Abstract
To elucidate the origins of the MHC-B-MHC-C pair and the MHC class I chain-related molecule (MIC)A-MICB pair, we sequenced an MHC class I genomic region of humans, chimpanzees, and rhesus monkeys and analyzed the regions from an evolutionary stand-point, focusing first on LINE sequences that are paralogous within each of the first two species and orthologous between them. Because all the long interspersed nuclear element (LINE) sequences were fragmented and nonfunctional, they were suitable for conducting phylogenetic study and, in particular, for estimating evolutionary time. Our study has revealed that MHC-B and MHC-C duplicated 22.3 million years (Myr) ago, and the ape MICA and MICB duplicated 14.1 Myr ago. We then estimated the divergence time of the rhesus monkey by using other orthologous LINE sequences in the class I regions of the three primate species. The result indicates that rhesus monkeys, and possibly the Old World monkeys in general, diverged from humans 27-30 Myr ago. Interestingly, rhesus monkeys were found to have not the pair of MHC-B and MHC-C but many repeated genes similar to MHC-B. These results support our inference that MHC-B and MHC-C duplicated after the divergence between apes and Old World monkeys.
Keywords: long interspersed nuclear element (LINE), MHC class I chain-related molecule, genomic evolution, primate evolution
With the progress of genome sequencing and analysis (1-8), various biological aspects have been revealed at the genomic level. One aspect is the notion that a genome is an evolutionary chimera; the genome of a species has been formed by incorporating a number of foreign pieces into the original structure during evolution. The foreign pieces include not only ordinary genes but also many types of repeated sequences, retroviruses, and genomic fragments. We now recognize that the genome of a species is not a “pure” entity, but is a hybrid of many pieces with different evolutionary origins.
For population genetics and molecular evolution, it has been shown that the majority of nucleotide substitutions observed so far are caused by neutral and selectively negative mutations (9-10), and a minor portion are attributable to selectively positive mutations (11). It can now be considered that evolution at the molecular level has been driven mainly by neutral, nearly neutral (12), and selectively negative mutations. On the other hand, the existence of genes evolving by selectively positive mutations has also clearly been demonstrated and confirmed at the genomic level for several cases, including the MHC (major histocompatibility complex) genes (13).
Another aspect that has been extensively verified at the genomic level is the theory of evolution by gene duplication postulated by Ohno in ref. 14. A number of genes have been shown to possess their duplicated counterparts in the yeast genome (15, 16), the human genome (17-19), and other genomes (20). Actually, in the case of the yeast genome, the duplicated genes are considered to be products of genome fragments duplication (GFD), rather than those of gene duplication (15, 16). The same is true for some cases of human duplicated genes that are not necessarily located on the same chromosome (18).
Recently, we have sequenced the complete MHC class I genome regions of humans, Homo sapiens (21), and chimpanzees, Pan troglodytes (22), and a part of the counterpart of rhesus monkeys, Macaca mulatta.∥ The genomic regions of the three species include a large number of orthologous (23) genes and of repeated sequences such as short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs). This feature is particularly prevalent in the MHC class I genomic region, and provides excellent materials for studying genome evolution.
In this paper we shall discuss the evolutionary origin and history of MHC-B and MHC-C and those of the ape MHC class I chain-related molecule (MIC)A and MICB with special reference to LINEs that have coevolved with the two pairs of genes. To verify our result, we also estimated the divergence time of rhesus monkey by using orthologous LINE sequences in the genomic regions of humans, chimpanzees, and rhesus monkeys. We shall also discuss the role of GFD in the expansion of a genome in evolution.
Origin of MHC-B and MHC-C
The human MHC-B and MHC-C were generated by GFD in evolution (24-28). This GFD was also confirmed by the genome sequencing of the whole human MHC gene complex (21). Therefore, our effort focused on elucidating the duplication or divergence time of the two duplicated genomic fragments, B and C, in which MHC-B and MHC-C were respectively located. Note that earlier reports on the same subject were based on the analyses of the MHC genes themselves or neighboring SINEs (29, 30). As mentioned earlier, MHC genes are good examples of selectively positive genes and, thus, are not guaranteed to have evolved constantly over time. SINEs are also considered to be subject to natural selection, because most of them are intact in the genome of a host species.
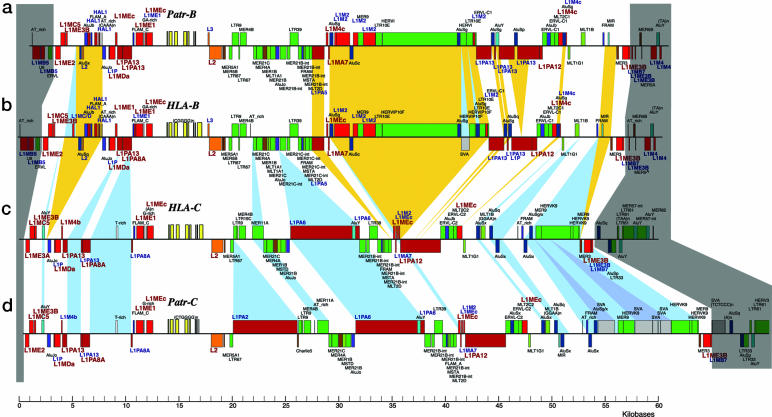
To estimate the duplication time with sufficient accuracy, we have to deal with genomic regions that have evolved constantly over time, such as neutral genes, nonfunctional genes, or noncoding sequences. We thoroughly analyzed the human B and C genomic fragments by using repeatmasker (www.repeatmasker.org, a service of the Genetic Information Research Institute), and we found that the region contained a large number of repeated sequences such as SINEs and LINEs that were paralogous (23) between the two duplicated fragments (Fig. 1 b and c). Three features were revealed regarding these repeated sequences. First, there were many pairs of paralogous LINE and SINE sequences in the duplicated fragments. Second, whereas all SINEs were complete, all LINEs were fragmented, indicating that the former were still functional and subject to natural selection, whereas the latter were free from natural selection. Third, fragmented LINE sequences were located in introns as well as intergenic regions, implying that the sequences have no functional interactions with neighboring genes. Based on these features, we chose the fragmented LINEs for our study, because our main purpose was to estimate the divergence times of the MHC genes in question. Accordingly, our next problem was to estimate the evolutionary rate of the LINE sequences in question.
Fig. 1.
Duplicated genomic fragments of MHC-B and MHC-C regions for humans and chimpanzees. Red, LINE fragmentary sequences used for the present analysis; blue, other sequences. Sequences on top of the line are located in 5′ to 3′ direction, whereas sequences underneath the line are in the 3′ to 5′ direction. Yellow square, an exon of the MHC gene. Other neighboring sequences are also presented to clarify that the LINEs used are paralogous within the species and orthologous between the species. Yellow area, insertion/deletion among the four fragmented sequences; gray area, a region outside of a duplicated fragment. Shown are MHC-B fragment of chimpanzees (a), MHC-B fragment of humans (b), MHC-C fragment of humans (c), and MHC-C fragment of chimpanzees (d).
Having sequenced the complete MHC class I region of chimpanzee, we proceeded to comparative analysis of LINE sequences of humans and chimpanzees, for which the divergence time was known. By scrutinizing and comparing the human and chimpanzee sequences, we found a number of possible orthologous LINE sequences in the B and C fragments of the two species (Fig. 1 a and b, for B fragment; c and d, for C fragment). All chimpanzee LINEs were also fragmented. We further examined whether these sequences were orthologous between the two species with respect to the order of locations, relationships with neighboring sequences, structure, 5′-3′ direction, and sequence homology.
We identified 31 orthologous LINE sequences in the B fragments and 23 orthologous LINE sequences in the C fragments of the two species. Among them 15 LINE sequences were shared by all four fragments (Fig. 1 a, b, c, and d). Using these 15 LINE sequences, we conducted pairwise alignments between the orthologous/paralogous LINE sequences and selected those with more than 100 aligned sites. As a result, we obtained 15 aligned sequences in the B fragments and 13 in the C fragments; these, respectively, included 6,941 and 9,910 nucleotide sites. We then used Kimura's two-parameter method (31) for computing the evolutionary distances between the aligned LINE sequences because the ratio of transition to transversion was ≈2.0 for each pair of the fragments. The results are given in Tables 1 and 2. We also tried Tamura's method (32), but the result was not significantly different from that given by Kimura's method, indicating that our estimation was not affected by the contents of the four bases.
Table 1. Comparison of MHC B and C regions: Identity percentage and number of sites (LINEs) compared.
| Chimp B | Human B | Human C | Chimp C | |
|---|---|---|---|---|
| Chimp B | — | 6,941 (15) | 6,233 (14) | 6,208 (13) |
| Human B | 97.8 | — | 6,309 (14) | 6,282 (13) |
| Human C | 92.1 | 92.1 | — | 9,910 (13) |
| Chimp C | 91.7 | 91.6 | 97.9 | — |
Identity (%) is located below and to the left of the diagonal; number of sites (LINEs) compared is located above and to the right of the diagonal.
Table 2. Comparison of MHC B and C regions: Evolutionary distances and divergence times.
| Chimp B | Human B | Human C | Chimp C | |
|---|---|---|---|---|
| Chimp B | — | 22.3 ± 1.0* | ||
| Human B | 0.0221 ± 0.0018 | — | ||
| Human C | 0.0839 ± 0.0039 | 0.0837 ± 0.0039 | — | 5.4† ± 0.4 |
| Chimp C | 0.0890 ± 0.0040 | 0.0897 ± 0.0040 | 0.0210 ± 0.0015 | — |
Evolutionary distance (substitutions per site), compared by Kimura's method (31), is located below and to the left of the diagonal; divergence time (Myr) is located above and to the right of the diagonal.
Calculated from an average of four evolutionary distances between B and C regions.
Taken from Olson and Varki (33) for calibration of divergence time.
For the divergence time between human and chimpanzee, we adopted an estimate of 5.4 million years (Myr) (33, 34). On the basis of this estimate, we obtained the evolutionary rate of the LINE sequences in the C fragments to be 3.89 × 10-9 substitutions per site per year. The rate of those in the B fragments was found not to be significantly different from that of the C fragments. Both rates were within the range of those of neutral genes (35). We thus applied the evolutionary rate to calibrating the distance between the LINE sequences in the B fragments and those in the C fragment, and we estimated the duplication time between MHC-B and MHC-C to be 22.3 ± 1.0 Myr ago (Tables 1 and 2). This result falls in the range of the estimate, 21-28 Myr ago, obtained by Pointkivska and Nei (29). On the other hand, Alvarez-Tejado et al. (36) argued that a New World monkey species, Saguinus oedipus, had an MHC gene similar to MHC-C. However, because they used incomplete data for the comparison of New World monkeys with gorillas and humans, their conclusion may not be valid.
Origin of MICA and MICB in Apes
Whereas the MHC class I genes have been extensively studied and well documented, the MIC genes have not, despite interesting features such as associations with diseases (37, 38). MIC genes are also responsible for an immune system that seems to be, on the whole, more primitive than that controlled by MHC genes (39, 40). Therefore, the question is whether the MIC immune system is really primitive and an evolutionarily predecessor of the MHC immune system or, conversely, the former is a simplified version of the latter.
There are seven MIC genes in the human genome (41), among which the pair of MICA and MICB has been studied and characterized more extensively than the others. We thus focused on the evolutionary origin of this pair in human and chimpanzee. To study the origin of the pair, we again analyzed the relevant sequences in the MHC genomic fragments of human and chimpanzee by using repeatmasker. The analysis revealed that, whereas the human MICA and MICB were intact, the chimpanzee MICA and MICB fused together by deletion of a downstream region of the former and an upstream region of the latter. The deletion occurred some time after the ape and human lineages diverged. The chimpanzee MICA and MICB may thus collectively be called MICAB, in which the upstream and downstream regions are respectively orthologous to the human MICA and MICB. This observation clearly indicates that the MICA and MICB duplicated earlier than the divergence of human and chimpanzee, but the question is how much earlier?
To answer this question, we similarly computed the evolutionary distance between the orthologous LINE sequences of the two species. In this case, the evolutionary rate turned out to be 3.74 × 10-9 substitutions per site per year. By applying the rate to the distance, we estimated that the duplication giving rise to MICA and MICB occurred 14.1 ± 0.9 Myr ago (Tables 3 and 4). Seo et al. (42) studied MIC genes in rhesus monkeys, and found no orthologous pair of MICA and MICB in monkeys. Their result is consistent with our estimate. On the basis of our estimate, we could predict that gorillas and orangutans also have an MIC pair orthologous to the human pair or one of the pair. In fact, Cattley et al. (43) published a strong possibility that orangutans have such orthologous pairs.
Table 3. Comparison of MIC regions: Identity percentage and number of sites (LINEs) compared.
| Human MICB | Chimp MICAB | Human MICA | |
|---|---|---|---|
| Human MICB | — | 4,462 (10) | 4,389 (10) |
| Chimp MICAB | 95.1 | — | 4,468 (10) |
| Human MICA | 94.8 | 98.0 | — |
Identity (%) is located below and to the left of the diagonal; number of sites (LINEs) compared is located above and to the right of the diagonal.
Table 4. Comparison of MIC regions: Evolutionary distances and divergence times.
| Human MICB | Chimp MICAB | Human MICA | |
|---|---|---|---|
| Human MICB | — | 14.1 ± 0.9* | |
| Chimp MICAB | 0.0512 ± 0.0035 | — | 5.4† ± 0.6 |
| Human MICA | 0.0546 ± 0.0036 | 0.0202 ± 0.0022 | — |
Evolutionary distance (substitutions per site), computed by Kimura's method (31), is located below and to the left of the diagonal; divergence time (Myr) is located above and to the right of the diagonal.
Calculated from an average of evolutionary distances between human MICB and chimp MICB regions and between human MICB and MICA regions.
Taken from Olson and Varki (33) for calibration of divergence time.
Our estimates for the MHC-B-MHC-C pair and the ape MICA-MICB pair indicate that the former is evolutionarily older than the latter. However, this result does not necessarily mean that the MHC immune system originated before the MIC immune system, because there are more MIC genes than ape MICA and MICB genes, as mentioned earlier. Those MIC genes should be analyzed to clarify the point at issue. It can be said at the present that even if the MIC immune system is evolutionarily older than the MHC system, new MIC loci could be generated by GFD or other evolutionary events.
Accuracy of Our Estimate in Reference to the Origin of Rhesus Monkeys
Our estimates of the evolutionary origin of MHC-B and MHC-C could be verified with more confidence by examining them against phylogenetic relationships that include not only human and chimpanzee but also more distantly related primate species. As we also sequenced a part of the MHC class I genomic region for rhesus monkey, we could use the sequence data to verify our estimates. One way to accomplish the verification would be to demonstrate accurately when the Old World monkey diverged from the lineage leading to human and chimpanzee and whether or not the monkey has an orthologous pair of MHC-B and MHC-C.
To estimate the divergence time, we used our own sequence data for human, chimpanzee, and rhesus monkey. From each of the three sequences we selected an orthologous genomic region stretching from the CDSN to FLOT1 loci and detected fragmented LINE sequences in the region by repeatmasker. We examined the detected LINE sequences to determine whether they were orthologous among the three species by the same criteria mentioned earlier and found that each region contained almost 100 fragmented LINE sequences, yielding ≈30,000 aligned sites in total (Tables 5 and 6). We then computed the Kimura distance (31) between each pair of the aligned and concatenated LINE sequences of the three species. Also by using the same divergence time between human and chimpanzee, we computed the evolutionary rate of the LINE sequences to be 2.31 × 10-9 substitutions per site per year, a value that again fell within the range of neutral mutation rates. This rate led us to an estimate that the divergence between rhesus monkeys and humans or chimpanzees occurred 27.0 ± 0.6 Myr ago (Tables 5 and 6).
Table 5. Comparison of neutral region (CDSN-FLOT1): Identity percentage and number of sites (LINEs) compared.
| Rhesus | Chimp | Human | |
|---|---|---|---|
| Rhesus | — | 29,344 (95) | 29,602 (98) |
| Chimp | 94.0 | — | 31,313 (98) |
| Human | 94.0 | 98.8 | — |
Identity (%) is located below and to the left of the diagonal; number of sites (LINEs) is located above and to the right of the diagonal.
Table 6. Comparison of neutral region (CDSN-FLOT1): Evolutionary distances and divergence times.
| Rhesus | Chimp | Human | |
|---|---|---|---|
| Rhesus | — | 27.0 ± 0.6* | |
| Chimp | 0.0629 ± 0.0015 | — | 5.4† ± 0.3 |
| Human | 0.0624 ± 0.0015 | 0.0125 ± 0.0006 | — |
Evolutionary distance (substitutions per site), computed by Kimura's method (31), is located below and to the left of the diagonal; divergence time (Myr) is located above and to the right of the diagonal.
Calculated from an average of evolutionary distances of neutral regions between humans and rhesus monkeys and between chimpanzees and rhesus monkeys.
Taken from Olson and Varki (33) for calibration of divergence time.
Recently, Steiper et al. (44) reported that humans and rhesus monkeys diverged 29.2 to 34.5 Myr ago, which is a little earlier than our estimate. This discrepancy is because of the differences in data, methods, and calibrations between the two approaches. If we adopt 6.0 Myr as the divergence time between humans and chimpanzees, as they did, our estimate would increase to 29.9 Myr, which is now in agreement with theirs. In either case, our result indicates that MHC-B and MHC-C originated by GFD in the lineage leading to humans and chimpanzees after apes diverged from the Old World monkeys.
We then scrutinized the MHC class I region of rhesus monkeys, and we found that monkeys possessed not the pair of MHC-B and MHC-C but 19 genes repeated in tandem. The repeated genes turned out to be similar to MHC-B rather than MHC-C. This finding clearly demonstrates that rhesus monkeys do not have an orthologous pair of MHC-B and MHC-C, confirming our inference that the duplication of MHC-B and MHC-C occurred after the divergence between apes and Old World monkeys.
Discussion
We would first like to emphasize the advantages of using fragmentary orthologous LINE sequences in the estimation of phylogenetic relationships and divergence times. First, fragmentary orthologous LINE sequences are guaranteed to have evolved constantly over time. Second, they can be aligned easily and securely. Third, they can be joined together with no discrimination to accumulate a sufficient number of base pairs for evolutionary analysis. Finally, because they are scattered over a genome, it is possible to conduct genomewide analysis.
The two pairs of MHC-B-MHC-C and MICA-MICB are respectively located in two pairs of duplicated genome fragments 45-55 kbp long. This observation raises a fundamental question regarding how each pair was duplicated. There are number of reports on the observation and the molecular mechanism of GFD (15, 16, 45-48). Koszul et al. (16) observed that the size of duplicated fragments ranges from 41 to 490 kbp in yeast. In some of these reports, Koszul et al. propose a two-step mechanism consisting of the generation of a chromosome breakage and a new replication induced by the breakage, in which the second step does not necessarily require a long sequence homology (see figure 1 of ref. 49 for details). This proposal implies that the existence of fragmentary LINE sequences or SINEs in a genome could trigger GFD and produce pairs of duplicated genome fragments, as in the present case. Because there are large numbers of LINEs and SINEs scattered over the genomes of human and other primate species, it is reasonable to speculate that duplicated genome fragments exist ubiquitously over their genomes, and there are lines of evidence for this speculation (e.g., see refs. 18 and 20). Therefore, GFD could be one of the major mechanisms common at least to the eukaryotes for expanding the size and function of the genome. GFD is, of course, an Epimethean event that keeps occurring unless natural selection or something else suppresses it (50). The force against GFD naturally operates in different manners in different species. Because rhesus monkeys have been shown to possess a number of repeated counterparts to MHC-B, the force is considered to operate less strongly on monkeys than on humans or chimpanzees.
Whereas the evolutionary rates of the fragmentary LINE sequences in the MHC and MIC regions are not significantly different from one another, the rate for the CDSN-FLOT1 region is somewhat slower. Although this difference does not seriously affect our estimates, some explanation is necessary. When we observed the distribution of genetic polymorphism over the entire MHC genomic region among human individuals, we found that the distribution was not uniform over the region, but showed a bell shape with a peak at each of the MHC-B and MHC-C loci (data not shown). This observation indicates that positive selection has operated not only on the MHC genes themselves but also on nearby regions. The same is possibly true for the counterpart sequences of the other two primate species. On the other hand, the influence of positive selection was not clearly observed in the CDSN-FLOTl region. These two distinct aspects could explain the difference in the evolutionary rates. Because we individually estimated the rate of the LINE sequences in the different genomic regions, our results are not affected by the difference in the rate.
It is interesting to ask which of the pair is closer to their ancestral gene, MHC-B or MHC-C. As we mentioned earlier, rhesus monkeys have repeated genes closer to MHC-B than MHC-C. It has also been reported that some orangutan individuals possess MHC-B but not MHC-C (51). Together, this report and our finding suggest that MHC-B is more ubiquitous than MHC-C and, thus, that the former is closer to their ancestral locus than the latter. In this regard, it is perhaps worthy of pointing out that the IMGT database (http://imgt.cines.fr:8104/) shows, as of May 2005, that the human MHC-B and MHC-C loci, respectively, have 661 and 190 alleles. More alleles at MHC-B locus than at MHC-C locus were observed also in chimpanzees (52).
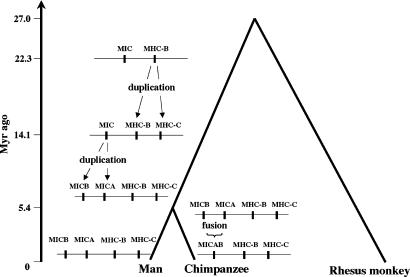
Having interpreted our results in reference to the theory of evolution by gene duplication postulated by Ohno in ref. 14, we propose a scenario regarding the origin and evolution of the MHC-B and MHC-C pair and the ape MICA and MICB pair. First, rhesus monkeys diverged from the lineage leading to apes ≈27 Myr ago; second, the ancestral MHC-B duplicated into MHC-B and MHC-C in the lineage leading to humans and chimpanzees 22.3 Myr ago; third, the ancestral ape MIC duplicated into the ape MICA and MICB in the same lineage, but later, 14.1 Myr ago; and, finally, the MICA and MICB fused by deletion in the lineage leading to chimpanzees (Fig. 2). Ohno mentioned that, by gene duplication, an organism acquires two advantages, new functions and diversified functions (14). The present case is one of the latter.
Fig. 2.
Duplications of the MHC-B-MHC-C pair and the MICA-MICB pair in reference to phylogeny of humans, chimpanzees, and rhesus monkeys. The vertical axis indicates time in Myr in ascending order. We cited the divergence time between humans and chimpanzees (5.4 Myr ago) to calibrate the divergence between rhesus monkeys and humans or chimpanzees and the two events of duplication, as depicted in the figure.
Acknowledgments
This work was supported in part by Ministry of Education, Science, Sports, and Culture of Japan grant (to K.F.-K., H.I., and Y.T.) and National Institute of Genetics Cooperative Research Program Grant 2004-41 (to K.F.-K.).
Author contributions: H.I. and Y.T. designed research; T.S., T.A., K.S., M.Y., and H.I. performed research; K.F.-K. and Y.T. analyzed data; and Y.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MIC, MHC class I chain-related molecule; LINE, long interspersed nuclear element; Myr, million years; GFD, genome fragments duplication; SINE, short interspersed nuclear element.
Footnotes
References
- 1.Goffeau, A., Barrell, B. G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J. D., Jacq, C., Johnston, M., et al. (1996) Science 274, 546-567. [DOI] [PubMed] [Google Scholar]
- 2.The C. elegans Sequencing Consortium (1998) Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- 3.Dunham, I., Shimizu, N., Roe, B. A., Chissoe, S., Hunt, A. R., Collins, J. E., Bruskiewich, R., Beare, D. M., Clamp, M., Smink, L. J., et al. (1999) Nature 402, 489-495. [DOI] [PubMed] [Google Scholar]
- 4.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 5.Hattori, M., Fujiyama, A., Taylor, T. D., Watanabe, H., Yada, T., Park, H. S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D. K., et al. (2000) Nature 405, 311-319. [DOI] [PubMed] [Google Scholar]
- 6.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs, R. A., Weinstock, G. M., Metzker, M. L., Muzny, D. M., Sodergren, E. J., Scherer, S., Scott, G., Steffen, D., Worley, K. C., Burch, P. E., et al. (2004) Nature 428, 493-521. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T. D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R., et al. (2004) Nature 429, 382-388. [DOI] [PubMed] [Google Scholar]
- 9.Kimura, M. (1968) Nature 217, 624-626. [DOI] [PubMed] [Google Scholar]
- 10.Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 11.Endo, T., Ikeo, K. & Gojobori, T. (1996) Mol. Biol. Evol. 13, 685-690. [DOI] [PubMed] [Google Scholar]
- 12.Ohta, T. (1973) Nature 246, 96-98. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L. & Nei, M. (1988) Nature 335, 167-170. [DOI] [PubMed] [Google Scholar]
- 14.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
- 15.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708-713. [DOI] [PubMed] [Google Scholar]
- 16.Koszul, R., Caburet, S., Dujon, B. & Fischer, G. (2004) EMBO J. 23, 234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasahara, M., Nakaya, J., Satta, Y. & Takahata, N. (1997) Trends Genet. 13, 90-92. [DOI] [PubMed] [Google Scholar]
- 18.Endo, T., Imanishi, T., Gojobori, T. & Inoko, H. (1997) Gene 205, 19-27. [DOI] [PubMed] [Google Scholar]
- 19.Shiina, T., Tamiya, G., Oka, A., Takishima, N., Yamagata, T., Kikkawa, E., Iwata, K., Tomizawa, M., Okuaki, N., Kuwano, Y., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 13282-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, W. H., Gu, Z., Cavalcanti, A. R. & Nekrutenko, A. (2003) J. Struct. Funct. Genomics 3, 27-34. [PubMed] [Google Scholar]
- 21.Aguado, B., Bahram, B., Beck, S., Campbell, R. D., Forbes, S. A., Geraghty, D., Guillaudeux, T., Hood, L., Horton, R., Inoko, H., et al. (1999) Nature 401, 921-923.10553908 [Google Scholar]
- 22.Anzai, T., Shiina, T., Kimura, N., Yanagiya, K., Kohara, S., Shigenari, A., Yamagata, T., Kulski, J. K., Naruse, T. K., Fujimori, Y., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7708-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitch, W. M. (1970) Syst. Zool. 19, 99-113. [PubMed] [Google Scholar]
- 24.Mizuki, N., Ando, H., Kimura, M., Ohno, S., Miyata, S., Yamazaki, M., Tashiro, H., Watanabe, K., Ono, A., Taguchi, S., et al. (1997) Genomics 42, 55-66. [DOI] [PubMed] [Google Scholar]
- 25.Kulski, J. K., Gaudieri, S., Bellgard, M., Balmer, L., Giles, K., Inoko, H. & Dawkins, R. L. (1997) J. Mol. Evol. 45, 599-609. [DOI] [PubMed] [Google Scholar]
- 26.Gaudieri, S., Giles, K. M., Kulski, J. K. & Dawkins, R. L. (1997) Hereditas 127, 37-46. [DOI] [PubMed] [Google Scholar]
- 27.Shiina, T., Tamiya, G., Oka, A., Yamagata, T., Yamagata, N., Kikkawa, E., Goto, K., Mizuki, N., Watanabe, K., Fukuzumi, Y., et al. (1998) Genomics 47, 372-382. [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki, M., Tateno, Y. & Inoko, H. (1999) J. Mol. Evol. 48, 317-327. [DOI] [PubMed] [Google Scholar]
- 29.Piontkivska, H. & Nei, M. (2003) Mol. Biol. Evol. 20, 601-609. [DOI] [PubMed] [Google Scholar]
- 30.Sawai, H., Kawamoto, Y., Takahata, N. & Satta, Y. (2004) Genetics 166, 1897-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura, M. (1980) J. Mol. Evol. 16, 111-120. [DOI] [PubMed] [Google Scholar]
- 32.Tamura, K. (1992) Mol. Biol. Evol. 9, 814-825. [DOI] [PubMed] [Google Scholar]
- 33.Olson, M. V. & Varki, A. (2003) Nat. Rev. Genet. 4, 20-28. [DOI] [PubMed] [Google Scholar]
- 34.Chen, F. C. & Li, W. H. (2001) Am. J. Hum. Genet. 68, 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, W.-H. & Graur, D. (1991) Fundamentals of Molecular Evolution (Sinauer, Sunderland, MA).
- 36.Alvarez-Tejado, M., Martinez-Laso, J., Garcia-de-la-Torre, C., Varela, P., Recio, M. J., Allende, L., Gomez-Casado, E. & Arnaiz-Villena, A. (1998) Eur. J. Immunogenet. 25, 409-417. [DOI] [PubMed] [Google Scholar]
- 37.Ghaderi, M., Nikitina Zake, L., Wallin, K., Wiklund, F., Hallmans, G., Lenner, P., Dillner, J. & Sanjeevi, C. B. (2001) Hum. Immunol. 62, 1153-1158. [DOI] [PubMed] [Google Scholar]
- 38.Gupta, M., Nikitina-Zake, L., Zarghami, M., Landin-Olsson, M., Kockum, I., Lernmark, A. & Sanjeevi, C. B. (2003) Hum. Immunol. 64, 553-561. [DOI] [PubMed] [Google Scholar]
- 39.Lanier, L. L. (1998) Annu. Rev. Immunol. 16, 359-393. [DOI] [PubMed] [Google Scholar]
- 40.Stephens, H. A. (2001) Trends Immunol. 22, 378-385. [DOI] [PubMed] [Google Scholar]
- 41.Bahram, S. (2000) Adv. Immunol. 76, 1-60. [DOI] [PubMed] [Google Scholar]
- 42.Seo, J. W., Bontrop, R., Walter, L. & Gunther, E. (1999) Immunogenetics 50, 358-362. [DOI] [PubMed] [Google Scholar]
- 43.Cattley, S. K., Longman, N., Dawkins, R. L., Gaudieri, S., Kulski, J. K. & Leelayuwat, C. (1999) Eur. J. Immunogenet. 26, 233-238. [DOI] [PubMed] [Google Scholar]
- 44.Steiper, M. E., Young, N. M. & Sukama, T. Y. (2004) Proc. Natl. Acad. Sci. USA 101, 17021-17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llorente, B., Durrens, P., Malpertuy, A., Aigle, M., Artiguenave, F., Blandin, G., Bolotin-Fukuhara, M., Bon, E., Brottier, P., Casaregola, S., et al. (2000) FEBS Lett. 487, 122-133. [DOI] [PubMed] [Google Scholar]
- 46.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397-408. [DOI] [PubMed] [Google Scholar]
- 47.Ira, G. & Haber, J. E. (2002) Mol. Cell. Biol. 22, 6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman, R. & Hughes, A. L. (2003) Mol. Biol. Evol. 20, 154-161. [DOI] [PubMed] [Google Scholar]
- 49.Hurles, M. (2004) PLoS Biol. 2, E206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohta, T. (2003) Genetica (The Hague) 118, 209-216. [PubMed] [Google Scholar]
- 51.Adams, E. J., Thomson, G. & Parham, P. (1999) Immunogenetics 49, 865-871. [DOI] [PubMed] [Google Scholar]
- 52.Adams, E. J., Cooper, S., Thomson, G. & Parham, P. (2000) Immunogenetics 51, 410-424. [DOI] [PubMed] [Google Scholar]