Abstract
Control of the bovine mastitis pathogen Streptococcus uberis requires sensitive and epidemiologically meaningful subtyping methods that can provide insight into this pathogen's epidemiology and evolution. Development of a multilocus sequence typing (MLST) scheme based on six housekeeping and virulence genes allowed differentiation of 40 sequence types among 50 S. uberis isolates from the United States (n = 30) and The Netherlands (n = 20). MLST was more discriminatory than EcoRI or PvuII ribotyping and provided subtype data with better epidemiological relevance, e.g., by discriminating isolates with identical ribotypes obtained from different farms. Phylogenetic analyses of MLST data revealed indications of reticulate evolution between genes, preventing construction of a core phylogeny based on concatenated DNA sequences. However, all individual gene phylogenies clearly identified a distinct pauA-negative subtaxon of S. uberis for which housekeeping alleles closely resembled those of Streptococcus parauberis. While the average GC content for five genes characterized was between 0.38 and 0.40, pauA showed a considerably lower GC content (0.34), suggesting acquisition through horizontal transfer. pauA also showed a higher nonsynonymous/synonymous rate ratio (dN/dS) (1.2) compared to the other genes sequenced (dN/dS < 0.12), indicating positive selection in this virulence gene. In conclusion, our data show that (i) MLST provides for highly discriminatory and epidemiologically relevant subtyping of S. uberis; (ii) S. uberis has a recombinatorial population structure; (iii) phylogenetic analysis of MLST data reveals an S. uberis subtaxon resembling S. parauberis; and (iv) horizontal gene transfer and positive selection contribute to evolution of certain S. uberis genes, such as the virulence gene pauA.
Streptococcus uberis is a major cause of bovine mastitis around the world (2, 29, 36, 38, 56). Udder infections can result from the cow-to-cow spread of the pathogen (38, 56) or, more likely, originate from environmental sources of S. uberis (2, 29, 53). Skin, bedding material, and feces can harbor S. uberis (1). For years, a vaccine has been sought after as a means to protect cows from S. uberis mastitis. Among other targets, proteins encoded by the housekeeping gene gapC and the virulence gene pauA have been the focus of vaccine studies (9, 30, 31). In addition to vaccine development, an enhanced understanding of the epidemiology and pathophysiology of S. uberis infections will allow for improvement of mastitis control programs (1, 38). Initial evidence indicates that the epidemiology and pathophysiology of S. uberis infections may be strain specific. For example, the average duration of udder infections in vivo differs between S. uberis strains (56), which may lead to differences in the ability of strains to spread between hosts, as described for Streptococcus pyogenes (22). In addition, in vitro adherence and invasion to mammary epithelium have been shown to differ between strains of S. uberis (27). Although it is not clear what role these processes play in vivo (29), they could potentially contribute to strain differences in chronicity and contagiousness of S. uberis infections (33, 56). Improved knowledge of strain characteristics associated with patterns of transmission, infection, or cure could contribute to improvement of herd- and cow-specific recommendations with respect to mastitis treatment and control, potentially reducing the current reliance on use of antimicrobials as the primary control strategy. Development of reliable, portable, and discriminatory subtyping methods for S. uberis represents a critical initial step to better define these strain differences and to provide diagnostic tools that can identify strains with different transmission and virulence characteristics.
Many phenotypic and DNA banding pattern-based typing methods have been used to characterize S. uberis, including bacteriocin typing (18), restriction endonuclease fingerprinting (18), rapidly amplified polymorphic DNA (RAPD) typing (37, 53, 56), repetitive element polymorphism-PCR (53), and pulsed-field gel electrophoresis (2, 38). While these methods have contributed to insights into the epidemiology of S. uberis mastitis, they have limitations in terms of typeability, discriminatory power, and reproducibility. For several streptococcal species, including S. agalactiae, S. pneumoniae, S. pyogenes, and S. suis, multilocus sequence typing (MLST) schemes have been developed (4, 6, 21, 26), which will likely supersede banding pattern-based typing methods in the near future. In contrast to banding patterns, DNA sequence data, such as those generated in MLST, are easy to standardize, store, and exchange electronically, facilitating analysis, interpretation, and diagnostic use of typing results for isolates from multiple sources, regions, and countries (4). In addition, while it is difficult to correlate band differences to genetic relatedness of strains (45), DNA sequence data can be used for phylogenetic and evolutionary analyses (7, 22, 41). Thus, MLST schemes provide more standardized and informative strain typing data than banding pattern-based methods. While the traditional MLST schemes are based on sequencing of multiple housekeeping genes (4), inclusion of virulence genes (41) or other hypervariable genes (35) in an MLST scheme can enhance discriminatory power. Because genetic variation accumulates slowly in housekeeping genes, MLST based on housekeeping genes is particularly suited for global epidemiological studies (4). Virulence genes may show more short-term variability, and sequencing of virulence genes may thus be more suited to study local epidemiology (57). Typing schemes that encompass housekeeping genes as well as virulence genes also allow for detection of parallel evolution and acquisition or loss of virulence genes (22, 41).
In order to develop a standardized DNA sequencing-based subtyping method for S. uberis, we designed an MLST scheme that includes housekeeping genes as well as virulence genes. A strain set that included a collection of 30 diverse S. uberis isolates from the United States as well as 20 S. uberis isolates from The Netherlands with well-defined epidemiological relatedness (56) was used for ribotyping as well as MLST typing to compare the discriminatory power of these subtyping methods as well as their abilities to reveal epidemiologically meaningful relationships. Phylogenetic analyses were conducted to provide insight into the population structure and evolution of S. uberis. These data will ultimately help us to better understand the evolution of strain-specific transmission and virulence characteristics in this important bovine pathogen and may help us to understand mechanisms behind emergence of new strains or shifts in mastitis epidemiology in response to control measures, including antibiotic treatment and vaccination.
MATERIALS AND METHODS
Bacterial isolates.
A total of 50 S. uberis isolates were selected from our strain collections to facilitate development and evaluation of an MLST scheme. A set of 30 isolates represented S. uberis obtained from bulk tank milk (BTM) collected from 30 different herds throughout New York State. These isolates were considered to be epidemiologically unrelated and should represent diverse sources, since BTM harbors S. uberis from mastitic cows as well as from the environment (19). Because previous reports have shown that the same S. uberis strain is rarely found in different herds (2, 34, 53), we also expected these isolates to be genetically unrelated. An additional 20 S. uberis isolates from The Netherlands, which originated from quarter milk samples from cows in two herds and represent well-defined epidemiological relationships (herd, cow, quarter, and time of isolation), were also included in our study to determine the ability of subtyping methods to provide epidemiologically relevant results; these isolates had previously been characterized by RAPD typing (56). In addition, one Streptococcus parauberis isolate was included in our study. Inclusion of this closely related species provided an outgroup for phylogenetic analyses and allowed us to quantify the degree of relatedness between S. uberis and S. parauberis, two species that are phenotypically indistinguishable (54).
BTM samples were collected and processed by Cornell University's Quality Milk Production Services following standard recommendations (36), and quarter milk samples from The Netherlands were collected and processed as described previously (56). Briefly, upon culture from milk, preliminary identification of isolates was based on colony morphology on blood-esculin agar, Gram stain, and catalase testing (36) followed by speciation with the API20STREP system as recommended by the manufacturer (BioMérieux, Inc., Hazelwood, MO). Isolates were stored at −80°C in brain heart infusion (BHI) broth with 15% glycerol. For ribotyping and additional analyses, streptococci were grown on BHI agar. Bacterial lysates for PCR were prepared as described by Furrer et al. (11). Species identity of all 50 S. uberis isolates and the S. parauberis isolate was confirmed by PCR using S. uberis or S. parauberis primers targeting species-specific parts of the 16S rRNA gene (13).
DNA banding pattern-based strain typing.
Ribotyping of 50 S. uberis isolates and one S. parauberis isolate was performed twice, once with restriction enzyme EcoRI and once with PvuII, using the RiboPrinter microbial characterization system (Qualicon, Wilmington, DE). RAPD typing of isolates from The Netherlands had been performed previously (56), using primer OPE-04 (5′-GTGACATGCC-3′) (37, 53). Simpson's index of discrimination (SID) (15) was calculated for ribotyping but not for RAPD typing, because RAPD results were not generated for BTM isolates.
MLST.
Six genes, including housekeeping genes, vaccine targets, and virulence genes (Table 1) were chosen for inclusion in the MLST scheme described here. For gapC and pauA, DNA sequence data from GenBank were used to design primers in Primer Select (DNAstar; Lasergene). Primers for amplification of cpn60, oppF, sodA, and tuf were taken from the literature (12, 24, 40, 43). Primer sequences, sources of sequence information, and reaction conditions for DNA amplification by PCR are listed in Table 2.
TABLE 1.
Target genes in multilocus sequence typing scheme for S. uberis
| Gene | Enzyme | Functional category |
|---|---|---|
| cpn60 | Chaperonin or heat shock protein 60 | Protein synthesis and metabolism (housekeeping) |
| gapC | Glyceraldehyde-3-phosphate dehydrogenase | Cell surface protein synthesis (housekeeping)a |
| oppF | Oligopeptide permease homolog | Amino acid acquisition (housekeeping/virulence)b |
| pauA | Plasminogen activator A | Cleavage of plasminogen (virulence gene)c |
| sodA | Superoxide dismutase | Antioxidant enzyme (housekeeping) |
| tuf | Elongation factor Tu | GTP binding protein (housekeeping) |
TABLE 2.
Primer sequences and source information, amplicon size, and thermocycling parameters for MLST of S. uberis
| Gene | Primersa | Reference or source | Size (nt) | PCR thermocycling parameters |
|---|---|---|---|---|
| cpn60 | GAI III GCI GGI GAY GGI ACI ACI AC | 12 | 600 | 94°C, 2:00; 20× (94°C, 1:00; TDb, 2:00; 72°C, 5:00); 20× (94°C, 1:00; 35°C, 2:00; 72°C, 5:00); 72°C, 10:00 |
| YKI YKI TCI CCR AAI CCI GGI GCY TT | ||||
| gapC | TTG GTA TTA ACG GTT TCG GTC | NCBIc | 906 | 94°C, 9:00; 35× (94°C, 1:00; 50°C, 1:00; 72°C, 1:00); 72°C, 5:00 |
| CAA GTT GAG CAG TGT AAG ACA TTT C | ||||
| oppF | GAA GCG AAG CTT TGG CT GG | 43 | 800 | 95°C, 4:00; 35× (95°C, 1:00; 55°C, 1:00; 72°C, 1:00); 72°C, 7:00 |
| GCA GCT TCT GCT TCT GTT GA | ||||
| pauA | TTC ACT GCT GTT ACA TAA CTT TGT G | NCBId | 976 | 94°C, 5:00; 35× (94°C, 1:00; 50°C, 1:00; 72°C, 1:00); 72°C, 5:00 |
| CCT TTG AAA GTG ATG CTC GTG | ||||
| sodA | CCI TAY ICI TAY GAY GCI YTI GAR CC | 40 | 480 | 95°C, 3:00; 35× (95°C, 0:30; 37°C, 2:00; 72°C, 1:30); 72°C, 10:00 |
| ARR TAR TAI GCR TGY TCC CAI ACR TC | ||||
| tuf | AAY ATG ATI ACI GGI GCI CAR ATG GA | 24 | 803 | 95°C, 3:00; 35× (95°C, 0:30; 55°C, 0:30; 72°C, 1:00); 72°C, 7:00 |
| AYR TTI TCI CCI GGC ATI ACC AT |
All primers are shown 5′ to 3′. For each gene, the first primer listed is the forward primer, and the second primer is the reverse primer. I = inosine; K = keto (G or T); R = purine (A or G); Y = pyrimidine (C or T).
Touchdown PCR from 45°C to 35°C with temperature decrease of 0.5°C per cycle.
PCR products were purified using the QIAquick 8 PCR purification kit (QIAGEN Inc., Valencia, CA). Sequencing was performed in both directions using PCR primers and Big Dye Terminator chemistry. Sequencing reactions were run on the Applied Biosystems automated 3730 DNA or ABI PRISM 3700 DNA analyzer.
Sequence data analysis.
Sequence data were proofread in SeqMan, and high-quality, double-stranded sequence data were used for further analysis. Sequences for each gene were aligned in MegAlign (DNAstar; Lasergene). Data for the full coding sequence were used for pauA, while partial coding sequences, read in frame, were used for housekeeping genes. DnaSP version 4.0 (42) was used for descriptive analyses, including allele assignment. Different alleles were assigned to gene sequences that differed from each other by one or more polymorphisms. Each unique combination of alleles was considered a sequence type (ST), and BURST analysis (based upon related STs) was used to identify single-locus variants (SLV), double-locus variants (DLV), and satellites (SAT) for each sequence type and to detect the presence of clonal complexes (http://fi-srvmlst1-ide.sm.med.ic.ac.uk/burst/ [last accessed 03/15/2004]).
DnaSP version 4.0 (42) was also used to calculate the G+C content of the sequenced genes as well as the number of polymorphic sites, average number of pair-wise differences between alleles, and average nonysynonymous/synonymous rate ratio (dN/dS). Calculations were performed twice: once for the set of all 50 S. uberis isolates, and once for a “core set” of 49 S. uberis isolates that did not include isolate FSL Z1-015. Isolate FSL Z1-015 had a highly divergent DNA sequence compared to the remaining S. uberis isolates (see Results) and strongly skewed the results of some analyses.
To asses whether the evolution of genes could be adequately represented by the classical (bi)furcating tree model, data were tested for reticulate evolution within genes (17, 44). Methods for detection of reticulate evolution within genes included (i) Rm, the minimum number of recombination events based on the four-gamete test (14) as implemented in DnaSP version 4.0; (ii) construction of split decomposition trees with 1,000 bootstrap replicates based on parsimony splits as implemented in SplitsTree 4.0 BETA 4 (16; http://www-ab.informatik.uni-tuebingen.de/software/splits/ [last accessed 12/06/2004]); and (iii) calculation of compatibility of informative sites within genes as implemented in Reticulate (17). Again, analyses were performed including and excluding the strongly divergent isolate FSL Z1-015.
The model of evolution best describing each gene was determined with ModelTest version 3.06 (39), using hierarchical likelihood ratio tests as the criterion. Settings for the best-fitting model were subsequently used in PAUP version 4.0 beta 10 to determine whether evolution under the specified model followed a molecular clock and to generate maximum likelihood (ML) trees with 100 bootstrap replicates. In these trees, S. parauberis was used as an outgroup. Finally, compatibility between genes was assessed through visual comparison of bootstrapped ML trees for individual genes and through analysis of concatenated sequence data for all genes in Reticulate (17).
Detection of positive selection.
Using codon substitution models that account for heterogeneous selection pressure at amino acid sites, positively selected sites can be specifically identified and the posterior probability of positive selection for each site can be calculated (55). The fit of nested models with and without positive selection can be compared using likelihood ratio tests. For genes with indications of positive selection based on average dN/dS, the codon substitution models 0, 1, 2, 3, 7, and 8, which are described in detail elsewhere (55), were used to test for evidence of amino acid sites under positive selection. Briefly, model 0 assumes that there is one dN/dS ratio for all sites. Model 1 assumes that a proportion of sites has dN/dS = 0 (purifying selection, conserved sites), while the remainder of sites in the gene have dN/dS = 1 (neutral selection). Model 2 assumes that dN/dS can take on one of three values, i.e., 0 (purifying selection), 1 (neutral selection), or a third value that is estimated by the model and that can either account for so-called “slightly deleterious” mutations or for positive selection but not for both at the same time. Model 3 is similar to model 2 but allows for multiple fixed levels of dN/dS. In model 7, levels for dN/dS are not fixed at discrete values but follow a beta distribution. Positive selection is not covered in model 7, because the beta distribution is restricted between dN/dS = 0 and dN/dS = 1. Model 8 combines the beta distribution from model 7 with an extra category for dN/dS, which does allow for positive selection (dN/dS > 1). Analyses were run in the codeml program of PAML version 3.13 (phylogenetic analysis using maximum likelihood), and likelihood ratio tests were performed for model 2 versus 1 (three versus two fixed levels of dN/dS), model 3 versus 0 (multiple versus one fixed level of dN/dS), and model 8 versus 7 (beta distribution of dN/dS with or without positive selection) (55).
In addition to PAML, a likelihood-based method for detection of positively selected sites, the parsimony-based ADAPSITE program was used to identify positively selected sites (47). The location of residues in the β-domain of the pauA-encoded plasminogen activator that were shown to have nonsynonymous mutations was visualized in Cn3D version 4.1 (http://www.ncbi.nih.gov/ [last accessed 04/28/2004]) using alignments with the homologous streptokinase sequence of Streptococcus dysgalactiae subsp. equisimilis (49).
RESULTS
Ribotyping.
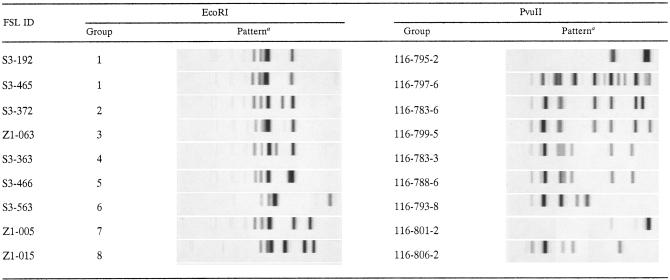
EcoRI ribotyping of 50 S. uberis isolates and one S. parauberis isolate yielded a limited number of bands that were difficult to group into distinct subtype patterns (Table 3). Ribotyping with PvuII resulted in 36 distinct subtype patterns (i.e., ribotypes), with 2 (e.g., FSL S3-192) to 16 bands (e.g., FSL S3-465) per pattern (Table 3). While the majority of ribotypes (n = 28) represented only a single isolate, six ribotypes were found among two isolates, and one ribotype each was found among four and seven isolates (Table 4). Of eight ribotypes that were detected more than once, three were detected on both continents. The SID for PvuII ribotyping of 50 S. uberis isolates was 0.973. RiboPrinter patterns for all isolates can be accessed online in PathogenTracker (www.pathogentracker.net) and are identified by a code (e.g., 116-799-5) consisting of the instrument identification (i.e., 116) and pattern identification (e.g., 799-5).
TABLE 3.
Examples of RiboPrint patterns of S. parauberis (FSL S3-563) and S. uberis (all other isolates) generated with the restriction enzymes EcoRI and PvuII
Full ribotyping results can be viewed at www.pathogentracker.net, including definitive grouping of isolates based on computer-assisted and visual interpretation of PvuII banding patterns. Grouping based on EcoRI patterns is tentative due to the limited number of bands per pattern.
TABLE 4.
Source information, strain assignment, and MLST results for 50 S. uberis isolates and one S. parauberis isolate from bovine milk
| Isolate | Allele
|
ST | PvuII ribotype | RAPD type | Countrya | Herd | Species | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cpn60 | gap | oppF | pauA | sod | tuf | |||||||
| A3-085 | 1 | 1 | 6 | 1 | 1 | 1 | 1 | 783-5 | NDb | USA | 1 | S. uberis |
| C1-308 | 2 | 2 | 1 | 2 | 1 | 2 | 7 | 783-4 | ND | USA | 2 | S. uberis |
| S3-182 | 3 | 3 | 2 | 3 | 1 | 3 | 18 | 795-1 | ND | USA | 3 | S. uberis |
| S3-192 | 2 | 4 | 3 | 4 | 2 | 4 | 12 | 795-2 | ND | USA | 4 | S. uberis |
| S3-197 | 13 | 8 | 3 | 2 | 1 | 4 | 41 | 783-3 | ND | USA | 5 | S. uberis |
| S3-198 | 2 | 4 | 2 | 1 | 2 | 1 | 10 | 795-4 | ND | USA | 6 | S. uberis |
| S3-259 | 4 | 1 | 1 | 5 | 1 | 5 | 19 | 795-6 | ND | USA | 7 | S. uberis |
| S3-286 | 2 | 4 | 3 | 1 | 1 | 4 | 11 | 795-7 | ND | USA | 8 | S. uberis |
| S3-298 | 1 | 4 | 4 | 1 | 2 | 4 | 4 | 783-3 | ND | USA | 9 | S. uberis |
| S3-301 | 2 | 4 | 1 | 4 | 1 | 4 | 8 | 795-8 | ND | USA | 10 | S. uberis |
| S3-304 | 5 | 4 | 1 | 4 | 1 | 4 | 22 | 790-6 | ND | USA | 11 | S. uberis |
| S3-311 | 1 | 4 | 1 | 6 | 1 | 6 | 3 | 793-3 | ND | USA | 12 | S. uberis |
| S3-314 | 1 | 4 | 5 | 0 | 1 | 4 | 6 | 790-2 | ND | USA | 13 | S. uberis |
| S3-328 | 5 | 4 | 4 | 1 | 2 | 4 | 25 | 783-3 | ND | USA | 14 | S. uberis |
| S3-363 | 5 | 4 | 1 | 7 | 6 | 4 | 23 | 783-3 | ND | USA | 15 | S. uberis |
| S3-372 | 1 | 1 | 6 | 1 | 1 | 4 | 2 | 783-6 | ND | USA | 16 | S. uberis |
| S3-391 | 6 | 4 | 7 | 1 | 1 | 4 | 32 | 797-5 | ND | USA | 17 | S. uberis |
| S3-396 | 7 | 4 | 8 | 1 | 2 | 4 | 34 | 783-3 | ND | USA | 18 | S. uberis |
| S3-406 | 2 | 4 | 4 | 1 | 1 | 7 | 13 | 790-4 | ND | USA | 19 | S. uberis |
| S3-415 | 5 | 6 | 3 | 4 | 3 | 7 | 29 | 790-6 | ND | USA | 20 | S. uberis |
| S3-423 | 5 | 4 | 4 | 8 | 1 | 6 | 26 | 783-6 | ND | USA | 21 | S. uberis |
| S3-451 | 2 | 4 | 1 | 4 | 1 | 4 | 8 | 788-4 | ND | USA | 22 | S. uberis |
| S3-457 | 2 | 4 | 1 | 4 | 1 | 4 | 8 | 788-5 | ND | USA | 23 | S. uberis |
| S3-465 | 5 | 4 | 1 | 4 | 1 | 4 | 22 | 797-6 | ND | USA | 24 | S. uberis |
| S3-466 | 1 | 4 | 4 | 1 | 9 | 1 | 5 | 788-6 | ND | USA | 25 | S. uberis |
| S3-474 | 5 | 1 | 1 | 4 | 3 | 7 | 21 | 788-7 | ND | USA | 26 | S. uberis |
| S3-483 | 6 | 4 | 2 | 6 | 1 | 6 | 31 | 797-7 | ND | USA | 27 | S. uberis |
| S3-513 | 2 | 2 | 1 | 2 | 1 | 2 | 7 | 783-4 | ND | USA | 28 | S. uberis |
| S3-525 | 5 | 4 | 1 | 9 | 2 | 4 | 24 | 793-5 | ND | USA | 29 | S. uberis |
| S3-549 | 6 | 7 | 1 | 9 | 1 | 7 | 33 | 793-6 | ND | USA | 30 | S. uberis |
| S3-563 | 8 | 5 | 0 | 0 | 4 | 8 | 35 | 793-8 | ND | USA | 31 | S. parauberis |
| Z1-004 | 2 | 7 | 11 | 1 | 5 | 9 | 15 | 795-2 | A | NL | 32 | S. uberis |
| Z1-005 | 5 | 4 | 14 | 1 | 1 | 4 | 28 | 801-2 | G | NL | 32 | S. uberis |
| Z1-006 | 5 | 4 | 9 | 9 | 6 | 4 | 27 | 806-5 | H | NL | 32 | S. uberis |
| Z1-008 | 2 | 9 | 10 | 1 | 6 | 7 | 17 | 801-3 | N | NL | 32 | S. uberis |
| Z1-014 | 5 | 1 | 1 | 1 | 7 | 10 | 20 | 783-3 | K | NL | 32 | S. uberis |
| Z1-015 | 9 | 10 | 0 | 0 | 10 | 11 | 36 | 806-2 | O | NL | 32 | S. uberis |
| Z1-040 | 2 | 7 | 11 | 1 | 5 | 9 | 15 | 790-2 | E | NL | 32 | S. uberis |
| Z1-063 | 10 | 4 | 1 | 10 | 1 | 4 | 37 | 799-5 | A | NL | 33 | S. uberis |
| Z1-075 | 2 | 8 | 3 | 4 | 1 | 4 | 16 | 800-4 | B | NL | 33 | S. uberis |
| Z1-082 | 2 | 8 | 3 | 4 | 1 | 4 | 16 | 795-5 | B | NL | 33 | S. uberis |
| Z1-086 | 11 | 11 | 1 | 10 | 1 | 4 | 39 | 799-5 | Q | NL | 33 | S. uberis |
| Z1-088 | 2 | 8 | 3 | 4 | 1 | 4 | 16 | 795-5 | I | NL | 33 | S. uberis |
| Z1-095 | 10 | 4 | 1 | 10 | 1 | 4 | 37 | 799-5 | A | NL | 33 | S. uberis |
| Z1-099 | 2 | 4 | 12 | 4 | 1 | 7 | 14 | 799-4 | L | NL | 33 | S. uberis |
| Z1-100 | 11 | 4 | 1 | 10 | 1 | 4 | 38 | 799-5 | P | NL | 33 | S. uberis |
| Z1-113 | 5 | 4 | 9 | 9 | 6 | 4 | 27 | 799-6 | J | NL | 33 | S. uberis |
| Z1-118 | 5 | 4 | 9 | 9 | 6 | 4 | 27 | 783-3 | G | NL | 33 | S. uberis |
| Z1-124 | 12 | 8 | 13 | 11 | 8 | 12 | 40 | 801-7 | A | NL | 33 | S. uberis |
| Z1-125 | 6 | 4 | 1 | 9 | 1 | 2 | 30 | 799-7 | D | NL | 33 | S. uberis |
| Z1-126 | 2 | 4 | 1 | 4 | 1 | 7 | 9 | 783-7 | M | NL | 33 | S. uberis |
NL, The Netherlands.
ND, not determined.
Multilocus sequence typing.
Typeability of S. uberis, i.e., the proportion of isolates yielding an amplicon with primers and conditions used (Table 2), was 100% for cpn60, gapC, sodA, and tuf, 98% for oppF (FSL Z1-015 not typeable), and 96% for pauA (FSL S3-314 and FSL Z1-015 not typeable). Sequence data for all isolates can be accessed on-line in PathogenTracker (www.pathogentracker.net). Between 9 and 14 alleles per gene were identified among the 50 S. uberis isolates (Table 5). Based on these allelic types, a total of 40 STs could be differentiated among the 50 S. uberis isolates characterized; the SID for MLST was 0.989. S. parauberis (isolate FSL S3-563) had unique alleles, which were not found among the 50 S. uberis isolates, for all four housekeeping genes. The S. parauberis isolate tested did not yield PCR amplicons for oppF and pauA.
TABLE 5.
Compositional characteristics of genes in MLST scheme for S. uberis
| Gene | Fragment size (bp) | No. of allelesa | % G+Ca | No. of polymorphic sites
|
No. of parsimony-informative sites
|
Pairwise nt difference
|
No. of substitutions
|
dN/dS
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonymous
|
Non- Synonymous
|
||||||||||||||
| Alla | Coreb | Alla | Coreb | Alla | Coreb | Alla | Coreb | Alla | Coreb | Alla | Coreb | ||||
| cpn60 | 537 | 12 | 0.39 | 93 | 19 | 13 | 11 | 7.3 | 4.1 | 94 | 20 | 5 | 0 | 0.01 | 0.00 |
| gapC | 843 | 10 | 0.40 | 67 | 11 | 7 | 7 | 4.1 | 1.7 | 55c | 8 | 9c | 3 | 0.07 | 0.12 |
| oppF | 723 | 14 | 0.40 | NA | 14 | NA | 9 | NA | 2.3 | NA | 12 | NA | 2 | NA | 0.05 |
| pauA | 861 | 11 | 0.34 | NA | 16 | NA | 9 | NA | 3.4 | NA | 4c | NA | 9c | NA | 1.20 |
| sod | 429 | 9 | 0.38 | 99 | 10 | 8 | 6 | 5.3 | 1.7 | 76 | 6 | 24 | 4 | 0.06 | 0.05 |
| tuf | 657 | 11 | 0.40 | 45 | 14 | 9 | 8 | 3.9 | 2.9 | 35 | 11 | 10 | 3 | 0.11 | 0.12 |
Based on all 50 S. uberis isolates, with the exception of oppF (no amplicon from FSL Z1-015) and pauA (no amplicon from FSL S3-314 and FSL Z1-015). NA, not applicable.
Based on core set of 48 (pauA) or 49 (other genes) S. uberis isolates. Isolate FSL Z1-015 with strongly divergent sequence was excluded from analysis to avoid skewing of results.
One complex codon (triplet of sites segregating for multiple codons; DnaSP version 4.0) was excluded from the analysis.
BURST analysis identified one group comprising 21 STs, four groups of 2 STs each, and nine singletons among 50 S. uberis isolates. Clonal complexes with ancestral types were not detected. Concordance between PvuII ribotypes and BURST grouping was poor; for five of eight ribotypes that were represented by multiple isolates, isolates were categorized into different BURST groups. Four isolates belonging to ribotype 116-799-5 and originating from one herd were SLVs or DLVs (Table 4).
Concordance of ribotyping, RAPD, and MLST.
RAPD data available for the 20 S. uberis isolates from The Netherlands (56), representing 15 RAPD types, were used to compare discriminatory power and epidemiological relevance of RAPD, ribotyping, and MLST. Three RAPD types, A, B and G, were represented by more than one isolate in the data set (Table 4). The one isolate with RAPD-A obtained from herd 32 differed in both its PvuII ribotype and its ST (Table 4) from the other three isolates with this RAPD type, which were from herd 33 (Table 4). Two of three RAPD-A isolates in herd 33 (Z1-063 and Z1-095), both collected in December 1997, shared the same ribotype (799-5) and ST (37). The third RAPD-A isolate (Z1-124) from herd 33, which had been collected in August 1998, had a different ribotype (801-7) and ST (40). Thus, ribotyping and MLST differentiated within RAPD-A in accordance with herd of origin and time of sample collection. For RAPD-B, two isolates (Z1-075 and Z1-082) were collected from two cows in herd 32 in May and June 1998 during an outbreak of S. uberis mastitis and were considered to be epidemiologically related (49). STs for these isolates were identical, while ribotype patterns differed by one band (Table 4). For RAPD-G, the two isolates collected from herd 32 and 33 (Z1-005 and Z1-118) differed in ribotype and ST, in accordance with the difference in herd of origin.
Ribotype 116-783-3 was represented by seven isolates, which originated from seven herds on two continents and were thus considered epidemiologically unrelated; these isolates were grouped into seven different STs. Four ribotypes (116-795-2, 116-783-6, 116-790-2, and 116-790-6) were represented by two isolates each, and within each of these ribotypes the isolates originated from different herds and belonged to different STs. Two other ribotypes were also represented by two isolates each. Isolates originated from one herd and two herds for ribotypes 116-795-5 and 116-783-4, respectively. Within each of these two ribotypes, isolates were assigned the same ST. Ribotype 116-799-5 was represented by four isolates from one herd. Among the four isolates, three RAPD types had been identified, and three STs were assigned in accordance with RAPD patterns. In summary, comparison of ribotyping, RAPD, and MLST results showed that MLST provided subtype discrimination within RAPD types or ribotypes, which is in accordance with the epidemiological origin of isolates.
Descriptive analysis of sequence data.
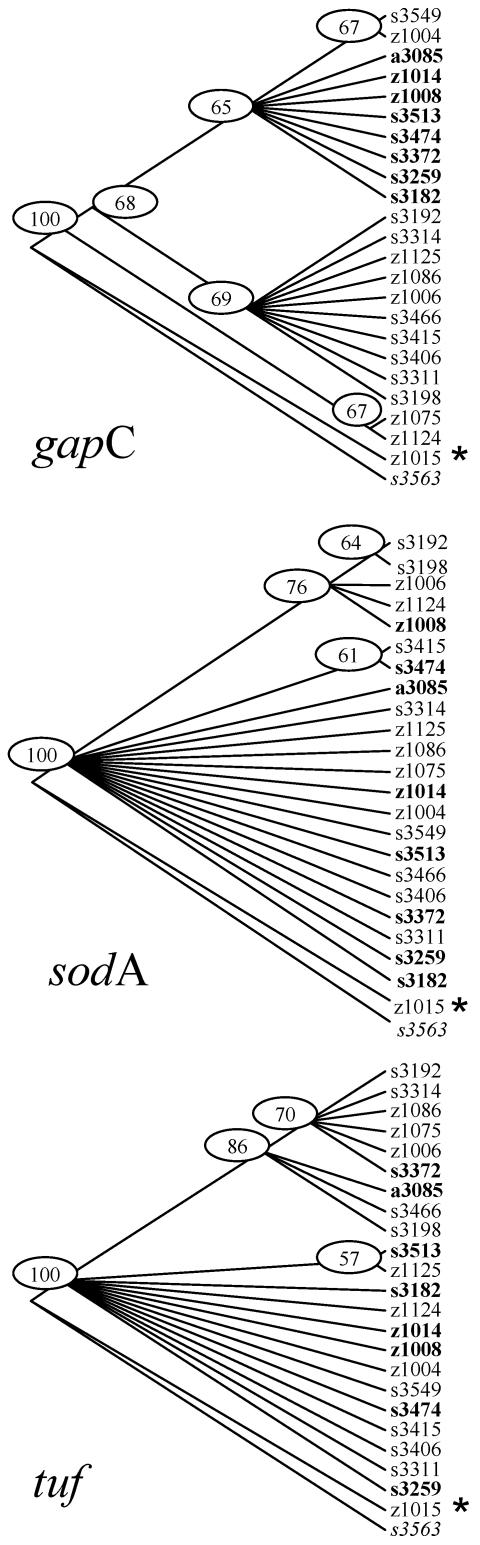
Sequence alignments and phylogenetic trees showed that the individual gene sequences for S. uberis isolate FSL Z1-015 were highly divergent from the consensus sequence for the other 49 S. uberis isolates (Fig. 1). For all housekeeping genes, sequences of FSL Z1-015 were more similar to the sequences for S. parauberis isolate FSL S3-563 and for other S. parauberis isolates obtained from GenBank (AF4855798, AF485799, and AF485800 for cpn60; AF421901 for gapC; AJ544723 for sodA; and AY267004 for tuf) than to the sequences for the other 49 S. uberis isolates. In bootstrap analysis of ML trees for housekeeping genes, FSL Z1-015 was separated from the remaining isolates with a bootstrap value of 100, while it was not separated from S. parauberis, which was included in the analysis as an outgroup (Fig. 1). To confirm the species identity of FSL Z1-015, partial sequencing of rnpB (encoding the RNA subunit of endoribonuclease P) and rpoB (encoding the beta subunit of RNA polymerase), two genes previously used to probe the phylogeny of streptococcal species, was performed as described elsewhere (3, 48). rnpB and rpoB sequences for FSL Z1-015 were identical to those obtained for six S. uberis isolates representing six EcoRI RiboPrinter patterns (Table 3, groups 1 to 5 and 7) and different from those for S. parauberis isolate FSL S3-563, confirming the classification of FSL Z1-015 as S. uberis. Analysis of sequence data after removal of FSL Z1-015, leaving a core set of MLST data for 49 S. uberis isolates, reduced the number of polymorphic sites per gene dramatically but had little impact on the number of parsimony-informative sites (Table 5). G+C content was approximately 39% for all genes, with the exception of pauA (34%) (Table 5). The average dN/dS was low (0.00 through 0.12) for all genes, again with the exception of pauA (dN/dS = 1.2). Exclusion of isolate FSL Z1-015 strongly reduced the number of synonymous and nonsynonymous substitutions, but the effect on dN/dS ratios was limited (Table 5).
FIG. 1.
Maximum-likelihood trees for gapC, sodA, and tuf based on sequence data for 50 S. uberis isolates. Sequences used represent all combinations of alleles for the three genes. Bootstrap values of >50 are shown. S. parauberis (S3-563) was used as an outgroup (italicized). Isolates from a major branch of the gapC tree are labeled (bold), and distribution of the isolates from this branch over sodA and tuf trees is shown. Bootstrap analysis clustered isolate FSL Z1-015 (star) together with S. parauberis and separate from other S. uberis isolates for all three genes.
Gene-specific reticulate and phylogenetic analysis.
The four-gamete test, splits decomposition analysis, and compatibility analysis all revealed limited recombination within cpn60, oppF, and pauA. Phylogenies of the housekeeping genes gapC, sodA, and tuf were all represented by bifurcating trees when isolate FSL Z1-015 was excluded from the alignments. In addition, the three genes followed the same evolutionary model (Hasegawa-Kishino-Yano model, i.e., variable base frequencies, variable transition and transversion frequencies, and molecular clock). Inclusion of FSL Z1-015 decreased within-gene compatibilities and affected either estimated variability in base frequencies or estimated transition or transversion ratios for each of the three genes (Table 6).
TABLE 6.
Evolutionary characteristics of genes in the MLST scheme for S. uberis
| Gene | Reticulate evolutiona,b
|
Tree-like evolutiona,c
|
|||
|---|---|---|---|---|---|
| Rm | SplitsTree | Compatibility | Basic model | Molecular clock | |
| cpn60 | 2 (3) | Net (net) | 0.891 (0.846) | TrN+G (TrN+G) | No (No) |
| gapC | 0 (2) | Tree (net) | 1.000 (0.905) | HKY (TVM) | Yes (Yes) |
| oppF | 2 (n.a.) | Net (NA) | 0.917 (NA) | TrN+G (NA) | Yes (NA) |
| pauA | 1 (n.a.) | Net (NA) | 0.944 (NA) | F81 (NA) | Yes (NA) |
| sodA | 0 (3) | Tree (net) | 1.000 (0.786) | HKY (K80) | Yes (Yes) |
| tuf | 0 (2) | Tree (tree) | 1.000 (0.917) | HKY (GTR) | Yes (Yes) |
Data represent a core set of 49 isolates excluding highly divergent isolate FSL Z1-015, which did not yield amplicons for oppF and pauA. Data for all 50 isolates are shown in brackets if applicable. NA, not applicable.
Rm, minimum number of recombination events based on four-gamete test (14). SplitsTree is the result of splits decomposition analysis, showing either treelike phylogeny (tree) or network (net) (16). Compatibility indicates the compatibility score of the parsimony-informative sites (17).
Basic model for tree-like phylogenies determined with MODEL TEST (39). For genes with reticulation, tree may not adequately represent evolution (17, 44). F81, Felsenstein 1981 model (variable base frequencies, all substitutions equally likely); K80, Kimura two-parameter model (equal base frequencies, variable transition and transversion frequencies); HKY, Hasegawa-Kishino-Yano model (variable base frequencies, variable transition and transversion frequencies); TrN, Tamura-Nei model (variable base frequencies, equal transversion frequencies, variable transition frequencies); TVM, transversion model (variable base frequencies, variable transversion frequencies, equal transition frequencies); GTR, general time-reversible model (variable base frequencies, symmetrical substitution matrix); G, gamma distribution (gamma-distributed site-to-site rate variation).
Reticulate evolution between genes.
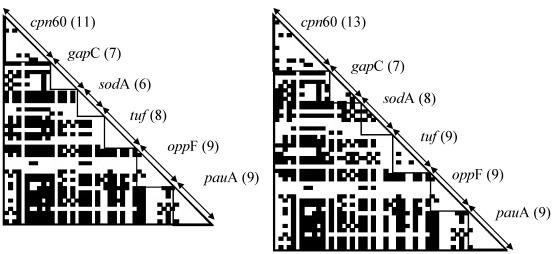
ML trees for gapC, sodA, and tuf, the three genes that showed tree-like evolution within the core set of 49 S. uberis isolates, are shown in Fig. 1. To simplify the figure, each combination of alleles that was observed for the three genes is represented by only one isolate. The major branches in the bootstrapped ML tree from any one gene were not in agreement with results for the two other genes. As an example, isolates from a branch in the gapC tree are bolded and their distribution over branches of the sodA and tuf trees is shown. These results indicate that there is reticulate evolution between genes and that there is no single consistent tree phylogeny that could adequately describe the evolution of all genes. This is in accordance with results from Reticulate, which showed significant clustering of compatible sites, both along the concatenated sequence as a whole (P < 0.01) and for individual genes (P ≤ 0.05 for each gene, with the exception of sodA in the analysis including FSL Z1-015). High compatibility of sites within genes (range, 89.1 to 100%; average, 95.9%) relative to compatibility of sites between genes (range, 35.4 to 72.9%; average, 55.9%) indicates that different genes in the core set of 49 isolates experienced distinct evolutionary histories. Visually, this is indicated by a large proportion of black squares for between-gene comparisons in the compatibility matrix (Fig. 2) with no black squares within genes. When all 50 isolates were included in the analysis, within-gene compatibility (range, 78.6 to 94.4%; average, 88.6%) was still much higher than compatibility between genes (43.6% to 76.4%; average, 59.9%), but incompatibilities were visible within each gene due to the inclusion of highly divergent isolate FSL Z1-015.
FIG. 2.
Compatibility matrix for concatenated sequence data of six genes of S. uberis. White squares and black squares indicate compatibility and incompatibility of sequence polymorphisms, respectively. The number of parsimony-informative sites for each gene is shown in parentheses after the gene designation. The right panel shows results for housekeeping genes of 50 isolates, while the left panel shows results for the core set of 49 isolates obtained after exclusion of highly divergent isolate FSL Z1-015. The core set shows perfect within-gene compatibility for gapC, sodA, and tuf as indicated by the white triangles. Results for oppF and pauA did not differ between panels because FSL Z1-015 did not yield amplicons for those genes, and sequence data were thus not included in either reticulogram.
Positive selection.
Log likelihood values obtained by PAML analysis of pauA sequence data indicated a better fit for models M2, M3, and M8, all of which allow for positive selection, than for models M0, M1, and M7, respectively, none of which allow for positive selection. Differences in model fit were not statistically significant. For comparison of M3 to M0 P was 0.10, and for other comparisons (for M2 versus M1 or for M8 versus M7) the observed chi-square value was 4.3, while the critical value for P values of <0.10 was 4.61. The proportion of positively selected sites was estimated at 7.8% in M3 and M8, and the dN/dS ratio was estimated to be 7.3 for sites under positive selection. However, due to the small number of polymorphic sites, reliable prediction of sites under positive selection and calculation of associated posterior probabilities was not possible. Inclusion of pauA sequences available from GenBank in the analysis did not improve performance of the model (results not shown). Similar calculational difficulties were encountered with parsimony-based detection of positively selected sites in ADAPTSITE. Because positively selected sites could not be identified accurately in pauA, which has a high average dN/dS, PAML or ADAPTSITE analysis was not undertaken for other genes.
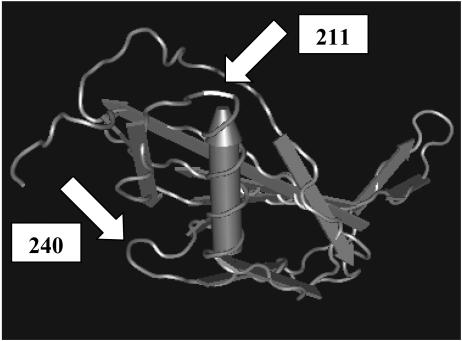
Amino acids encoded by nonsynonymous sites in pauA are shown in Table 7. Codon 211 was highly variable and contained synonymous as well as nonsynonymous mutations. It encodes Arg, Leu, or Ser in the three most common alleles of pauA, detected 15, 13, and 6 times, respectively, among the 49 S. uberis isolates, with each allele detected among isolates from the United States as well as Europe (Table 7). Residue 211 is located at the tip of an α-helix in the β-domain of streptokinase (Fig. 3). Codon 240 encodes a residue located at the tip of a loop between β5 and β6 strands which connects two β-folded sheets (49) (Fig. 3). In the three most common alleles of pauA, codon 240 codes for glycine, a small, hydrophilic amino acid that allows for flexibility in stereochemical conformation, or proline, a hydrophobic amino acid that limits structural flexibility (Table 7).
TABLE 7.
Amino acids encoded by codons with nonsynonymous mutations in plasminogen activator gene pauA of S. uberis
| Allele | No. of isolates | Sourcea | Codonb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 23 | 40 | 73 | 82 | 115 | 124 | 211 | 240 | 242 | |||
| 1 | 15 | USA (10), NL (5) | Gly | Val | Pro | Asp | Ala | Arg | Arg | Arg | Pro | Asp |
| 2 | 3 | USA | Gly | Val | Pro | Asn | Ala | Arg | Arg | Arg | Pro | Asp |
| 3 | 1 | USA | Gly | Val | Pro | Asp | Ala | Arg | Arg | Arg | Pro | Asp |
| 4 | 13 | USA (8), NL (5) | Gly | Val | Pro | Asp | Ala | Gln | Gln | Leu | Gln | His |
| 5 | 1 | USA | Ala | Val | Pro | Asn | Ala | Arg | Arg | Leu | Pro | Asp |
| 6 | 2 | USA | Gly | Val | Pro | Asn | Ala | Arg | Arg | Argc | Pro | Asp |
| 7 | 1 | USA | Gly | Val | Pro | Asp | Thr | Arg | Arg | Arg | Pro | Asp |
| 8 | 1 | USA | Gly | Ile | Pro | Asp | Ala | Arg | Arg | Arg | Pro | Asp |
| 9 | 6 | USA (2), NL (4) | Gly | Val | Pro | Asp | Ala | Arg | Arg | Ser | Pro | Asp |
| 10 | 4 | NL | Gly | Val | Pro | Asp | Ala | Arg | Arg | Leu | Pro | Asp |
| 11 | 1 | NL | Gly | Val | His | Asp | Ala | Arg | Arg | Leu | Gln | Asp |
Isolates originated from bovine milk and were collected from bulk tank milk in the United States (USA; n = 29) and from quarter milk samples in The Netherlands (NL; n = 19). Two additional isolates, one from the United States and one from The Netherlands, did not produce pauA amplicons.
Amino acids differing from the majority are bolded.
Arg is encoded by CGA in allele 6 and by CGC in other alleles with an Arg codon.
FIG. 3.
β-Domain of streptokinase from S. dysgalactiae subsp. equisimilis (49), showing a single α-helix (tube-shaped arrow) and β-folded sheets consisting of multiple strands (flat gray arrows). White arrows indicate the localization of the codons homologous to codons 211 and 240 in the S. uberis PauA sequence.
DISCUSSION
Using MLST analysis based on four housekeeping genes (cpn60, gapC, sodA, and tuf), a putative virulence gene (oppF), and a known virulence gene (pauA), we have shown that (i) MLST provides for highly discriminatory and epidemiologically relevant subtyping of S. uberis; (ii) S. uberis has a recombinatorial population structure; (iii) phylogenetic analysis of MLST data reveals an S. uberis subtaxon with high genetic similarity to S. parauberis in terms of presence of virulence genes and DNA sequence of housekeeping genes; and (iv) horizontal gene transfer and positive selection contribute to evolution of certain S. uberis genes, e.g., the virulence gene pauA.
MLST provides for highly discriminatory and epidemiologically relevant subtyping of S. uberis.
While selection of genes to be included in an MLST scheme is facilitated by the availability of partial and/or complete genome sequences for the organism of interest, no genome sequences for S. uberis were available at the initiation of our project. Target genes were thus selected based on availability of sequence data and/or PCR primer sequences. Housekeeping genes, virulence genes, and genes encoding vaccine targets were included in our MLST scheme to allow studies on population genetics as well as the evolution of virulence and vaccine target genes in S. uberis. The housekeeping genes cpn60 (chaperonin or heat shock protein gene), sodA (superoxide dismutase), and tuf (elongation factor Tu) were chosen because these genes are conserved across bacterial species, allowing us to use previously described degenerate primers for amplification of these genes (12, 24, 40). These primers can thus be used for DNA sequence-based bacterial speciation and for subtyping schemes, such as MLST. For example, primers for these genes have been successfully used in our laboratory for species identification and characterization of the mastitis pathogens S. agalactiae and S. dysgalactiae subsp. dysgalactiae and for enterococcal species isolated from milk and dairy farm environments. In addition to the three housekeeping genes described above, we also included two genes whose products have been the target of vaccine studies, i.e., gapC (glyceraldehyde-3-phosphate dehydrogenase), a housekeeping gene (9), and pauA (plasminogen activator A), a virulence gene (31), in our MLST scheme. Finally, oppF, encoding an oligopeptide permease homolog, was included because this gene is essential for the growth of S. uberis in milk (43). Due to its potential role in infection, this gene may also be classified as a putative virulence gene. Analysis of dN/dS ratios showed that all genes, except pauA, appeared under purifying selection, indicating that the five genes other than pauA provide appropriate information for probing the population genetics of S. uberis, although oppF cannot be amplified from all S. uberis isolates. While the relative location of the selected MLST genes on the S. uberis genome could not be mapped, the distinct similarity patterns for different genes suggest that the target genes represent sufficiently diverse locations to be appropriate for MLST.
Our results show that MLST provides for more discriminatory subtype differentiation than banding pattern-based methods (ribotyping and RAPD). While similar results have previously been obtained for other human and animal streptococcal pathogens (4, 6, 26), our data further show that development of a sensitive MLST-based subtyping scheme is possible even with limited a priori sequence information (i.e., without the availability of a genome sequence). Our data thus also indicate the feasibility of developing MLST-based subtyping methods for economically less important streptococcal pathogens, such as S. dysgalactiae subsp. dysgalactiae and S. parauberis, for which complete genome sequences may not be available in the near future. The fact that three of the primers used in the MLST scheme described here also allow for amplification of genes from these species further supports the feasibility of developing a core MLST scheme for all streptococcal mastitis pathogens, which could be supplemented with sequencing of species-specific virulence genes. In addition to the discriminatory capability of the MLST scheme described here, we also showed that MLST provides epidemiologically meaningful subtype information, e.g., by differentiating epidemiologically unrelated isolates from different herds, which share identical ribotypes or RAPD patterns. MLST will thus provide a better tool to detect farm-to-farm S. uberis transmission and pathogen reemergence on a given farm. Finally, as discussed in more detail below, MLST, unlike banding pattern-based subtyping methods, allows for meaningful phylogenetic and evolutionary analyses (5), which can provide important insight into S. uberis biology, population genetics, and transmission patterns, providing information that could help in the design of improved mastitis control strategies.
S. uberis has a recombinatorial population structure.
Our data show that while within-gene recombination is limited, considerable reticulate evolution occurs between genes. Thus, a species tree or core phylogeny for S. uberis based on concatenated sequence data could not be constructed (44). Our results are consistent with previous reports, which showed that most streptococcal species generally show a highly recombinatorial population structure. For example, in S. pyogenes, group C streptococci (GCS) and group G streptococci (GGS) reticulate evolution between genes has been shown to play an important role in overall sequence diversity, while only a limited number of housekeeping genes show evidence of within-gene reticulate evolution (23). In some species, for example, in the highly pathogenic species S. pyogenes, clonal groups are preserved despite widespread occurrence of reticulate evolution. These clonal complexes may be niche specific, as shown for S. pyogenes (22) and S. suis (26). In other streptococcal species, such as GCS and GGS, clonal groups have not been detected. Interestingly, for streptococcal species (23) as well as for other pathogens, e.g., Mycobacterium spp. (10) and Listeria monocytogenes (35), increasing evidence is emerging that lineages or species with a commensal lifestyle show higher levels of recombination, possibly facilitating rapid adaptation to different environments, while host-adapted lineages or species often show highly clonal population structures. Results from our current analysis did not identify any clones of S. uberis that could be associated with increased pathogenic potential or targeted through specific mastitis control programs. Rather, a nonclonal population structure was detected, similar to results for commensal streptococcal species (GCS, GGS). This is in accordance with the concept that S. uberis is predominantly an opportunistic environmental pathogen and not a host- or organ-adapted pathogen with specific virulence characteristics. Future studies using a larger number of S. uberis isolates from a variety of sources, including from fecal samples, environmental sources, and animals with clinical S. uberis mastitis, will be required, though, to comprehensively probe the population genetics of S. uberis.
Phylogenetic analysis of MLST data reveals an S. uberis subtaxon genetically resembling S. parauberis.
Advantages of MLST over banding pattern-based methods include the ability to use MLST data to infer phylogenetic relationships and thus reliably define bacterial species and species-like subgroups. Recently, Lan and Reeves (28) specifically suggested that MLST data provide a better approach for defining bacterial species than 16S rRNA analysis or DNA-DNA hybridization. In our study, MLST data for four housekeeping genes allowed us to specifically define S. uberis isolate FSL Z1-015 as a unique subtaxon which is only distantly related to the remaining S. uberis isolates. While ribotyping and RAPD fingerprinting assigned unique banding patterns to this isolate, they failed to reveal the extent of the differences between FSL Z1-015 and other S. uberis isolates, some of which also had unique banding patterns. Interestingly, isolate FSL Z1-015 also lacks the virulence gene pauA as well as oppF, encoding an oligopeptide permease that plays a role for bacterial growth in milk, two genes also absent in S. parauberis. Since S. parauberis cannot be differentiated from S. uberis based on cultural morphology or biochemical properties, it was originally considered a subtype of S. uberis, and only 16S rRNA sequence data allowed for separation of the species S. uberis and S. parauberis (54). Previously described species-specific 16S rRNA PCR assays (13) and rnpB and rpoB, two genes previously used to study the phylogeny of streptococcal species (3, 48), clearly suggest that FSL Z1-015 belongs to the species S. uberis, but sequencing data from housekeeping genes showed that FSL Z1-015 represents a subtaxon distinct from both S. uberis and S. parauberis. Isolate FSL Z1-015 originated from a milk sample in The Netherlands. Meanwhile, ongoing work in our laboratory has identified at least four additional isolates originating from environmental samples in the United States that also belong to this subtaxon, as determined by analysis of sequence data for cpn60, gapC, sodA, and tuf. Like FSL Z1-015, these isolates did not yield amplicons for oppF and pauA (R. N. Zadoks and M. Wiedmann, unpublished results).
The evolution of pauA differs from the evolution of housekeeping genes.
Absence of a pauA amplicon was observed in 2 of 50 isolates in our study (4.0%) and in 4 of 130 S. uberis isolates (3.1%) in a study from Germany (25). Diagnostic methods such as multiplex PCR that target pauA for detection of S. uberis will thus fail to diagnose a small proportion of S. uberis-positive samples. In addition to lower prevalence, pauA displayed a lower GC content than the other five genes sequenced. These findings suggest that S. uberis may have acquired this virulence gene by horizontal transfer. PauA, or plasminogen activator A, is a streptokinase, and transfer of streptokinase genes between streptococcal species has been described before, specifically, between S. pyogenes in humans and GCS or GGS (22).
pauA also represented the only gene that showed strong positive selection, as evidenced by an average dN/dS above 1. Our observation of positive selection in pauA is noteworthy, because PauA has been studied as a potential vaccine target (8, 31) and epitopes that contain positively selected sites may be less suitable as vaccine targets than epitopes that consist exclusively of negatively selected amino acid sites (46). Although expression of PauA is not essential for infection of the mammary gland, as indicated by the isolation of pauA-negative isolates from mastitic cows and by experimental studies (50), PauA is still considered to play a critical role in the pathogenesis of mastitis (52). Some substitutions in pauA are inconsequential. For example, substitutions for alanine by glycine (residue 15) or valine by isoleucine (residue 23) are unlikely to have a functional impact because the amino acids substituting each other have very similar sizes and properties. In addition, they are located in a signal peptide that is not part of the functional domains of streptokinase (20). Other nonsynonymous substitutions on the other hand may have an effect on the conformation and/or functionality of the protein, such as the substitution for arginine by leucine or serine (residue 211), or substitution for glycine by proline (residue 240). The specified residues are located in the β-domain of streptokinase and differ considerably in stereochemical properties, which could potentially affect protein folding and activity. Differences between S. uberis isolates in plasminogen activity have been reported (20) but not correlated to specific amino acid polymorphisms. A recent study on regions of PauA critical to recruitment and activation of plasminogen did not identify any of the codons listed in Table 7 as such (51). Thus, while positive selection clearly plays a role in the evolution of pauA, its impact on pathogenesis is as yet unclear.
In S. pyogenes both positive selection and horizontal transfer of streptokinase genes contribute to streptokinase gene diversity (22). Transfer of streptokinase genes is not just a historic event but can take place between S. pyogenes and S. dysgalactiae subsp. equisimilis when they occupy the same niche (22). The origin of streptokinase genes in S. uberis is unknown in terms of time or donor species. Observations for S. pyogenes and S. uberis, both of which show evidence for horizontal gene transfer and diversifying selection, suggest that there is potential for further evolution of S. uberis streptokinases in response to selective pressures such as vaccination. Vaccine candidates other than PauA are currently being investigated (32). For genes encoding novel vaccine targets, as for streptokinase genes, continued monitoring of gene diversity and evolution, particularly in vaccine studies or applications, may be advised.
Conclusion and outlook.
We developed an MLST scheme for the bovine udder pathogen S. uberis based on four housekeeping genes, a putative virulence gene (oppF), and an established virulence gene (pauA). While previous reports on development of MLST-based subtyping methods have used various numbers of isolates from as few as 28 (57) to as many as 294 (26), we show that use of a well-defined smaller collection of isolates can allow for initial development and validation of an MLST scheme, particularly in the case of an organism like S. uberis, which was previously known to be highly diverse (29, 34, 53). MLST-based subtyping of S. uberis was superior to banding pattern-based methods in terms of discriminatory ability, concordance with epidemiological data, and quantitative information regarding relatedness of isolates. Inclusion of three housekeeping genes that can be amplified from a variety of streptococcal and enterococcal species provides a first step towards development of a “multispecies” mastitis pathogen MLST scheme. pauA-negative isolates, which go undetected by pauA-based diagnostic methods, constitute a small proportion of S. uberis isolates from milk and may represent a new subtaxon of S. uberis that is genetically closely related to S. parauberis. Reticulate evolution contributes to a limited extent to genetic variability within genes but plays a major role in overall sequence variability. The nonclonal structure of the S. uberis population is in accordance with the notion that S. uberis is an environmental opportunist rather than a host-adapted pathogen. Evolution of virulence genes, specifically pauA, differed from the evolution of housekeeping genes, and routine inclusion of housekeeping as well as (additional) virulence genes in MLST schemes for S. uberis should be considered. Horizontal gene transfer and positive selection may contribute to acquisition or evolution of new allelic types for pauA or alternative plasminogen activators and could affect the long-term efficacy of a vaccine based on such virulence genes. In the development and evaluation of S. uberis vaccines, creation of subunit vaccines targeting conserved epitopes and monitoring of target gene diversity and evolution should be considered.
Acknowledgments
We thank Mary Carson, Alphina Ho, Alana Jonat, and Katy Windham for technical assistance and Kendra Nightingale, Sharinne Sukhnanand, and Qi Sun for helpful discussions and suggestions regarding phylogenetic analyses.
This publication was developed with help of the Cornell University Center for Biotechnology, a New York State Center for Advanced Technology supported by New York State and industrial partners. The project was funded in part by the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (USDA-CSREES) award 2004-35204-14220.
REFERENCES
- 1.Bramley, A. J. 1984. Streptococcus uberis udder infection—a major barrier to reducing mastitis incidence. Br. Vet. J. 140:328-335. [DOI] [PubMed] [Google Scholar]
- 2.Douglas, V. L., S. G. Fenwick, D. U. Pfeiffer, N. B. Williamson, and C. W. Holmes. 2000. Genomic typing of Streptococcus uberis isolates from cases of mastitis, in New Zealand dairy cows, using pulsed-field gel electrophoresis. Vet. Microbiol. 75:27-41. [DOI] [PubMed] [Google Scholar]
- 3.Drancourt, M., V. Roux, P. E. Fournier, and D. Raoult. 2004. rpoB gene sequence-based identification of aerobic gram-positive cocci of the genera Streptococcus, Enterococcus, Gemella, Abiotrophia, and Granulicatella. J. Clin. Microbiol. 42:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 5.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finch, J. M., A. Winter, A. W. Walton, and J. A. Leigh. 1997. Further studies on the efficacy of a live vaccine against mastitis caused by Streptococcus uberis. Vaccine 15:1138-1143. [DOI] [PubMed] [Google Scholar]
- 9.Fontaine, M. C., J. Perez-Casal, X. M. Song, J. Shelford, P. J. Willson, and A. A. Potter. 2002. Immunisation of dairy cattle with recombinant Streptococcus uberis GapC or a chimeric CAMP antigen confers protection against heterologous bacterial challenge. Vaccine 20:2278-2286. [DOI] [PubMed] [Google Scholar]
- 10.Frothingham, R. 1999. Evolutionary bottlenecks in the agents of tuberculosis, leprosy, and paratuberculosis. Med. Hypotheses 52:95-99. [DOI] [PubMed] [Google Scholar]
- 11.Furrer, B., U. Candrian, C. Hoefelein, and J. Luethy. 1991. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 70:372-379. [DOI] [PubMed] [Google Scholar]
- 12.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan, A. A., I. U. Khan, A. Abdulmawjood, and C. Lammler. 2001. Evaluation of PCR methods for rapid identification and differentiation of Streptococcus uberis and Streptococcus parauberis. J. Clin. Microbiol. 39:1618-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson, R. R., and N. L. Kaplan. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 17.Jakobsen, I. B., and S. Easteal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. 12:291-295. [DOI] [PubMed] [Google Scholar]
- 18.Jayarao, B. M., S. P. Oliver, J. R. Tagg, and K. R. Matthews. 1991. Genotypic and phenotypic analysis of Streptococcus uberis isolated from bovine mammary secretions. Epidemiol. Infect. 107:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayarao, B. M., and D. R. Wolfgang. 2003. Bulk-tank milk analysis. A useful tool for improving milk quality and herd udder health. Vet. Clin. North Am. Food Anim. Pract. 19:75-92. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen, L. B., K. Poulsen, M. Kilian, and T. E. Petersen. 1999. Purification and cloning of a streptokinase from Streptococcus uberis. Infect. Immun. 67:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalia, A., and D. E. Bessen. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J. Bacteriol. 186:110-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalia, A., M. C. Enright, B. G. Spratt, and D. E. Bessen. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 69:4858-4869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Ke, D., M. Boissinot, A. Huletsky, F. J. Picard, J. Frenette, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Evidence for horizontal gene transfer in evolution of elongation factor Tu in enterococci. J. Bacteriol. 182:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan, I. U., A. A. Hassan, A. Abdulmawjood, C. Lammler, W. Wolter, and M. Zschock. 2003. Identification and epidemiological characterization of Streptococcus uberis isolated from bovine mastitis using conventional and molecular methods. J. Vet. Sci. 4:213-224. [PubMed] [Google Scholar]
- 26.King, S. J., J. A. Leigh, P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lammers, A., C. J. van Vorstenbosch, J. H. Erkens, and H. E. Smith. 2001. The major bovine mastitis pathogens have different cell tropisms in cultures of bovine mammary gland cells. Vet. Microbiol. 80:255-265. [DOI] [PubMed] [Google Scholar]
- 28.Lan, R., and P. R. Reeves. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 29.Leigh, J. A. 1999. Streptococcus uberis: a permanent barrier to the control of bovine mastitis? Vet. J. 157:225-238. [DOI] [PubMed] [Google Scholar]
- 30.Leigh, J. A. 2002. Immunisation of dairy cattle with recombinant Streptococcus uberis GapC or chimeric CAMP antigen confers protection against heterologous bacterial challenge (comment on M.C. Fontaine et al. 2002). Vaccine 20:3047. [DOI] [PubMed] [Google Scholar]
- 31.Leigh, J. A., J. M. Finch, T. R. Field, N. C. Real, A. Winter, A. W. Walton, and S. M. Hodgkinson. 1999. Vaccination with the plasminogen activator from Streptococcus uberis induces an inhibitory response and protects against experimental infection in the dairy cow. Vaccine 17:851-857. [DOI] [PubMed] [Google Scholar]
- 32.Leigh, J. A., P. N. Ward, and T. R. Field. 2004. The exploitation of the genome in the search for determinants of virulence in Streptococcus uberis. Vet. Immunol. Immunopathol. 100:145-149. [DOI] [PubMed] [Google Scholar]
- 33.Matthews, K. R., R. A. Almeida, and S. P. Oliver. 1994. Bovine mammary epithelial cell invasion by Streptococcus uberis. Infect. Immun. 62:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDougall, S., T. J. Parkinson, M. Leyland, F. M. Anniss, and S. G. Fenwick. 2004. Duration of infection and strain variation in Streptococcus uberis isolated from cows' milk. J. Dairy Sci. 87:2062-2072. [DOI] [PubMed] [Google Scholar]
- 35.Meinersmann, R. J., R. W. Phillips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Mastitis Council. 1999. Laboratory handbook on bovine mastitis. National Mastitis Council, Madison, Wis.
- 37.Oliver, S. P., B. E. Gillespie, and B. M. Jayarao. 1998. Detection of new and persistent Streptococcus uberis and Streptococcus dysgalactiae intramammary infections by polymerase chain reaction-based DNA fingerprinting. FEMS Microbiol. Lett. 160:69-73. [DOI] [PubMed] [Google Scholar]
- 38.Phuektes, P., P. D. Mansell, R. S. Dyson, N. D. Hooper, J. S. Dick, and G. F. Browning. 2001. Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J. Clin. Microbiol. 39:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 40.Poyart, C., P. Berche, and P. Trieu-Cuot. 1995. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol. Lett. 131:41-45. [DOI] [PubMed] [Google Scholar]
- 41.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 42.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 43.Smith, A. J., A. J. Kitt, P. N. Ward, and J. A. Leigh. 2002. Isolation and characterization of a mutant strain of Streptococcus uberis, which fails to utilize a plasmin derived beta-casein peptide for the acquisition of methionine. J. Appl. Microbiol. 93:631-639. [DOI] [PubMed] [Google Scholar]
- 44.Smouse, P. E. 2000. Reticulation inside the species boundary. J. Classification 17:165-173. [Google Scholar]
- 45.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, Y. 2004. Negative selection on neutralization epitopes of poliovirus surface proteins: implications for prediction of candidate epitopes for immunization. Gene 328:127-133. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki, Y., T. Gojobori, and M. Nei. 2001. ADAPTSITE: detecting natural selection at single amino acid sites. Bioinformatics 17:660-661. [DOI] [PubMed] [Google Scholar]
- 48.Tapp, J., M. Thollesson, and B. Herrmann. 2003. Phylogenetic relationships and genotyping of the genus Streptococcus by sequence determination of the RNase P RNA gene, rnpB. Int. J. Syst. Evol. Microbiol. 53:1861-1871. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., J. Tang, B. Hunter, and X. C. Zhang. 1999. Crystal structure of streptokinase beta-domain. FEBS Lett. 459:85-89. [DOI] [PubMed] [Google Scholar]
- 50.Ward, P. N., T. R. Field, C. D. Rapier, and J. A. Leigh. 2003. The activation of bovine plasminogen by PauA is not required for virulence of Streptococcus uberis. Infect. Immun. 71:7193-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward, P. N., T. R. Field, E. L. Rosey, A. B. bu-Median, R. A. Lincoln, and J. A. Leigh. 2004. Complex interactions between bovine plasminogen and streptococcal plasminogen activator PauA. J. Mol. Biol. 342:1101-1114. [DOI] [PubMed] [Google Scholar]
- 52.Ward, P. N., and J. A. Leigh. 2004. Genetic analysis of Streptococcus uberis plasminogen activators. Indian J. Med. Res. 119(Suppl.):136-140. [PubMed] [Google Scholar]
- 53.Wieliczko, R. J., J. H. Williamson, R. T. Cursons, S. J. Lacy-Hulbert, and M. W. Woolford. 2002. Molecular typing of Streptococcus uberis strains isolated from cases of bovine mastitis. J. Dairy Sci. 85:2149-2154. [DOI] [PubMed] [Google Scholar]
- 54.Williams, A. M., and M. D. Collins. 1990. Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp. nov. J. Appl. Bacteriol. 68:485-490. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zadoks, R. N., B. E. Gillespie, H. W. Barkema, O. C. Sampimon, S. P. Oliver, and Y. H. Schukken. 2003. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol. Infect. 130:335-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, W., B. M. Jayarao, and S. J. Knabel. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]