Abstract
The accepted limitations associated with classic culture techniques for the diagnosis of invasive fungal infections have lead to the emergence of many non-culture-based methods. With superior sensitivities and quicker turnaround times, non-culture-based methods may aid the diagnosis of invasive fungal infections. In this review of the diagnostic service, we assessed the performances of two antigen detection techniques (enzyme-linked immunosorbent assay [ELISA] and latex agglutination) with a molecular method for the detection of invasive Candida infection and invasive aspergillosis. The specificities for all three assays were high (≥97%), although the Candida PCR method had enhanced sensitivity over both ELISA and latex agglutination with values of 95%, 75%, and 25%, respectively. However, calculating significant sensitivity values for the Aspergillus detection methods was not feasible due to a low number of proven/probable cases. Despite enhanced sensitivity, the PCR method failed to detect nucleic acid in a probable case of invasive Candida infection that was detected by ELISA. In conclusion, both PCR and ELISA techniques should be used in unison to aid the detection of invasive fungal infections.
Advances in medical intervention provide a growing number of compromised patients who are at risk from opportunistic fungal infections. The prophylactic use of azole antifungal drugs has led to the emergence of an increasing number of azole-resistant fungal strains. Diagnostic tests, such as blood culture, show poor sensitivity for the detection of Candida and especially Aspergillus spp. (10). It is therefore necessary to develop and evaluate non-culture-based methods for the detection of systemic fungal infections.
The use of PCR in the detection of systemic fungal infection has been extensively published and provides potential in terms of sensitivity and specificity. A range of PCR targets have been used including cytochrome P450 genes (25), heat shock protein genes (1), and pH regulation genes (17), although primers that amplify the rRNA genes (18S, 28S, and 5.8S rRNA genes) are the most frequently used due to their universal nature and large copy number. A variety of postamplification methods have been used to exploit the variable regions within the rRNA amplicons and identify the genus or species causing infection. These include nested PCR (32, 40), restriction fragment length polymorphism (RFLP) (26), PCR-enzyme-linked immunosorbent assay (ELISA) (20), single-strand confirmation polymorphism (38), hybridization with specific probes (10), and sequencing (8), all of which increase complexity. The most promising PCR technique utilizes fluorescently labeled specific probes and real-time PCR. This removes the need for postamplification handling, reducing both turnaround time and the potential for contamination, and provides a species/genus level of identification, depending on the design of the probe.
Alternatively, serological assays for the detection of invasive fungal infection are commercially available, but they still need to be evaluated on a large scale. Methods have been developed for the detection of both circulating antibodies and antigens, but the usefulness of antibody detection may be limited when the patients under investigation are immunosuppressed and/or heavily colonized but uninfected (42). However, the use of both antigen and antibody detection improves the chances of early detection of systemic candidal infection (43).
Among the various antigens present in the blood of patients suffering from invasive fungal infection, carbohydrates, particularly cell wall components, are favored for the diagnosis of the infection. For the detection of candidal infections, the polysaccharide mannan is a major marker, as it contributes over 7% of the dry weight of the yeast Candida albicans (11), is noncovalently bound to the cell wall and is extremely immunogenic. For the detection of Aspergillus infections, the presence of the carbohydrate galactomannan (GM) is determined. Aspergillus fumigatus releases large quantities of GM into culture media, but it has not been proven whether circulating GM in body fluids is the same as detected in vitro (18). There have been problems with false positives when trying to detect circulating GM. These have been attributed to cross-reactivity with cyclophosphamide (13), adsorption of dietary GM from damaged gut endothelium (19), and β-lactam antibiotic usage (34), but they could also represent subclinical infection or recovery of the patient before confirmation of diagnosis. Many of these problems can be overcome by the screening of multiple sequential samples from the same patient.
Early commercial serological tests utilized antibody-coated latex agglutination, but research evaluating these assays reported that less than 12% of positive cases were detected from single serum samples (12). This unacceptable sensitivity is principally due to the fluctuating levels of antigenemia and the swift removal of mannan and GM from the circulation (12). Improved detection levels for both Candida and Aspergillus have been achieved by the introduction of an ultrasonication step, this concentrates the latex/antigen particles and, in doing so, enhances the sensitivity a reported 250- and 500-fold for mannan and GM, respectively (12).
A more promising serological method is the commercially available sandwich ELISA, which is available for the detection of both Candida and Aspergillus. Again, the assays detect circulating mannan and GM with sensitivity limits of <0.25 ng per ml of serum (28) and 1.0 ng per ml of serum (33), respectively. In terms of specificity, both assays seem accurate, whereas in terms of sensitivity, the performance of these assays varies greatly. The Candida antigen ELISA has a reported sensitivity of 40% when not combined with the antimannan antibody ELISA (31), whereas the Aspergillus assay has been reported to have sensitivities of over 90% for patients with confirmed aspergillosis (histopathology and/or positive blood culture) (22).
This retrospective review evaluated three of the systems, namely latex agglutination, ELISA, and PCR in terms of sensitivity, specificity, simplicity of use (turnaround time), and finally, cost in a high-risk patient group.
MATERIALS AND METHODS
Latex agglutination.
Pastorex kits (Bio-Rad, United Kingdom) were used for the detection of the relevant Candida and Aspergillus antigens. Positive and negative controls were included in the kit and used in every assay. The positive controls were the respective antigens mannan and GM (75 ng/ml) prepared from C. albicans and A. fumigatus, and the negative control was glycine buffer (pH 8.2). An additional negative control in the form of serum from a healthy donor was included in every run.
The method was modified to include an ultrasonication step as described by Grundy et al. (12). Briefly, 300 μl of serum was mixed with 100 μl of treatment reagent and heated to 100°C for 3 min to dissociate immune complexes and to minimize nonspecific reactions. The treated serum was centrifuged for 10 min at 10,000 × g. Forty microliters of the supernatant was mixed gently with 10 μl of the antibody-coated latex particles and taken up into a 2-mm capillary by capillary action. The capillary was positioned in the chamber of the ultrasonication apparatus and exposed to ultrasound for 5 min with an applied transducer voltage of 50 V, peak to peak. After sonication, the reaction mix was eluted and vigorously mixed to detach any aggregates (not actually agglutinated) and examined under a microscope.
Enzyme-linked immunosorbent assay.
Platelia kits (Bio-Rad, United Kingdom) were used for the detection of the relevant Candida and Aspergillus antigens (Ags). Positive and negative controls were included in the kit, but the Platelia Aspergillus Ag also used a threshold serum and the Platelia Candida Ag used a positive-standard serum (mannan, 2 ng/ml), which were used to construct a standard curve. An additional negative control in the form of serum from a healthy donor was included in every run.
The method was as described in the supplier's manual. Briefly, the serum was heat-treated as described above, and then 50 μl of the supernatant was mixed with 50 μl of the peroxidase-labeled anti-GM (mannan) monoclonal antibody in the anti-GM (mannan) antibody-sensitized wells, which were sealed and incubated for 90 min at 37°C. The plates were washed five times and dried before 200 μl of the substrate-chromogen reaction solution was added. The reaction was allowed to develop for 30 min in darkness, before 100 μl of 1.5 N sulfuric acid was added to stop the reaction. The optical density of each well was read at 450/620 nm (OD450/620).
Galactomannan results were determined using a threshold index (OD450/620 of sample/OD450/620 of threshold serum), where an index value above 1.5 corresponded to a positive result. When determining Platelia Candida results, an OD450/620 corresponding to a mannan concentration of 0.5 ng/ml of serum or above was considered positive.
DNA extraction.
DNA was extracted from EDTA-whole blood as described by Einsele et al. (10) using recombinant lyticase (Sigma-Aldrich, United Kingdom) and the QIAmp tissue kit (QIAGEN, United Kingdom) with a minor modification. When precipitating DNA with ethanol, the solution was incubated on ice for 30 min; this was found to increase the yield of the DNA. Positive (10 CFU/ml C. albicans/A. fumigatus)- and negative-control bloods were included in all extraction procedures, and all reagents were filter sterilized through 0.2-μm filters before use.
Primer and probe design.
Panfungal primers (L18F, 5′-CTC GTA GTT GAA CCT TGG; L18R, 5′-GCC TGC TTT GAA CAC TCT) and the Candida-specific probe (Candida, 5′-TTT TGA TGC GTA CTG GAC CCT GT) were designed as described by White et al. (39).
Light-Cycler PCR amplification.
PCR amplification was carried out as described by White et al. (39). The lower limit of sensitivity was 5 CFU/ml, and the Candida probe was specific for Candida albicans, Candida dubliniensis, Candida kefyr, Candida krusei, Candida glabrata, Candida parapsilosis, and Candida tropicalis. The PCR was completed within 45 min.
Aspergillus RFLP digestion.
The PCR amplicons were digested using the restriction enzyme ScaI as described previously (39). In total, 7 species of Aspergillus (Aspergillus candidus, A. fumigatus, Aspergillus flavus, Aspergillus niger, Aspergillus nidulans, Aspergillus oryzae, and Aspergillus terreus) were tested, and when digested, they gave three representative fragments (150, 100, and 50 bp). The PCR amplicons of a further 10 Candida species, Cryptococcus neoformans, and Saccharomyces cerevisiae were also tested, but these did not contain the necessary recognition sequence for ScaI and remained undigested.
Contamination control.
In controlling contamination, minor modifications were applied to the protocol described previously (39). Before any stage of the experiment was performed, all work services including cabinets, pipettes, racks, and microcentrifuges, including rotors and adaptors, were wiped down with Microsol (Anachem, United Kingdom) and DNAzap (Ambion, United Kingdom).
Each stage was carried out in separate laboratories that were independently equipped, including laboratory coats. To prevent DNA carryover, personnel performing the DNA extraction were not allowed to carry out the PCR setup on the same day.
All PCR reagents were aliquoted into single-use sterile tubes in a UV-treated clean cupboard that was also used for PCR master mix setup. The aliquoted master mix was transferred to another room for the addition of the template. After the addition of the DNA, the PCR mix was transferred to the amplification room.
Before any transfer racks were returned to their original rooms, they were soaked in Microsol and wiped down with DNAzap.
Samples.
Clinicians requesting fungal PCR or antigen testing sent 4 ml of EDTA and clotted whole-blood specimens to the laboratory within 1 week of requesting other diagnostic assays (e.g., blood cultures).
Patients.
A group of 105 patients considered to be at high risk for invasive fungal infection were screened using ultrasound enhanced latex agglutination (UELA), ELISA, and PCR. The main body of patients (77) were suffering from hematological malignancies, with 39 patients suffering from leukemia, 19 patients suffering from lymphoma, 5 patients suffering from myeloma, and a further 14 patients receiving bone marrow transplantation. All of the hematology patients were neutropenic (<500 neutrophils per μl of blood) and pyrexial (body temperature of >38°C) and were not responding to antibacterial antibiotics. Other patients, most of whom had at least one of the above host factors, were receiving treatment for a variety of complaints, including respiratory failure, esophageal rupture, cystic fibrosis, diabetes, pneumonia, premature birth, renal failure, and cirrhosis, and two patients were postoperative following cardiac and abdominal surgery.
Using the classifications defined in the EORTC guidelines, the 77 hematology patients were categorized as to the degree of their fungal infection. Proven cases (n = 2; 1 Candida infection and 1 invasive Aspergillus [IA] infection) were defined by histological evidence and/or culture from blood or other sterile sites. Probable cases (n = 15; 13 Candida infections and 2 IA infections) were defined by host factors plus computed tomography/radiological evidence with supporting microbiological signs. The presence of a significant bacteremia caused the probable diagnosis to be reclassified as possible if the organism was resistant to the antibacterial antibiotics prescribed and provided only one organism was isolated from the blood (mixed cultures could represent an infection with more than one causative agent, of which fungi could be included).
Possible cases (n = 18; 14 Candida infections and 4 IA infections) were defined by host factors in the absence of any clinically significant bacteremias plus either microbiological or computed tomography/radiological evidence. Patients originally defined as having a possible infection but with a clinically significant bacteremia were reclassified as at high risk for a fungal infection. All patients with host factors, with or without a bacteremia and no evidence of an invasive fungal infection, were also defined as high risk. In total, 42 hematology patients were categorized as being at high risk for a fungal infection.
For the 28 nonhematological patients, all but one of which were critical care patients, proven cases (n = 1 candidemia) were defined by the isolation of Candida from sterile sites (excluding drain fluids and urine) and by histopathology for IA. Probable invasive Candida infections (n = 5) were classified by the presence of risk factors (surgery, antibiotic use, prematurity, venous catheters, parenteral nutrition, and immunosuppression) with clinical signs of infection (fever unresponsive to antibacterial antibiotics) and Candida colonization at multiple (>2) noncontiguous anatomical sites. Furthermore, in neonates, persistent candiduria was considered significant. Possible invasive Candida infections (n = 4) were defined by clinical signs of infection linked to predisposing conditions such as necrotizing pancreatitis, anastomotic breakdown, or recurrent perforation of the bowel or spontaneous esophageal rupture with no evidence of a clinically significant bacteremia. In the nonhematological patient group, no categories for probable and possible IA were included, and patients who could not be classified proven were considered high risk. Patients with no evidence of fungal colonization, predisposing conditions, or risk factors with or without a clinically significant bacteremia were categorized as high risk. In total, 18 nonhematological patients were categorized as high risk.
RESULTS
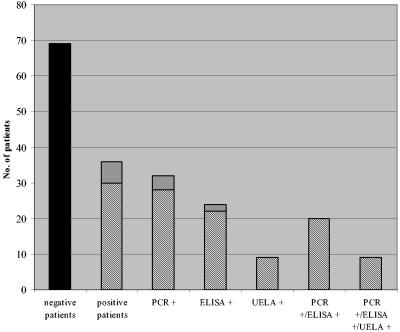
A total of 214 EDTA-whole-blood and serum samples from 105 patients were available for analysis. In total, 36 patients were determined positive by at least 1 of the 3 techniques evaluated. Thirty-two patients were positive by PCR, 24 patients were positive by ELISA, and 9 patients were positive by UELA (Fig. 1). Twenty patients were positive by both PCR and ELISA (Fig. 1). There were 10 PCR-positive-ELISA-negative patients, and a further 2 cases provided samples that were unsuitable for ELISA. Of the 4 ELISA-positive-PCR-negative patients, 2 were caused by unsuitable samples and 2 were genuinely ELISA positive and PCR negative. All samples that were UELA positive were also positive by the other techniques (Fig. 1).
FIG. 1.
Distribution of results for the 105 patients tested for invasive fungal infection by PCR, ELISA, and UELA. Black, negative; light gray, positive for Aspergillus; dark gray, positive for Candida.
Invasive Candida infections.
The majority of the positive patients were positive for Candida (30/36) (Fig. 1). Twenty-eight patients were positive for Candida by PCR, 22 were positive by ELISA, 9 were positive by UELA, and 20 were positive by both PCR and ELISA (Fig. 1 and Table 1).
TABLE 1.
Comparison of PCR and ELISA results for detection of invasive Candida infections in the 105 patients studieda
| PCR result (n) | No. of patients with ELISA result (n)
|
|
|---|---|---|
| Positive (22) | Negative (83) | |
| Positive (28) | 20 | 8 |
| Negative (77) | 2 | 75 |
95% confidence limits, −0.4 to 12.1%.
When these results were collated with the predicted degree of systemic fungal infection for each patient, both patients that had proven candidemias were positive by PCR, whereas ELISA only detected 1 case of proven candidemia and UELA failed to detect any cases (Table 2). There were 18 cases of probable invasive Candida infection, with 17 patients positive by PCR, 14 patients positive by ELISA, and 5 patients positive by UELA. Thirteen patients were positive by both PCR and ELISA (Table 2).
TABLE 2.
Comparison of PCR and ELISA results for detection of invasive Candida infections in the 105 patients categorized by predicted degree of invasive fungal infection
| Predicted degree of infection and PCR result (n) | No. of patients with ELISA result
|
95% confidence limits | |
|---|---|---|---|
| Positive | Negative | ||
| Proven (2) | 1 | 1 | −27.3 to 90.6 |
| Positive (2) | 1 | 1 | |
| Negative (0) | 0 | 0 | |
| Probable (18) | 14 | 4 | −8.9 to 40.7 |
| Positive (17) | 13 | 4 | |
| Negative (1) | 1 | 0 | |
| Possible (18) | 5 | 13 | −12.1 to 32.7 |
| Positive (7) | 4 | 3 | |
| Negative (11) | 1 | 10 | |
| High risk (67) | 2 | 65 | −5.8 to 5.8 |
| Positive (2) | 2 | 0 | |
| Negative (65) | 0 | 65 | |
There were 18 cases of possible invasive Candida infection, with 7 patients positive by PCR, 5 patients positive by ELISA, and 4 patients positive by both of these methods (Table 2). Four patients were positive when tested by UELA. Of the 67 patients with a suspected IA or at high risk for a fungal infection, only 2 were positive for Candida by both PCR and ELISA (Table 2).
The sensitivity of the Candida PCR assay was 95.0% (99.1 to 76.4%, 95% confidence limits) compared to a sensitivity of 75.0% (88.8 to 55.1%, 95% confidence limits) for the Candida ELISA. However, the 20% difference in sensitivities between PCR and ELISA was not statistically different (95% confidence intervals, 42.8 to −4.6%). The specificities of the Candida PCR and ELISA were the same at 97.0% (89.8 to 99.2%, 95% confidence limits). The positive and negative predictive values (PPV and NPV, respectively) for the Candida PCR were 90.5% and 98.5%, respectively, and similar values (88.2% PPV, 92.8% NPV) were calculated for the Candida ELISA. The Candida UELA showed the worst sensitivity (25%), with 95% confidence intervals (−41.3 and −84.4%) confirming that UELA was less sensitive than PCR. However, the Candida UELA was specific (100%).
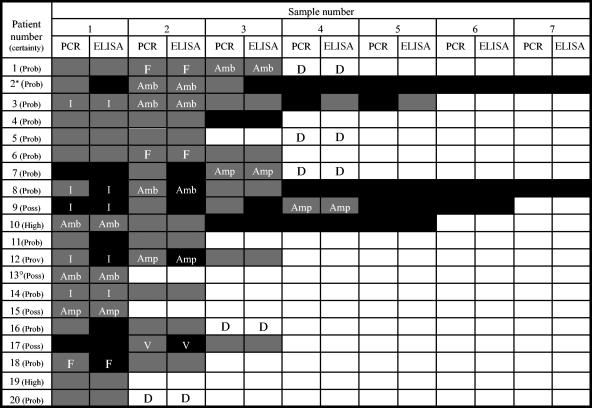
After analysis of the 20 patients that were both PCR and ELISA positive for Candida (1 proven, 13 probable, 4 possible, 2 high risk), it was noted that, in 45% of these patients, the PCR method detected the infection earlier than the ELISA method (Fig. 2). In 3 patients, PCR was positive a mean of 6.3 (±2.9) days prior to the ELISA, and in 6 patients, PCR was positive a mean of 2.2 (±1.8) days ahead of the ELISA (Fig. 2). The other 11 patients became PCR and ELISA positive at the same time. Patients 4, 11, and 19 were positive by both PCR and ELISA, and patient 11 was also positive for Candida by UELA. However, they appeared to be false-positive results, as all recovered without antifungal therapy.
FIG. 2.
Comparison of PCR and ELISA results for the 20 patients who tested positive for invasive Candida infection by both techniques. Black, negative sample; gray, positive sample; white, no sample received; F, fluconazole treatment started; Amp, amphotericin B treatment started; Amb, AmBisome treatment started; I, itraconazole treatment started; V, voriconazole treatment started; D, patient died; Prov, proven candidemia; Prob, probable invasive Candida infection; Poss, possible invasive Candida infection; High, high risk for fungal infection; *, in addition to AmBisome, patient 2 received amphotericin B (nonliposomal) and itraconazole (prophylactic); °, in addition to AmBisome, patient 13 received amphotericin B (nonliposomal), fluconazole, and itraconazole.
Invasive aspergillosis.
Only seven patients had any clinical or microbiological evidence of IA. Four patients were positive by PCR and two patients were positive by ELISA, but no samples were positive for Aspergillus by both techniques (Fig. 1). No samples were positive for Aspergillus by UELA (Fig. 1). Under present European guidelines, the permitted GM cutoff index used to define positivity is 1.5. However, recent research suggests that an index of 0.5 is a more definitive threshold. Reinterpretation of the Aspergillus ELISA results using the 0.5-threshold did not change the overall results, and all negative samples had an index below 0.5.
The PCR detected the one proven case, the two probable cases, and one high-risk case, whereas the ELISA only detected two possible cases. Calculating significant sensitivity values for the Aspergillus assays tested is not plausible due to the small number of proven/probable cases; however, the specificities of the PCR assay and ELISA are high (≥99%).
Bacteremias.
On completion of the fungal study, bacterial blood culture (BC) data from each patient were gathered. Of the 105 patients in the study, 56 had no evidence of bacterial blood-borne infection, while 49 had at least one bacterial BC-positive result, although 24 of these were suspected coagulase-negative staphylococcus skin contaminants and were considered skin contaminants if only a single isolate was obtained. Of the 69 patients negative by all three techniques, 21 (30.4%) had significant bacterial BC-positive isolates, with the remaining 48 patients having no significant isolates. From the 36 patients positive by at least one of three techniques, only 4 (11.1%) had significant positive BC isolates.
When these data were collated with the predicted degree of systemic fungal infection, none of the proven cases of systemic fungal infection had a significant bacterial BC. Only 4 of the 20 probable cases of systemic fungal infection had positive bacterial blood culture, whereas none of the 22 possible cases of systemic fungal infection had a significant bacterial BC. Twenty-one of the 60 high-risk cases had significant positive bacterial BC.
Independent of the predicted certainty of fungal infection, the percentage of clinically significant bacterial BC-positive results was always greater in the cases that were negative by all three techniques than in the cases that were positive by at least one technique, and this was confirmed by 95% confidence intervals (2.2 to 32.8%).
DISCUSSION
This research presents a retrospective comparison of two commercially available antigen detection tests and an in-house PCR assay for the detection of invasive Aspergillus and Candida infections. As only a small number of invasive Aspergillus infections were available, the relevance of the results, in terms of comparison with other Aspergillus studies, is minimal.
However, the benefits of the Aspergillus PCR-RFLP assay are highlighted by the fact that it did detect the only proven case of invasive aspergillosis that was consistently negative by both antigen detection methods and had no ante mortem Aspergillus culture evidence. The case was only proven on autopsy where histopathology discovered septate branching hyphae, vegetation on the mitral valve and papillary muscle, and a well-demarcated lesion in the left lung (24). Conversely, other works (7) have reported that GM ELISA is as sensitive or even more sensitive than the PCR method used. However, as they were describing a rare case of primary digestive aspergillosis, it is difficult to compare the cases.
This highlights the problems when trying to evaluate published research designed to compare ELISA and PCR methodologies. Many groups use different specimen types, with serum rather than EDTA-whole blood often targeted (3, 4, 9, 14, 16, 29, 40, 41). The use of serum over EDTA-whole blood allows the same specimen to be tested by both ELISA and PCR and thus provides the opportunity to perform a direct comparison. Testing serum specimens by PCR relies on the detection of free circulating Aspergillus DNA, as any Aspergillus spores/hyphae present in blood could either be phagocytosed or simply trapped by the clot formation and removed by subsequent centrifugation. The use of EDTA-whole-blood samples allows the detection of all free circulating DNA, spores, or hyphal fragments that have found their way into the bloodstream (36, 37). This could explain why many groups find ELISA more sensitive than PCR when testing serum specimens (4, 7, 9), but this group and others (6, 15) find PCR more sensitive when testing EDTA-whole-blood samples. Alternatively, sample volume could be an issue. As the volumes of serum used for DNA extraction tend to be less than the volumes of EDTA-whole blood, it has been argued that larger blood volumes may increase the sensitivity of the assay due to the higher fungal yield (10). Other research (30) using PCR and GM ELISA to diagnose invasive pulmonary aspergillosis used bronchoalveolar lavage specimens. Both tests showed high rates of sensitivity, and it was suggested that a combined use of the methods might improve diagnosis of invasive pulmonary aspergillosis. However, the clinical relevance of bronchoalveolar lavage specimens is unclear, as false-positive samples due to contamination or colonization may arise (5) and serial testing is difficult due to the invasive nature of the sampling procedure.
Comparing the ELISA versus PCR studies is complicated further by the fact that the majority use an amplification system that is familiar to (usually developed by) them. It would be desirable to evaluate the most widely used PCR protocols by way of multiple consensus studies and in doing so decide on the best system(s) that have been extensively validated. This optimal PCR procedure can then be used in multiple comparison studies with the various antigen tests and a principal method deduced.
Of the 105 patients in the study, 23 were categorized as having a proven/probable invasive fungal (Candida or Aspergillus) infection, 22 were categorized as having a possible infection, and the remaining 60 were thought to be at high risk for a fungal infection. Comparing this research with many of the other published articles, we found that the PCR sensitivity and specificity values were comparable to previous methods (8). Like other groups (16), we found the ELISAs to be less sensitive than PCR; however, they did show good specificity, a detail that has been noted previously in detecting IA by PCR and ELISA (15, 27). Furthermore, the Candida ELISA sensitivity and specificity values presented here compare with those published by Sendid et al. (31), who reported sensitivities and specificities of 80 and 93%, respectively, for the detection of mannanemia when combined with antibody detection. Other groups (2, 35) have reported high sensitivity values (90 to 100%) when using ELISA for the detection of Aspergillus GM. The difference in sensitivity between the Candida PCR and ELISA was deemed to be statistically insignificant. However, clinically, the difference is of value, as the ELISA failed to detect 1 case of proven and 4 cases of probable invasive Candida infection that were detected by PCR. Conversely, the PCR only failed to detect 1 case of probable invasive Candida infection that was detected by the ELISA. Nevertheless, taking the serious nature of these infections into consideration, it is recommended that, if possible, both PCR and ELISA should be utilized when trying to diagnose cases of fungemia.
The Candida UELA assay provided a poor sensitivity (25%), although both the Aspergillus and Candida UELA assays did provide high degrees of specificity (>90%), a trait that has been highlighted previously by other groups (23, 31). The PPVs were similar for the Candida PCR and ELISA, although the NPVs were slightly better for the PCR method. The NPVs for the Candida UELA were always lower than those for the PCR and ELISA; however, the PPVs were comparable with the other methods.
Twenty of the patients studied were positive for an invasive Candida infection by both PCR and ELISA. It was noted that 9 of these patients became PCR positive a mean of 3.5 (±2.9) days prior to becoming ELISA positive, a figure that is comparable to the research of Kami et al. (15), who reported PCR-positive results 2.8 (±4.1) days prior to ELISA positives for the detection of IA. Of the three patients (4, 11, and 19) who appeared to be falsely positive for invasive Candida infection, two (4 and 11) were probable cases. Both had high levels of Candida colonization (Candida was cultured from central line tips, wound swabs, nondirected bronchial lavage fluid, urine, and skin swabs). This high level of candidal colonization could explain why these patients were positive by PCR and ELISA.
The objective of this retrospective study was to evaluate the performances of PCR, ELISA, and UELA in detecting invasive Candida and IA infections. In terms of sensitivity (Candida assays only), the PCR achieved the highest value, although when compared with ELISA, this was not statistically significant. The specificities of all of the assays were similar. In terms of simplicity, i.e., number of steps/time to result, the UELA was the best, taking less than 15 min to complete. The ELISA followed, taking 2 to 3 h, and finally, the PCR method that took 1 day to complete, due to an extensive extraction procedure. The overall cost per sample (excluding the necessary controls and labor costs) is similar at £3.13 to 3.54 and £3.83 for the ELISA and UELA, respectively. The PCR assay is also comparable, with a cost of £3.50 per sample, again excluding labor costs. Introducing a wastage factor (20%) for the use of controls and the labor costs necessary to analyze 10 samples, then the UELA assay becomes the cheapest costing <£6 per sample, followed by the ELISA at £7 and the PCR at >£10.
This PCR assay is a sensitive and specific method for the detection of IA and invasive Candida infection. However, because of the additional concerns over contamination (21), it should only be performed in designated diagnostic molecular biology laboratories. It should be used in parallel with, but not replace, ELISA (9) and has the potential to improve invasive fungal infection diagnosis.
REFERENCES
- 1.Arancia, S., S. Sandini, A. Cassone, F. De Bernardis, and R. La Valle. 1997. Construction and use of PCR primers from a 70 kDA heat shock protein gene for identification of Candida albicans. Mol. Cell. Probes 11:329-336. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. J., S. de Marie, D. Willemse, H. A. Verbrugh, and I. A. J. M. Bakker-Woudenberg. 2000. Quantitative galactomannan detection is superior to PCR in diagnosing and monitoring invasive pulmonary aspergillosis in an experimental rat model. J. Clin. Microbiol. 38:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bialek, R., D. Moshous, J. L. Casanova, S. Blanche, and C. Hennequin. 2002. Aspergillus antigen and PCR in bone marrow transplanted children. Eur. J. Med. Res. 7:177-180. [PubMed] [Google Scholar]
- 4.Bretagne, S., J.-M. Costa, E. Bart-Delabesse, N. Dhédin, C. Rieux, and C. Cordonnier. 1997. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 5.Bretagne, S., J.-M. Costa, A. Marmorat-Khuong, F. Poron, C. Cordonnier, M. Vidaud, and J. Fleury-Feith. 1995. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J. Clin. Microbiol. 33:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchheidt, D., C. Baust, H. Skladny, J. Ritter, T. Suedhoff, M. Baldus, W. Seifarth, C. Leib-Moesch, and R. Hehlmann. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428-435. [DOI] [PubMed] [Google Scholar]
- 7.Chambon-Pautas, C., J.-M. Costa, M-T. Chaumette, C. Cordonnier, and S. Bretagne. 2001. Galactomannan and polymerase chain reaction for the diagnosis of primary digestive aspergillosis in a patient with acute myeloid leukaemia. J. Infect. 43:213-214. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. C. A., C. L. Halliday, and W. Meyer. 2002. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med. Mycol. 40:333-357. [DOI] [PubMed] [Google Scholar]
- 9.Costa, C., J.-M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Einsele, H., H. Herbart, G. Roller, J. Löffler, I. Rotherhöfer, C. A. Müller, R. A. Bowden, J.-A. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukazawa, Y. 1989. Antigenic structure of Candida albicans. Immunochemical basis of the serological specificity of the mannans in yeasts. Immunol. Ser. 47:37-62. [PubMed] [Google Scholar]
- 12.Grundy, M. A., R. A. Barnes, and W. T. Coakley. 1995. Highly sensitive detection of fungal antigens by ultrasound-enhanced latex agglutination. J. Med. Vet. Mycol. 33:201-203. [DOI] [PubMed] [Google Scholar]
- 13.Hashiguchi, K., Y. Niki, and R. Soejima. 1994. Cyclophosphamide induces false-positive results in detection of Aspergillus antigen in urine. Chest 105:975-976. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, A., Y. Yamakami, P. Kamberi, E. Yamagata, R. Karashi. H. Nagaoka, and M. Nasu. 1998. Comparison of PCR, (1→3)-beta-D-glucan and galactomannan assays in sera of rats with experimental invasive aspergillosis. J. Clin. Lab. Anal. 12:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 16.Kawamura, S., S. Maesaki, T. Noda, Y. Hirakata, K. Tomono, T. Tashiro, and S. Kohno. 1999. Comparison between PCR and detection of antigen in sera for diagnosis of pulmonary aspergillosis. J. Clin. Microbiol. 37:218-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurzai, O., W. J. Heinz, D. J. Sullivan, D. C. Coleman, M. Frosch, and F. A. Mühlschlegel. 1999. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J. Clin. Mircobiol. 37:1587-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lescher-Bru, V., A. Cavalier, E. Pernot-Marino, H. Koenig, D. Eyer, J. Waller, and E. Condolfi. 1998. Aspergillus galactomannan antigen detection with Platelia® Aspergillus: multiple positive antigenemia without Aspegillus infection. J. Mycol. Med. 8:112-113. [Google Scholar]
- 20.Löeffler, J., H. Herbart, S. Sepe, U. Schumacher, T. Klingebiel, and H. Einsele. 1998. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med. Mycol. 36:275-279. [DOI] [PubMed] [Google Scholar]
- 21.Löeffler, J., H. Herbart, R. Bialek, L. Hagmeyer, D. Schmidt, F. P. Serey, M. Hartmann, J. Eucker, and H. Einsele. 2000. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 38:3830-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maertens, J., J. Van Eldere, J. Verhaegen, E. Verbeken, J. Verschakelen, and M. Boogaerts. 2002. Use of circulating galactomannan screening for early diagnosis of invasive aspergillosis in allogeneic stem cell transplant recipients. J. Infect. Dis. 186:1297-1306. [DOI] [PubMed] [Google Scholar]
- 23.Manso, E., M. Montillo, G. De Sio, S. D'Amico, G. Discepoli, and P. Leoni. 1994. Value of antigen and antibody detection in the serological diagnosis of invasive aspergillosis in patients with haematological malignancies. Eur. J. Clin. Microbiol. Infect. Dis. 13:756-760. [DOI] [PubMed] [Google Scholar]
- 24.McCracken, D., R. Barnes, C. Poynton, P. L. White, N. Isik, and D. Cook. 2003. Polymerase chain reaction aids in the diagnosis of an unusual case of Aspergillus niger endocarditis in a patient with acute myeloid leukaemia. J. Infect. 47:344-347. [DOI] [PubMed] [Google Scholar]
- 25.Morace, G., M. Sanguinetti, B. Posteraro, G. Lo Cascio, and G. Fadda. 1997. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J. Clin. Microbiol. 35:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morace, G., L. Pagano, M. Sanguinetti, B. Posteraro, L. Mele, F. Equitani, G. D'amore, G. Leone, and G. Fadda. 1999. PCR-restriction enzyme analysis for detection of Candida DNA in blood from febrile patients with haematological malignancies. J. Clin. Microbiol. 37:1871-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinel, C., H. Fricker-Hidalgo, B. Lebeau, F. Garban, R. Hamidfar, P. Ambroise-Thomas, and R. Grillot. 2003. Detection of circulating Aspergillus fumigatus galactomannan: value and limits of the Platelia test for diagnosing invasive aspergillosis. J. Clin. Microbiol. 41:2184-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson, M. D., and M. H. Kokki. 1999. New Perspectives in the diagnosis of systemic fungal infections. Ann. Med. 31:327-333. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, T., K. Ikegami, E. Yoshinaga, R. Uesugi-Hayakawa, and A. Wakizaka. 2000. Rapid, sensitive and simple detection of Candida deep mycosis by amplification of 18S ribosomal RNA gene; comparison with assay of serum beta-D-glucan level in clinical samples. Tohoku J. Exp. Med. 190:119-128. [DOI] [PubMed] [Google Scholar]
- 30.Sanguinetti, M., B. Posteraro, L. Pagano, G. Pagliari, L. Fianchi, L. Mele, M. La Sorda, A. Franco, and G. Fadda. 2003. Comparison of real-time PCR, conventional PCR and galactomannan antigen detection by enzyme-linked immunosorbent assay using bronchoalveolar lavage fluid samples from hematology patients for diagnosis of invasive pulmonary aspergillosis. J. Clin. Microbiol. 41:3922-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sendid, B., M. Tabouret, J. L. Poirot, D. Mathieu, J. Fruit, and D. Poulain. 1999. New enzyme immunoassays for sensitive detection of circulating Candida albicans mannan and antimannan antibodies: useful combined test for diagnosis of systemic candidiasis. J. Clin. Microbiol. 37:1510-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skladny, H., D. Buchheidt, C. Baust, F. Krieg-Schneider, W. Seifarth, C. Leib-Mösch, and R. Hehlmann. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stynen, D., J. Sarfati, F. Symoens, A. Goris, N. Nolard and J-P. Latge. 1991. Rat monoclonal antibodies against exocellular carbohydrate antigens of Aspergillus and dermatophytes, p. 181-193. In J.-P. Latge and D. Boucias (ed.), Fungal cell wall and immune response. Springer-Verlag, Berlin, Germany.
- 34.Sulahian, A., S. Touratier, and P. Ribaud. 2003. False positive test for Aspergillus antigenaemia related to concomitant administration of piperacillin and tazobactam. N. Engl. J. Med. 349:2366-2367. [DOI] [PubMed] [Google Scholar]
- 35.Swanink, C. M. A., J. F. G. M. Meis, A. J. M. M. Rijs, J. P. Donnelly, and P. E. Verweij. 1997. Specificity of a sandwich enzyme-linked immunosorbent assay for detecting Aspergillus galactomannan. J. Clin. Microbiol. 35:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Burik, J., D. Myerson, R. Schreckhise, and R. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verweij, P. E., J. Figueroa, J. van Burik, M. D. Holdom, E. Del-Cas, B. L. Gómez, and M. J. Mendes-Giannini. 2000. Clinical applications of non-culture based methods for the diagnosis and management of opportunistic and endemic mycoses. Med. Mycol. 38(Suppl. 1):161-171. [PubMed] [Google Scholar]
- 38.Walsh, T. J., A. Francesconi, M. Kasai, and S. J. Chanock. 1995. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J. Clin. Microbiol. 33:3216-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, P. L., A. Shetty, and R. A. Barnes. 2003. Detection of seven Candida species using the Light-Cycler system. J. Med. Microbiol. 52:229-238. [DOI] [PubMed] [Google Scholar]
- 40.Williamson, E. C. M., J. P. Leeming, H. P. Palmer, C. G. Steward, D. Warnock, D. I. Marks, and M. R. Millar. 2000. Diagnosis of invasive aspergillosis in bone marrow transplant recipients by polymerase chain reaction. Br. J. Haematol. 108:132-139. [DOI] [PubMed] [Google Scholar]
- 41.Yamakami, Y., A. Hashimoto, E. Yamagata, P. Kamberi, R. Karashima, H. Nagai, and M. Nasu. 1998. Evaluation of PCR for detection of DNA specific for Aspergillus species in sera of patients with various forms of pulmonary aspergillosis. J. Clin. Microbiol. 36:3619-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo, S. F., and B. Wong. 2002. Current status of non-culture methods for diagnosis of invasive fungal infections. Clin. Microbiol. Rev. 15:465-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yera, H., B. Sendid, N. Francois, D. Camus, and D. Poulain. 2001. Contribution of serological tests and blood culture to the early diagnosis of systemic candidiasis. Eur. J. Clin. Microbiol. Infect. Dis. 20:864-870. [DOI] [PubMed] [Google Scholar]