutes with UNG to eliminate potential carryover; (ii) reverse transcription at 53℃ for 10 minutes; (iii) initial denaturation at 95℃ for 2 minutes; (iv) amplification through 45 cycles of denaturation at 95℃ for 3 seconds and annealing/extension at 60℃ for 30 seconds. Subsequently, all the crude PCR products were sent out for Sanger sequencing.
The NGS library was prepared with amplicon methodology using the CleanPlex SARS-CoV-2 FLEX Research and Surveillance Panel (Cat#. 918010, Paragon Genomics, CA) or Illumina COVIDSeq Test (FDA EUA approved, cat# 20049393) and sequenced on the MiSeq instrument (Illumina, CA). All the raw data were analyzed by ARTIC workflow on Galaxy (https://github.com/galaxyproject/SARS-CoV-2), using a reference sequence from the NCBI database (NC_045512.2) (46–48). All tools were used with default parameters.
RESULTS
XNA enhances the ability to distinguish viral variants from wild type in RT-qPCR
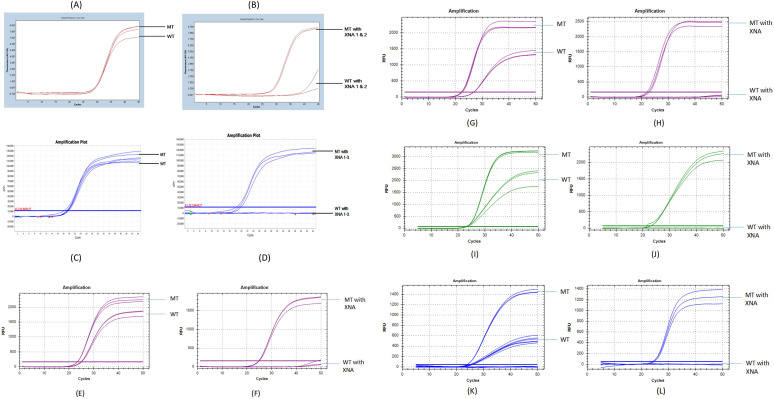
In order to test whether XNA clamping of the wild-type sequences enhances mutant detection, we compared the RT-qPCR results with and without XNA. An amplification curve of the D614G mutant vs wild type without XNA showed that it is difficult to distinguish the mutant and the wild-type virus (Fig. 2A and C). However, with XNA, the differentiation of the mutant from wild-type SARS-CoV-2 (Fig. 2B and D) was clear (ΔCt, ~13–20). Figure 2E to L showed L452R, T478K, K417T, and K417N detection with or without XNA. To optimize the assay, we tested different concentrations of XNA. Representatively, the optimization results of D614G and N501Y XNAs are shown in Tables S4 and S5, respectively. The XNA that generated the largest ΔCt between the wild type and the mutant, within its series, was selected for use in RT-qPCR. For example, 8 µM D614G XNA001 resulted in the largest ΔCt 15.9, and 0.25 µM N510Y XNA003 resulted in the largest ΔCt 32.7. Through the optimization process, we were able to select D614 XNA001, N501 XNA003, T478 XNA001, L452 XNA003, and K417 XNA001 as the optimal XNAs (their confirmed structural or mass spectra data are shown in Fig. S1 and Table S3a and b).
Fig 2.
qPCR test with and without XNA. For D614G mutation detection: (A) amplification curve of D614G mutant (MT) vs WT without XNA. Both wild-type and mutant had ~30 Ct. It is difficult to distinguish mutant and wild type. (B) Amplification curve of D614G mutant vs wild type with XNA. Wild-type amplification was blocked and had high Ct ~ 40, but mutant was enhanced and had Ct ~ 27, making the differentiation of WT/MT highly reliable, easy, and accurate. For N501Y mutation detection: (C) N501Y mutant (MT) vs WT without XNA. Both wild-type and mutant had ~21 Ct . It is difficult to distinguish the mutant and the wild type. (D) N501Y mutant vs wild type with XNA. Wild-type amplification was blocked and had high Ct > 40, but mutant was enhanced and had Ct ~ 20, making the distinction apparent and interpretation intuitive. For L452R mutation detection: (E) L452R mutant (MT) vs WT without XNA. The ∆Ct was around 2. (F) L452R MT vs WT with XNA. Wild-type amplification was blocked and had high Ct > 40, made ∆ Ct to 25. For T478K mutation detection: (G) T478K MT vs WT without XNA (H) T478K MT vs WT with XNA. The ∆ Ct between MT and WT increased from 3 to 27. For K417T mutation detection: (I) K417T MT vs WT without XNA (J) K417T MT vs WT with XNA. The ∆ Ct between MT and WT increased from 0.5 to 28 and wild-type amplification was fully blocked. For K417N mutation detection: (K) K417N MT vs WT without XNA (L) K417N MT vs WT with XNA. The ∆ Ct between MT and WT increased from 1 to 25, and wild-type amplification was fully blocked.
Analytical sensitivity and cross-reactivity evaluations
In order to determine the analytical sensitivity of the assay, we performed the QuantiVirus SARS-CoV-2 variant detection test on a Bio-Rad CFX384. We diluted SARS-CoV-2 RNA control 16/17/23 which contains the N501Y, D614G, L452R, T478K, K417T, and K417T mutants (Twist Bioscience, CA) from 800 copies/mL down to 25 copies/mL (viral copy number per milliliter of transport media) and tested each level dilution repeatedly in 24 replicates (Table 1; Table S10). The LoD was determined to be 100 copies/mL for each mutant with 95% CI 76.9%–99.8%.
TABLE 1.
LoD of the multiplex RT-qPCR for SARS-CoV-2 variant detectionb
| Target | Copies/mL | Avg Ct | SD | CV | Overall testsa (N) |
Detection rate (95% CI) |
|---|---|---|---|---|---|---|
| N501Y | 200 | 33.76 | 0.29 | 0.86% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 34.43 | 0.29 | 0.83% | 23/24 | 95.83% (76.9%–99.8%) | |
| 50 | 36.89 | 0.27 | 0.73% | 22/24 | 91.67% (71.5%–98.5%) | |
| 25 | 37.94 | 0.29 | 0.77% | 21/24 | 87.50% (66.5%–96.7%) | |
| D614G | 200 | 32.42 | 0.26 | 0.79% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 34.28 | 0.34 | 1.00% | 23/24 | 95.83% (76.9%–99.8%) | |
| 50 | 37.20 | 0.25 | 0.67% | 20/24 | 83.33% (61.8%–94.5%) | |
| 25 | 38.92 | 0.27 | 0.69% | 19/24 | 79.17% (57.3%–92.1%) | |
| L452R | 200 | 34.95 | 0.27 | 0.78% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 35.78 | 0.30 | 0.84% | 23/24 | 95.83% (76.9%–99.8%) | |
| 50 | 36.84 | 0.24 | 0.66% | 22/24 | 91.67% (71.5%–98.5%) | |
| 25 | 37.90 | 0.20 | 0.53% | 20/24 | 83.33% (61.8%–94.5%) | |
| K417T | 200 | 32.32 | 0.33 | 1.01% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 33.49 | 0.30 | 0.90% | 24/24 | 100.00% (82.8%–100%) | |
| 50 | 34.56 | 0.28 | 0.80% | 23/24 | 95.83% (76.9%–99.8%) | |
| 25 | 36.36 | 0.32 | 0.87% | 22/24 | 91.67% (71.5%–98.5%) | |
| K417N | 200 | 32.88 | 0.29 | 0.89% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 33.71 | 0.30 | 0.90% | 24/24 | 100.00% (82.8%–100%) | |
| 50 | 34.81 | 0.28 | 0.82% | 22/24 | 91.67% (71.5%–98.5%) | |
| 25 | 36.69 | 0.25 | 0.68% | 22/24 | 91.67% (71.5%–98.5%) | |
| T478K | 200 | 33.32 | 0.27 | 0.82% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 34.20 | 0.27 | 0.80% | 24/24 | 100.00% (82.8%–100%) | |
| 50 | 35.49 | 0.21 | 0.60% | 23/24 | 95.83% (76.9%–99.8%) | |
| 25 | 37.32 | 0.30 | 0.81% | 22/24 | 91.67% (71.5%–98.5%) | |
| ORF | 200 | 33.32 | 0.28 | 0.83% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 33.93 | 0.28 | 0.82% | 24/24 | 100.00% (82.8%–100%) | |
| 50 | 34.99 | 0.23 | 0.65% | 22/24 | 91.67% (71.5%–98.5%) | |
| 25 | 36.32 | 0.28 | 0.76% | 22/24 | 91.67% (71.5%–98.5%) | |
| RP | 200 | 34.17 | 0.29 | 0.85% | 24/24 | 100.00% (82.8%–100%) |
| 100 | 34.78 | 0.28 | 0.81% | 24/24 | 100.00% (82.8%–100%) | |
| 50 | 36.84 | 0.29 | 0.79% | 23/24 | 95.83% (76.9%–99.8%) | |
| 25 | 38.04 | 0.32 | 0.84% | 23/24 | 95.83% (76.9%–99.8%) |
Each concentration was tested repeatedly by 24 replicated tests. For details, see Table S9.
CV, coefficient of variation; ORF, SARS-CoV-2 open reading fragmes; RP, human RNase P gene.
We evaluated the cross-reactivity of the assay to a variety of other pathogens. The results are summarized in Table S6 and indicate that there is no cross-reactivity in our XNA-based qPCR assay among the SARS-CoV-2 variants and any of the organisms tested including MERS-CoV.
Use of XNA-based RT-qPCR to confirm rapid surge of Alpha variant in San Francisco Bay Area in early 2021
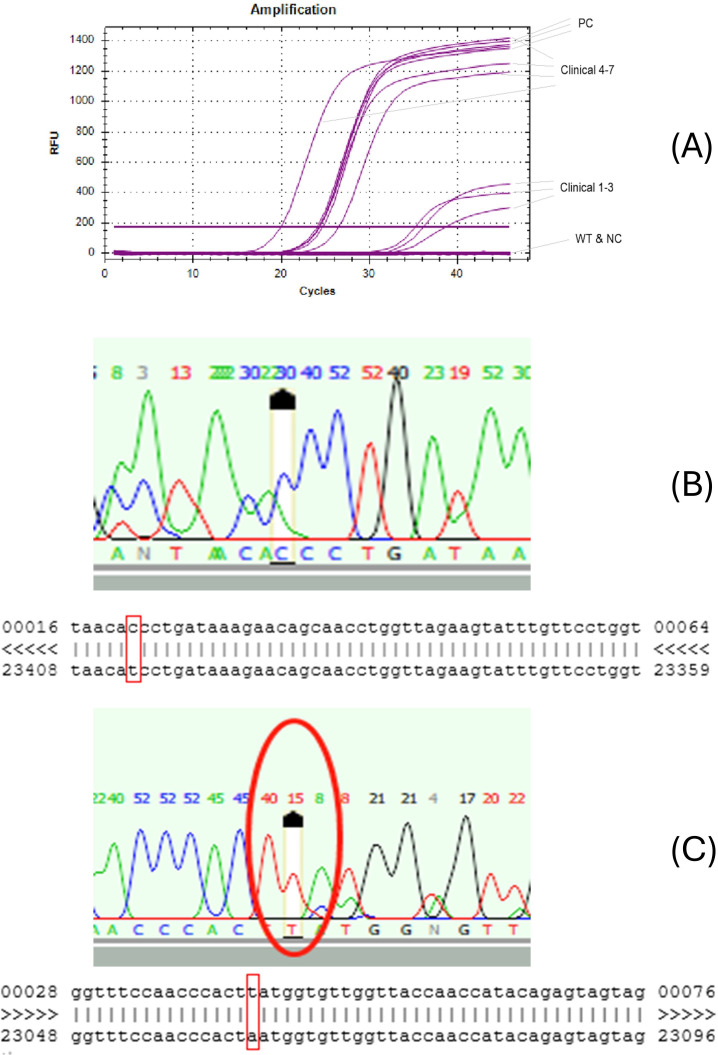
In order to determine the ability of the QuantiVirus SARS-CoV-2 Variants Detection kit to detect the Alpha variant, 374 confirmed positive samples that were collected between January and March 2021 in the San Francisco Bay Area were analyzed. Among the 139 positive specimens sampled from mid-January 2021, 58 (41.7%) were positive for the D614G mutation but not the N501Y (Tables 2 and 3). None of the samples were positive for both N501Y and D614G mutations, a defining characteristic of the Alpha variant (B1.1.7). However, of the 139 positive specimens sampled from late February and the 96 positive specimens collected in March 2021, there were 7 (5.04%) and 10 (10.42%) specimens, respectively, that were positive for both mutations. The data suggested an increase in the Alpha variant frequency in Northern California, from 0% in January to above 10% in March 2021. Additionally, the multiplex RT-qPCR test showed high specificity, i.e., amplification in non-N501Y samples and negative controls was completely blocked by the XNA in the RT-qPCR, while amplification was present in all N501Y positive clinical samples and positive controls (Fig. 3A).
TABLE 2.
Summary of SARS-CoV-2 alpha variant detection in January to March 2021a
| Sample collection time | RT-qPCR test | N501Y | D614G | ORF1abb | RNase P gene |
|---|---|---|---|---|---|
| January 2021 | Positive | 0 | 58 | 132 | 139 |
| Total (N) | 139 | 139 | 139 | 139 | |
| Alpha variant detection rate (%) | 0 | 41.7 | 95.0 | 100.0 | |
| February 2021 | Positive | 7 | 78 | 139 | 139 |
| Total | 139 | 139 | 139 | 139 | |
| Alpha variant detection rate (%) | 5.0 | 56.1 | 100.0 | 100.0 | |
| March 2021 | Positive | 10 | 41 | 96 | 96 |
| Total | 96 | 96 | 96 | 96 | |
| Alpha variant detection rate (%) | 10.4 | 42.7 | 100.0 | 100.0 |
Three hundred seventy-four clinical samples collected in middle January, late February, and March 2021 in the California Bay area.
ORF1ab gene was targeted for wild-type SARS-CoV-2.
TABLE 3.
| SARS-CoV-2 | Patient samples (N) | Quantivirus SARS-COV-2 variant detection | Variant identification | Sensitivity (95% CI) | Specificity (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant detection | Positive | Negative | Alpha | Early mutation/wild type | Negative | ||||||
| Positive | 374 | N501Y | 17 | 357 | 17 | 160/197 | 102 | 100% (0.77–1.00) | 100% (0.99–1.00) | 100% (0.77–1.00) | 100% (0.99–1.00) |
| D614G | 177 | 197 | 100% (0.97–1.00) | 100% (0.98–1.00) | 100% (0.97–1.00) | 100% (0.98–1.00) | |||||
| ORF1ab | 374 | 0 | 100% (0.99–1.00) | 100% (0.96–1.00) | 100% (0.99–1.00) | 100% (0.96–1.00) | |||||
| Rp | 476 | 0 | |||||||||
| Negative | 102 | ||||||||||
Bio-Rad CFX384 qPCR instrument was used for this test; all of 374 positive samples were detected ORF1ab gene (wild type) positive; all variants data were confirmed with Sanger sequencing.
Positive predictive value (PPV) and negative predictive value (NPV) were calculated by comparing qPCR data with Sanger sequencing data for each sample.
Fig 3.
(A) Representative RT-qPCR amplification curves of SARS-CoV-2 variant N501Y. PC, positive control; WT, wild type; NC, negative control. Clinical, clinical samples 1–7. (B and C) Confirmation of D164G and N501Y mutations by Sanger sequencing. (B) Sanger sequencing peaks (C in the red circle: D614G mutant). Sequence alignment of D614G, red squares indicate D164G mutant sequence T < C. (C) Sanger sequencing peaks (T in the red circle: N501Y mutant). Sequence alignment of N501Y. Red squares indicate N501Y mutant sequence A < T.
To verify that the D614G and the D614G/N501Y mutations in these mutant samples were detected correctly, we amplified the mutant target by PCR, and the amplicons from these samples were analyzed by Sanger sequencing. All 160 D614G positive samples, 17 D614G/N501Y double positive samples, and 197 wild-type samples, as determined by the Quantivirus SARS-CoV-2 Variants Detection kit, were confirmed by Sanger sequencing indicating that the specificity of the test was 100%. The Sanger sequencing peaks showed the target mutations in the viral cDNA, T < C in the case of D614G (Fig. 3B), and A < T in the case of N501Y (Fig. 3C).
As summarized in Table 2 and Table 3, the XNA-based RT-qPCR has a positive predictive value (PPV) of 100.0% (95% CI: 0.99–1.00) and negative predictive value (NPV) of 100% (95% CI: 0.96–1.00) for detecting the Alpha variant by comparing its Sanger Sequencing data.
Use of XNA-based RT-qPCR to detect breakthrough COVID-19 cases caused by Delta variant and Omicron variant
Despite widespread vaccination, a significant number of breakthrough cases were seen in late 2021 and early 2022 due to emergence of two new SARS-CoV-2 variants, Delta and Omicron. To examine the ability of the Quantivirus SARS-CoV-2 Delta Plus Detection kit to detect these variants, we analyzed 39 NPS that were initially collected from symptomatic COVID-19-positive individuals from Franciscan University in Ohio (Fall 2021) who had been vaccinated (“breakthrough” infection). In addition, six NPS samples collected from healthy donors were analyzed as well. All 36 breakthrough patient samples tested positive for the SARS-CoV-2 Delta variant as they displayed the T478K mutation with the wild-type ORF1ab and three Delta plus variants as K417N and T478K mutant detectable, whereas all six samples collected from healthy donors tested negative for SARS-CoV-2 (Table S7) and an additional 44 negative samples were confirmed negative (Table 4). The data were compared with Sanger sequence and determined that the assay’s PPV for Delta detection was 100% (95% CI: 0.89–1.0), and its NPV was 100% (95% CI: 0.91–1.0).
TABLE 4.
Summary of Delta variant detection for breakthrough patient samples from October to November 2021
| qPCR kit | Patient sample (N) | Mutant target | Positive | Negative | Variant identification | Comparing with Sanger sequence | |||
|---|---|---|---|---|---|---|---|---|---|
| Delta | Delta+ | Negative | PPV (%) | NPV (%) | |||||
| Quantivirus SARS-CoV-2 delta plus | 89 | K417N | 3 | 86 | 36 | 3 | 50 | 100 (95% CI: 0.888–1.0) | 100 (95% CI: 0.911–1.0) |
| T478K | 39 | 50 | |||||||
| Orf1ab | 39 | 50 | |||||||
| RP | 89 | 0 | |||||||
To assess the ability of QuantiVirus SARS-CoV-2 Variants Detection kit to detect the Omicron variant, we tested an additional 37 NPS samples, 34 of which were collected from vaccinated symptomatic patients in early January 2022. All 34 samples collected from vaccinated symptomatic patients tested positive, while the other three samples tested negative for SARS-CoV-2 by the FDA EUA-approved kit (QuantiVirus SARS-COV-2 Multiplex Test). All 34 positive samples further tested positive for the K417N, T478K, N501Y, and D614G mutations, characteristic of the Omicron variant, using the XNA-based RT-qPCR assay in this kit (Table S8). We also tested 50 negative confirmed samples. The data were compared with Sanger Sequence and determined that the assay’s PPV for Omicron detection was 100% (95% CI: 0.87–1.00), and its NPV was 100% (95% CI: 0.92–1.00; Table 5).
TABLE 5.
Summary of Omicron variant detection for breakthrough patients during early 2022
| qPCR kit | Patient sample (N) | Mutant target | Positive | Negative | Variant identification | Comparing with Sanger sequence | ||
|---|---|---|---|---|---|---|---|---|
| Omicron | Negative | PPV (%) | NPV (%) | |||||
| Quantivirus SARS-CoV-2 variant detection | 87 | D614G | 34 | 53 | 34 | 53 | 100 (95% CI: 0.87–1.0) | 100 (95% CI: 0.92–1.0) |
| N501Y | ||||||||
| K417N | ||||||||
| T478K | ||||||||
| Orf1ab | ||||||||
| RP | 87 | 0 | ||||||
Summary of XNA-based RT-qPCR assay in detecting SARS-CoV-2 variants and comparing with NGS
In total, 649 samples were screened using the kit, including 447 SARS-CoV-2 positive samples and 202 negative samples (Table 6). Among the 447 positive samples, the unique combination sets of detected viral S-gene mutations suggest the following categorization: 17 cases of the Alpha variant, 5 cases of the Beta variant, 36 cases of the Delta variant, 3 cases of the Delta Plus variant (AY.4.2), and 34 cases of the Omicron variant were detected. Notably, there were 160 cases which were only positive for D614G mutation alone (not yet acquiring the N501Y mutation as the Alpha variant) and 192 cases of “wild type” (no mutations) among the 447 COVID-positive samples, mostly from early 2021. In summary, 21.3% (95/447) of the samples were identified to be SARS-CoV-2 variants, and their prevalence over time followed the sequential emergence of the variants in North America. At least 57% (255/447) of the positive samples had at least one mutation. All the data were confirmed by Sanger Sequencing.
TABLE 6.
| Sample total (N) | Status (n) pos/neg | Variant (n) | ORF1ab | D614G | N501Y | T478K | L452R | K417T | K417N | Variant identity |
|---|---|---|---|---|---|---|---|---|---|---|
| 649 | 447 (positive) | 34 | x | x | x | x | x | Omicron | ||
| 36 | x | x | x | x | Delta | |||||
| 3 | x | x | x | x | x | Delta plus | ||||
| 5 | x | x | x | x | Beta | |||||
| 17 | x | x | x | Alpha | ||||||
| 160 | x | x | Early mutation | |||||||
| 192 | x | Wild type | ||||||||
| 202 (negative) | 202 | Negative |
All data were confirmed by comparing Sanger sequencing data.
"X" mean positive detection for each mutation.
To further validate the XNA-based RT-qPCR assay in detecting SARS-CoV-2 variants, a random subset of eight positive variant samples, one wild type, and two SARS-CoV-2 negative samples from early 2021 were tested by this new assay and NGS. Out of the nine samples that tested positive using the kit, eight variants were found to be N501Y/D614G positive, while the remaining one sample was identified as the wild type without any detected mutations. On the other hand, the two SARS-CoV-2 negative samples tested negative using the same kit. All 11 samples were then sequenced by NGS using the CleanPlex SARS-CoV-2 FLEX kit, and all eight positive samples were confirmed as Alpha variants, while the one wild type was confirmed as wild type (Lineage 19A, Nextstrain clad), and the two negative samples were confirmed as negative (Table S9). We additionally sequenced and analyzed 13 positive samples (vaccinated breakthrough cases) and two negative samples collected in early 2022 using both the QuantiVirus SARS-CoV-2 Variants Detection Kit and the Illumina COVIDSeq Test. Again, the results were all congruent (Table S9). The data demonstrated that our RT-qPCR platform gave the same result as the Illumina COVIDSeq Test with 100% concordance.
DISCUSSION
We have described the successful development of a multiplex RT-qPCR testing platform for the rapid detection of SARS-CoV-2 variant strains using XNA-based molecular-clamping technology. In order to develop a sensitive and specific molecular-clamping assay, it is not only necessary to select a suitable set of primers and probe for a given mutant gene target, but it is also imperative to choose an XNA with appropriate sequences and desired performance characteristics. Our XNA selection process included a twofold approach. First, during the sequence design, we excluded those improper sequences with problematic features (e.g., sequences that were too long or too short, had too high or too low Tm values, high purine content, long purine stretch, or unwanted self-complementarity within or between XNA molecules). Second, for each mutation assay, we designed and synthesized multiple XNAs and compared their qPCR clamping robustness and clamping specificity (higher delta Ct difference between wild-type gene and mutation gene amplification). Notably, across the XNA groups, the XNA with a Tm of nearly 80℃ stood out in the selection process, which is likely related to the established RT-qPCR temperature-cycling conditions (49).
Using this assay, we were able to track the changes in SARS-CoV-2 variants over time. Due to its higher transmissibility compared to the original SARS-CoV-2 strain, the D614G mutant of SARS-CoV-2 became the dominant variant in the beginning of 2021 in the United States. Its sub-clade, the N501Y mutation, independently emerged in the UK and South Africa and subsequently spread to North America during the first half of 2021. Our data indicated that the spread of this Alpha variant in northern California likely started in February 2021 because patient samples collected in January 2021 were all negative for the N501Y mutation, whereas the N501Y mutation was detectable in up to 5% of the samples collected in late February 2021. Although the U.S. CDC predicted that, according to an epidemiology model (50) at that time, the B.1.1.7 or Alpha variant would quickly dominate soon, the Alpha variant was soon largely replaced around the middle of 2021 by the even more potent and infectious Delta variant. The Delta variant in turn was soon replaced by the more contagious Omicron variants in late 2021 to early 2022. Our RT-qPCR data, confirmed by Sanger or NGS sequencing, were consistent with the sequential surges of COVID-19 cases caused by the D614G mutant and the Alpha, Delta, and Omicron variants in the United States from late 2020 to early 2022.
Availability of rapid and accurate testing platforms is critical for tackling the challenge of emerging SARS-CoV-2 variants, especially to understand breakthrough cases and provide appropriate treatments. Among the many detection methods, a qPCR-based platform could serve as a rapid, cost-effective, and practical testing tool for monitoring SARS-CoV-2 evolution.
A few biotechnology companies and academic institutions have been developing or have reported variant detection methods using qPCR. These methods can be categorized into one of the following: (i) mutant gene-specific or allele-specific primers and probe; (ii) spike gene target failure; (iii) E gene target failure; and (iv) ORF gene deletion (51–54). Some of the methods still require NGS to confirm the results, whereas other platforms require pre-testing by regular SARS-CoV-2 RT-qPCR before carrying out the variant assay. While these assays can be useful in certain circumstances (55), they are limited by the effectiveness and accuracy of the primers and/or probes used. This issue can limit the use of these variant testing methodologies. For example, the allele-specific PCR (AS-PCR) method quickly attracted attention for mutation detection by some test developers (56). However, AS-PCR has two inherent shortcomings: (i) due to the fixed 3′ end of the allele-specific primer, it is not always feasible to choose optimal primers for PCR amplification; and (ii) high purity DNA/RNA is essential as low-quality or crude DNA/RNA samples are prone to providing inconclusive results (57).
However, XNA-based qPCR can overcome these aforementioned shortcomings because XNA sequences can be designed with more flexibility compared to restrictive primer sequence design (58), and XNA-based qPCR works better with samples of low-concentration DNA/RNA and samples with high-background mutant gene targets. Noteworthy is that the XNAs have been made with practically appealing cost and high efficiency—the unit cost of XNA is between the oligo primer and the Taqman PCR probe. Compared to AS-PCR, as demonstrated in our studies, the XNA-based RT-qPCR assay can achieve a lower LoD, about 100 copies/mL for variant detection.
Of particular note, the determined LoD 100 copies/mL here refers to the original, collected patient sample, not the extracted concentrated RNA sample. Based on the described procedure of virus RNA extraction, we can calculate the copy number per PCR (10 µL/reaction) to be 2.8–3.7 copies per reaction, this result was well consistent with the theoretical considerations in the MIQE guidelines (59). The molecular-clamping technology used in this study increases the sensitivity and specificity of conventional qPCR as wild-type background amplification is minimized by this method. This XNA clamping-based RT-qPCR assay can also improve the multiplexing of targets, as we were able to differentiate the wild-type SARS-CoV-2 (ORF1ab gene for wild-type detection) and its variants in a single run. In addition to detecting the existing SARS-CoV-2 mutations D614G and N501Y, the assay described here also detected L452R, T478K, K417N, and K417T. The presence or absence of these major mutations can be used to suggest and identify all five VOCs based on their unique mutation profiles VOCs (60) (Table S1). As a result, this assay covers almost all SARS-CoV-2 variants (both VOC and VOI), including Delta, “Delta Plus,” and Omicron, all of which have the potential to breakthrough vaccine-induced protective immunity (60). Of note, it is natural to consider an additional E484A mutation to help detect Omicron, but we finally opted to use the unique combination of N501Y, T478K, and K417N mutations to detect Omicron by using the already-available K417 XNA and K417N primers/probe to maximally speed up the variants assay efforts.
This strategy can provide an effective, rapid, and easily adoptable testing platform for known and emerging SARS-CoV-2 variants in the future, and this technology can be readily adopted by clinical laboratories that perform routine SARS-CoV-2 RT-qPCR testing. Given the established stages of the COVID-19 pandemic, future new variant(s) with increased transmissibility could potentially lead to an exponential increase in cases and/or deaths (13). Therefore, timely and convenient detection of the concerning variants is of high importance.
There are 15 mutations in the RBD of Omicron variant, including Q393R, K417N, T478K, S477N, E484A, G496S, Q498R, Y505H, G446S, N501Y, N440K, S75F, S373P, S371L, and G339D (29). This poses a significant challenge for RT-qPCR-based assays as this high number of mutations can render some primers dysfunctional. Interestingly, the N501Y/D614G primer, which works well for Alpha, Beta, and Delta variant detection, had a lower signal for N501Y/D614G detection in Omicron variants due to the fact that the original N501Y/D614G primer was located in Omicron high mutant region (data in Tables S8 and S9). To avoid this issue in the future, we have re-designed a new set of “degenerate” primers and probes to better fit the mutant area in Omicron, while still covering the original N501Y/D614G region in Alpha, Beta, and Delta variants. Evaluation of this revised N501Y/D614G assay design showed that it is effective in distinguishing these variants effectively. This illustrates how this method can be rapidly adapted to test new and emerging variants while still successfully detecting all known variants.
Despite this issue, our XNA-based RT-qPCR assay kit was able to identify the Omicron variant because of the presence of three mutations N501Y/D614G/T478K/K417N and the absence of L452R mutation, a combination unique to the Omicron variant. Indeed, we were able to detect and differentiate Omicron and Delta variants and successfully detected the Omicron variant in patients who had breakthrough SARS-CoV-2 infections in early January 2022.
Noteworthy, compared to the NGS method in existing SARS-CoV-2 variant detection, the cost (time, reagents, and other resources) of using RT-PCR is considerably cheaper. However, NGS (when WGS is used) is the Gold-Standard method in discovering the emerging unknown variants.
Furthermore, compared to the basic RT-qPCR method, our PCR method using XNA molecular-clamping technology offers a more straightforward and easier interpretation of experimental data, thus saving time and resources; meanwhile, the design and synthesis of XNA molecular clampers were optimized so as to merely add a small extra cost (lower than the synthetic cost of well-known Taqman probe).
The development of mutation-specific RT-qPCR, combined with wild-type gene blockers, has utilized various techniques. For instance, Chaintoutis et al. introduced a method using locked nucleic acids (LNA)-modified oligonucleotides to distinguish target mutations from wild-type genes. This method incorporated LNA modifications into TaqMan probes and non-extendable blocking oligonucleotides to enhance the Tm, thereby improving the test’s specificity and sensitivity. However, the same LNA modifications that increased the Tm also posed limitations. LNA-modified probes could potentially hybridize non-specifically, and the non-extendable oligonucleotide blockers, which had a lower Tm than the LNA probes, might not fully block the wild-type genes (61, 62).
In addition to RT-qPCR and genotyping methods, electrophoresis-based fragment analysis presents an alternative for detecting COVID-19 variants. Clark et al. designed a multiplex fragment analysis technique called CoVarScan, which distinguishes PCR target variants by size and fluorescent color. This method targets eight mutation hotspots in SARS-CoV-2 VOCs, with fragments analyzed via capillary electrophoresis (ABI 3730XL). However, the sensitivity of CoVarScan does not match that of RT-qPCR, and the requirement for capillary electrophoresis constrains its broader application (63).
In contrast to this novel XNA-assisted RT-qPCR method for RNA detection of SARS-CoV-2 variants, there have been no studies reported to our best knowledge to utilize the molecular clampers like PNAs in RT-qPCR SARS-CoV-2 variant detection and identification. One close example was to use PNA clampers in digital PCR for variant detection reported most recently (64). Additionally, other studies using PNA clampers have developed a non-PCR method, such as a biosensor technique (65) and loop-mediated isothermal amplification based on SARS-CoV-2 variants detection (66). These also reflect the wide applications, flexibility, and vast potential of applying molecular clampers like XNAs in clinically relevant molecular diagnosis.
There are a few limitations in this study: (i) relatively limited number of samples (n < 1,000); (ii) less attention to newer variants or subvariants due to our main focus on the earlier, most threatening VOCs from Alpha to Delta variants prior to the Omicron variant in the pandemic; (iii) multiple tubes instead of a single tube is included in this kit due to the detection of >4 targets and also consideration of minimizing the assay interference between close targets (e.g., L452 and T478).
In the further development of viral variant detection methods, the following directions may be worthy of attention: PCR probe improvement with even higher specificity; a higher degree of multiplexing; artificial-intelligence-assisted assay design for rapid multiplexing optimization; more integration of multiple techniques/platforms to better supplement each other (isothermal, qPCR, digital PCR, and NGS) for different purposes of variant detection (monitoring, screening, point-of-care, etc.).
In summary, we have developed a multiplex RT-qPCR testing platform for the rapid detection of SARS-CoV-2 variants using the XNA-based molecular-clamping technology. This testing platform can be adopted by laboratories that perform routine SARS-CoV-2 PCR testing, providing a rapid and cost-effective method in lieu of NGS-based assays, for detecting, differentiating, and monitoring SARS-CoV-2 variants. This assay is easily scalable to any new variant(s) should it emerge.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to DiaCarta Clinical Laboratory colleagues Yulia Loginova and Eric Abbott for their help in obtaining clinical samples. We also thank the UCSF DeGrado Lab for using MALDI-TOF mass spectrometry instrument.
Data collection, analysis, and interpretation: S.S., M.J., A.Y.F., J.L. L.P. C.Z., M.Y.S., C.M.L., M.R., M.K., D.K., J.N., and I.A. Chemical synthesis, purification, and analysis: A.Y.F. and D.C. Clinical sample processing: J.L., S.S., L.P., and J.A.P. Writing of original draft: S.S., M.J., A.Y.F., and M.Y.S. Revision and editing: A.Y.F., M.Y.S., S.S., D.K., J.A.P., and C.M.L. Conceptualization: M.Y.S., A.Y.F., and Q.S. Project planning and administration: M.S. and A.Z.
S.S., A.Y.F., M.J., J.L., Z.C., L.P., D.C., Q.S., A.Z., and M.Y.S. have been employed by DiaCarta Inc. during the period of this work.
AFTER EPUB
[This article was published on 16 October 2024 with a typo in the keywords. The first keyword was corrected in the current version, posted on 5 November 2024.]
Contributor Information
Michael Y. Sha, Email: mikesha168@gmail.com.
Leiliang Zhang, Shandong First Medical University, Jinan, Shandong, China.
Wayne Xianding Deng, University of California San Francisco, San Francisco, California, USA.
Junping Peng, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China.
DATA AVAILABILITY
The data generated and analyzed during this study are available from the corresponding author upon reasonable request.
ETHICS APPROVAL
Relevant portions of the study were approved by the institutional review board (IRB) at University of California at San Francisco (UCSF) (UCSF IRB #11-05207) as a no-subject contact study with waiver of consent and as exempt under category 4 and by the IRB at Franciscan University (FUS IRB #2021-07). All experiments were performed in accordance with relevant guidelines and regulations.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.04248-23.
Tables S1 to S10; Fig. S1.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. COVID-19 cases . WHO COVID-19 dashboard. Available from: https://data.who.int/dashboards/covid19/cases. Retrieved 13 Feb 2023.
- 2. CDC . 2020. COVID data tracker. Centers for Disease Control and Prevention. Available from: https://covid.cdc.gov/covid-data-tracker. Retrieved 13 Feb 2023. [Google Scholar]
- 3. Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E, on behalf of COVID-19 Genomics Consortium UK . 2020. Preliminary genomic characterization of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 4. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, et al. 2021. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature New Biol 592:438–443. doi: 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 5. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D da S, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, et al. 2021. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372:815–821. doi: 10.1126/science.abh2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. 2021. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA 325:1324–1326. doi: 10.1001/jama.2021.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, et al. 2021. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 184:3426–3437. doi: 10.1016/j.cell.2021.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll T, Fox D, van Doremalen N, Ball E, Morris MK, Sotomayor-Gonzalez A, Servellita V, Rustagi A, Yinda CK, Fritts L, Port JR, Ma Z-M, Holbrook MG, Schulz J, Blish CA, Hanson C, Chiu CY, Munster V, Stanley S, Miller CJ. 2022. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog 18:e1009914. doi: 10.1371/journal.ppat.1009914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. WHO . 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. Available from: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. Retrieved 13 Feb 2023.
- 10. Tracking SARS-CoV-2 variants. 2024. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants. Retrieved 10 Jun 2024.
- 11. CDC . 2020. COVID data tracker. Centers for Disease Control and Prevention. Available from: https://covid.cdc.gov/covid-data-tracker. Retrieved 10 Jun 2024. [Google Scholar]
- 12. SARS-CoV-2 variants of concern as of 31 May 2024. 2024. Available from: https://www.ecdc.europa.eu/en/covid-19/variants-concern. Retrieved 10 Jun 2024.
- 13. Wise J. 2020. COVID-19: new coronavirus variant is identified in UK. BMJ:m4857. doi: 10.1136/bmj.m4857 [DOI] [PubMed] [Google Scholar]
- 14. ECDC . 2023. Risk of spread of new SARS-CoV-2 variants of concern in the EU/EEA - first update - 21 January 2021. ECDC Stockh [Google Scholar]
- 15. CDC . 2020. US COVID-19 cases caused by variants. Centers for Disease Control and Prevention. Available from: https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html. Retrieved 13 Feb 2023. [Google Scholar]
- 16. Helix . The Helix COVID-19 surveillance dashboard. Available from: https://www.helix.com/pages/helix-covid-19-surveillance-dashboard. Retrieved 13 Feb 2023.
- 17. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) Consortium, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. doi: 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. 2021. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26:2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. CDC . 2020. COVID data tracker. Centers for Disease Control and Prevention. Available from: https://covid.cdc.gov/covid-data-tracker/#variant-summary. Retrieved 13 Feb 2023. [Google Scholar]
- 20. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827. doi: 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH III, Leist SR, Schäfer A, Nakajima N, Takahashi K, et al. 2020. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370:1464–1468. doi: 10.1126/science.abe8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, et al. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184:64–75. doi: 10.1016/j.cell.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagen A. SARS-CoV-2 variants vs vaccines. ASM.org. Available from: https://asm.org:443/Articles/2021/February/SARS-CoV-2-Variants-vs-Vaccines. Retrieved 25 May 2024. [Google Scholar]
- 24. Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. 2021. SARS-CoV-2 variants of concern are emerging in India. Nat Med 27:1131–1133. doi: 10.1038/s41591-021-01397-4 [DOI] [PubMed] [Google Scholar]
- 25. Konings F, Perkins MD, Kuhn JH, Pallen MJ, Alm EJ, Archer BN, Barakat A, Bedford T, Bhiman JN, Caly L, et al. 2021. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat Microbiol 6:821–823. doi: 10.1038/s41564-021-00932-w [DOI] [PubMed] [Google Scholar]
- 26. Expert reaction to cases of variant B.1.617 (the “Indian variant”) being investigated in the UK. Science Media Centre. Available from: https://www.sciencemediacentre.org/expert-reaction-to-cases-of-variant-b-1-617-the-indian-variant-being-investigated-in-the-uk. Retrieved 25 May 2024. [Google Scholar]
- 27. Li B, Deng A, Li K, Hu Y, Li Z, Shi Y, Xiong Q, Liu Z, Guo Q, Zou L, et al. 2022. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat Commun 13:460. doi: 10.1038/s41467-022-28089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Callaway E. 2021. Heavily mutated Omicron variant puts scientists on alert. Nature New Biol 600:21–21. doi: 10.1038/d41586-021-03552-w [DOI] [PubMed] [Google Scholar]
- 29. European Centre for Disease Prevention and Control . 2021. Implications of the emergence and speed of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron), for the EU/EEA. Stockholm: ECDC [Google Scholar]
- 30. UK Health Security Agency . 2022. SARS-CoV-2 variants of concern and variants under investigation in England: technical briefing 37. London: UK Health Security Agency [Google Scholar]
- 31. Yelagandula R, Bykov A, Vogt A, Heinen R, Özkan E, Strobl MM, Baar JC, Uzunova K, Hajdusits B, Kordic D, Suljic E, Kurtovic-Kozaric A, Izetbegovic S, Schaeffer J, Hufnagl P, Zoufaly A, Seitz T, Födinger M, Allerberger F, Stark A, Cochella L, Elling U, VCDI . 2021. Multiplexed detection of SARS-CoV-2 and other respiratory infections in high throughput by SARSeq. Nat Commun 12:3132. doi: 10.1038/s41467-021-22664-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Breakthrough Technologies . 2022. MIT technology review. Available from: https://www.technologyreview.com/2022/02/23/1045416/10-breakthrough-technologies-2022. Retrieved 25 May 2024.
- 33. Bedotto M, Fournier P-E, Houhamdi L, Levasseur A, Delerce J, Pinault L, Padane A, Chamieh A, Tissot-Dupont H, Brouqui P, Sokhna C, Azar E, Saile R, Mboup S, Bitam I, Colson P, Raoult D. 2021. Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant. J Clin Virol 139:104814. doi: 10.1016/j.jcv.2021.104814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Annavajhala MK, Mohri H, Wang P, Nair M, Zucker JE, Sheng Z, Gomez-Simmonds A, Kelley AL, Tagliavia M, Huang Y, Bedford T, Ho DD, Uhlemann A-C. 2021. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature New Biol 597:703–708. doi: 10.1038/s41586-021-03908-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Q, Chen L, Long Y, Tian H, Wu J. 2013. Molecular beacons of xeno-nucleic acid for detecting nucleic acid. Theranostics 3:395–408. doi: 10.7150/thno.5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kyger EM, Krevolin MD, Powell MJ. 1998. Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal Biochem 260:142–148. doi: 10.1006/abio.1998.2687 [DOI] [PubMed] [Google Scholar]
- 37. Sun Q, Pastor L, Du J, Powell MJ, Zhang A, Bodmer W, Wu J, Zheng S, Sha MY. 2021. A novel xenonucleic acid-mediated molecular clamping technology for early colorectal cancer screening. PLoS One 16:e0244332. doi: 10.1371/journal.pone.0244332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee B, Chern A, Fu AY, Zhang A, Sha MY. 2024. A highly sensitive XNA-based RT-qPCR assay for the identification of ALK, RET, and ROS1 fusions in lung cancer. Diagn (Basel) 14:488. doi: 10.3390/diagnostics14050488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen S, Brown R, Kim D, Pastor L, Fu AY, Kaur P, Babu A, Liu W, Zhang A, Tanaka H, Sha MY. 2024. Abstract 7295: a novel XNA technology-based assay to detect JAK2 V617F mutation by real-time PCR. Cancer Res 84:7295–7295. doi: 10.1158/1538-7445.AM2024-7295 [DOI] [Google Scholar]
- 40. Liu W, Brown R, Fu A, Shen S, Sha M, Zhang A. 2023. Abstract 6681: XNA increases assay sensitivity in sanger sequencing, qPCR, NGS and CRISPR mutant screening. Cancer Res 83:6681–6681. doi: 10.1158/1538-7445.AM2023-6681 [DOI] [Google Scholar]
- 41. Anosova I, Kowal EA, Dunn MR, Chaput JC, Van Horn WD, Egli M. 2016. The structural diversity of artificial genetic polymers. Nucleic Acids Res 44:1007–1021. doi: 10.1093/nar/gkv1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun Q, Li J, Ren H, Pastor L, Loginova Y, Madej R, Taylor K, Wong JK, Zhang Z, Zhang A, Lu CM, Sha MY. 2021. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLoS One 16:e0243183. doi: 10.1371/journal.pone.0243183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pinheiro VB, Holliger P. 2014. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol 32:321–328. doi: 10.1016/j.tibtech.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. FDA EUA-approved DiaCarta QuantiVirus SARS-CoV-2 test kit. 2020. Available from: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2. Retrieved 13 Feb 2023.
- 45. Fernandes JD, Hinrichs AS, Clawson H, Gonzalez JN, Lee BT, Nassar LR, Raney BJ, Rosenbloom KR, Nerli S, Rao AA, Schmelter D, Fyfe A, Maulding N, Zweig AS, Lowe TM, Ares M, Corbet-Detig R, Kent WJ, Haussler D, Haeussler M. 2020. The UCSC SARS-CoV-2 genome browser. Nat Genet 52:991–998. doi: 10.1038/s41588-020-0700-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544. doi: 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blankenberg D, Johnson JE, Team TG, Taylor J, Nekrutenko A. 2014. Wrangling Galaxy’s reference data. Bioinformatics 30:1917–1919. doi: 10.1093/bioinformatics/btu119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 20:8. doi: 10.1186/s13059-018-1618-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giesen U, Kleider W, Berding C, Geiger A, Orum H, Nielsen PE. 1998. A formula for thermal stability (Tm) prediction of PNA/DNA duplexes. Nucleic Acids Res 26:5004–5006. doi: 10.1093/nar/26.21.5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. 2021. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-January 12, 2021. MMWR 70:95–99. doi: 10.15585/mmwr.mm7003e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harper H, Burridge A, Winfield M, Finn A, Davidson A, Matthews D, Hutchings S, Vipond B, Jain N, Edwards K, Barker G, COVID-19 Genomics UK (COG-UK) Consortium . 2021. Detecting SARS-CoV-2 variants with SNP genotyping. PLoS One 16:e0243185. doi: 10.1371/journal.pone.0243185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, Larsen BB, Anderson C, White S, Cassens T, et al. 2021. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184:2587–2594. doi: 10.1016/j.cell.2021.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, Meex C, Melin P, Hayette M-P, Bours V, Durkin K. 2020. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol 58:e01598-20. doi: 10.1128/JCM.01598-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. COVID-19 test makers are adapting for variants. 2021. BioWorld. Available from: https://www.bioworld.com/articles/503479-covid-19-test-makers-are-adapting-for-variants. Retrieved 25 May 2024. [Google Scholar]
- 55. De Pace V, Bruzzone B, Orsi A, Ricucci V, Domnich A, Guarona G, Randazzo N, Stefanelli F, Battolla E, Dusi PA, Lillo F, Icardi G. 2022. Comparative analysis of five multiplex RT-PCR assays in the screening of SARS-CoV-2 variants. Microorganisms 10:306. doi: 10.3390/microorganisms10020306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brito-Mutunayagam S, Maloney D, McAllister G, Dewar R, McHugh M, Templeton K. 2022. Rapid detection of SARS-CoV-2 variants using allele-specific PCR. J Virol Methods 303:114497. doi: 10.1016/j.jviromet.2022.114497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Myakishev MV, Khripin Y, Hu S, Hamer DH. 2001. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res 11:163–169. doi: 10.1101/gr.157901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oligo design tools. 2023. Available from: www.thermofisher.com/us/en/home/life-science/oligonucleotides-primers-probes-genes/custom-dna-oligos/oligo-design-tools.html. Retrieved 13 Feb 2023.
- 59. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 60. Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. 2021. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 372:n579. doi: 10.1136/bmj.n579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaintoutis SC, Chassalevris T, Balaska S, Mouchtaropoulou E, Tsiolas G, Vlatakis I, Tychala A, Koutsioulis D, Argiriou A, Skoura L, Dovas CI. 2021. A novel real-time RT-PCR-based methodology for the preliminary typing of SARS-CoV-2 variants, employing non-extendable LNA oligonucleotides and three signature mutations at the spike protein receptor-binding domain. Life (Basel) 11:1015. doi: 10.3390/life11101015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Owczarzy R, You Y, Groth CL, Tataurov AV. 2011. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry 50:9352–9367. doi: 10.1021/bi200904e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clark AE, Wang Z, Ostman E, Zheng H, Yao H, Cantarel B, Kanchwala M, Xing C, Chen L, Irwin P, Xu Y, Oliver D, Lee FM, Gagan JR, Filkins L, Muthukumar A, Park JY, Sarode R, SoRelle JA. 2022. Multiplex fragment analysis for flexible detection of all SARS-CoV-2 variants of concern. Clin Chem 68:1042–1052. doi: 10.1093/clinchem/hvac081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang L, Parvin R, Lin S, Chen M, Zheng R, Fan Q, Ye F. 2024. Peptide nucleic acid clamp-assisted photothermal multiplexed digital pcr for identifying SARS-CoV-2 variants of concern. Adv Sci (Weinh) 11:e2306088. doi: 10.1002/advs.202306088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Y, Zhao S, Xu Z, Qiao X, Li M, Li Y, Luo X. 2023. Peptide nucleic acid and antifouling peptide based biosensor for the non-fouling detection of COVID-19 nucleic acid in saliva. Biosens Bioelectron 225:115101. doi: 10.1016/j.bios.2023.115101 [DOI] [PubMed] [Google Scholar]
- 66. Iijima T, Sakai J, Kanamori D, Ando S, Nomura T, Tisi L, Kilgore PE, Percy N, Kohase H, Hayakawa S, Maesaki S, Hoshino T, Seki M. 2023. A new method to detect variants of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification combined with a bioluminescent assay in real time (RT-LAMP-BART). Int J Mol Sci 24:10698. doi: 10.3390/ijms241310698 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S10; Fig. S1.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The data generated and analyzed during this study are available from the corresponding author upon reasonable request.