Abstract
Loss of immune function and an increased incidence of myeloid leukemia are two of the most clinically significant consequences of aging of the hematopoietic system. To better understand the mechanisms underlying hematopoietic aging, we evaluated the cell intrinsic functional and molecular properties of highly purified long-term hematopoietic stem cells (LT-HSCs) from young and old mice. We found that LT-HSC aging was accompanied by cell autonomous changes, including increased stem cell self-renewal, differential capacity to generate committed myeloid and lymphoid progenitors, and diminished lymphoid potential. Expression profiling revealed that LT-HSC aging was accompanied by the systemic down-regulation of genes mediating lymphoid specification and function and up-regulation of genes involved in specifying myeloid fate and function. Moreover, LT-HSCs from old mice expressed elevated levels of many genes involved in leukemic transformation. These data support a model in which age-dependent alterations in gene expression at the stem cell level presage downstream developmental potential and thereby contribute to age-dependent immune decline, and perhaps also to the increased incidence of leukemia in the elderly.
Keywords: leukemia, microarray, ontogeny, lineage potential
Studies from various systems have suggested that the physiologic process of aging is perhaps best characterized as a failure to maintain appropriate tissue homeostasis or to return to a homeostatic condition after exposure to stress or injury, and that homeostatic failure of organs and tissues ultimately underlies age-associated decline. Consistent with this assertion, many of the phenotypes observed in patients with progeroid syndromes, and in the animals that model them, suggest an imbalance between cell loss and cell renewal. Given that homeostasis in adult tissues is largely maintained by tissue-specific stem cells, it has been suggested that age-dependent stem cell dysfunction may play a central role in the aging process (1).
The major sites of hematopoiesis change during murine ontogeny, beginning with the yolk sac [7.5-10 days postcoitum (dpc)], the aorta/gonad/mesonephros (8.5-11 dpc), followed by the fetal liver (10-16 dpc), and finally the bone marrow (BM) (16 dpc-adulthood). Although hematopoietic stem cells (HSCs) function throughout the lifetime of an organism to give rise to all cells of the blood, the phenotypic and functional properties of HSCs change during ontogeny. For example, it is well documented that the repopulating activity and lymphoid potential of fetal liver HSCs exceeds that of BM-derived HSCs (2). Additionally, we have previously demonstrated that during early embryonic T cell development, there is only a narrow time window in which stem cells are capable of producing Vγ3+ and Vγ4+ T cells (3). In a similar manner, CD5+ B1 B cells with VH11 rearrangements are produced only during fetal development (4). The precise timing of these developmental changes suggests the existence of molecular switches that are tightly regulated during ontogeny.
Advanced age is also accompanied by a variety of changes in the hematolymphoid system including age-dependent deficiencies in T and B lymphocyte production that combine to reduce the competence of the adaptive immune system in the elderly. The most clinically significant aspect of age-dependent hematopoietic dysfunction, however, is the dramatically increased incidence of leukemias and other hematological diseases that accompany aging. Interestingly, whereas pediatric leukemias predominantly involve lymphoid lineages, the leukemias that manifest in old age are largely myeloid in origin, suggesting that the malignant capacity of different hematopoietic progenitors changes with age. Although the timing of these age-dependent changes is less precise than the developmental changes that take place during early ontogeny, their pervasiveness argues that there may also be an underlying “programmed” component to their genesis (in addition to the stochastic events that certainly contribute to their manifestation). Indeed such components may simply reflect the natural course of developmental processes that extend beyond adolescence into midlife and even old age.
Historically, the impact of aging on primitive hematopoietic cells has been studied by using numerous assays including colony-forming unit-spleen (CFU-S) activity (5), cobblestone area-forming cell (CAFC) activity (9), BM transplantation (6, 7), and serial transplantation (8). These studies, however, often lead to conflicting conclusions with CFU-S and CAFC activities, suggesting considerable age-dependent differences (5, 9), whereas single or serial transplantation experiments of BM suggest that aging had little impact on stem cell function (7, 8). The discrepant conclusions of these studies, however, could be partly caused by differences in mouse strains used, because strain-dependent increases or decreases in primitive hematopoietic cell frequency and function with age have been reported (9).
The advent of techniques permitting the prospective isolation and purification of HSCs (10) prompted a reexamination of the impact of aging on HSCs (2, 11, 12). The most striking observation made in these studies was that aging was accompanied by a steady-state increase in the frequencies of primitive hematopoietic cells (2, 12), a finding that stressed the necessity of using purified stem cells to assay age-dependent functional changes on a per-cell basis. Although some studies have attempted to map age-dependent characteristics to specific chromosomal loci (reviewed in ref. 13), the question of whether or not there is a wider intrinsic molecular program underlying the age-dependent changes of the hematopoietic system remains relatively unexplored.
To further elucidate the cellular and molecular changes underlying HSC aging, we evaluated the intrinsic functional and molecular properties of highly purified HSCs and progenitor cells in young and old mice and generated a genomewide expression profile of aging stem cells by microarray analysis. Our findings suggest that many of the features that underlie aging of the hematopoietic system result directly from intrinsic changes that occur at the level of long-term HSCs.
Materials and Methods
Mice. All young (2-3 months), middle-aged (12 months), and old (22-24 months) mice used in this study were C57BL/6. Old and middle-aged mice were obtained from the National Institute of Aging (Bethesda), and young mice were obtained from the Stanford University Laboratory Animal Facility.
Purification of Cells. Long-term HSCs (LT-HSCs) were isolated by lineage depletion of whole BM using unconjugated antibodies (CD3, CD4, CD5, CD8, IL7Rα, B220, Ter119, Gr1, and Mac1) and Dynabeads M-450 beads (Dynal, Oslo). Depleted cells were next c-kit-enriched by using streptavidin-conjugated magnetic beads (Miltenyi, Bergisch Gladbach, Germany) followed by staining with ScaI, c-kit, CD34 (Pharmingen), and flk2 (eBioscience, San Diego). All antibodies were prepared in the Weissman laboratory at Stanford unless otherwise noted. Cells were sorted on FACSVantage or FACS Aria sorters (Becton Dickinson). For side population analysis, lineage-depleted BM cells were stained with Hoechst 33342 as described (14), followed by cell surface staining. All cells used for functional and microarray evaluations were double-sorted for purity. Cells were maintained on ice when possible through all procedures.
Transplantation Experiments. Competitive reconstitution was performed by using the congenic CD45.1/CD45.2 mouse system as described (2). Evaluations of BM residing progenitors regenerated after transplant were performed as described (15, 27) with the addition of staining against flk2 for common lymphoid progenitors (CLPs). Samples were costained for CD45.2 to reveal their donor origin. All flow cytometry and FACS data were analyzed with flowjo software (Treestar, Ashland, OR).
Microarray Analysis and Identification of Age-Regulated Genes. A total of 20,000-30,000 LT-HSCs from young and old mice were double-sorted, with the second sort done directly into Trizol. BM cells from 35-40 mice were used to isolate LT-HSCs for each young sample, and 5-10 mice were used to isolate LT-HSCs for each old sample. RNA was isolated and twice amplified with a RiboAmp RNA amplification kit (Arcturus Engineering, Mountain View, CA). Amplified cRNA was streptavidin-labeled, fragmented, and hybridized to Affymetrix 430-2.0 arrays as recommended by the manufacturer (Affymetrix, Santa Clara). Arrays were scanned with a Gene Chip Scanner 3000 (Affymetrix) running gcos 1.1.1 software. Scanned data were exported to dchip software for normalization. A perfect-match/mismatch (PM/MM) model was used for the calculation of expression values (16). To determine whether transcripts were absent, the sample population was divided into young (n = 3) and old (n = 5) subgroups, with the criterion for an absent transcript being a majority of absent calls in both subgroups. Probe sets found to be nonexpressed in mouse LT-HSCs were eliminated from further analyses. Of the original 45,101 probe sets screened, 25,788 probe sets were determined to be present. Expression data were analyzed by using statistical analysis of microarray software, with a calculated median false discovery rate of ≈10%. Heat mapping was done with heatmap builder, version 1.0 software. The categorization of genes into lymphoid and myeloid groupings was done by comprehensive evaluation of the relevant literature for all of the age-regulated genes. All array data are available at the Stanford Microarray Database (http://genomewww5.stanford.edu).
Results
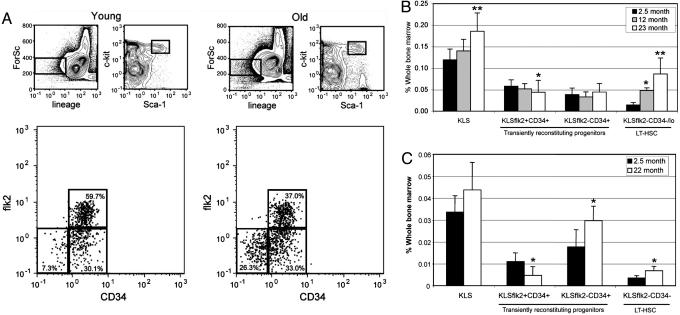
The ability of HSCs to permanently reconstitute myeloablated recipients in all blood cell lineages is the most rigorous criteria for evaluating HSC activity. In the mouse, most, if not all, of the long-term reconstituting HSCs reside within the c-kit-positive, lineage-negative, and Sca-1-positive (KLS) fraction of cells in the BM (10, 17). However, KLS cells are heterogeneous with only a small fraction of these cells possessing long-term repopulating ability (18). To isolate a more purified population of long-term reconstituting cells from the KLS population, we used two additional markers, CD34 (18) and flk2 (19, 20), which we and others have shown can be used to isolate HSCs. By FACS we were able to isolate three populations (KLSflk2+CD34+, KLSflk2-CD34+, and KLSflk2-CD34-) from the BM of young (2-3 months of age) and old (22-24 months of age) mice (Fig. 1A). These populations were transplanted into young congenic recipients to determine which were capable of long-term multilineage reconstitution and whether the cell surface phenotype of such cells would change over time. Peripheral blood analysis of transplanted recipients at 4, 12, 20, and 28 weeks posttransplant demonstrated that regardless of donor age, the KLSflk2-CD34+ and KLSflk2+CD34+ populations gave rise only to transient lympho-myeloid reconstitution (Table 1 and Fig. 4, which are published as supporting information on the PNAS web site). This finding is consistent with two recent reports demonstrating that KLSflk2-CD34+ cells are capable only of short-term multilineage reconstitution (21), whereas KLSflk2+CD34+ cells are primarily responsible for lymphoid reconstitution and possess only limited myeloid potential (22). In contrast, all long-term multilineage repopulating activity was found exclusively within the KLSflk2-CD34- population (Table 1). These data establish that BM cells with the surface phenotype of KLSflk2-CD34- are the only cells within the KLS fraction that possess long-term multilineage reconstituting ability (LT-HSC), and that the cell surface phenotype of LT-HSCs as defined by these markers is not altered with age.
Fig. 1.
Increased self-renewal of LT-HSCs as a consequence of age. (A) Representative FACS profiles of young and old BM cells showing HPCs and multipotent progenitor cells. (B) BM frequencies of KLS cell subsets from young (n = 16), middle-aged (n = 5), and old mice (n = 17). (C) BM frequencies of donor-derived KLS cell subsets after transplantation of BM from young or old donors into young recipients 4 months posttransplant (n = 4 young, n = 3 old). Recipient mice were transplanted with 5-fold more young cells to approximate stem cell equivalents. Statistically significant differences (P < 0.05) are indicated by *. ** in B indicate statistical significance to both of the other age groups.
Previous studies have reported an age-dependent expansion in steady-state frequencies of HSCs in C57BL/6 strains of mice (2, 12), although the question of whether or not this expansion was a cell intrinsic property of aging HSCs, or a consequence of the aging BM microenvironment, has not been addressed. We therefore examined the BM of young, middle-aged (12 months), and old mice to evaluate the impact of aging on the steady-state frequencies of each of the KLSflk2CD34 populations and found that whereas the frequency of the KLSflk2-CD34+ population did not appreciably change with age, the frequency of the KLSflk2+CD34+ population was significantly diminished in old mice (1.4-fold, P < 0.015). In contrast, the frequency of the LT-HSC subset was significantly increased in the BM of aged mice (6.0-fold, P < 0.001), whereas middle-aged animals showed an intermediate phenotype, suggesting that these changes progress steadily with age (Fig. 1B). A similar age-dependent increase in HSC frequency was observed when using side population activity (23) as a criterion for isolating HSC (Fig. 5, which is published as supporting information on the PNAS web site). To address whether the changes in stem cell frequencies is a cell autonomous property of aging HSCs, we transplanted whole BM from young and old donors into young congenic recipients and assayed recipient BM for donor-derived stem and progenitor cell frequencies 4 months posttransplant (Fig. 1C). This analysis revealed that whereas the frequency of donor-derived KLSflk2+CD34+ cells was reduced in recipients transplanted with old BM (2.3-fold, P < 0.04), the frequency of donor-derived LT-HSCs was significantly increased in recipients transplanted with old BM (1.9-fold, P < 0.02). These results demonstrate that the age-dependent expansion of LT-HSCs is a transplantable cell autonomous property of these cells, a finding that establishes a link between aging and increased LT-HSC self-renewal.
To gauge the qualitative performance of the aged LT-HSCs, cells from young and old donors were sorted by FACS and transplanted into young congenic recipients in a competitive setting (24). Peripheral blood analysis of donor-derived B, T, and myeloid cells at 4, 12, 20, and 28 weeks posttransplant revealed that total donor-derived reconstitution was significantly lower (P < 0.05) in mice transplanted with old LT-HSCs at all time points measured (Fig. 2A Left and Fig. 4). This analysis also revealed a significant reduction in the ability of old LT-HSCs to give rise to peripheral B lymphocytes at all time points measured (P < 0.05), with a corresponding trend toward increased myelopoiesis and T lymphopoiesis (Fig. 2A Right). These findings were independently confirmed in experiments in which we isolated and competitively transplanted HSCs isolated by either a KLSflk2- phenotype (data not shown) or a combination of KLSCD34- surface marking and the isolation of cells with the highest capacity to efflux Hoechst 33342 (14) (Fig. 5). They were also confirmed in limit dilution experiments that evaluated the lineage potential of individual LT-HSC clones (Table 2, which is published as supporting information on the PNAS web site).
Fig. 2.
Aging is associated with stem cell intrinsic alterations in lineage and progenitor potential. (A) Multilineage reconstitution (Left) and lineage potential (Right) from transplant experiments of LT-HSCs from young or old mice. Fifty KLSflk2-CD34- cells purified from young or old CD45.2 donors were transplanted into congenic (CD45.1) young recipients against 3 × 105 competitor cells. Peripheral blood analysis was done at the indicated time points, and donor repopulating ability is presented as the percent of total white blood cells. Donor contribution to lymphoid and myeloid lineages is presented as the percent of B, myeloid, and T cells. (B) Aged BM microenvironment does not affect the lineage distribution of adoptively transferred LT-HSCs long term. Ninety LT-HSCs from young mice were transplanted into congenic young or old recipients along with 3 × 105 competitor BM cells from old donors. Peripheral blood analysis was done at the indicated time points, and donor-derived contribution to B cells and myeloid cells is presented as the percent of T cell receptor (TCR) β-negative cells so as not to bias the B cell and myeloid cell data by negating the contribution of the involuted thymus in the old recipients. The contribution to T cells was looked at separately by gating on total donor-derived contribution to all lineages before assessing T cell contribution (Right). (C) BM frequencies of CMPs, GMPs, and MEPs from young (n = 10) and old mice (n = 9). (D) BM frequencies of CLP in young (n = 8), middle-aged (n = 5), and old mice (n = 7). (E) BM frequencies of donor-derived myeloid progenitors from either young or old donors into young recipient mice 4 months posttransplant (n = 4 young, n = 3 old). (F) BM frequencies of donor-derived CLPs from either young or old donors into young recipient mice 4 months posttransplant (young n = 9, old n = 4). Statistically significant differences are indicated by *; ** indicates statistical significance to both age groups.
It has been suggested that the BM microenvironment of aged animals can negatively modulate B cell output (25). To address whether this assertion was applicable to adoptively transferred LT-HSCs, we transplanted purified young LT-HSCs into young and old recipients and evaluated their lineage potential by peripheral blood analysis at multiple time points post-transplant. Although the lineage distribution of transplanted LT-HSCs suggested that the old BM microenvironment adversely impacted B cell production short term, long-term analyses revealed that the ability of young stem cells to generate mature B cells was unaffected by the age of the BM microenvironment (Fig. 2B Left). In contrast, the development of peripheral T cells was severely affected by the aging microenvironment, presumably because of thymic involution of the aged recipients (Fig. 2B Right). Taken together, these results demonstrate that aging is associated with a cell intrinsic reduction in the ability of LT-HSCs to give rise to B lymphocytes that is independent of the aging BM microenvironment.
LT-HSCs give rise to mature blood cells by differentiating through a succession of increasingly committed downstream progenitor cells (26). The altered capacity of old LT-HSCs to generate mature cells in the competitive transplant experiments prompted us to examine the frequency of committed myeloid and lymphoid progenitors in aged animals. Analysis of committed myeloid progenitors (15) from young and old mice by flow cytometry (Fig. 6, which is published as supporting information on the PNAS web site) revealed that whereas the frequencies of common myeloid progenitors (CMPs) and megakaryocyte-erythrocyte progenitors (MEPs) were unaffected by age, old animals exhibited a significant increase (1.7-fold, P < 0.001) in granulocyte-macrophage progenitor (GMP) cells (Fig. 2C). We next examined the impact of aging on the frequency of CLPs by using a variation of an earlier published protocol (27) that takes advantage of additional cell surface markers to further purify this subset (Fig. 6 and H. Karsunky and I.L.W., unpublished work). These analyses showed that old mice exhibited a significant decrease in CLP frequency compared with young mice (2.3-fold, P < 0.001) (Fig. 2D). The CLP frequency in middle-aged animals was found to be intermediate between young and old, suggesting that CLP frequencies progressively decline with age.
To address whether the differences in steady-state frequencies of committed progenitor cells were an intrinsic property of aged LT-HSCs, we transplanted whole BM from young and old donors into young recipients and assayed for donor-derived progenitor cell contribution 4 months posttransplant. We found that donor-derived myeloid progenitor frequencies were comparable regardless of donor age (Fig. 2E), suggesting that the expansion of GMPs that we observed at steady state in old animals (Fig. 2C) was, to some extent, contingent on the aging of the BM microenvironment. In contrast, donor-derived CLP frequencies were significantly reduced (3-fold, P < 0.004) in recipient animals transplanted with BM from old donors (Fig. 2F). In combination with the cell intrinsic reduction of the KLSflk2+CD34+ cells (Fig. 1D), these results indicate that aging leads to a cell autonomous reduction in the ability of LT-HSCs to generate the earliest lymphoid progenitors, while leaving their ability to generate myeloid progenitors intact.
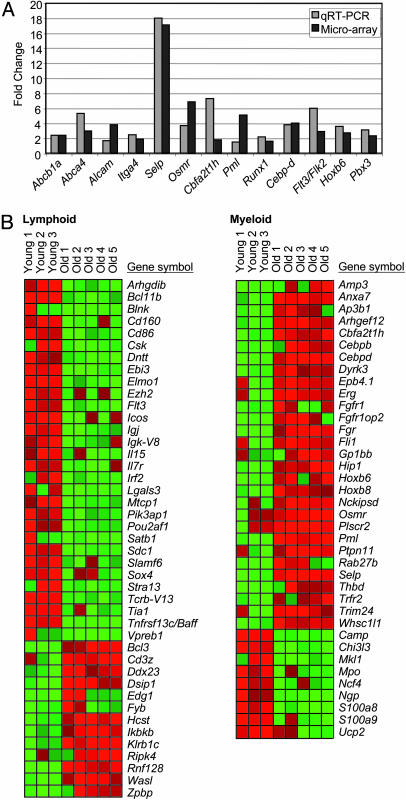
To maintain a steady pool of stem cells while giving rise to differentiated effector cells, a balance must be struck between stem cell self-renewal and differentiation. The cell autonomous nature of LT-HSC aging regarding increased self-renewal (Fig. 1), diminished lymphoid potential (Fig. 2), and differential capacity to generate committed lymphoid and myeloid progenitors (Fig. 2) prompted us to examine the changes in gene expression underlying these age-dependent functional changes. To this end, we performed microarray analysis on highly purified LT-HSCs isolated from young and old mice by using Affymetrix whole-genome arrays containing 45,101 probe sets corresponding to ≈34,000 genes. Array data (eight arrays total: three young and five old) was analyzed using statistical analysis of microarray software to identify age-regulated genes by using stringent parameters corresponding to a calculated median false discovery rate of ≈10%. This analysis identified 907 age-regulated genes in LT-HSCs (Tables 3 and 4, which are published as supporting information on the PNAS web site), of which 59% (532/907) were characterized and annotated genes, whereas the remaining 41% (375/907) were uncharacterized transcripts (Fig. 7, which is published as supporting information on the PNAS web site). To validate the array data by independent means, we performed quantitative RT-PCR (qRT-PCR) on 25 genes, 24 of which were found to be in close agreement with the microarray data (Fig. 3A).
Fig. 3.
Down-regulation of lymphoid genes and the up-regulation of myeloid genes in aged LT-HSCs. (A) Validation of microarray data by qRT-PCR. Fold change of expression of several age-regulated genes as determined by qRT-PCR (light gray) and our microarray analysis (dark gray) is shown. qRT-PCR fold change is presented as the average of three independent experiments. (B) Heat map showing the expression of all of the age-regulated genes identified as lymphoid-specific (Left) and myeloid-specific (Right) in aging LT-HSCs. Gene up-regulation in old LT-HSCs is presented in red and gene down-regulation is in green.
Because gene expression analysis using microarray technology allows for the simultaneous measurement of the expression of thousands of genes, approaches to gain insight into global patterns of gene expression have been developed. To this end, the Gene Ontology (GO) classification system has annotated and assigned genes to common biological processes, molecular functions, and cellular components (28). We therefore analyzed our microarray data by using the GO annotations linked to the Affymetrix platform and gostat software (29) to determine whether specific patterns of gene regulation accompanied LT-HSC aging. This analysis revealed significant statistical overrepresentation of several GO categories in the age-regulated gene set (Table 5, which is published as supporting information on the PNAS web site). Of these, genes involved in signal transducer activity (P ≪ 0.001), and receptor activity (P ≪ 0.001) were found to be highly overrepresented in aging LT-HSC (Table 5). The majority of the genes within these categories fell into several subcategories with correspondingly significant representation including genes involved in transmembrane receptor activity (P ≪ 0.001), G protein-coupled receptor binding (P < 0.02), transmembrane receptor protein tyrosine kinase activity (P < 0.002), and protein tyrosine phosphatase activity (P < 0.01) (Table 5). Thus the differential expression of genes involved in cell signaling is a central underlying molecular feature of LT-HSC aging, a finding that is consistent with the vital role that such regulatory pathways play in HSC self-renewal, lineage commitment, and disease (30). It was also noteworthy that several of the functional categories that have previously been reported to accompany aging of other tissues such as DNA repair (31, 32), stress response (31-34), and inflammation (31-33) were not found to be differentially regulated in aging LT-HSCs (Table 5).
Malignant transformation of normal cells into cancerous cells is believed to require multiple mutagenic hits that alter several aspects of a cell's normal physiology (reviewed in ref. 35). One crucial change that must take place in the transformation of a normal cell is the acquisition of self-renewal capability. It has been proposed that stem cells might represent ideal targets for malignant transformation because they already have the molecular machinery in place for self-renewal. Furthermore, by virtue of their longevity, stem cells have a greater chance than many other cell types of acquiring the succession of mutations and epigenetic changes necessary for transformation (36). In this context, it was remarkable that of the 317 characterized genes up-regulated in aged stem cells, 16 (5.0%) have been implicated in the genesis of various subtypes of human leukemias. Of these, Runx1/AML1, Cbfa2t1h/ETO, Pml, Nckipsd, Arhgef12, Trim24, Erg, Fgfr1, Whsc1/1, Fgfr1op2, Hip1, Dab2ip, and Fgfr3 are translocated in the majority of human myeloid leukemias, whereas Bcl3, Pbx1, and Maf are involved primarily in lymphoid leukemias (Table 3). Although it should be emphasized that the products of many of these genes play roles in normal hematopoiesis, including self-renewal, it is tempting to speculate that the increased expression of leukemia-associated genes with old age might facilitate leukemic transformation by rendering these loci more susceptible to translocation, perhaps in a manner similar to the transcription-dependent chromosomal rearrangements that accompany Ig class switching recombination (reviewed in ref. 37).
An examination of the genes identified in our microarray analysis of LT-HSCs revealed expression of numerous genes whose products are known to function in more committed progenitor cells. This finding is consistent with previous studies using expression arrays (38) or single-cell PCR (39, 40) that have suggested that such “promiscuous” transcription of lineage-associated transcripts in stem cells precedes lineage commitment and is required to prime primitive hematopoietic cells for differentiation toward more committed downstream progenitor cells. In light of this notion, it was remarkable that many such lineage-associated genes were differentially expressed in LT-HSCs. These included 43 genes whose products are critical determinants of lymphoid specification and function, the vast majority of which (70%, 30/43) were found to be down-regulated in old LT-HSCs (Fig. 3B). We also observed age-dependent deregulation of 38 genes involved in mediating myeloid specification and function, the majority of which, in striking contrast to the lymphoid genes, were found to be up-regulated in old LT-HSCs (76%, 29/38) (Fig. 3B). These data demonstrate that differential expression of genes governing lineage specification and function is an intrinsic underlying molecular property of LT-HSC aging and strongly suggest that the age-dependent down-regulation of genes mediating lymphoid specification and function and up-regulation of genes mediating myeloid specification and function combine in a concerted program to skew the lineage potential of LT-HSCs from lymphopoiesis toward myelopoiesis with age.
Discussion
Although not widespread, some studies have undertaken global approaches to address the changes in gene expression that accompany aging in mammalian tissues. Such studies have reported that aging is frequently characterized by up-regulation of genes involved in stress responses (31-34), inflammatory responses (31-33), and DNA repair (31, 32), suggestive of an increased need to cope with the accumulation of macromolecular damage that is believed to accompany aging. By virtue of their longevity, it might have been predicted that similar gene expression changes would be observed in an aging stem cell profile. Our analysis of highly purified LT-HSCs does not, however, indicate that such processes are markedly altered as these cells age. The divergence of these aging expression profiles might reflect the fact that although most tissues are comprised of multiple cell types with divergent expression profiles, our experiments using highly purified stem cells were designed to address the age-dependent changes of a single cell type. And because aging is known to be accompanied by changes in the cellular composition of tissues, including age-dependent increases in lymphocytic infiltration, studies measuring the age-dependent gene expression profile of whole tissues are likely to reflect alterations in cellular composition in addition to cell intrinsic age-regulated changes per se.
Alternatively, these data may reflect more fundamental differences in the way in which stem cells and differentiated cells and tissues age. For example, one reason LT-HSC aging may not be associated with an up-regulation of genes involved in stress response or DNA repair could reflect the fact that while many tissues are largely comprised of postmitotic cells with high metabolic activities, LT-HSCs are mitotic cells that reside largely in the metabolically inactive Go phase of the cell cycle (41). Thus it is conceivable that these cells are exposed to lower levels of damage-inducing metabolic side products and reactive oxygen species than most cell types present in metabolically active tissues, which would abrogate the need to up-regulate cellular stress response pathways. Furthermore, stem cells may be uniquely equipped to handle damage in ways that many differentiated cell types are not. This concept has precedent in stem cell biology and is typified by high levels of expression of many ABC/MDR transporter genes in stem cells, whose products play key physiological roles in cytoprotection. In this regard, it is noteworthy that elevated levels of three ABC transporters, Abca4, Abcb1a, and Abcc1, accompanied LT-HSC aging (Table 3), an observation that contrasts the tissue-associated age-dependent decreases in ABC transporter expression that were reported to accompany aging across species (42). Whether these observations will extend to other adult stem cell populations is an issue of great interest.
Many studies have described the diminished capacity for lymphopoiesis that accompanies aging (reviewed in ref. 25). Although thymic involution, which begins at around the time of puberty, is believed to be the major determinant underlying reductions in T lymphocyte production, the mechanisms leading to age-dependent reductions in B lymphocyte production remain elusive. Studies have suggested that the age-dependent decrease in B cell potential results from changes that occur within the aging BM microenvironment, as a result of defects in committed B lineage progenitor cells, or are caused by an inability of the newly made B cells to migrate to peripheral compartments (reviewed in ref. 25). In this study, we report that the diminished capacity to generate mature peripheral B lymphocytes with age can be traced to the level of the LT-HSC, a finding that is consistent with previous studies (11, 12). Furthermore, our experiments transplanting young LT-HSCs into young or old recipients indicated that although the aging BM microenvironment seemed to adversely impact mature B cell production in the short term, the long-term effects of the aging BM microenvironment on B cell production were not significant.
A recent report demonstrated that CLP cells were significantly decreased in old mice (43). This report raised the possibility of an alternative underlying mechanism for the diminished B cell output distinct from the previously reported age-related deficiencies in cells already committed to a B cell fate (reviewed in ref. 25). By isolating a more purified population of CLPs we confirmed the observations of Miller and Allman (43) that showed that CLP frequencies were dramatically reduced with age. Additionally, we observed age-dependent decreases in the transiently reconstituting KLSCD34+flk2+ cells, which lie upstream of CLP and have recently been shown to contribute primarily to lymphoid reconstitution while possessing only limited myeloid potential (22). Importantly, the diminished potential to generate both CLPs and KLSflk2+CD34+ cells was found to be a cell intrinsic property of aging LT-HSCs. In contrast, myeloid progenitors were found at normal (MEP and CMP) or elevated (GMP) frequencies in the steady state of old animals and were generated just as readily by old and young LT-HSCs after transplantation. These findings argue that the changes in lineage potential that accompany aging are caused by intrinsic differences in the capacity of LT-HSCs to give rise to lymphoid and myeloid progenitor cells. Moreover, the striking finding that LT-HSC aging is accompanied by systematic down-regulation of genes mediating lymphoid specification and function, and up-regulation of genes mediating myeloid specification and function, strongly suggests that the changes in lineage potential that accompany hematopoietic aging are underwritten by age-dependent changes in gene expression at the stem cell level.
The cell intrinsic nature of many of the properties of LT-HSC aging including increased self-renewal, altered lineage potential, and differential capacity to generate lymphoid and myeloid progenitors indicates that profound perturbations in homeostatic control underlie LT-HSC aging. These findings are particularly noteworthy in the context of recent studies demonstrating the involvement of HSCs in leukemic development (44), which represents the extreme endpoint of disrupted homeostatic control in the hematopoietic system. Furthermore, the age-dependent up-regulation of the principal protooncogenes in myeloid leukemogenesis, combined with the increased propensity of aged LT-HSCs to commit toward myeloid lineages, provides a cellular and molecular rationale for the increased incidence of myeloid leukemias in the elderly.
Supplementary Material
Acknowledgments
We thank L. Jerabek for laboratory management, C. Richter for antibody production, L. Hildalgo for animal care, and the Stanford Genome Technology Center for use of the facility. This work was supported by National Institutes of Health Grant 5 R01 CA86065 (to I.L.W.). D.J.R. is supported by The Damon Runyon Cancer Foundation. D.B. is supported by a Swedish Medical Research Council scholarship.
Abbreviations: HSC, hematopoietic stem cell; LT-HSC, long-term HSC; BM, bone marrow; CLP, common lymphoid progenitor; KLS, c-kit positive, lineage negative, and Sca-1 positive; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; GMP, granulocyte-macrophage progenitor; qRT-PCR, quantitative RT-PCR.
References
- 1.Schlessinger, D. & Van Zant, G. (2001) Mech. Ageing Dev. 122, 1537-1553. [DOI] [PubMed] [Google Scholar]
- 2.Morrison, S. J., Wandycz, A. M., Akashi, K., Globerson, A. & Weissman, I. L. (1996) Nat. Med. 2, 1011-1016. [DOI] [PubMed] [Google Scholar]
- 3.Ikuta, K., Kina, T., MacNeil, I., Uchida, N., Peault, B., Chien, Y. H. & Weissman, I. L. (1990) Cell 62, 863-874. [DOI] [PubMed] [Google Scholar]
- 4.Hardy, R. R., Wei, C. J. & Hayakawa, K. (2004) Immunol. Rev. 197, 60-74. [DOI] [PubMed] [Google Scholar]
- 5.Albright, J. W. & Makinodan, T. (1976) J. Exp. Med. 144, 1204-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micklem, H. S., Ford, C. E., Evans, E. P., Ogden, D. A. & Papworth, D. S. (1972) J. Cell Physiol. 79, 293-298. [DOI] [PubMed] [Google Scholar]
- 7.Harrison, D. E. & Doubleday, J. W. (1975) J. Immunol. 114, 1314-1317. [PubMed] [Google Scholar]
- 8.Ogden, D. A. & Micklem, H. S. (1976) Transplantation 22, 287-293. [DOI] [PubMed] [Google Scholar]
- 9.de Haan, G., Nijhof, W. & Van Zant, G. (1997) Blood 89, 1543-1550. [PubMed] [Google Scholar]
- 10.Spangrude, G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241, 58-62. [DOI] [PubMed] [Google Scholar]
- 11.Kim, M., Moon, H. B. & Spangrude, G. J. (2003) Ann. N.Y. Acad. Sci. 996, 195-208. [DOI] [PubMed] [Google Scholar]
- 12.Sudo, K., Ema, H., Morita, Y. & Nakauchi, H. (2000) J. Exp. Med. 192, 1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang, Y. & Van Zant, G. (2003) Curr. Opin. Hematol. 10, 195-202. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki, Y., Kinjo, K., Mulligan, R. C. & Okano, H. (2004) Immunity 20, 87-93. [DOI] [PubMed] [Google Scholar]
- 15.Akashi, K., Traver, D., Miyamoto, T. & Weissman, I. L. (2000) Nature 404, 193-197. [DOI] [PubMed] [Google Scholar]
- 16.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikuta, K. & Weissman, I. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1502-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osawa, M., Hanada, K., Hamada, H. & Nakauchi, H. (1996) Science 273, 242-245. [DOI] [PubMed] [Google Scholar]
- 19.Christensen, J. L. & Weissman, I. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14541-14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adolfsson, J., Borge, O. J., Bryder, D., Theilgaard-Monch, K., Astrand-Grundstrom, I., Sitnicka, E., Sasaki, Y. & Jacobsen, S. E. (2001) Immunity 15, 659-669. [DOI] [PubMed] [Google Scholar]
- 21.Yang, L., Bryder, D., Adolfsson, J., Nygren, J., Mansson, R., Sigvardsson, M. & Jacobsen, S. E. (2004) Blood 105, 2717-2723. [DOI] [PubMed] [Google Scholar]
- 22.Adolfsson, J., Månsson, R., Buza-Vidas, N., Hultquist, A., Liuba, K., Jensen, C., Bryder, D., Yang, L., Borge, O. J., Thoren, L., et al. (2005) Cell 121, 295-306. [DOI] [PubMed] [Google Scholar]
- 23.Goodell, M. A., Brose, K., Paradis, G., Conner, A. S. & Mulligan, R. C. (1996) J. Exp. Med. 183, 1797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison, D. E. (1980) Blood 55, 77-81. [PubMed] [Google Scholar]
- 25.Linton, P. J. & Dorshkind, K. (2004) Nat. Immunol. 5, 133-139. [DOI] [PubMed] [Google Scholar]
- 26.Kondo, M., Wagers, A. J., Manz, M. G., Prohaska, S. S., Scherer, D. C., Beilhack, G. F., Shizuru, J. A. & Weissman, I. L. (2003) Annu. Rev. Immunol. 21, 759-806. [DOI] [PubMed] [Google Scholar]
- 27.Kondo, M., Weissman, I. L. & Akashi, K. (1997) Cell 91, 661-672. [DOI] [PubMed] [Google Scholar]
- 28.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beissbarth, T. & Speed, T. P. (2004) Bioinformatics 20, 1464-1465. [DOI] [PubMed] [Google Scholar]
- 30.Phillips, R. L., Ernst, R. E., Brunk, B., Ivanova, N., Mahan, M. A., Deanehan, J. K., Moore, K. A., Overton, G. C. & Lemischka, I. R. (2000) Science 288, 1635-1640. [DOI] [PubMed] [Google Scholar]
- 31.Lu, T., Pan, Y., Kao, S. Y., Li, C., Kohane, I., Chan, J. & Yankner, B. A. (2004) Nature 429, 883-891. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C. K., Weindruch, R. & Prolla, T. A. (2000) Nat. Genet. 25, 294-297. [DOI] [PubMed] [Google Scholar]
- 33.Kayo, T., Allison, D. B., Weindruch, R. & Prolla, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 5093-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, C. K., Klopp, R. G., Weindruch, R. & Prolla, T. A. (1999) Science 285, 1390-1393. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57-70. [DOI] [PubMed] [Google Scholar]
- 36.Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. (2001) Nature 414, 105-111. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhuri, J. & Alt, F. W. (2004) Nat. Rev. Immunol. 4, 541-552. [DOI] [PubMed] [Google Scholar]
- 38.Akashi, K., He, X., Chen, J., Iwasaki, H., Niu, C., Steenhard, B., Zhang, J., Haug, J. & Li, L. (2003) Blood 101, 383-389. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto, T., Iwasaki, H., Reizis, B., Ye, M., Graf, T., Weissman, I. L. & Akashi, K. (2002) Dev. Cell 3, 137-147. [DOI] [PubMed] [Google Scholar]
- 40.Hu, M., Krause, D., Greaves, M., Sharkis, S., Dexter, M., Heyworth, C. & Enver, T. (1997) Genes Dev. 11, 774-785. [DOI] [PubMed] [Google Scholar]
- 41.Cheshier, S. H., Morrison, S. J., Liao, X. & Weissman, I. L. (1999) Proc. Natl. Acad. Sci. USA 96, 3120-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarroll, S. A., Murphy, C. T., Zou, S., Pletcher, S. D., Chin, C. S., Jan, Y. N., Kenyon, C., Bargmann, C. I. & Li, H. (2004) Nat. Genet. 36, 197-204. [DOI] [PubMed] [Google Scholar]
- 43.Miller, J. P. & Allman, D. (2003) J. Immunol. 171, 2326-2330. [DOI] [PubMed] [Google Scholar]
- 44.Warner, J. K., Wang, J. C., Hope, K. J., Jin, L. & Dick, J. E. (2004) Oncogene 23, 7164-7177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.