Abstract
RNA molecules with internal 2′,5′-branches are intermediates in RNA splicing, and branched RNAs have recently been proposed as retrotransposition intermediates. A broadly applicable in vitro synthetic route to branched RNA that does not require self-splicing introns or spliceosomes would substantially improve our ability to study biochemical processes that involve branched RNA. We recently described 7S11, a deoxyribozyme that was identified by in vitro selection and has general RNA branch-forming ability. However, an important restriction for 7S11 is that the branch-site RNA nucleotide must be a purine (A or G), because a pyrimidine (U or C) is not tolerated. Here, we describe the compact 6CE8 deoxyribozyme (selected using a 20 nt random region) that synthesizes 2′,5′-branched RNA with any nucleotide at the branch site. The Mn2+-dependent branch-forming ligation reaction is between an internal branch-site 2′-hydroxyl nucleophile on one RNA substrate with a 5′-triphosphate on another RNA substrate. The preference for the branch-site nucleotide is U > C ≅ A > G, although all four nucleotides are tolerated with useful ligation rates. Nearly all other nucleotides elsewhere in both RNA substrates allow ligation activity, except that the sequence requirement for the RNA strand with the 5′-triphosphate is 5′-pppGA, with 5′-pppGAR (R = purine) preferred. These characteristics permit 6CE8 to prepare branched RNAs of immediate practical interest, such as the proposed branched intermediate of Ty1 retrotransposition. Because this branched RNA has two strands with identical sequence that emerge from the branch site, we developed strategies to control which of the two strands bind with the deoxyribozyme during the branch-forming reaction. The ability to synthesize the proposed branched RNA of Ty1 retrotransposition will allow us to explore this important biochemical pathway in greater detail.
INTRODUCTION
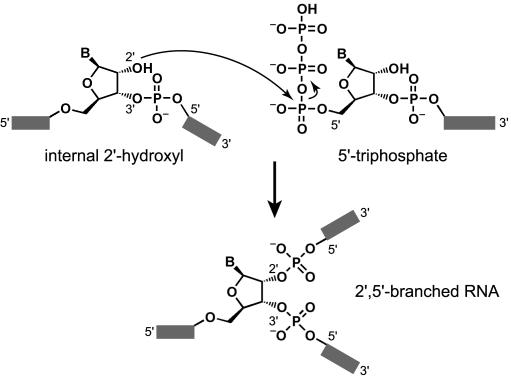
RNA molecules with 2′,5′-branched linkages (Figure 1) are well-recognized intermediates in RNA splicing by both group II introns and the spliceosomes of higher organisms (1–3). More recently, branched RNAs have been proposed during Ty1 retrotransposition, where they may facilitate the key strand-transfer step (4,5), although recent experiments suggest that this hypothesis is likely to be incorrect (6). A general approach for preparing branched RNA will allow investigation of such biochemical processes and may also enable construction of topologically novel nucleic acids (7–9) that incorporate covalently branched backbones. Others have reported efforts toward 2′,5′-branched RNA based on solid-phase synthesis (10–14), but these methods often inherently limit the products that may be formed. For example, in some solid-phase approaches, two of the strands that emerge from the branch site must have identical sequences (11,12). In addition, solid-phase RNA synthesis has an intrinsic length limit of <100 nt (15,16), which is much shorter than many natural branched RNAs of biological interest. Finally, solid-phase synthesis of branched RNA requires special nucleoside monomers; therefore, this approach is not readily accessible by many laboratories that lack either the facilities for synthetic organic chemistry or familiarity with the associated procedures.
Figure 1.
Formation of 2′,5′-branched RNA by reaction of an internal 2′-hydroxyl group with a 5′-triphosphate (B = nucleobase).
We have previously applied in vitro selection to identify deoxyribozymes (DNA enzymes) (17–20) that ligate RNA (21–32). These deoxyribozymes function in one of two ways: they mediate the reaction of a hydroxyl group nucleophile on one substrate with a 2′,3′-cyclic phosphate on the second substrate (21–26), or they catalyze the reaction of a hydroxyl group on one substrate with a 5′-triphosphate on the second substrate (27–32). The latter reaction forms 2′,5′-branched RNA when the nucleophilic hydroxyl group is at an internal 2′-position of the substrate (Figure 1). Although our previously reported branch-forming deoxyribozymes have been both interesting and useful (20), they have varying tolerance for specific RNA substrate sequences. For example, consider the 7S11 DNA enzyme, which is currently our most general branch-forming deoxyribozyme with regard to the RNA nucleotides other than the branch site (29,30). Despite its overall generality for most RNA substrate nucleotides, 7S11 requires a branch-site adenosine or—with reduced ligation rate—a branch-site guanosine. Significantly, 7S11 does not tolerate branch-site pyrimidine nucleotides, giving <5% ligation yield with these substrates (29,30). This limitation prevents the preparation of certain branched RNAs, such as the proposed Ty1 retrotransposition intermediate, which has a branch-site uridine nucleotide (4).
Subsequent to the identification of 7S11, we reported a different selection effort that initially used RNA substrate sequences derived from two natural sources, a spliceosomal substrate and a group II intron (32). In the collection of resulting deoxyribozymes, many different nucleotides of the left-hand RNA substrate were able to provide a 2′-hydroxyl group to react with a 5′-triphosphate (each deoxyribozyme catalyzed one particular ligation reaction). This suggested little inherent preference for specific branch points, at least for these RNA substrate sequences. These experiments used N40 random DNA regions, which have been used in nearly all of our previous studies. Here, we performed an analogous selection using a much smaller N20 random region; an explicit goal was the identification of very compact deoxyribozymes with useful catalytic activity. We find that one of the new deoxyribozymes, 6CE8, can use any branch-site nucleotide with both reasonable generality for the surrounding RNA sequences and useful catalytic parameters. As a demonstration of its practical utility, we show that 6CE8 is capable of synthesizing the proposed Ty1 branched intermediate, which will facilitate direct biochemical assessment of the role of branched RNA in retrotransposition.
MATERIALS AND METHODS
RNA and DNA oligonucleotides
RNA oligonucleotides were prepared either by solid-phase synthesis (Dharmacon, Inc., Lafayette, CO or HHMI–Keck Laboratory, Yale University, New Haven, CT) or by in vitro transcription using T7 RNA polymerase with an appropriate double-stranded DNA template (33,34). DNA oligonucleotides were prepared at IDT (Coralville, IA). The sequence of the 6CE8 deoxyribozyme is 5′-CCGCGCTAGAACATGGCACTCAGAGCGCACGGCGAGTACATGAGACTTCC-3′, where the underlined regions are the DNA binding arms (complementary to the original RNA substrate sequences as shown in Figures 5 and 6) and the intervening non-boldface sequence is the enzyme region. The boldface AG dinucleotide corresponds to the 2 nt shown explicitly in the DNA strand in the summary image of Figure 7 (for any particular RNA substrate sequence, the second of these 2 nt was chosen following the guidelines depicted in that figure). The DNA binding arms were chosen to maintain complementarity to the RNA substrates for any specific experiment. All RNA and DNA oligonucleotides were purified by denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid, 2 mM EDTA, pH 8.3) as described previously (21,28).
Ty1 RNA sequences
The putative Ty1 branch was proposed previously (4). We derived the complete Ty1 sequences (shown partially in Figure 8 and fully in the Supplementary Material) from the information provided previously (35). See the Supplementary Material for sequences pertinent to the hairpin and disruptor ligation approaches of Figure 9.
In vitro selection procedure
The in vitro selection effort denoted CE used the same procedure as described for the previously reported CA selection, except that an N20 random region was used in place of the N40 random region (32). During selection, the left-hand (L) RNA substrate sequence was 5′-GGAAGUCUCAUGUACUAUCG-3′, and the right-hand (R) RNA substrate sequence was 5′-GAAUGUUCUAGCGCGGA-3′. The key selection step in which the two RNA substrates were joined used incubation conditions of 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl and 20 mM MnCl2 at 37°C for 1 h.
Kinetic assays
The general assay methods have been reported in detail (21,24,27,28). Here, a 6 μl sample containing 5′-32P-radiolabeled left-hand RNA substrate (L), deoxyribozyme (E) and right-hand RNA substrate (R) in 5 mM HEPES, pH 7.5, 15 mM NaCl and 0.1 mM EDTA was annealed by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 10 μl with total concentrations of 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl and 20 mM MnCl2. In the final sample, the ratio L:E:R was equal to 1:3:6 with the concentration of E ≈ 0.15–0.50 μM. When a variable amount of disruptor oligonucleotide (D) was included for the strategy of Figure 9B, the ratio L:E:R:D was 1:3:6:(1 or 3 or 6), as described in Supplementary Material. After the addition of Mn2+ as the last component, the sample was incubated at 37°C. Aliquots were withdrawn at desired timepoints and quenched onto stop solution (80% formamide, 1× TB, 50 mM EDTA, 0.025% each bromophenol blue and xylene cyanol), followed by PAGE and exposure to a PhosphorImager screen. Values of kobs and final yield were obtained by fitting the yield versus time data directly to first-order kinetics, i.e. yield = Y·(1 − e−kt), where k = kobs and Y = final yield.
Partial alkaline hydrolysis assays
The assays of Figure 4B used branched RNA that was prepared using the 6CE8 deoxyribozyme with a 5′-32P-radiolabeled L substrate. Each sample comprised 10 pmol of L (of which 1 pmol was 5′-32P-radiolabeled, and the remainder was cold-phosphorylated), 30 pmol of E and 60 pmol of R in a 20 μl reaction volume. The branched RNA product was purified by 20% PAGE. A portion of this sample (∼100 fmol) was then incubated in 10 μl of 50 mM NaHCO3, pH 9.2 for 10 min and quenched with 10 μl of stop solution; 4 μl was taken for PAGE. To provide a standard alkaline hydrolysis ladder, the same assay was performed on the L substrate. The ladder was calibrated by RNase T1 digestion of the L substrate, using ∼100 fmol of L and 1 U of RNase T1 (Ambion) for 5 min at room temperature in the manufacturer's buffer.
Debranching enzyme assays
The branched RNA product (∼15–50 fmol, prepared as described above) was incubated with 75 ng yeast debranching enzyme Dbr that was previously purified by glycerol gradient gel electrophoresis. The estimated mole ratio of Dbr to RNA was 30–100:1. The Dbr was a gift from Scott W. Stevens (University of Texas at Austin). The incubation with Dbr was in 10 μl containing 20 mM HEPES, pH 7.5, 0.5 mM MgCl2, 125 mM KCl and 1 mM DTT at 30°C for up to 30 min. In all the cases, the cleavage kobs increased by <2-fold when the Dbr:RNA ratio was increased from 30:1 to 100:1.
Hairpin and disruptor approaches to synthesize the proposed Ty1 branched RNA
The two reaction steps of Figure 9C were performed as follows. For the 6CE8 ligation step, branched RNA was prepared using the 6CE8 deoxyribozyme with a 5′-32P-radiolabeled L substrate, which is 335 or 338 nt in length for the hairpin and disruptor approaches, respectively. Each sample comprised 20 pmol of L (of which 2 pmol was 5′-32P-radiolabeled, and the remainder was cold-phosphorylated), 60 pmol of E and 120 pmol of R in a 40 μl reaction volume. For the disruptor approach only, RQ1 DNase (4 μl of 1 U/μl; Promega) was added and the sample was incubated at 37°C for 10 min to digest the disruptor oligonucleotide, which induced smearing of all bands on PAGE if not removed (data not shown). The branched RNA product (537 or 540 nt in length) was purified by 6% PAGE. For the subsequent 10–23 cleavage step, the purified branched product was cleaved with an appropriate 10–23 deoxyribozyme (see sequences in Supplementary Material) in a 10 μl incubation with 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl and 5 mM MnCl2 at 37°C. The 10–23 cleavage reaction using 60 mM MgCl2 instead of Mn2+ was at least 5-fold slower, and the final cleavage yield was also slightly lower (∼65 versus >90%).
RESULTS
In vitro selection of compact RNA ligase deoxyribozymes
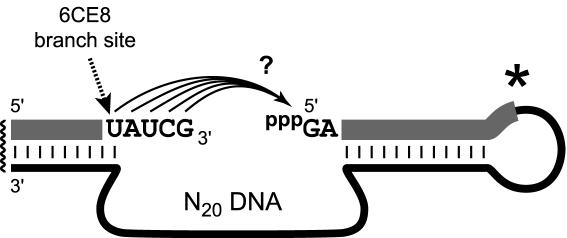
In a previous report, we described two selections for branch-forming deoxyribozymes that used N40 random DNA regions (32). Here, we performed an analogous selection starting with a smaller N20 random region, using a selection strategy whose key step is depicted in Figure 2. Consistent with our laboratory's ongoing selection nomenclature, this new effort was designated ‘CE’. The CE selection is directly analogous to the CA selection of our previous report (32), in which the identities of the RNA substrates were derived from the sequence of the ai5γ group II intron (1,36). Reducing the length of the random region to 20 nt means that all of potential DNA enzyme ‘sequence space’ is covered here using a 200 pmol initial pool (200 pmol × 1014 molecules compared with 420 = 1012 possible sequences; therefore ∼100× coverage of sequence space). This contrasts with only ∼10−10 × coverage of sequence space with the previously used N40 pool. The key selection step of the procedure (Figure 2) was performed with incubation conditions of 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl and 20 mM MnCl2 at 37°C for 1 h. The selection procedure was iterated for multiple rounds until the ligation activity (i.e. fraction of pool reacting in a particular selection round) leveled off at ∼30% at round 6.
Figure 2.
The key step of in vitro selection, in which the deoxyribozyme (lower strand) catalyzes the RNA ligation reaction of Figure 1. The covalent loop marked with an asterisk is present during the selection procedure, but the loop is not required for practical ligation of two RNA substrates. The internal nucleotide marked with an arrow is the branch site for the 6CE8 deoxyribozyme, which is the focus of this report.
A preliminary analysis of the RNA products from the uncloned 6CE pool indicated a mixture of branched and linear linkages (data not shown). This is similar to our observation for the N40-containing CA selection before cloning. Individual 6CE deoxyribozymes were cloned and surveyed, both to determine their ligation activities (i.e. rates and yields) and to pinpoint the sites of the newly formed linkages. One particular deoxyribozyme used the internal uridine 2′-hydroxyl indicated in Figure 2 as the nucleophile to attack the 5′-triphosphate (see data below). This deoxyribozyme was named 6CE8, reflecting its origin in the 6th round of the CE selection, clone number 8. Because 6CE8 uses a branch-site pyrimidine, which is an activity that is generally lacking by our other deoxyribozymes such as 7S11 (29,30), 6CE8 was pursued in greater detail as the primary focus of this report. The mfold-predicted secondary structure (37,38) of this deoxyribozyme features a stem–loop region (Figure 3A), but we have not probed this structure experimentally.
Figure 3.
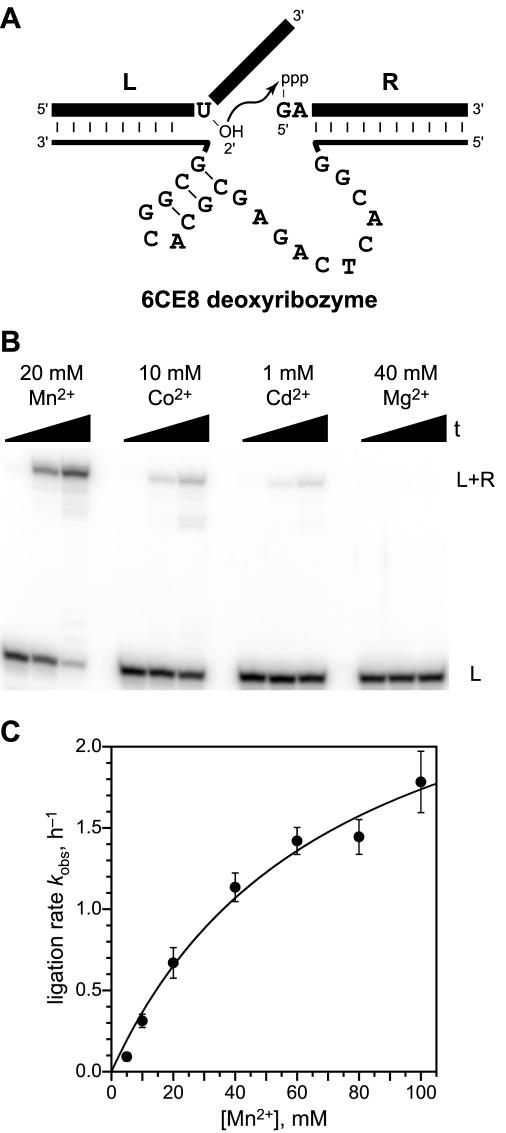
The 6CE8 deoxyribozyme and characterization of its ligation activity. (A) Mfold-predicted (37,38) secondary structure of 6CE8 (thin bottom strand, with the 20 nt enzyme region shown explicitly) bound intermolecularly to its two RNA substrates, L and R (thick top strands). The branch-site nucleotide is shown as U but may be any nucleotide; see Figure 7 for a more complete depiction of the substrate sequence requirements. (B) Ligation of RNA substrates in 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM KCl, and the indicated divalent metal ion at 37°C (see Figure 4 for kinetic plots using Mn2+). L and R denote the left-hand and right-hand RNA substrates, respectively. Timepoints were taken at 0, 1 and 6 h for Mn2+ and 0, 6 and 24 h for the other ions. Each of Co2+ and Cd2+ were tested at 100 μM, 1 mM and 10 mM in a similar fashion; only the concentrations shown here led to detectable ligation activity (≥0.1%; data not shown). Other ions that were examined but showed no detectable ligation activity at any tested concentration (100 μM–10 mM) were Ca2+, Ni2+, Zn2+ and Cu2+. (C) Determination of the Kd,app for Mn2+. The fit value of Kd,app is 71 ± 3 mM.
Characterization of 6CE8 RNA ligation activity: rate, yield and metal dependence
The overall ligation activity of 6CE8 was characterized in the trimolecular RNA:RNA:DNA format of Figure 3A. Under the standard incubation conditions (20 mM Mn2+, pH 7.5, 37°C) and using the same RNA substrate sequences that were used during selection, kobs = 0.67 ± 0.09 h−1 (n = 22) with ∼90% ligation yield (Figure 3B, Mn2+ lanes). Higher pH cannot be used owing to oxidation of Mn2+, and the kobs was substantially lower at pH 7.0 or pH 6.5 (data not shown). Buffer compounds other than HEPES led to no improvement in rate or yield (e.g. Tris; data not shown). Several other metal ions were tested in place of Mn2+ (Figure 3B). Aside from Cd2+ and Co2+, which showed only small amounts of ligation upon extended incubation (8–11% in 24 h), no activity was detected with other ions, including Mg2+. The Kd,app for Mn2+ is ∼70 mM (Figure 3C). For all assays described below, the original 20 mM Mn2+ incubation conditions were employed.
Tolerance of 6CE8 for different branch-site nucleotides
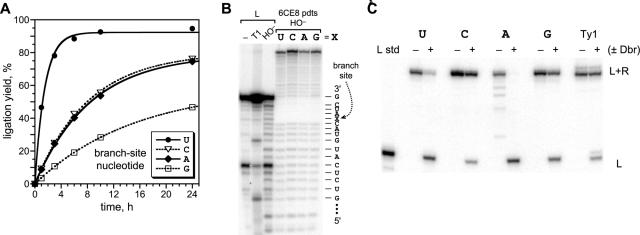
The initial characterization experiments demonstrated that 6CE8 has useful rate and yield for practical RNA ligation. The most important remaining question with regard to the practical application of 6CE8 is its generality for various RNA substrate sequences. This generality was determined experimentally, focusing first on the key branch-site RNA nucleotide position. The tolerance of 6CE8 for different nucleotides at the branch site was assessed with RNA substrates for which the branch-site nucleotide was changed, but all others were kept identical to those used during selection. Unlike previous branch-forming deoxyribozymes such as 7S11, substantial ligation activity was observed for 6CE8 with any of the four nucleotides at the branch site (Figure 4A). For any branch-site nucleotide other than U, the kobs was lower by 5–10-fold, but substantial ligation activity was still observed. The overall kobs trend of U > C ≅ A > G contrasts sharply with the analogous trend for 7S11, which is A ≫ G with C and U essentially inactive (29).
Figure 4.
Successfully changing the branch-site RNA nucleotide while retaining 6CE8 ligation activity. (A) Kinetic data for RNA ligation by 6CE8 for RNA substrates with one of the four possible branch-site nucleotides. The DNA nucleotide opposite the branch site was maintained as A in all cases (see text). Incubation conditions were the same as for Figure 3A (20 mM Mn2+). krel values relative to branch-site U were branch-site C 0.19, A 0.18 and G 0.10. (B) Assessment of the branch-site location in the 6CE8 products using partial alkaline hydrolysis (HO−). L is the left-hand RNA substrate, for which the sequence is marked along the side of the gel image; T1 is RNase T1 digestion (G-specific) for ladder calibration. The spurious cleavage band seen here at a particular C nucleotide for all samples, including the no treatment (−) lane, was not reproducibly observed (see Supplementary Material). (C) Cleavage of the branched RNA products using debranching enzyme (Dbr), where + denotes 30 min incubation. The first four sets of lanes are the same 6CE8 products from (B), with the branch-site nucleotides shown above the gel. The fifth set of lanes is the core of the proposed Ty1 branched RNA, which has a branch-site uridine (see Figure 8A). For each set of lanes, the debranched product migrates at the position of the appropriate L substrate standard, which is shown on the far left for the first four assays (the L standard is not shown for the Ty1 product). From timepoints taken between 2 and 30 min, the kobs and cleavage yield values were as follows: U, 0.18 min−1 and 82%; C, 0.19 min−1 and 33%; A, >5 min−1 and 95%; G, 0.30 min−1 and 50%; Ty1, 0.16 min−1 and 25%.
In these 6CE8 assays, the DNA nucleotide opposite the branch-site RNA nucleotide was maintained as A, regardless of the branch-site RNA nucleotide identity. When this DNA nucleotide was instead changed to G and the assays with all four branch-site RNA nucleotides were repeated, the ligation activity was nearly abolished for all four RNA substrates (<1% yield; data not shown). Therefore, although the initial arrangement of RNA and DNA (Figure 2) suggests that these two nucleotides might form an RNA:DNA base pair, the experimental data indicate otherwise, because an A is required in the DNA regardless of the RNA nucleotide's identity.
For each of the 6CE8 ligation products, the precise branch-site location was verified using partial alkaline hydrolysis of the purified branched RNA product. This experiment leads to a ladder with a gap for 2′,5′-branched RNA, where the gap location indicates the site of branching (27,39). Regardless of the substrate sequence, the branch formed by 6CE8 is found at the RNA nucleotide marked in Figure 2 (Figure 4B). We further verified the nature of the new branched linkage using yeast debranching enzyme Dbr, which specifically cleaves 2′,5′-branched linkages (40,41). The 6CE8 branched product with a branch-site A was cleaved rapidly and completely by Dbr, whereas the products that incorporate any of the other three RNA branch-site nucleotides were cleaved less efficiently (Figure 4C). This sequence dependence is roughly consistent with the known RNA substrate preferences of Dbr (40), although these preferences have not been delineated systematically.
Tolerance of 6CE8 for nucleotide changes in the left-hand (L) binding arm
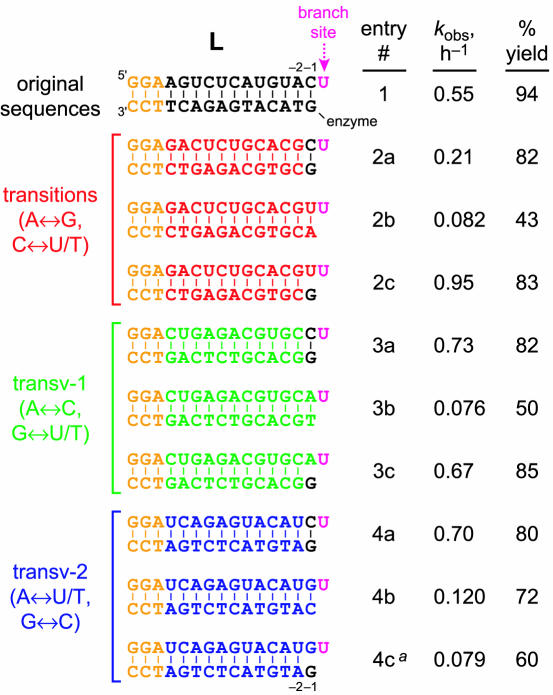
The tolerance of 6CE8 for any branch-site nucleotide (Figure 4) is promising for its generality. However, truly broad applicability would require that this deoxyribozyme also tolerates a wide range of RNA substrate sequences away from the branch-site nucleotide. The effect of changing the nucleotide sequences in the binding arms was assessed comprehensively, beginning with the left-hand (L) RNA substrate in the ‘binding arm’ region where the DNA binds via Watson–Crick base pairs (Figure 5). For convenience and also for consistency with the natural RNA splicing convention (39), the branch-site nucleotide position of the L substrate is designated as position +1. Nucleotides to its 3′-side are designated +2 and higher, whereas nucleotides to its 5′-side are designated −1, −2 and so on; the first nucleotide to the 5′-side of the branch is position −1. As we have done elsewhere (30), systematic changes to the RNA nucleotides relative to the original sequence (as used during selection) are denoted as transitions (A↔G and C↔U); as ‘transversions of type 1’ (transv-1; A↔C and G↔U); or as ‘transversions of type 2’ (transv-2; A↔U and G↔C). Analogous nomenclature for positions and mutations applies for changes to the DNA nucleotides.
Figure 5.
Effects on 6CE8 ligation activity of changes to the L substrate nucleotides for RNA positions in the DNA-binding region. Incubation conditions were the same as for Figure 3A (20 mM Mn2+). The RNA substrate sequence is shown on top (5′–3′), and the complementary DNA is shown below the RNA. The leading 5′-GGA sequence (orange) was kept constant to permit transcription of each substrate. The nucleotide opposite the branch-site uridine (pink) was A in all the cases and is omitted from these images because the data suggest that there is not a Watson–Crick base pair at this position (see text). In addition, the AUCG nucleotides of the L substrate to the 3′-side of the branch-site U are omitted for clarity. The yield is from an exponential curve fit to the yield versus time data unless kobs < 0.15 h−1, in which case the yield is from the 24 h data point. See Supplementary Material for plots of the datasets. aThe same experiment except with A instead of G in the DNA at position −1 gave much lower rate and yield (data not shown).
As a first test for varying L, all RNA nucleotides of the L substrate up to and including position −2 were systematically changed by any of the transitions, transv-1 or transv-2, with corresponding DNA changes to maintain Watson–Crick base pairing. In all the cases, the effects on 6CE8 ligation activity were modest or negligible (Figure 5, compare entries 2a, 3a and 4a with entry 1). This indicates substantial generality for the L sequence. However, analogous experiments with the RNA changes extending 1 nt further to include position −1 led uniformly to significant decreases in activity, with negative effects on both rate and yield (entries 2b–4b versus entry 1). To determine the extent to which the RNA nucleotide at position −1 could be changed, additional experiments were performed with the RNA changes again extending to position −1, but now with the DNA nucleotide at position −1 maintained as G. Any of C, U or A at RNA position −1 led to successful ligation, but in contrast, G at RNA position −1 led to a low kobs (entries 2c–4c versus entry 1).
In summary of the Figure 5 data, all RNA positions of the L substrate up to and including position −2 can be any nucleotide while permitting high 6CE8 ligation rate and yield. To achieve this, the DNA nucleotides of the binding arm up to and including position −2 must be changed to maintain straightforward Watson–Crick complementarity with the RNA. The RNA nucleotide at position −1 can be any nucleotide except G, with the DNA nucleotide at position −1 maintained as G (regardless of the RNA nucleotide identity) for best rate and yield. However, if the RNA nucleotide at position −1 is G, then the best choice for the opposing DNA nucleotide is C. In this case, although the ligation rate is substantially lowered, useful ligation yield is still observed at long incubation times.
Tolerance of 6CE8 for changes in the L overhanging (unpaired) RNA nucleotides
We assessed the effects of changes to the unpaired nucleotides in the 3′-tail of the L RNA substrate. These nucleotides are not base paired with DNA according to the selection design (Figure 2), and we have observed specific sequence requirements for analogous overhanging nucleotides in other branch-forming DNA enzymes (28). When the overhanging RNA nucleotide at position +2 (i.e. one position to the 3′-side of the branch-site uridine) was changed from A in the parent sequence to each of the other three nucleotides, the ligation activity decreased in the order A ≈ U > G > C. This reflects a modest sequence requirement at position +2 (data not shown; C still permitted 44% ligation yield in 23 h, with krel = 0.16 compared with A at position +2). The maintenance of the branch site at position +1 for the RNA substrate with U at +2 was verified using partial alkaline hydrolysis, analogous to Figure 4B (see Supplementary Material). Therefore, the potentially ambiguous uridines at adjacent positions +1 and +2 (as well as +3) do not induce branching at the ‘wrong’ nucleotide.
For overhanging nucleotides further removed from the branch site (i.e. positions +3 and beyond), the substrate sequences …UAUCG-3′ (parent sequence, where U is the branch site) and …UAACA-3′ [available from a previous selection effort (32)] gave nearly identical ligation activities (data not shown). Although we have not systematically examined changes at positions +3 and beyond, the available data do not indicate any strict requirements, and our subsequent experiments with the considerably different Ty1 substrate sequences confirmed this generality (see below).
We also examined changes to the length of the L substrate 3′-end. When the 3′-end of the L substrate was extended by 16 nt, kobs increased by ∼3-fold (data not shown, and see long Ty1 substrates below). Thus, 6CE8 appears to benefit from a long, unpaired 3′-tail on the L substrate. When this tail was instead shortened from a 4 nt overhang (UAUCG) by 2 nt to UAU, no decrease in activity was observed (see Supplementary Material). Shortening the tail further to UA led to a modest (<2-fold) drop in rate with maintenance of high yield. Therefore, even a 1 nt branch may be synthesized by 6CE8. However, when the tail was removed entirely (i.e. the ‘branch-site’ U was the 3′-terminal nucleotide of L, leading in this case to linear RNA), kobs decreased ∼12-fold, and the maximal ligation yield was only ∼30%. It is likely that the resulting linear RNA has a 2′–5′ linkage, as was the case for the branch-forming 9F7 deoxyribozyme with a similar tail-less L substrate (28), although this has not been tested explicitly for 6CE8.
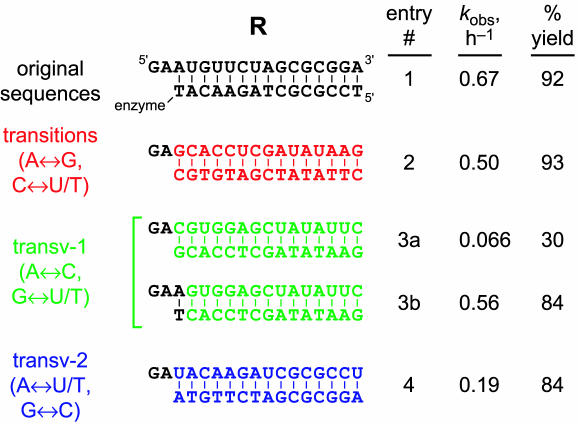
Tolerance of 6CE8 for nucleotide changes in the right-hand (R) substrate
The right-hand (R) RNA substrate has just two unpaired nucleotides in the original selection design (Figure 2). Because the 5′-terminus of R bears a triphosphate, only the 5′-adenosine analog is readily prepared by T7 RNA polymerase transcription (42,43). When the R substrate was 5′-pppA instead of 5′-pppG with no other nucleotide changes, no ligation activity was observed at all (data not shown). Similarly, when the second nucleotide was changed from A to any of the other 3 nt, no activity was observed (data not shown). Thus, the strict sequence requirement 5′-pppGA exists at these 2 nt positions.
In contrast to these observations, changes were tolerated further along the R substrate. All nucleotides starting with the third were changed systematically by either transitions, transv-1 or transv-2 as described above for the L substrate. Although transitions in R were tolerated with little effect, transv-1 and transv-2 substitutions lowered kobs by 10- and 4-fold, respectively (Figure 6, compare entries 1, 2, 3a and 4). The origin of this effect was traced solely to the third nucleotide, because an R substrate beginning 5′-pppGAA… but continuing with the transv-1 changes was ligated with rate and yield almost identical to the parent sequence (entry 3b). Thus, we conclude that the R substrate has a strict requirement for sequence 5′-pppGA and also a preference for 5′-pppGAR (R = purine), with no other requirements. These findings are supported by the Ty1 sequence experiments reported below, in which the substrates have a large number of changes relative to the parent sequences. A summary of all of the sequence requirements for 6CE8 is provided in Figure 7.
Figure 6.
Effects on 6CE8 ligation activity of changes to the R substrate nucleotides for RNA positions in the DNA-binding region. Incubation conditions were the same as for Figure 3A (20 mM Mn2+). The RNA substrate sequence is shown on top (5′–3′), and the complementary DNA is shown below the RNA. The yield is from an exponential curve fit to the yield versus time data, except for entry 3a, for which the yield is from the 20 h data point. When the DNA sequence is changed to remove the Watson–Crick base pairing at the third RNA nucleotide, ligation activity is reduced or abolished (data not shown).
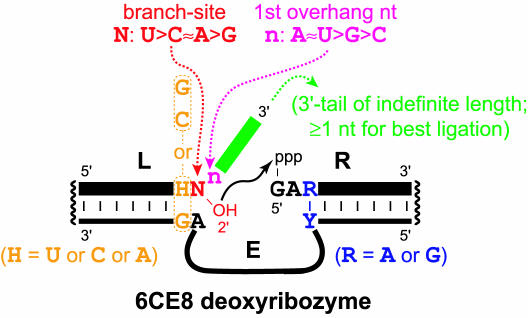
Figure 7.
Summary of sequence requirements for the 6CE8 deoxyribozyme. The preference for a purine (R, blue) at the third position of the right-hand substrate is not absolute (Figure 6); U is tolerated reasonably well, and C supports diminished ligation activity. The 6CE8 sequence in the enzyme region between the indicated Y (i.e. T or C to pair with the purine in the RNA substrate) and the indicated AG is 5′-GGCACTCAGAGCGCACGGCG-3′, as shown in Figure 3A. The parallel vertical lines denote regions of Watson–Crick complementarity between the RNA substrates (thick bars) and DNA binding arms (thin bars).
Synthesis of the proposed Ty1 branched RNA by the 6CE8 deoxyribozyme
As a practical branch-forming application, we sought to use 6CE8 for ligation of RNA substrate sequences that are largely unrelated to those used during selection. For this purpose, we used 6CE8 to synthesize the proposed Ty1 branched RNA intermediate, which has a branch-site uridine (Figure 8A) (4,5) and thus cannot be prepared with the 7S11 deoxyribozyme. We first established that the short Ty1 RNA sequences that interact directly with the DNA binding arms of 6CE8 are indeed tolerated by 6CE8 (Figure 8B, first set of lanes). This experiment used short RNA substrate arms to bind with the DNA and also a short 3′-unpaired tail on the L substrate. The kobs was 3.4 h−1, which is ∼5-fold higher than observed with the original selection substrates, and the ligation yield was >75%. Partial alkaline hydrolysis demonstrated that the correct branch-site uridine was used (see Supplementary Material), and Dbr-catalyzed cleavage of the branched product proceeded rather inefficiently, as expected for U-branched RNA (Figure 4C).
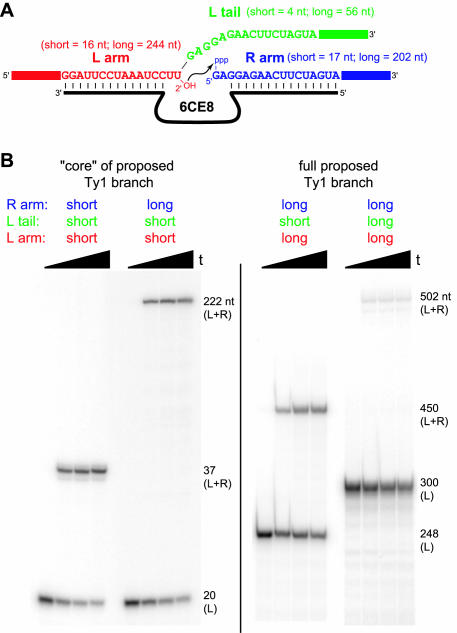
Figure 8.
Synthesis of the proposed Ty1 branched RNA by the 6CE8 deoxyribozyme. (A) Diagram and partial sequences of the proposed Ty1 branched RNA. In the specific set of long RNAs examined in this study, the L substrate is 300 nt (243 nt to the 5′-side of the branch site and 56 nt to the 3′-side of the branch site), and the R substrate is 202 nt. The minimal ‘core’ of this branched RNA consists solely of those nucleotides that bind directly to the 6CE8 DNA binding arms (shown explicitly here), plus 4 nt of the L tail. See Supplementary Material for complete RNA sequences and their correspondence to the various regions of the natural Ty1 RNA (U3, R, U5 and PBS). (B) Ligation by 6CE8 to form the minimal Ty1 core using various combinations of short and long L arm, L tail and R arm. For the L arm and R arm, ‘short’ refers to just the sequences shown explicitly in (A), and ‘long’ refers to the full sequences. For the L tail, ‘short’ refers to the 4 nt GAGG tail, and ‘long’ refers to 56 nt of sequence identical to the R arm. For all experiments, the L substrate was 5′-32P-radiolabeled; timepoints were taken at t = 0, 10, 20 and 30 min. The first two sets of lanes are 20% PAGE and the last two sets of lanes are 6% PAGE. Values of kobs and yield at 30 min timepoint: first set of lanes, 3.4 h−1 and 77%; second set of lanes, 3.2 h−1 and 61%; third set of lanes, 2.2 h−1 and 29% (50% at 90 min, data not shown); fourth set of lanes, ∼0.05 h−1 and 3%. For the first three sets of lanes, analogous experiments except with the short L tail sequence of AUCG instead of GAGG gave essentially equivalent results (data not shown).
Synthesis of the entire proposed Ty1 branched RNA is much more challenging because both the R arm and the L tail inherently have identical sequences for the 56 nt closest to the ligation site (Figure 8A, note the identical blue and green sequences) (4,5). This sequence identity complicates proper annealing of the RNA to the deoxyribozyme. More precisely, successful branch formation by 6CE8 requires that the L tail does not compete with the R arm for annealing to the DNA binding arm. However, when the 5′-end of L is properly annealed to its own DNA binding arm, the L tail can readily anneal to the other DNA binding arm in an intramolecular sense, thereby preventing the R arm from interacting properly. Indeed, when both R and the full L substrate (containing a long 3′-tail with the same sequence as R) were incubated with 6CE8, little ligation product was formed (Figure 8B, last set of lanes). Because each of the other arm and substrate combinations in Figure 8 were successfully ligated, the lack of activity in the fourth set of lanes is specifically owing to competition for the DNA between the R arm and L tail RNA sequences.
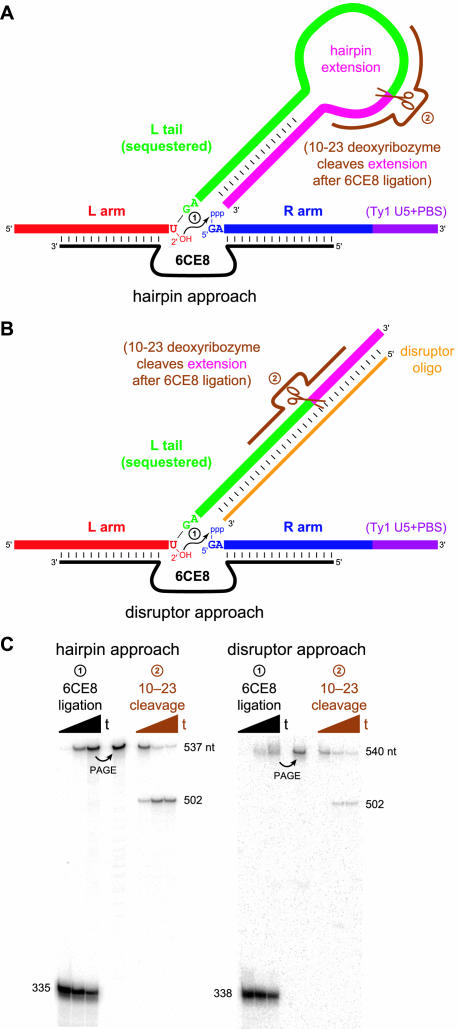
We sought to surmount this binding problem in two ways. In the ‘hairpin’ approach (Figure 9A), the L tail was sequestered in an intramolecular hairpin secondary structure by extending the L substrate sequence. The large hairpin loop included a 10–23 deoxyribozyme cleavage site, such that 6CE8-catalyzed branch formation can be followed by 10–23 cleavage of the unwanted 3′-portion of the hairpin. Alternatively, in the ‘disruptor’ approach (Figure 9B), a DNA disruptor oligonucleotide with substantial complementarity to the L tail was added before initiating ligation. Again, 6CE8 ligation is followed by 10–23 cleavage of the extension sequence.
Figure 9.
The hairpin and disruptor approaches for synthesizing the full proposed Ty1 branched RNA. (A) Schematic diagram of the hairpin approach. The L tail (green) is extended with appropriate RNA sequence (pink) such that the tail is sequestered in a hairpin, thereby allowing the R arm (blue, with the same sequence as the L tail) to bind to the DNA (black). Following 6CE8-catalyzed branch formation, the extension sequence is cleaved by a 10–23 deoxyribozyme (brown). (B) Schematic diagram of the disruptor approach. The L tail is extended with arbitrary sequence (pink) that is different from the 3′-end of the R arm (purple). A disruptor DNA oligonucleotide (orange) then sequesters the L tail, and following 6CE8 ligation, the extension sequence is cleaved by a 10–23 deoxyribozyme (brown). See Supplementary Material for complete sequence information for both approaches, including several variants of the disruptor oligonucleotide; the optimal disruptor is not entirely complementary to the L tail region. (C) Experimental data that demonstrate successful application of both the hairpin and disruptor approaches. Sizes in nucleotides are shown along the side of each gel; see Supplementary Material for detailed sequence information. Timepoints were taken at t = 0, 5 and 30 min for 6CE8 ligation and 0, 10 and 30 min for 10–23 cleavage. For ligation using the hairpin approach, kobs = 2.6 h−1 with 44% yield at 30 min. For ligation using the disruptor approach, kobs = 2.3 h−1 with 25% yield at 30 min. The 10–23 cleavage yields at 30 min were 86% (hairpin approach) and 57% (disruptor approach).
Both the hairpin and disruptor approaches successfully allowed preparation of the full proposed Ty1 branched RNA (Figure 9C). The long substrates were particularly susceptible to non-specific degradation; therefore, the incubation times were carefully optimized. The hairpin approach led to ∼41% yield for ligation to form branched RNA. In a second step after ligation, the hairpin extension was readily cleaved with the 10–23 deoxyribozyme. Separately, the disruptor approach was optimized by tuning the strength of the disruptor:L tail interaction via judicious inclusion of mismatches and wobble pairs (see Supplementary Material for full details). In summary of these optimization experiments, the best disruptor was fully complementary to the L tail both near the ligation site and for the entire length of the extension sequence that is different from the R arm, whereas a substantial number of mismatches and wobble pairs were optimal in the intervening region. The best disruptor allowed ∼25% ligation yield. Subsequent to ligation, the L extension sequence was successfully removed with the 10–23 deoxyribozyme, as was done for the hairpin approach.
DISCUSSION
Deoxyribozyme-catalyzed formation of branched RNA with any branch-site nucleotide
Using in vitro selection, we previously established the ability of deoxyribozymes to synthesize branched RNA (27–30). However, on the basis of our earlier reports, certain branched RNA sequences could not be created in useful yield by any of our deoxyribozymes. Here, we sought to expand the utility of branch-forming deoxyribozymes in the context of a particular goal: synthesis of the branched RNA that has been proposed as a Ty1 retrotransposition intermediate (4,5). The new Mn2+-dependent 6CE8 deoxyribozyme (Figure 3) has useful kinetic parameters and can synthesize branched RNA with any branch-site nucleotide including a pyrimidine (Figure 4), which matches the branch-site uridine of the proposed Ty1 intermediate. In addition, 6CE8 tolerates a wide range of nucleotides further from the branch site on both RNA substrates (Figures 5 and 6). Only the right-hand RNA substrate has a strict sequence requirement, and this extends over only 2 nt (Figure 6, 5′-pppGA). Because of its wide substrate tolerance as summarized in Figure 7, 6CE8 can indeed synthesize the proposed Ty1 branched RNA (Figures 8 and 9), thereby achieving the goal set forth at the outset of this study.
Despite its utility, the 6CE8 deoxyribozyme is not a perfect reagent. For example, Mn2+ is not an optimal cofactor for a preparative deoxyribozyme owing to concerns about non-specific metal-dependent RNA degradation. In practice, we suppress non-specific degradation to the greatest possible extent by controlling the pH and incubation time. We were pleased to find that the proposed Ty1 branch was prepared with substantially higher kobs than the branch from the selection substrates themselves, which helps to avoid degradation of the inherently long substrates. Other changes relative to the selection substrates also increase kobs, such as changing the nucleotide immediately before the branch-site U (e.g. UU instead of CU, Figure 5) or extending the L substrate 3′-tail. In future applications, 6CE8 should be tested on a small scale with any specific combination of RNA substrate sequences to determine if sufficient activity exists for preparing the desired branched RNA in the necessary quantity.
For branches that correspond to many group II intron and spliceosomal sequences, the 5′-pppGA sequence requirement of 6CE8 may be problematic, because a typical 5′-splice site RNA sequence instead begins with 5′-GU. Although 6CE8 itself cannot be used in such cases, appropriate deoxyribozymes could likely be obtained by new selection efforts or by re-directing the outcome of the original CE selection (e.g. by changing the relevant RNA substrate sequences during the procedure). Nevertheless, despite its limitations, 6CE8 is directly useful to prepare the putative Ty1 branch, and 6CE8 is also able to synthesize certain well-studied group II intron RNAs, such as ai5γ (1,36). Indeed, we are actively pursuing experiments with synthetic branched RNA for both of these systems. The 502 nt branched RNA whose synthesis is shown in Figures 8 and 9 is large enough to encompass all key elements of the proposed Ty1 branch (44); therefore, relevant biochemical tests may immediately be performed. In particular, we intend to test the synthetic branched RNA as a template for cDNA synthesis. This is similar to previous assays that successfully used separate linear 5′ and 3′ Ty1 RNAs for integration into a nucleoprotein complex and subsequent reverse transcription assays (44).
Overcoming special challenges in synthesizing the large proposed Ty1 branch
During the synthesis of the proposed Ty1 branch, we addressed the demanding question of how to control which of two nearly identical substrate strands anneals to the deoxyribozyme (Figure 8A). The data in Figure 8B indicate that when both the L tail and R arm of the substrates have identical sequences, the L tail competitively anneals to the DNA binding arm, thereby preventing proper annealing of the R arm. Because certain targets, such as the proposed Ty1 branch, intrinsically have this type of structural self-similarity, an approach to thwart this annealing problem was required.
The hairpin and disruptor approaches shown in Figure 9A and B each were successful for this purpose. The hairpin approach sequesters the L tail, allowing the R arm to bind to the deoxyribozyme. We presume that this succeeds because an intramolecular hairpin within the L tail is favored over the alternative structure in which the L tail extension binds to the R arm sequence, leaving the L tail itself to bind to the DNA. The disruptor approach also sequesters the L tail, but in this case the L tail and R arm are distinguished by appending onto the L tail arbitrary nucleotides that differ in sequence from those naturally present at the end of the R arm. Therefore, the disruptor oligonucleotide preferentially targets the L tail plus its extension, after optimizing the precise disruptor composition (see Supplementary Material). For both the hairpin and disruptor approaches, substantial (albeit incomplete) branch formation is observed. Furthermore, the extension sequence is readily removed with a 10–23 deoxyribozyme (Figure 9C).
Mechanistic implications of 6CE8 branch-site tolerance and small size
The observation that 6CE8 tolerates any branch-site RNA nucleotide implies that this deoxyribozyme does not make critical contacts with the branch-site nucleobase. Other than this straightforward inference, it is difficult to draw specific mechanistic conclusions. The 6CE8 deoxyribozyme is one of the smallest RNA ligase deoxyribozyme that we have reported to date. Its 20 nt enzyme region approaches the size of the RNA-cleaving deoxyribozymes such as 10–23, 8–17 and related enzymes (45–47) that are now standard tools for the in vitro manipulation of RNA (48). Because we have not yet attempted to minimize the structure or sequence of 6CE8, we do not know whether all 20 nt of the enzyme region are required for its activity. The identification of such a small yet highly functional deoxyribozyme provides further encouragement that deoxyribozymes will continue to emerge as useful reagents for bioorganic and biochemical synthesis (20).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Scott W. Stevens (University of Texas at Austin) for the generous gift of purified yeast debranching enzyme Dbr. The authors also thank members of the Silverman laboratory for discussions. This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the March of Dimes Birth Defects Foundation (Research Grant No. 5-FY02-271), the National Institutes of Health (GM-65966), the American Chemical Society Petroleum Research Fund (38803-G4) and the UIUC Department of Chemistry (all to S.K.S.). S.K.S. is the recipient of a fellowship from The David and Lucile Packard Foundation. E.D.P. was supported by an NIH Cell & Molecular Biology Training Grant. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Peebles C.L., Perlman P.S., Mecklenburg K.L., Petrillo M.L., Tabor J.H., Jarrell K.A., Cheng H.L. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- 2.Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987;50:17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- 3.Burge C.B., Tuschl T., Sharp P.A. In: The RNA World. 2nd edn. Gesteland R.F., Cech T.R., Atkins J.F., editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 4.Cheng Z., Menees T.M. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science. 2004;303:240–243. doi: 10.1126/science.1087023. [DOI] [PubMed] [Google Scholar]
- 5.Perlman P.S., Boeke J.D. Ring around the retroelement. Science. 2004;303:182–184. doi: 10.1126/science.1093514. [DOI] [PubMed] [Google Scholar]
- 6.Coombes C.E., Boeke J.D. An evaluation of detection methods for large lariat RNAs. RNA. 2005;11:323–331. doi: 10.1261/rna.7124405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeman N.C. Biochemistry and structural DNA nanotechnology: an evolving symbiotic relationship. Biochemistry. 2003;42:7259–7269. doi: 10.1021/bi030079v. [DOI] [PubMed] [Google Scholar]
- 8.Seeman N.C. At the crossroads of chemistry, biology, and materials. Structural DNA nanotechnology. Chem. Biol. 2003;10:1151–1159. doi: 10.1016/j.chembiol.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Seeman N.C. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 10.Damha M.J., Ogilvie K.K. Synthesis and spectroscopic analysis of branched RNA fragments: messenger RNA splicing intermediates. J. Org. Chem. 1988;53:3710–3722. [Google Scholar]
- 11.Damha M.J., Zabarylo S. Automated solid-phase synthesis of branched oligonucleotides. Tetrahedron Lett. 1989;30:6295–6298. [Google Scholar]
- 12.Damha M.J., Ganeshan K., Hudson R.H., Zabarylo S.V. Solid-phase synthesis of branched oligoribonucleotides related to messenger RNA splicing intermediates. Nucleic Acids Res. 1992;20:6565–6573. doi: 10.1093/nar/20.24.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganeshan K., Tadey T., Nam K., Braich R., Purdy W.C., Boeke J.D., Damha M.J. Novel approaches to the synthesis and analysis of branched RNA. Nucleosides Nucleotides. 1995;14:1009–1013. [Google Scholar]
- 14.Reese C.B., Song Q. A new approach to the synthesis of branched and branched cyclic oligoribonucleotides. Nucleic Acids Res. 1999;27:2672–2681. doi: 10.1093/nar/27.13.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaringe S.A. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 16.Marshall W.S., Kaiser R.J. Recent advances in the high-speed solid phase synthesis of RNA. Curr. Opin. Chem. Biol. 2004;8:222–229. doi: 10.1016/j.cbpa.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Breaker R.R., Joyce G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y. DNAzymes—a new class of enzymes with promise in biochemical, pharmaceutical, and biotechnological applications. Chem. Eur. J. 2002;8:4589–4596. [Google Scholar]
- 19.Emilsson G.M., Breaker R.R. Deoxyribozymes: new activities and new applications. Cell. Mol. Life Sci. 2002;59:596–607. doi: 10.1007/s00018-002-8452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silverman S.K. Deoxyribozymes: DNA catalysts for bioorganic chemistry. Org. Biomol. Chem. 2004;2:2701–2706. doi: 10.1039/B411910J. [DOI] [PubMed] [Google Scholar]
- 21.Flynn-Charlebois A., Wang Y., Prior T.K., Rashid I., Hoadley K.A., Coppins R.L., Wolf A.C., Silverman S.K. Deoxyribozymes with 2′–5′ RNA ligase activity. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 22.Flynn-Charlebois A., Prior T.K., Hoadley K.A., Silverman S.K. In vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′–5′ to 2′–5′. J. Am. Chem. Soc. 2003;125:5346–5350. doi: 10.1021/ja0340331. [DOI] [PubMed] [Google Scholar]
- 23.Ricca B.L., Wolf A.C., Silverman S.K. Optimization and generality of a small deoxyribozyme that ligates RNA. J. Mol. Biol. 2003;330:1015–1025. doi: 10.1016/s0022-2836(03)00654-5. [DOI] [PubMed] [Google Scholar]
- 24.Prior T.K., Semlow D.R., Flynn-Charlebois A., Rashid I., Silverman S.K. Structure–function correlations derived from faster variants of a RNA ligase deoxyribozyme. Nucleic Acids Res. 2004;32:1075–1082. doi: 10.1093/nar/gkh263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semlow D.R., Silverman S.K. Parallel selections in vitro reveal a preference for 2′–5′ RNA ligation by deoxyribozyme-mediated opening of a 2′,3′-cyclic phosphate. J. Mol. Evol. 2005 doi: 10.1007/s00239-004-0326-y. in press. [DOI] [PubMed] [Google Scholar]
- 26.Hoadley K.A., Purtha W.E., Wolf A.C., Flynn-Charlebois A., Silverman S.K. Zn2+-dependent deoxyribozymes that form natural and unnatural RNA linkages. Biochemistry. 2005;44 doi: 10.1021/bi050146g. doi:10.1021/bi050146q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Silverman S.K. Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 2003;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Silverman S.K. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 29.Coppins R.L., Silverman S.K. A DNA enzyme that mimics the first step of RNA splicing. Nature Struct. Mol. Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- 30.Coppins R.L., Silverman S.K. A deoxyribozyme that forms a three-helix-junction complex with its RNA substrates and has general RNA branch-forming activity. J. Am. Chem. Soc. 2005;127:2900–2907. doi: 10.1021/ja044881b. [DOI] [PubMed] [Google Scholar]
- 31.Coppins R.L., Silverman S.K. Rational modification of a selection strategy leads to deoxyribozymes that create native 3′–5′ RNA linkages. J. Am. Chem. Soc. 2004;126:16426–16432. doi: 10.1021/ja045817x. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Silverman S.K. Directing the outcome of deoxyribozyme selections to favor native 3′–5′ RNA ligation. Biochemistry. 2005;44:3017–3023. doi: 10.1021/bi0478291. [DOI] [PubMed] [Google Scholar]
- 33.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 35.Boeke J.D., Garfinkel D.J., Styles C.A., Fink G.R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 36.Bonitz S.G., Coruzzi G., Thalenfeld B.E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrome oxidase. J. Biol. Chem. 1980;255:11927–11941. [PubMed] [Google Scholar]
- 37.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu V.T., Adamidi C., Liu Q., Perlman P.S., Pyle A.M. Control of branch-site choice by a group II intron. EMBO J. 2001;20:6866–6876. doi: 10.1093/emboj/20.23.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nam K., Hudson R.H., Chapman K.B., Ganeshan K., Damha M.J., Boeke J.D. Yeast lariat debranching enzyme. Substrate and sequence specificity. J. Biol. Chem. 1994;269:20613–20621. [PubMed] [Google Scholar]
- 41.Ooi S.L., Dann C., III, Nam K., Leahy D.J., Damha M.J., Boeke J.D. RNA lariat debranching enzyme. Methods Enzymol. 2001;342:233–248. doi: 10.1016/s0076-6879(01)42548-1. [DOI] [PubMed] [Google Scholar]
- 42.Coleman T.M., Wang G., Huang F. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 φ2.5 promoter. Nucleic Acids Res. 2004;32:e14. doi: 10.1093/nar/gnh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang F., Wang G., Coleman T., Li N. Synthesis of adenosine derivatives as transcription initiators and preparation of 5′ fluorescein- and biotin-labeled RNA through one-step in vitro transcription. RNA. 2003;9:1562–1570. doi: 10.1261/rna.5106403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cristofari G., Bampi C., Wilhelm M., Wilhelm F.X., Darlix J.L. A 5′–3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoro S.W., Joyce G.F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 47.Cruz R.P.G., Withers J.B., Li Y. Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Pyle A.M., Chu V.T., Jankowsky E., Boudvillain M. Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol. 2000;317:140–146. doi: 10.1016/s0076-6879(00)17012-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.