Abstract
Many systems have been designed for the detection of SARS-CoV-2, which is the virus that causes COVID-19. SARS-CoV-2 is readily transmitted, resulting in the rapid spread of disease in human populations. Frequent testing at the point of care (POC) is a key aspect for controlling outbreaks caused by SARS-CoV-2 and other emerging pathogens, as the early identification of infected individuals can then be followed by appropriate measures of isolation or treatment, maximizing the chances of recovery and preventing infectious spread. Diagnostic tools used for high-frequency testing should be inexpensive, provide a rapid diagnostic response without sophisticated equipment, and be amenable to manufacturing on a large scale. The application of these devices should enable large-scale data collection, help control viral transmission, and prevent disease propagation. Here we review functional nanomaterial-based optical and electrochemical biosensors for accessible POC testing for COVID-19. These biosensors incorporate nanomaterials coupled with paper-based analytical devices and other inexpensive substrates, traditional lateral flow technology (antigen and antibody immunoassays), and innovative biosensing methods. We critically discuss the advantages and disadvantages of nanobiosensor-based approaches compared to widely used technologies such as PCR, ELISA, and LAMP. Moreover, we delineate the main technological, (bio)chemical, translational, and regulatory challenges associated with developing functional and reliable biosensors, which have prevented their translation into the clinic. Finally, we highlight how nanobiosensors, given their unique advantages over existing diagnostic tests, may help in future pandemics.
Keywords: biosensors, low-cost diagnostics, point of care, rapid testing, nanomaterials, SARS-CoV-2, COVID-19, future pandemics
Graphical abstract
1. INTRODUCTION
In response to a pandemic threat, the rapid identification and detection of disease-causing pathogens are critical public health measures. Technologies capable of rapid detection would help protect uninfected individuals against viral transmission while accelerating the response to epidemics or pandemics, such as that caused by SARS-CoV-2. Public health interventions aimed at controlling disease spread include physical distancing, evaluating the severity of the disease, and treating patients accordingly.1,2 In the case of SARS-CoV-2, the protocol recommended by the Centers for Disease Control and Prevention (CDC) and other health organizations is the accurate detection of SARS-CoV-2 nucleic acids present in oropharyngeal/nasopharyngeal (OP/NP) fluids by using real-time reverse transcription-polymerase chain reaction (RT-PCR).2 Indeed, several RT-PCR-based diagnostic kits for SARS-CoV-2 detection are already available; however, they have several drawbacks. These kits are costly and time-consuming to operate. Furthermore, advanced equipment and skilled staff are needed to process test results.1 The high cost of diagnosis and the scarcity of test kits, especially in developing countries, hinder the continual monitoring of community transmission and their widespread use.
The case of SARS-CoV-2 is particularly problematic because this virus can be transmitted from people who are presymptomatic, symptomatic, or asymptomatic; thus, identifying infected subjects based on symptoms alone (and then isolating them) is insufficient to stop the virus from spreading. Population-wide screening tests to detect infected individuals are the most effective way to break the chain of transmission and stop the pandemic.3 The development of advanced and accessible POC devices to test both symptomatic and asymptomatic individuals is required to quickly limit viral transmission in human populations. These diagnostic tools should be inexpensive and easy to use, allowing frequent testing, i.e., multiple tests per week.2,4
Inexpensive tests with rapid outcomes are of the utmost importance in areas where there are accelerated rates of transmission of COVID-19 (or other infectious diseases) because they allow massive and periodic community testing in decentralized testing sites and can be used frequently. Infections can be detected with such tests in a timely manner, so that even if the tests do not meet the benchmark analytic limit of detection, infections can be contained by steps that can be taken immediately.4 Additionally, rapid and cost-effective testing can minimize the economic effects of the pandemic, because there is less disruption of usual activities.5 In addition to the low market cost, if such testing devices are portable and easy to manufacture at a large scale, they are more likely to be widely used.
The COVID-19 outbreak challenged the scientific community to respond rapidly by developing and improving rapid diagnostics, vaccines, medicines, and functional protective materials.6–8 Advances in the fields of materials science, microfabrication, and sensing have enabled the development of several diagnostic methods that hold promise for future pandemics. These technologies overcome the limitations of traditional methods by providing fast clinical outcomes, reduced cost, easy operation, reproducibility, and precision. Optical, mainly colorimetric, and electrochemical biosensors stand out as effective options because of their well-established fabrication and functionalization techniques, such as screen printing, inkjet printing, roll-to-roll, and 3D printing. Additionally, the use of low-cost and accessible materials, such as paper substrates, allows portability and easy disposal of the test after use.9 These devices can be miniaturized and require only a small volume (microliters) of biofluids to perform accurate bioassays with readout directly on a smart device, such as a cell phone.10–14 Functional materials can be combined to enable catalytic responses with enhanced sensitivity and large surfaces, on which biological receptors (enzymes, antibodies, and aptamers) can be immobilized. Such materials provide adequate selectivity and analytical parameters for diagnostic tests.
Thus, in this review, we focus on the use of cost-effective nanomaterial-based optical and electrochemical approaches for the development of accessible diagnostic tests for COVID-19 with the idea that these technologies may be applicable in future pandemics. We describe nanomaterials coupled with paper-based analytical devices and other convenient platforms with emphasis on the use of traditional lateral flow technology (antigen and antibody immunoassays) and innovative biosensing methods. In summary, nanomaterials enhance the biosensing response (signal transduction) and, consequently, method sensitivity. The main benefits of nanomaterials are their higher surface area/volume ratio, which increases the detectability of the assay compared to bulk equivalents, as well as their unique optical, plasmonic, and electrical properties.15,16 However, using nanomaterials presents several challenges in developing diagnostics in terms of scalability, mass production, and sensitivity. Therefore, we have included critical discussions about the need for controlled synthesis, strategies for the biofunctionalization of nanomaterials (i.e., mainly metallic nanoparticles), and the importance of evaluating their stability and homogeneity in order to build reproducible and reliable biosensing diagnostic tests.
We highlight diagnostic methods that meet the ASSURED criteria, i.e., that have the following features: Affordable, Sensitive, Specific, User-friendly, Rapid and robust, sophisticated Equipment-free, and Delivered to the end-users.17,18 Currently, there is not a consensus or well-established cutoff value in the literature for considering a diagnostic test low-cost. The World Health Organization (WHO), through the ASSURED criteria, suggests typical values of US $1.00 to US $10.00 per test to be considered affordable, varying from rapid antigen tests to molecular assays and depending on the disease evaluated.18 In 2020, a global partnership guided by WHO established efforts to provide affordable tests for COVID-19 at prices lower than US $5.00 per test aiming for applications in low- to middle-income countries.19 In addition, a modeling study comparing different testing scenarios, showed that the frequent use (weekly test) of US $5.00 per test is more cost-effective than the status-quo strategy of symptom-based testing and isolation.20 Therefore, for point-of-care applications, we estimate the cutoff value as US $5.00 per biosensor test to consider it a low-cost solution. However, it is important to highlight that few studies have thus far described the estimated costs of diagnostics tests. Moreover, reported costs are not standardized and vary depending on the manufacturer, scale, and factors such as labor intensity, fabrication, and commercialization costs. We also provide recommendations, key challenges, and future directions needed to translate biosensors into rapid and reliable diagnostic tests that are scalable and widely used in the clinic and that may help in upcoming pandemics and outbreaks.
2. SARS-COV-2 STRUCTURE AND BIOMARKERS FOR COVID-19 DIAGNOSIS
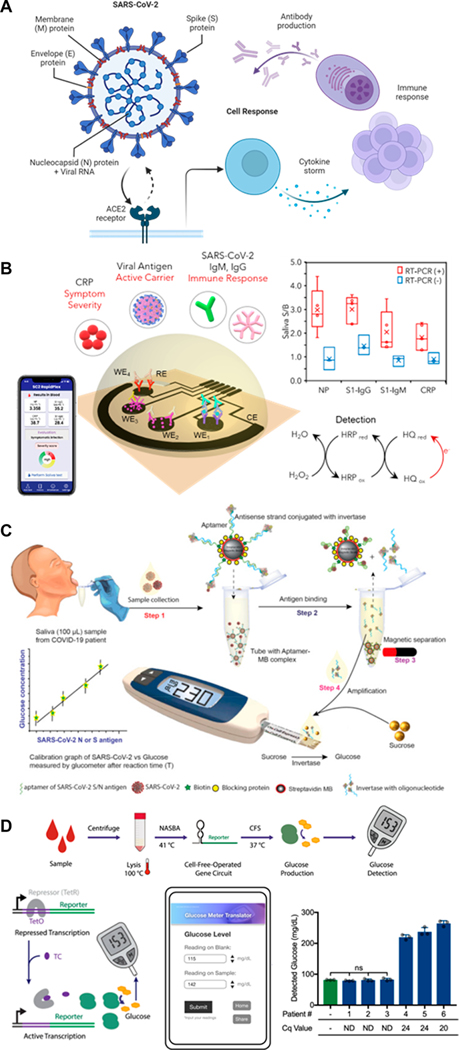
SARS-CoV-2 is an enveloped positive-sense single-stranded RNA virus with nanometric dimensions (80–120 nm diameter)21 whose genetic material is wrapped within lipid bilayers. SARS-CoV-2 RNA encodes four major structural proteins: the spike protein (S protein), which binds to the host receptor resulting in the consequent entry of the virus to the host cell; the nucleocapsid protein (N protein), which encapsulates the viral RNA; and the membrane (M) and envelope (E) proteins that enable morphogenesis of the viral envelope (Figure 1A).21,22 Thus, S, M, and E are readily available, whereas N is accessible after the viral particle is lysed; i.e., methods for detecting N protein require sample treatment prior to the measurements.
Figure 1.
Enzyme-linked signal transduction for SARS-CoV-2 detection. (A) SARS-CoV-2 structure and downstream biomarkers of SARS-CoV-2 infection. Created with BioRender.com. (B) Laser-engraved graphene-based immunosensor for the rapid, multiplexed detection of SARS-CoV-2 antigen, antibodies, and C-reactive protein as demonstrated in saliva. Reproduced with permission from ref 31. Copyright 2020 Matter. (C) Aptamer-based detection of SARS-CoV-2 antigen in saliva with invertase-facilitated signal amplification and glucometer measurement. Reproduced with permission from ref 74. Copyright 2021 Elsevier. (D) A cell-free gene circuit sensor combining synthetic biology and an off-the-shelf glucometer for the detection of SARS-CoV-2 nucleic acids. Reproduced with permission from ref 75 under CC BY 4.0. Copyright 2021 Springer Nature.
The genomic material and structural proteins of SARS-CoV-2 have been investigated as antigenic biomarkers for detection by nanomaterial-based biosensors. The most common biomarkers for targeted diagnosis are antigens, antibodies, and receptors, each of which represents specific responses to the virus by the human host. Molecules derived from inflammatory processes (e.g., C-reactive protein, serum amyloid A, interleukin 6, lactate dehydrogenase, cardiac troponin, and D-dimer) or that are expressed or suppressed in tissues and organs such as blood, kidneys, heart, and liver, have also been explored for disease diagnosis.12,23 The main antigens, antibodies, and receptors used as biomarkers for the detection of SARS-CoV-2 and for diagnosing COVID-19 are given in Table 1.
Table 1.
Examples of Biomarkers Used for the Detection of SARS-CoV-2 by Nanomaterials-Based Biosensorsa
| biological target | technique | time (min) | reference(s) |
|---|---|---|---|
| S and N genes | PN | 10 | 24 |
| DPV | 120 | 25 | |
| ORF1ab and N genes | LSPR | 2 | 26 |
| RdRp | LSPR | 2 | 26 |
| S protein | EIS | 4 | 27,28 |
| SWV | 6.5 | 29 | |
| PN | 5 | 30 | |
| N protein | DPV and OCP-EIS | 10 | 31 |
| Immunoglobulins G and M antibodies | LFIA DPV and OCP-EIS |
15 10 |
32,33 31 |
| SWV | 45 | 34 |
PN, plasmonic nanoparticles; DPV, differential pulse voltammetry; LSPR, localized surface plasmon resonance; EIS, electrochemical impedance spectroscopy; SWV, square wave voltammetry; OCP-EIS, open-circuit potential-electrochemical impedance spectroscopy; LFIA, lateral flow immunoassay. RdRp, RNA-dependent RNA polymerase.
Nanomaterials developed for the selective detection of SARS-CoV-235 provide the basis for a multitude of POC devices for COVID-19 diagnosis.36 The effectiveness of POC biosensors, as well as their testing time, depends on identifying specific molecular biomarkers and using adequate receptors. Examples of biomarker-based nanomaterials used in biosensors are described below.
2.1. Protein Biomarkers.
The S protein is the biomarker that has been the most extensively leveraged for SARS-CoV-2 detection. It has 66 N-glycosylation sites37 and comprises three subunits: S1, S2, and a single transmembrane anchor.38 The S protein selectively binds to human angiotensin-converting enzyme 2 (ACE2), a cellular surface receptor, through its receptor-binding domain (RBD).39 This binding is essential for membrane fusion between the virus and the host cell.40 Numerous SARS-CoV-2 variants have mutations in the S protein, affecting the interaction of the virus with ACE2. However, variation in the S protein often leads to increased binding affinity, thus, not altering the ability of ACE2-based biosensors to accurately detect the viral infection.41,42 Therefore, native ACE227,29,30 and synthetic variants43–46 have been used to trap or selectively bind to SARS-CoV-2 viral particles on the surface of nanomaterials used to coat POC devices.27,29,30,47,48 The ACE2 receptor (or a variant thereof), in the form of a purified protein, is often covalently anchored onto the nanomaterial surface through amide or thiol bonds. As the binding of the S protein is highly dependent on the receptor’s conformational integrity, strategies for stabilizing ACE2, such as generating stable constrained derivatives or using ACE2-based molecules with increased binding affinity compared to native ACE2, are among the most common ways to construct nanomaterial-based biosensors that detect SARS-CoV-2. These biosensors, which consist of functionalized nanoparticles, nanofilms, or electrodes,39,49 have been applied to POC devices that detect S protein by techniques ranging from basic colorimetric assays to more complex and more sensitive electrochemical methods, e.g., electrochemical impedance spectroscopy and square-wave voltammetry.
The N protein is strongly immunogenic, and its sequence is highly conserved. These features allow its use as a diagnostic biomarker for screening for SARS-CoV-2 using nanomaterial-based biosensors.39,50 Nucleocapsid antibodies, similarly to the binding of the ACE2 receptor to protein S, have been widely used for highly specific POC devices. Like ACE2 and its variants, the nucleocapsid antibody is usually anchored onto the nanomaterial surface via chemical modification. Activation of the responsive surface makes it possible to accurately detect very small amounts of virus, corresponding to a limit of detection (LOD) of ≈ fg mL−1.39
Antibodies such as immunoglobulins G (IgG) and M (IgM) have been widely used for a large variety of COVID-19 tests. However, immunoglobulins often do not appear immediately in the patient’s blood, and early infections can be misdiagnosed. More sensitive POC devices for IgG and IgM detection are commonly manufactured with complementary SARS-CoV-2 antigens and antihuman immunoglobulins functionalized onto nanoparticles and used in colorimetric assays. For instance, a lateral flow immunoassay (LFIA) was developed by using antigen-decorated gold nanoparticles (AuNPs) that detected IgM and IgG antibodies against SARS-CoV-2 with a clinical detection sensitivity of 88.66% and a specificity of 90.63%.32 The diagnostic device was made by immobilizing antihuman-IgM and antihuman-IgG on nitrocellulose membrane-based test strips, which captured AuNP-S protein-IgM and AuNP-S protein-IgG conjugates from the flowing blood sample. Detection of SARS-CoV-2 with this device took 15 min.
2.2. Nucleic Acid Biomarkers.
Nucleic acid testing is the basis of RT-PCR diagnostic methods, which are performed in well-equipped and relatively automated clinical settings.51 For POC applications, several approaches have been developed to translate PCR-based diagnostic methods to more suitable biosensor technologies to enable decentralized and frequent COVID-19 diagnostics. The detection of SARS-CoV-2 RNA requires specific and highly sensitive assays, such as LFIA, colorimetric, and multifunctional assays, which are mainly associated with amplification techniques, such as isothermal amplification. Microfluidic systems have many applications in molecular diagnostics, with an increasing number of solutions relying on microfluidic-related platforms with nucleic acid amplification stages and testing.52,53
Alternative strategies for viral detection include SARS-CoV-2 RNA-based nanomaterials, which have been used as biosensor-coating elements that combine a plasmonic photothermal (PPT) effect and a localized surface plasmon resonance (LSPR) effect.26 The biosensor was designed such that a thermally dewetted gold nanofilm self-assembles onto a BK7 glass surface. Two-dimensional gold nanoisland (AuNI) chips were incorporated and then functionalized with the complementary DNA receptors, leading to nucleic acid hybridization and resulting in the sensitive detection of SARS-CoV-2. To enhance the sensing properties, the laser beam was positioned such that the plasmonic resonances of PPT and LSPR were excited at two different wavelengths. This in situ PPT enhancement, by improving the hybridization kinetics, improved the specific nucleic acid detection; in fact, gene sequences in the pmol L−1 concentration range could be accurately discriminated.26
One challenge in using nucleic acid biomarkers is their degradation during the storage and transport of samples or swabs. RNA, unlike DNA, is easily degraded. As a result, the sample storage, handling, and RNA isolation stages must be optimized to minimize RNA loss owing to degradation at each step.54 Another important point is that viruses continually mutate, necessitating protocol adaptations of diagnostic methods. Recent studies on the primers and probes used for RT-PCR revealed that certain genetic mutations hampered the sensitivity of the reverse primer for RNA-dependent RNA polymerase (RdRp) and the forward primer targeting the N gene.51,55,56 Sensitivity can be maximized by utilizing conserved regions of the nucleic acids wherever feasible. The accurate tracking of primer and probe changes may make it easier to develop tests as well as to redesign and adjust existing ones. The development of platforms that are adaptable and that can target multiple biomarkers of the virus should be prioritized.51
Recent advances have been made in the detection of SARS-CoV-2 through nucleic acid sensing. For example, CRISPR-Cas-based diagnostics have emerged as robust next-generation platforms. Combining CRISPR with isothermal techniques and a lateral flow readout, CRISPR-based diagnostic systems can match the accuracy of RT-qPCR and the speed of currently available lateral flow rapid tests, while overcoming the limitations of both platforms.57 Additionally, interferon-stimulated genes (ISG)-based COVID-19 diagnostics have been developed to achieve higher sensitivity and specificity compared to RT-qPCR. This technique is less likely to produce false negatives, is flexible regarding the type of sample (e.g., compatible with saliva, blood, and respiratory specimens), and can detect the virus at lower concentrations.58
3. BIOFLUIDS: PROS AND CONS
OP/NP biofluid samples have been largely used for the detection of COVID-19, mainly for RT-PCR as the gold standard diagnostic protocol. Nevertheless, the large-scale, repetitive monitoring of viral loads by collecting OP/NP biofluid samples is not effective for tracking infection levels in populations.59 Healthcare staff involved in the collection of OP/NP specimens risk accidental transmission because of contact with infected patients during specimen collection and the likelihood of exposure to virus particles transmitted by sneezing, coughing, or gag reflex. Furthermore, the collection of specimens may be painful for patients or result in bleeding, especially for those with thrombocytopenia.59–61 The use of OP/NP swabs by patients themselves may lead to misdiagnosis,28 and so biofluid collection would ideally be restricted to trained operators. Other biofluids, such as sputum, transtracheal aspirates, or bronchoalveolar lavage fluid samples, are commonly assessed in clinical practice for respiratory disease monitoring; however, these samples are unsuitable for SARS-CoV-2 diagnostic evaluation because sample collection is invasive and very low viral titers are collected from asymptomatic subjects.61,62 In general, viral shedding in throat swabs and sputum samples is at a maximum 5 to 6 days after symptoms begin.2
Blood sample analyses are of limited use for SARS-CoV-2 detection because sample collection is invasive, sample manipulation is laborious (samples are also processed for use as serum or plasma), and storage and pretreatment are required before diagnostic tests can be performed. Thus, for COVID-19-related tests, blood samples have been used mainly to test for antibodies to SARS-CoV-2, i.e., serological testing, which is usually performed after full recovery from COVID-19. Antibody tests involve evaluating antibody levels using enzyme-linked immunosorbent assays (ELISAs) and LFIA technologies.63
Saliva is a convenient biofluid for less invasive diagnosis and presents several advantages in terms of sample collection (e.g., samples can be taken repeatedly), manipulation, processing, safety, longitudinal monitoring, application to large populations, and transportation.59 Saliva, unlike blood, does not coagulate and remains stable for 24 h at room temperature and up to 1 week at 4 °C, so sampling can be done at home or at the POC. Saliva specimens can be obtained by asking patients to spit into a sterile collection tube, a procedure that is neither invasive nor painful.61 Furthermore, such sampling procedures minimize the chance of exposing healthcare workers to infected samples at hotspot test points. Additionally, a recent study demonstrates that the mean SARS-CoV-2 titers (virus copies mL−1) can be as much as five times higher in saliva compared with nasopharyngeal swabs,64 highlighting the practicality of saliva for high-frequency diagnosis. However, it is important to establish a protocol to collect saliva prior to the ingestion of liquids and foods to avoid dilution and biofluid contamination.
4. ELECTROCHEMICAL METHODS
Electrochemical methods have emerged as pivotal tools for biosensing applications, offering exceptional sensitivity and selectivity, mainly associated with nanomaterials and biological receptors. Leveraging the interactions between biological molecules and functionalized conductive surfaces, these methods enable the conversion of biochemical events to measurable electrical signals. The principles underlying electrochemical biosensors encompass a diverse array of techniques including amperometry, potentiometry, voltammetry, and impedance spectroscopy. With the ability to provide rapid, label-free, and real-time monitoring, electrochemical biosensing enables innovative solutions to some of the most pressing challenges in health, i.e., cost-effective and decentralized diagnostics.
4.1. Nanomaterials for Electrochemical Sensing Platforms.
Nanomaterials have been applied to SARS-CoV-2 electrochemical sensing schemes to efficiently transduce the signal generated upon the selective biorecognition of SARS-CoV-2 biomarkers. Nanostructured materials overcome some of the limitations of electrochemical sensing by enhancing the signal-to-noise ratio. Nanomaterials may improve the electrochemical performance in part because they have large specific surface areas and high charge carrier mobility.
Nanostructures may also impart antifouling properties to the electrode, improving performance in complex biological media.65 Gold nanostructures may be formed via bottom-up or top-down synthesis for improved electrochemical performance. Zero-dimension (0D) AuNPs may be used to create additive nanostructures on the electrode surface. AuNPs may be deposited onto relatively inert conductive materials to provide a chemically reactive surface for functionalization via Au-thiol binding. For example, ACE2-coated AuNPs were anchored onto inexpensive graphite electrodes to develop a Low-cost Electrochemical Advanced Diagnostic (LEAD) for robust SARS-CoV-2 sensing.29 In contrast, spongelike nanoporous gold is fabricated subtractively by etching gold–metal alloys. By alteration of the fabrication parameters, tunable porosity and behavior may be achieved. Beyond increasing the electroactive surface area, it has also been found that nanopore structures alter surface charge screening, limiting the Debye effect and enhancing electron transfer at small pore sizes.66 High-throughput laser interference lithography was used by Yoon et al. to fabricate nanoporous gold electrode arrays for their SARS-CoV-2 nucleic acid sensor.67
Carbon-based nanomaterials are an inexpensive alternative to noble metals and have valuable physicochemical properties for biosensing. Carbon allotropes exhibit distinct properties at each level: 1D, carbon nanotubes; 2D, graphene; and 3D, graphite. As a 2D nanomaterial, graphene demonstrates exceptional conductivity with a high surface-to-volume ratio. Graphene is a highly scalable nanomaterial synthesized by chemical vapor deposition, including CO2 laser engraving.31
Graphene may be functionalized through covalent interactions at planar edges and defects, which at high quantities may adversely impact conductivity. Instead, graphene functionalization relies on π–π stacking and van der Waals interactions, with polymer deposition introducing desired functional groups for covalent biomolecule immobilization.68 Graphene remains one of the most common nanomaterials used in electrochemical sensing. Given its low cost, high performance, and mass-manufacturability, graphene is an advantageous nanomaterial for SARS-CoV-2 biosensor design. One example of its use is the field-effect transistor (FET)-based device to detect SARS-CoV-2 in human nasopharyngeal swabs.69 This device was developed by transferring graphene onto a SiO2/Si substrate by the wet-transfer method. Subsequently, the graphene surface was coated with a specific antibody for the S protein, resulting in the highly sensitive detection of SARS-CoV-2 (from 1 to 100 fg mL−1).
MXenes are 2D nanomaterials composed of transition-metal elements (Mn+1) incorporated into carbon or nitrogen elements (Xn). MXenes are typically produced via chemical etching of a silicon- or aluminum-based precursor, resulting in the formation of various chemically reactive terminal groups (e.g., oxygen, hydroxyl, or fluorine). MXene is an attractive nanomaterial as it has high conductivity and forms stable covalent bonds via reactive surface functional groups. Given these properties, MXene may improve signal-to-noise ratios in comparison to graphene. MXenes, however, are typically made up of small flakes (approximately 3 μm laterally), resulting in increased discontinuities. Li et al.70 improved the continuity of the structure by using a thin film composite of graphene and MXene, producing a high-performance electrochemical transduction material within the transistor they designed.
4.2. Point-of-Care Electrochemical Sensing Platforms.
Electrochemical signal transduction is a rapid and highly quantitative method that can be incorporated into COVID-19 diagnostic platforms. SARS-CoV-2 is primarily transmitted via respiratory droplets and aerosols; therefore, monitoring the virus in exhaled breath would be valuable for assessing infection rates and preventing transmission. Breath analysis, specifically the detection of volatile organic compounds (VOCs), is a quick, convenient, and portable way to detect SARS-CoV-2. As infection with this virus alters some metabolic pathways, a portable breathalyzer was developed for COVID-19 screening that detects the volatile biomarker nitric oxide.71 In this device, a room-temperature ionic liquid was used to modify the electrode surface, allowing fast and selective diffusion of NO to the sensor surface for chronoamperometric detection. An important feature of breathalyzers is that sensing may be expanded to detect a breath “signature” using a panel of VOCs rather than a single type of molecule. For example, a VOC sensing array was fabricated with eight chemiresistors made of gold nanoparticles modified with diverse organic ligands drop-casted onto circular interdigitated platinum electrodes.72 Changes in the electrical resistance with respect to the baseline represented features in the breath signal as a function of time. Discriminant analysis could then be used to distinguish the breath signature of COVID-19-infected individuals from that of either healthy controls or those with other lung infections with 76% and 95% accuracy in the test sets, respectively, suggesting the utility of the VOC sensing array as a portable screening tool.
Bioreceptor–ligand binding is transduced to an electrochemical signal often through: (1) a redox reaction facilitated by an enzyme, (2) changes in the presence of a redox tag or indicator at the surface of the electrode, or (3) inherent impedance changes detected in a transistor setup. Nanomaterials are used, as described previously, for the efficient transfer of electrons. Charge transfer may be measured as changes in electrical current using electrochemical sensing techniques, such as chronoamperometry and voltammetry. Redox reactions may be driven by applying a potential close to the indicator’s standard potential using cyclic voltammetry (CV), square wave voltammetry (SWV), differential pulse voltammetry (DPV), or other techniques. These techniques are all compatible with portable potentiostat designs: signal transduction to an electrical readout allows for simple onboard integration, data processing, and data transfer using wireless Bluetooth modules for immediate diagnostic interpretation and reporting. Additionally, advances in low-power integrated microsystems allow these diagnostic systems to be miniaturized so that they are highly portable and can be used for POC, screening, and telemedicine applications. In this section, several electrochemical sensing platforms for portable POC SARS-CoV-2 detection are described.
4.2.1. Enzyme-Based Signal Transduction.
Enzyme-facilitated redox reactions can be used for robust signal transduction; the presence of an excess of the chemical mediator allows for built-in signal amplification. Electrochemical signals are sensed via the chronoamperometric detection of the conjugated horseradish peroxidase (HRP) activity of the detector antibody. As an example, a multiplexed electrochemical immunosensing platform, SARS-CoV-2 RapidPlex, offers a low-cost, rapid way to simultaneously detect several SARS-CoV-2 biomarkers (Figure 1B).31 In SARS-CoV-2 RapidPlex, four laser-engraved graphene electrodes coated with 1-pyrenebutyric acid were individually decorated with capture antibodies and proteins for the selective detection of the SARS-CoV-2 N protein, antiprotein S IgG and IgM antibodies, and C-reactive protein. SARS-CoV-2 RapidPlex was able to quantitatively assess the infection status, immune response, and severity of the infection in less than 10 min from the addition of serum or saliva.
One drawback to immunosensing schemes is the requirement for multistep reagent addition, but this can be addressed by using microfluidic solutions. For example, a capillary-flow immunoassay was designed to automate the sequential delivery of reagents.73 In this assay, a microfluidic circuit was created by using stacked layers of hydrophilic polyester and double-sided adhesive films. This circuit directed fluid flow from inlets and preloaded reagent pads to a nitrocellulose membrane sensing region. A screen-printed carbon electrode (SPCE), positioned parallel to the nitrocellulose membrane, measured the HRP oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) for the detection of IgG antibodies selective for the SARS-CoV-2 N protein.
Commercial off-the-shelf glucose meters offer a well-established and portable enzymatic signal transduction method that can be cleverly utilized for the indirect detection and signal amplification of SARS-CoV-2 viral products. An off-the-shelf glucose meter facilitated signal transduction and amplification for picomolar-level detection of SARS-CoV-2 viral antigen in saliva (Figure 1C).74 Upon viral antigen aptamer binding, the complementary antisense strand of the aptamer, conjugated with invertase, was released from the aptamer strand and isolated via magnetic separation. The antigen-binding signal was amplified via the conversion of sucrose to glucose by invertase for glucometer detection. Another RNA sensor was developed using a synthetic biology-based approach, specifically, a toehold switch gene circuit with a downstream trehalase reporter enzyme that ultimately produces glucose, which correlates with viral RNA binding (Figure 1D).75 To eliminate the problem of the background signal associated with endogenous glucose in patient samples, three methods were explored: nucleic acid purification, sample dilution, and programmable glucose reduction using glucose dehydrogenase coupled with NAD. The RNA biosensor selectively detected as few as 100 copies of SARS-CoV-2 RNA. Thus, a compact, portable viral RNA measurement system was developed by utilizing the simple transduction method of a glucose meter.
4.2.2. Redox Indicators.
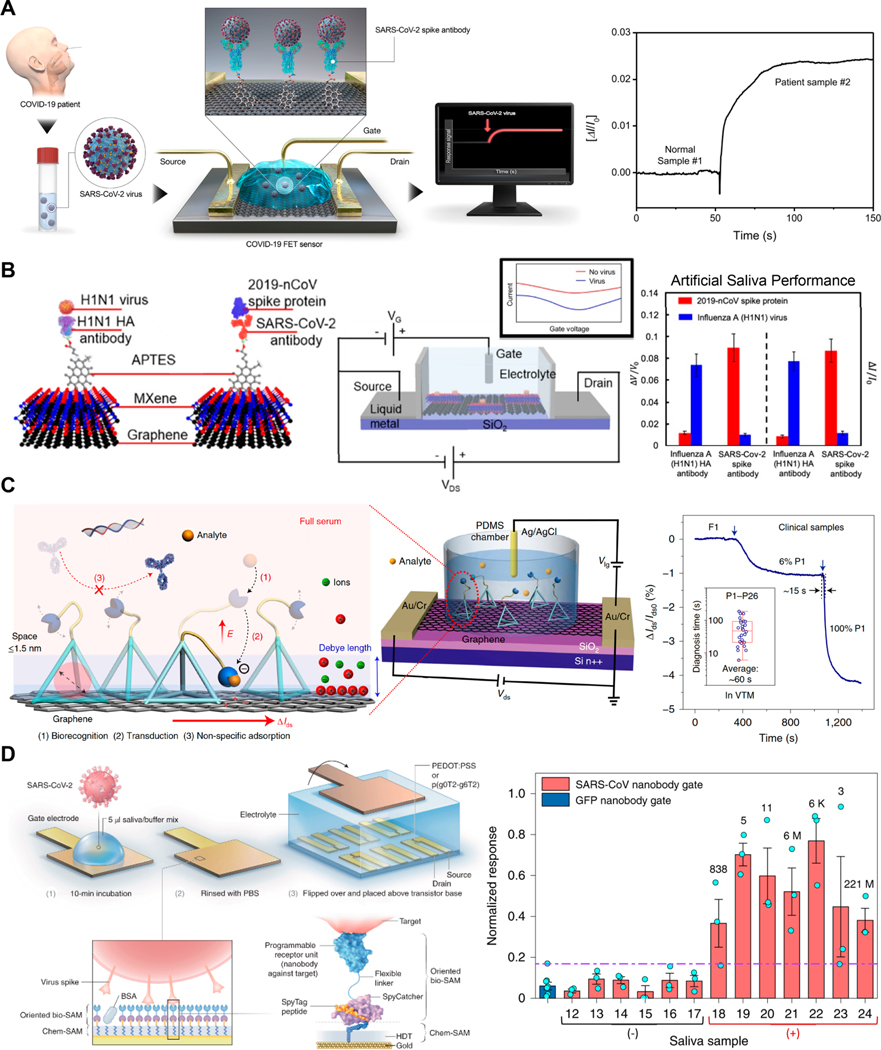
Electroanalytical methods may be used to drive and measure the electron transfer in a redox reaction. Voltammetric techniques can selectively measure redox agents according to their redox peak intensity and location. CV was used to detect SARS-CoV-2 RNA and single point mutations via metal-ion intercalation (Figure 2A).67 Mg2+ and Ag+ ions were intercalated at site-specific mismatches between target RNA and SARS-CoV-2 sensing probes immobilized on a nanoporous Au electrode array, resulting in the ultrasensitive multiplexed quantification of SARS-CoV-2 RNA mutations without requiring conventional tagging or amplification processes. DPV and SWV enhance the detection of charge transfer reactions by isolating the Faradaic current from the charging current at an applied poteproduces glucose, which correlates with viral RNA bindingntial pulse. In the presence of a redox indicator such as [Fe(CN)6]3−/4−, DPV and SWV can sensitively evaluate changes in charge transfer at the sensing surface in the event of the analyte binding to the electrode. Immunoassay and receptor–ligand binding signal transduction may be simplified this way for the label-free detection of SARS-CoV-2 antigen76 and antibodies.34,77 Combining sample collection and antibody detection steps, a cotton-tipped immunosensor swab was designed with a competitive binding scheme on a carbon nanofiber-modified screen-printed electrode (SPE).77 Nucleic acid binding may be assessed via DPV, but detection often requires amplification. In one study, prior to DPV detection, target SARS-CoV-2 genes were amplified in a portable on-body patch using isothermal recombinase polymerase.78 Molecularly imprinted polymer (MIP)-based sensors are also being evaluated by using pulse voltammetry to measure target binding to “imprinted” target cavities in the polymer-coated sensor surface. MIPs have been designed for the rapid detection of the SARS-CoV-2 N50 and S protein79 in nasopharyngeal swab samples (Figure 2B).
Figure 2.
Electrochemical detection of SARS-CoV-2 using redox indicators. (A) Detection of SARS-CoV-2 RNA and single-point mutations based on metal ion intercalation sensed via CV. Reproduced with permission from ref 67. Copyright 2022 ACS. (B) Molecularly imprinted polymer for SARS-CoV-2 antigen detection as demonstrated in control, spiked, and positive nasopharyngeal samples. Reproduced with permission from ref 79. Copyright 2022 Elsevier. (C) Ferrocene-tagged DNA molecular pendulum conjugated with an antibody for the selective detection of SARS-CoV-2 S protein and intact virus. Reproduced with permission from ref 84. Copyright 2021 ACS.
A disposable paper-based biosensor integrated with a smartphone-assisted Sensit Smart potentiostat for COVID-19 diagnosis was reported by Lomae and coauthors.80 The working electrode was modified with a pyrrolidinyl peptide nucleic acid (acpcPNA), a biological recognition element to capture the target complementary DNA (cDNA) (in this case, the SARS-CoV-2 N gene). Hybridization of the cDNA with acpcPNA hindered the redox conversion of [Fe(CN)6]3−/4− redox probe, leading to a decrease in amperometric response correlated with SARS-CoV-2 concentration. The method presented a linear response ranging from 0.1 to 200 nmol L−1 and was applied to 10 clinical samples. Rocha and coauthors81 used low-cost and eco-friendly materials such as polyester sheets, graphite, and natural resin to create stencil-printed sensors. The nanobiosensor developed comprises gold nanoparticles stabilized with cysteamine, glutaraldehyde, anti-SARS-CoV-2 S protein monoclonal antibody as the biological receptor, and bovine serum albumin as the protective layer. The diagnostic test was carried out using a SWV measurement of [Fe(CN)6]3−/4− after sample incubation. The method presented a linear response in the concentration range from 250 pg mL−1 to 20 μg mL−1 of S protein, with a limit of detection of 36.3 pg mL−1. The rapid test was applied with 100% accuracy to a panel of 44 undiluted swab samples.
DPV and SWV may also be used to evaluate the charge transfer rates of an incorporated redox moiety, such as methylene blue (MB). The charge transfer rate varies with the distance of the redox moiety from the electrode surface, and charge transfer becomes a diffusion-limited process when measurements are performed at a low frequency. A binding-induced conformational change in the MB-labeled nucleic acid aptamer probe elicits a measurable change in the SWV peak current. A high-affinity aptamer-based sensor detected the SARS-CoV-2 S protein at the picomolar level.82 In contrast to a binding-induced conformation change, SWV can also measure the change in the resonant motion of a redox-tagged probe that occurs in response to the binding.83 Peptide probes specific to SARS-CoV-2 protease, protein S, and chemokine receptor 5 (CCR5) were used to detect SARS-CoV-2 and measure symptom severity.83 In another study, a molecular pendulum scheme was used to evaluate a change in redox-tagged DNA free motion (Figure 2C).84 When a positive potential is applied, negatively charged DNA is electrostatically attracted to the surface. An anti-S protein antibody is tethered to the linker DNA for the capture of S protein and SARS-CoV-2 viral particles, which upon binding elicit a size-dependent drag force.84 The molecular pendulum thus senses bound SARS-CoV-2 viral particles by tracking changes in the transit time of the redox probe to the electrode surface.
4.2.3. Functionalized Transistors.
Functionalized transistors offer an ultrasensitive, label-free method for electrochemical sensing. Transistor-based sensors can provide miniaturized biosensing, requiring minimal sample volumes and eliciting a large change in current upon small parameter changes. Antibody-decorated FETs for the detection of SARS-CoV-2 S respond immediately upon antigen binding and subsequent changes in charge distribution.69,70 Both graphene69 and MXene-graphene70 FET immunosensors achieved LODs of 1 fg mL−1 (Figure 3A,B). FET immunosensors were multiplexed for the selective detection of H1N1 influenza and SARS-CoV-2 viruses using distinct microfluidic sensing reservoirs.70 Another device was designed to address the problem of reduced sensitivity of microscale transducers in complex biofluids, which is caused by interference from nonspecific biomolecules. To address this issue, a graphene FET was built, consisting of a molecular electromechanical system (MolEMS) composed of a complementary probe linked to a flexible ss-DNA with a stiff tetrahedral ds-DNA base; this device directly detected SARS-CoV-2 RNA in nasopharyngeal samples (Figure 3C).85 The tetrahedral base acts as a spacer that limits the interference of background noise beyond the Debye length while still allowing the bound probe to bend close to the graphene surface. Wang et al. demonstrated the use of selective aptamer probes to sense additional proteins, small molecules, and trace metals.85
Figure 3.
Functionalized transistors for SARS-CoV-2 detection. (A) Graphene FET for the ultrasensitive detection of SARS-CoV-2 antigen in nasopharyngeal samples. Reproduced with permission from ref 69. Copyright 2020 ACS. (B) MXene-graphene FET for the multiplexed detection of SARS-CoV-2 and H1N1 antigens in artificial saliva samples. Reproduced with permission from ref 70. Copyright 2021 ACS. (C) Graphene FET for the MolEMS-based detection of unamplified SARS-CoV-2 RNA in nasopharyngeal samples. Reproduced with permission from ref 85. Copyright 2022 Springer Nature. (D) OECT capable of single molecule detection of SARS-CoV-2 virus using a high-density nanobody self-assembled monolayer with demonstrated use in saliva. Reproduced with permission from ref 87. Copyright 2021 Springer Nature.
Organic electrochemical transistors (OECTs) have also been used to transduce and amplify biological binding events in complex biofluids.86,87 A POC OECT platform achieved a LOD for SARS-CoV-2 IgG of 10 fmol L−1 in saliva and serum samples.86 Applying voltage pulses on gate electrodes during IgG incubation enhanced ligand binding and reduced the required incubation time to less than 5 min. OECT detection of SARS-CoV-2 antigen was enhanced down to single molecule sensitivity by using (1) p(g0T2-g6T2) as a polymer instead of the commonly used conductive polymer, poly(3,4-ethylenedioxythiophene) polystyrenesulfonate (PEDOT:PSS), and (2) nanobodies for high-density, well-oriented functionalization (Figure 3D).87
5. OPTICAL METHODS
Optical methods being extensively explored for the development of diagnostic assays include spectrophotometric analysis, Raman spectroscopy, surface-enhanced Raman spectroscopy (SERS), surface plasmon resonance (SPR), and other techniques.26,88–91 However, few of these methods provide adequate portability, reduced analysis cost, or rapid results, which are required for decentralized and high-frequency testing. Thus, in this review, we focus mainly on the use of colorimetric and fluorescent biosensing approaches for the diagnosis of COVID-19. The use of nanomaterials coupled with paper-based analytical devices and other convenient platforms will be presented with an emphasis on the use of traditional lateral flow technology (antigen and antibody immunoassays) and innovative biosensing methods. Particular attention is paid to diagnostic methods that meet the ASSURED criteria.
5.1. Nanomaterials for Optical-Sensing Platforms.
As a zero-dimensional nanomaterial, AuNPs constitute the labeling system most widely used for lateral flow tests. Colloidal suspensions of metallic nanoparticles are widely used for colorimetric approaches in solution because they exhibit localized surface plasmon resonance (LSPR). LSPR induces the nanoparticles, in the presence of the target analyte, to aggregate, causing a color change in the solution that is visible to the naked eye.92,93 A challenge for the use of AuNPs and other metallic nanoparticles in colorimetric tests is that they tend to aggregate in environments having high ionic strength or containing impurities. Controlling this aggregation requires a balance of the interparticle attractive and repulsive forces with the use of adequate functional or stabilizing agents.93 To provide selective recognition and accurate diagnosis, metallic nanoparticles can be functionalized with specific recognizing elements or receptors for the target biomolecule. For example, biorecognition elements with terminal thiol groups can be conjugated to AuNPs, via adsorption processes or gold–thiol surface chemistry, to improve stability and recognition.94
Other metallic nanoparticles, such as silver (AgNP) and copper (CuNP), have also been used for colorimetric (bio)sensors. Color changes observed with these sensors are associated with the dispersion-aggregation state of the nanoparticles in colloidal solution and lateral flow assay (LFA) technology. However, these nanoparticles are less stable than AuNPs, and their instability impedes synthesis and storage.95 Metal oxide nanoparticles, such as Fe3O4 and CeO2, and carbon nanotubes catalyze the reaction of a peroxidase substrate or have intrinsic peroxidase activity. These classes of nanomaterials are also being used in colorimetric approaches.93
Light-emitting materials (fluorophores) are being used in fluorescence-based optical biosensors, such as LFA, for quantitative or semiquantitative detection. Alternatives to AuNPs include inorganic semiconductor quantum dots (QDs), carbon dots (CDs), graphene nanostructures, organic conjugated polymer nanoparticles, fluorescent microspheres, and lanthanide nanoparticles.93,94,96 QDs (colloidal semi-conductor nanocrystals) have unique optical and electronic properties and are only 1–10 nm in size. The emission wavelengths of QDs can be tuned from ultraviolet (UV) to near-infrared (NIR) by taking advantage of the quantum confinement effects. QDs have several qualities that represent improvements over small-molecule organic dyes, including better quantum yield, Stokes shift, emission wavelength tunability, photostability, and absorption and emission profiles.93,97 The excellent optical characteristics of lanthanide-doped nanoparticles include a long luminescence lifetime, upconversion luminescence in the NIR to the visible range, and sharp emission peaks. These characteristics make it possible to achieve a high signal-to-noise ratio, which is useful for the development of sensitive and selective fluorescent diagnostic tests.94
In general, the optical properties and stability of these nanomaterials are largely impacted by their size, defects, and functionalization. Although several synthesis and functionalization protocols have been reported in the literature, an important consideration is the scale at which the nanomaterials can be produced. Large-scale production (or mass production) of these nanomaterials for rapid tests or assays may affect their reproducibility, uniformity, or stability. Therefore, careful characterization of nanomaterial functionalization and bioconjugation with receptors is crucial for the development of reproducible, robust, and accurate diagnostic tests.
5.2. Lateral Flow Assays (LFAs) and ELISA-Based Methods.
LFAs are widely used to detect pathogens and various biomarkers. The pregnancy test is a familiar example of an LFA technology13 that is commercially available in the form of a paper-based diagnostic device. LFA devices can be divided into nucleic acid lateral flow assays, which detect amplicons generated during amplification methods such as PCR, and LFIAs that use antibodies as recognition elements.98 LFIAs have been applied for the rapid screening of SARS-CoV-2 infection. Typically, LFIAs have labeled antibodies that bind to a biomolecule and emit a colorimetric or fluorescent readout.99,100 LFIAs are cost-efficient, portable, and simple to use and give rapid results, qualities that are not all found in other conventional detection methods, such as ELISA or polymerase chain reaction (PCR). LFIAs use the same design principle as ELISA, consisting of an immobilized capture antibody or antigen that is bound to a solid-phase nitrocellulose (NC) membrane rather than a plastic well.13,101 Unlike multiple-step ELISA, LFIAs are one-step assays. Biofluid is introduced into the LFIA such that capillary action causes the sample to flow through a paper substrate up to the testing line, where the immobilized bioreceptor provides target recognition and reports its presence.
Whereas a drop of blood is usually required for antibody tests, antigen tests more commonly make use of saliva or OP/NP samples. Antigen tests provide an early identification of infected individuals and may be useful for the early detection of viruses in asymptomatic individuals. Antigens are less likely than RNA to degrade during transport and storage, because they are more stable. During the second and third weeks following the initial infection with SARS-CoV-2, antibody levels typically rise, so antibody testing is not effective for the prompt diagnosis of COVID-19. However, levels of IgG and IgM antibodies may be helpful indicators for analyzing the progress of the disease post-illness or the effectiveness of immunization.94
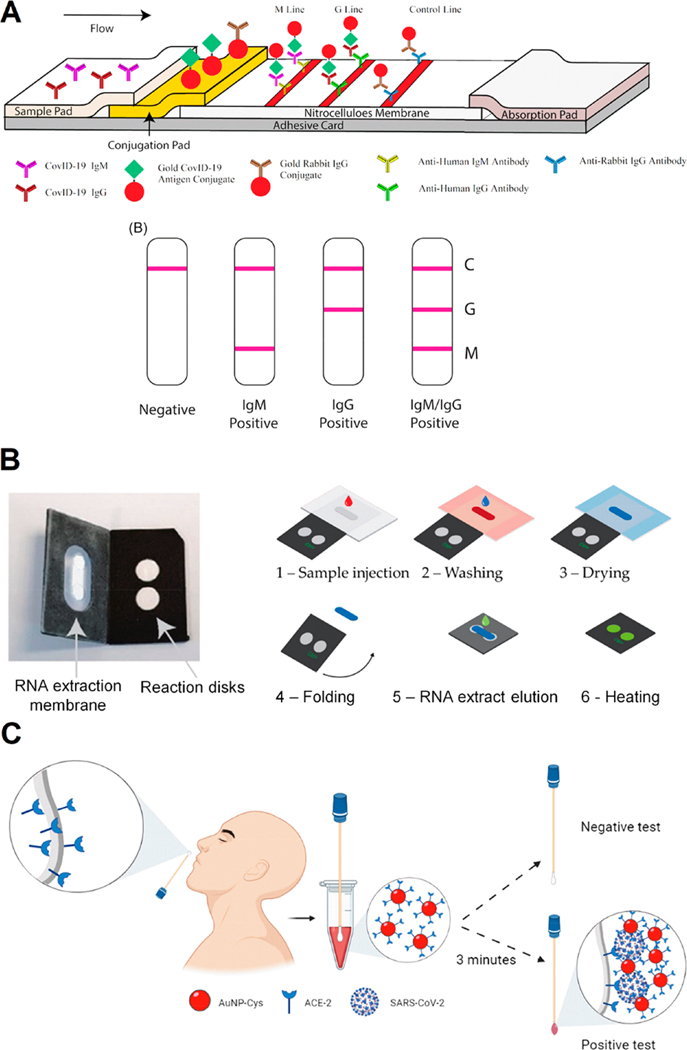
In LFAs for viral antigens, when the sample solution is introduced into the sample pad, particles functionalized with specific antibodies that identify the viral membrane proteins bind to the virus. The particles are AuNPs, AgNPs, or microparticles (latex beads). The nanoparticle-virus complexes are then captured by secondary antibodies that bind to SARS-CoV-2; these secondary antibodies have been immobilized on the test line, causing changes in color (positive test result; Figure 4A).99
Figure 4.
Colorimetric approaches for COVID-19 diagnosis. (A) Schematic illustration of rapid SARS-CoV-2 IgM-IgG combined antibody test with an illustration of different testing results; C, means control line; G, means IgG line; M, means IgM line. Reproduced from ref 32. Copyright 2020 Li et al. Journal of Medical Virology Published by Wiley Periodicals, Inc. (B) POC LAMP-based device and workflow. (1) The sample (with lysed virus) is injected onto the extraction membrane. (2) Washing. (3) Drying of the extraction membrane. (4) Folding of the device so the extraction membrane comes into contact with the two reaction disks. The LAMP mix and primers (COVID-19 and human 18S RNA for the test and control disk, respectively) are freeze-dried on both disks, permitting reverse transcription and amplification. (5) Elution of the RNA from the capture membrane to the reaction disks, subsequently sealing with PCR tape. (6) Heating at 65 °C. Read-out in real-time with intercalating agent SYTO82. Adapted with permission under a Creative Commons CC BY 4.0 License from ref 113. Copyright 2021 Garneret et al. (C) Schematic representation of the use of cotton swab colorimetric biosensor using AuNPs functionalized with ACE2. The colored cotton swab indicates a positive result for SARS-CoV-2. Reproduced with permission from ref 30. Copyright 2021 ACS.
The concentration of SARS-CoV-2 virions is often extremely low in specimens and difficult to detect directly using lateral flow technology. As a result, LFAs have been combined with nucleic acid amplification methods such as RT-PCR and isothermal amplification to enhance their sensitivity, or used as a readout of clustered regularly interspaced short palindromic repeats (CRISPR).102,103 Amplification approaches can be combined with LFIA by using functionalized primers to generate dual hapten-labeled amplicons that bind both to the test strip and a control strip.104 While these methods are often highly sensitive, the potential for nonspecific amplification may result in false-positives. Other approaches reported for nucleic acid detection in LFA involve amplification through methods such as displacement amplification or rolling circle amplification.104,105 Combining LFAs with amplification techniques is likely to simplify and improve traditional assays because amplified nucleic acids would enhance the sensitivity of LFAs so that diagnostic results could be directly visualized.100 However, the development of POC nucleic acid detection devices has been slowed down by the need to prepare high-purity templates for sample treatment and to select amplification reactions that ensure sufficient accumulation of target amplicons.
LFAs have been extensively developed and commercialized for the detection of COVID-19 antibodies and antigens. Table 2 summarizes some contributions regarding the LFAs and other optical-based strategies for COVID-19 testing. LFAs provide a short testing time (typically 10–20 min) but are not highly sensitive, which can lead to false negative results. Recent studies have focused on enhancing the sensitivity of these devices since poor sensitivity is their primary drawback.100 Nanomaterials, functionalization strategies, amplification approaches, and capturing agents are the most promising parameters to enhance the colorimetric or fluorescent readout of these tests.94 For further information and relevant discussion about LFAs focused on COVID-19 diagnosis, the readers can check these important review articles.100,104,106,107
Table 2.
Comparison of Optically Based Tests Applied for COVID-19 Diagnosis
| method | target | sensitivity (%) | specificity (%) | time (min) | ref. |
|---|---|---|---|---|---|
| Fluorescent LFIA using latex microsphere | N protein | 67.15 | 100 | 15 | 124 |
| Colorimetric LFIA using red latex beads | N protein | – | – | 20 | 125 |
| Colorimetric LFIA using AgNP | N protein | 57.6 | 99.5 | 15 | 126 |
| Fluorescent LFIA using silica-core@dual QD-shell nanocomposites | IgM and IgG | 97.37 | 95.54 | 15 | 127 |
| Colorimetric LFIA using AuNP | IgM | 100 | 93.3 | 15 | 128 |
| Fluorescent LFIA using selenium nanoparticles | IgM and IgG | 93.33 | 97.34 | 5 | 129 |
| Fluorescent LFIA using Lanthanide-doped polystyrene nanoparticles | IgG | 100 | 93 | 10 | 130 |
| Colorimetric paper-based RT-LAMP | N gene, RdRp gene, and orf1ab | 97 | 100 | 60 | 131 |
| Fluorescent paper-based RT-LAMP | RdRp gene | – | – | 60 | 113 |
| Fluorescent 3D-printed cartridge RT-LAMP | gene Orf 1a, gene S, gene Orf 8, and gene N | 100 | 100 | <40 | 114 |
| Colorimetric colloidal solution of AuNPs | S, E, and M proteins | 96 | 98 | – | 118 |
| Colorimetric cotton swabs modified with ACE2 and AuNPs | S protein | 96 | 84 | 5 | 30 |
| Colorimetric AuNPs capped with thiol-modified antisense oligonucleotides | N gene | – | – | 10 | 24 |
Antibodies, antigens, proteins, and glycoproteins in biological samples are commonly measured by ELISA, an immunological assay. ELISA has been applied to evaluate the levels of anti-SARS-CoV-2 antibodies. However, conventional ELISA is labor-intensive and time-consuming, and immuno-binding usually takes hours, limiting the potential application of ELISA for POC testing.108 As a consequence, some miniaturized high-throughput ELISAs have recently been developed. Gong et al.109 described a microfluidic platform that isolates and collects serum samples from whole blood by a pulling-force spinning top and then uses a paper-based microfluidic ELISA for quantitative IgA/IgM/IgG measurements. This approach, which does not require any sophisticated instruments, seems promising for POC applications because it is simple and inexpensive and because its consumption of reagents is ten times lower than conventional ELISA. Liu et al.108 proposed a strategy for regulating pressure for controllable lab-on-a-chip reciprocating-flowing (RF) immunobinding. This system enabled the antibodies in the fluid to repeatedly contact the corresponding immobilized antigens on the substrate during continuous repeated forward and backward flow, achieving adequate immunobinding within 60 s. RF-immunobinding was combined with ELISA as an RF-ELISA for the serological detection of anti-SARS-CoV-2 nucleoprotein, with a testing time of around five min for each assay, enabling efficient result turnaround for frequent testing.
5.3. RT-LAMP-Based Methods.
Loop-mediated isothermal amplification (LAMP) is a DNA amplification method used for the rapid and sensitive detection of a gene of interest.110,111 LAMP combined with reverse transcription (reverse transcription loop-mediated isothermal amplification; RT-LAMP) has been used to detect respiratory RNA viruses including SARS-CoV-2. Given its high specificity, sensitivity, cost-effectiveness, and fast turnaround time (typically 30 min), RT-LAMP offers a powerful alternative to RT-PCR. The detection of viral RNA by RT-LAMP is potentially simple, scalable, and broadly applicable. RT-LAMP assays require incubation at a constant temperature but, unlike RT-PCR–based methods, do not require sophisticated instruments.111,112 A portable paper-based microfluidic method has been described that extracts SARS-CoV-2 RNA, amplifies it isothermally by RT-LAMP, and detects it with intercalating dyes or fluorescent probes (Figure 4B). This method presented a LOD similar to that of RT-PCR (1 genome copy per microliter of the clinical sample). For POC applications, the authors created a portable, inexpensive device (US $2–4) to accommodate the foldable paper-based system and integrate the analytic steps.113 Another isothermal RT-LAMP nucleic acid–based detection of SARS-CoV-2 was described by Ganguli et al.,114 consisting of a portable POC device that detects SARS-CoV-2 nucleic acid from viral transport medium (VTM) samples using an additively manufactured three-dimensional cartridge and a smartphone as detector. This device discriminated positive SARS-CoV-2 samples among the 10 clinical samples tested in 30 min.
5.4. Hyperspectral Imaging.
Hyperspectral imaging (HSI) is a noninvasive imaging technique used for disease diagnosis. Also termed imaging spectroscopy, HSI has the ability to acquire multiple wavelengths across a continuous spectrum of light. Hence, in contrast to conventional RGB images with three distinct intensities per pixel, each pixel’s value in HSI represents a continuous spectrum. Analyzing the data captured by the HSI system, one can visualize it as a hypercube by examining a spectrum of light rather than assigning primary colors.15 The degree of contrast in hyperspectral imaging is directly associated with the maximum analyte concentration.115 For example, Alafeef and coauthors116 described a hyperspectral sensor based on hafnium nanoparticles (HfNPs) for ultrasensitive detection of SARS-CoV-2. The authors applied the method to 100 COVID-19 clinical samples with a 100% specificity. Density functional theoretical calculations have shown that HfNPs exhibit greater shifts in their absorption wavelength and light scattering when they bind to the target SARS-CoV-2 RNA sequence compared to gold nanoparticles.116 This property contributes to achieving a detection limit as low as the yoctomolar levels (10−24 mol L−1).
It is worth noting that despite the promise of the HSI method for COVID-19 diagnosis, it still presents significant drawbacks for POC applications, primarily its cost and complexity despite the possibility of using smartphones for practical applications. Currently, to effectively analyze hyperspectral data for frequent, decentralized testing applications, high-performance computing, sensitive detectors, and substantial data storage capacities and advances are still necessary.
5.5. Other Colorimetric and Fluorescent Biosensors.
Biosensors that incorporate colorimetric nanomaterial-based bioassays do not require a sophisticated apparatus for their operation or the interpretation of results; furthermore, these sensors have an easy-to-read visual output.24 The outstanding optical properties of biosensors based on functional metal nanoparticles make them appropriate for rapid colorimetric diagnostic tests for POC applications.117,118 As a result of localized SPR coupling among nanoparticles (i.e., AuNPs), the color of the colloidal suspension changes from red to blue in the presence of the target analyte.118 Interactions between the target species and the receptor immobilized on the nanoparticle surface induce AuNP aggregation.
Moitra et al.24 reported a colorimetric assay that involved capping AuNPs with thiol-modified antisense oligonucleotides (ASOs) specific for the N-gene of SARS-CoV-2. In the presence of their target RNA sequence, the functionalized AuNPs agglomerate selectively, and a change occurs in their SPR, so that within ten min, positive COVID-19 samples can be identified from isolated RNA samples by the naked eye. Taking a similar approach, Ventura et al.118 described a colorimetric biosensor in which AuNPs were functionalized with antibodies targeting the spike, envelope, and membrane proteins of SARS-CoV-2. The interaction of the viral particles with a colloidal solution of the functionalized AuNPs produced a color change in a few minutes. The authors used optical density measurements to detect viral particles in nasal and throat swabs. For high viral loads (Ct < 15), the color change from red to purple could be seen by the naked eye.
Another interesting approach, reported by Ferreira et al.,30 was the development of a colorimetric biosensor fabricated on cotton swabs using AuNPs modified with ACE2. This biosensor costs $0.15 to produce and was able to detect SARS-CoV-2 within 5 min using a smartphone camera as a detector (Figure 4C). The cotton swabs were first functionalized with the ACE2 receptor for the selective and rapid recognition of the SARS-CoV-2 S protein, while the swab was incubated in the clinical sample. Then, the cotton swab to which the SARS-CoV-2 S protein had bound was soaked in a dispersion of AuNPs functionalized with ACE2 for 3 min. This step caused the nanoparticles to aggregate onto the swab surface (positive COVID-19 test), which formed a molecular sandwich, visibly dyeing the cotton surface. This cotton swab-based biosensor is a convenient and accessible method for high-frequency tests since the color change is immediately visible.
Nguyen et al. designed a wearable face mask with a lyophilized CRISPR sensor for the noninvasive colorimetric detection of SARS-CoV-2.119 The SARS-CoV-2 face mask sensor detects the virus as it accumulates on the inside of the mask when the individual coughs, talks, or breathes normally. Although this shelf-stable device has a long testing time (∼90 min), it also has several distinct advantages: it does not require a power source; it operates autonomously at near-ambient temperature; there are no liquids involved; and it provides a visual output and weighs only 3 g. The LOD was reported to be 500 copies (17 aM) of SARS-CoV-2 in vitro transcribed RNA.120
LFAs, ELISA, and other approaches commonly use fluorescence detection. Numerous fluorescent probes, including high-quality optical fibers and suitable optical instruments, are sold commercially at a low cost. Fluorescence-based sensing has good sensitivity and is noninvasive, and biocompatible imaging agents are readily available. These devices can be used with smartphones as simple detectors and processors for POC tests.93 For example, Celiker et al.121 used a synthetic approach to produce a graft fluorescent copolymer PPy-g-PCL conjugated to SARS-CoV-2 antibodies, yielding photoinduced step-growth polymerization of pyrene (Py) and ring-opening polymerization of ε-caprolactone (PCL). After the fluorescent graft copolymer was functionalized with antibodies recognizing SARS-CoV-2, this platform was adapted with a paper-based (dot-blot) bioassay system, which was tested using human nasopharyngeal samples. The sensitivity of the device was reported as 93.33%, and the testing time was 15–20 min.
An interesting biosensor for COVID-19 diagnosis was described by Zheng et al.,122 the test consists of a nanometer-scale fluorescent biosensor that couples CdSe-ZnS quantum dots (QDs) with peptides and the B-cell epitopes of SARS-CoV-2. This system identifies the corresponding antibody with a limit of detection of 100 pmol L−1, provides a rapid response (5 min), and has high sensitivity for detecting COVID-19 antibodies (92.3 to 98.1%, depending on the peptides used). Lin et al.123 proposed a cholesteric liquid crystal (CLC) biosensor for the one-step, wash-free, fast detection of SARS-CoV-2 with little sample preprocessing. Immunocomplexes formed by target ligand binding to the antibody-modified LC surface disturb the organized arrangement of the CLC molecules and shift the CLC from a flat state to a focal cone state. Facilitated by alignment layers of N, N-dimethyl-n-octadecyl-3-aminopropyltrimethoxysilyl chloride, this optical method allows for SARS-CoV-2 S protein concentrations to be observed using the naked eye by apparent textural changes of the CLCs. Accurate S-protein quantification was performed using a microfiber spectrometer and image recognition software.123
In Table 2, we compare the performance of some colorimetric and fluorescent approaches applied to COVID-19 diagnosis, highlighting analytical and clinical advantages, mainly in terms of high-throughput analyses. Another important point is their cost, as a lower cost facilitates more frequent testing in limited-resource countries.
6. TRANSLATING BIOSENSOR TECHNOLOGIES INTO COMMERCIAL TESTS: CHALLENGES AND FUTURE DIRECTIONS
Well-established diagnostic technologies, such as RT-PCR and LFA, have been approved for use in humans by regulatory agencies such as the FDA, and are being widely used to diagnose infectious agents, including SARS-CoV-2. Despite their excellent detection accuracy, PCR methods have limited scalability, which is crucial for widespread testing and disease prevention. In addition, PCR tests require specialized personnel and equipment as well as specific reagents that may not be readily available in many parts of the world. This gap between testing requirements and the availability of these materials poses a challenge to the effective deployment of PCR, particularly in low-resource settings. Additionally, PCR experiments are time-consuming and often centralized, making them less amenable for data collection and testing, extending the time needed to execute and share diagnostic results.132,133 Conversely, LFA tests possess the portability, accessibility, and cost-effectiveness required for high-frequency testing but usually lack the sensitivity and selectivity of PCR that is needed to yield a reliable diagnosis. These limitations of existing methods have led the research community to develop alternative technologies displaying advantages over the established approaches. Biosensors are one such technology, and biosensor prototypes that are capable of diagnosing COVID-19 and other infectious agents at high throughput are emerging. Despite their promise, biosensor technologies have still not been translated into the clinic. The most common barriers include their limited long-term stability, low test reproducibility, sensitivity, and selectivity issues, as well as the need to resolve the complex regulatory hurdles intrinsic to deploying a diagnostic device in different countries.132,134
These obstacles have so far prevented the commercial translation of biosensor-based tests. Despite their promise, biosensor tests do not yet represent a substantial share of the total number of diagnostic technologies being translated into the clinic. Among FDA-authorized at-home over-the-counter (OTC) COVID-19 diagnostic tests for self-testing, most tests are LFA biosensor technologies using different amplification strategies.135 The commercial test prices usually range from US $5 to US $50 and the main bottlenecks to reducing the biosensor costs are the high costs of natural bioreceptors, which can be reduced by biological engineering production or the use of synthetic receptor mimetics; and the use of expensive sensing platforms, which has been overcome with the use of inexpensive substrates, such as paper-based platforms. Despite the plethora of emerging affordable electrochemical biosensors for COVID-19 diagnosis, to the best of our knowledge, no low-cost electrochemical diagnostic test has been approved for POC applications and is commercially available on the market. As will be further discussed, issues regarding the sensor stability and reproducibility should be solved and the biosensing signal still needs to be improved.136
In addition to guidance from regulatory agencies such as the FDA, several steps are still needed to advance biosensor technology: improving the analytical parameters of existing prototypes (sensitivity, stability, reproducibility, and accuracy), improving clinical parameters (test sensitivity and selectivity, as well as long-term stability), and assessing biosensors in operational settings, i.e., beyond routine laboratory-scale optimizations and testing. The stages involved in developing a diagnostic device are formally indicated by the Technology Readiness Level (TRL), a scoring system that identifies critical stages in the development of diagnostic devices. TRL increases from 1 (evaluation of device technology’s fundamental principles) to 9 (a real system tested in an operative environment).99
As the device progresses from TRL 1 to TRL 9, there are five main bottlenecks that may be encountered; briefly: (i) the feasibility of the approach has not been demonstrated, e.g., there is insufficient information about sensitivity and selectivity for proof of concept because such information can only be obtained by analyzing a wide range of clinical samples; (ii) the proposed detection method is too slow, too cumbersome (multistep), or too costly (i.e., it requires expensive equipment) to be easily used at the point of need; (iii) the shelf life of the device is short, or sophisticated storage conditions are needed, limiting its decentralized use; (iv) regulatory approval has not been obtained from the appropriate regulatory agency to ensure that the proposed diagnostic test has been assessed sufficiently for analytical and clinical validity, as well as for safe and effective use by the final user; and (v) the proposed fabrication and functionalization technologies are not scalable enough for market launch.99
The stability and reliability of biosensors depend on the quality of the transducer biofunctionalization and the storage conditions, which can either shorten or extend the shelf life. The storage conditions of the bioreceptors are critical for maintaining their activity. Freeze–thaw cycles can induce protein aggregation or denaturation, impairing protein stability, and can ultimately affect biosensor performance.137 Experiments to establish long-term stability, selectivity, and reliability can take a long time, as significant statistical data need to be collected in order to validate results and determine the adequate conditions of use and storage.
To biofunctionalize nanomaterials or transducers, a variety of surface modification approaches have been devised, including physical adsorption, chemical adsorption, covalent binding, and affinity-based interactions.138,139 The use of antifouling agents as permeable polymeric coatings to minimize nonspecific binding is also important to improve the selectivity and robustness of the biosensors.140,141 Adequate functionalization of the nanostructured transducer surface with the bioreceptor is essential to provide sensitive, stable, and reproducible operation of biosensors since the efficacy of the sensor depends on the orientation and activity of the biological receptor. The recognition or interaction sites need to be available (steric-free) for the target analyte to bind or interact with, and for these events to be translated into a readable signal by the transducer. For example, the human and engineered ACE2 receptor has been largely used in the development of biosensor technologies for SARS-CoV-2 detection.142 The interaction between the receptor-binding domain of S protein and ACE2 involves 20 residues in the N-terminal helix of the receptor.40,143 Thus, this moiety needs to be sterically available after biofunctionalization to create a sensitive biosensor response.
Appropriate large-scale manufacturing methods and biosensor reliability are still critical aspects that need to be overcome to facilitate the commercialization and widespread use of biosensors. Challenges facing the large-scale synthesis of nanomaterials include obtaining controlled size distribution and defects and stabilizing the nanomaterials at a low cost. Lowering the cost of nanomaterial manufacturing, as well as advances in functionalization, will make it possible to design and use biofunctionalized nanomaterials to develop simple, sensitive, and cost-effective methods for detecting SARS-CoV-2 and its associated biomarkers as well as other infectious agents.
In addition to controlling the synthesis and functionalization of nanomaterials on a large scale, manufacturing reproducible biosensors can be improved by the use of automation because manual approaches can produce biosensors with uneven performance. However, biosensors can also vary from batch to batch because the biological reagents used are commonly commercialized based on their purity without examining their reactivity.99,134 For example, enzymes acquired from different sources may have different activities, adding variability to experiments with biosensors. Thus, continuously assessing reagent reactivity is important in biosensor development and use. Another concern is maintaining the optimal activity of the biological receptors since the long-term stability of the biosensor device will deeply impact the likelihood of commercial implementation of these devices. To achieve stability, current biofunctionalization strategies incorporate monolayer membranes, three-dimensional constructions, and other interfaces to create antifouling structures and favorable physical and chemical environments (i.e., control of pH, ionic strength, humidity, light protection, and other specific conditions), which are key to prolonging bioreceptor stability and biosensor shelf life.144–146
Besides the use of unusual materials or preparation methods for improving the robustness of the biosensors, some strategies rely on the use of synthetic receptors that selectively bind to the target analyte.147 Mimetic materials can provide superior stability for operational use compared to biological materials. For COVID-19 diagnostics, tailored chemical receptors based on engineered versions of enzymes and antibodies have been described; the use of molecularly imprinted polymers potentially offers solutions to the limited stability of biological receptors.148,149
Streamlining biosensor technologies would also facilitate their widespread use. Often, biosensors use sophisticated analytical instruments and software that can be difficult to operate by nonexperts, preventing their translation to POC settings.134 Future studies should focus on test procedures, ideally creating methodologies that do not require lab-based materials, multiple steps, or skilled operators to translate these technologies to clinical and commercial use. Microfluidic systems, for example, offer an excellent alternative to integrate preanalysis steps, allowing delivery of the sample to the sensing region while performing separation, preconcentration, and derivatizations, among other steps. All POC applications require the integration of an accessible detector with a user-friendly interface to facilitate and accelerate interpretation of the results. Future work is needed in these areas to bring biosensors to the market.
It is worth mentioning the importance of evaluating the safety of different biosensor technologies during their use in testing sites, providing adequate training, and establishing clear guidance for accurate testing. Given the diverse range of healthcare professionals (e.g., physicians, nurses, and medical technologists, among others) and workflows involved in POC testing, there are additional risks related to the use of such technologies, i.e., electrical shock, chemical, and biological contamination during simplified protocol applications (sometimes without personal protective equipment), as well as the importance of adequate sample inactivation and disposal protocols after use to diminish the risk of operator’s exposure and contamination.150
For the widespread deployment and mass manufacturing of single-use POC tests, sustainability must be considered. Electronic readers and signal processing components should be reused and disposable; single-use components should be minimized. Greener and sustainable approaches using less toxic and biodegradable materials (paper,9 cellulose151) should be prioritized during sensor development.
The use of diagnostic technologies also raises ethical issues, such as concerns over data confidentiality, ownership, and privacy.134,152 Combining diagnostic devices with contact tracing apps, while valuable for surveillance, may not be trusted by users who are concerned with confidentiality and with data acquisition by third parties for purposes unrelated to COVID-19 surveillance.99 Indeed, Bhalla et al. discuss how consumers and patients are less likely to read the policies and terms described in self-testing biosensors and mobile health applications than in other circumstances.134 Nonmandatory contact-tracing apps that protect privacy while balancing the demands and aims of users, policymakers, and developers warrant future inquiry and development.99
7. CONSIDERATIONS AND LESSONS LEARNED TO COMBAT FUTURE PANDEMICS
The properties of functional nanomaterials (high conductivity, electromagnetic field enhancement, quantum confinement, high surface area for functionalization with biological receptors, and catalytic properties) and nanotechnology, more generally, enhance the sensitivity, stability, and selectivity of biosensing devices. Advances in nanotechnology (synthesis, functionalization, and bioconjugation with receptors) and fabrication methods that enable reproducible, robust, and low-cost testing devices for operation at the POC are crucial in combating COVID-19 and will also be fundamental in controlling future endemic and pandemic diseases.
In the past two decades, response times for estimating and tracking infected patients have accelerated. For example, the response time was substantially quicker during the COVID-19 pandemic compared to the SARS (Severe Acute Respiratory Syndrome) and MERS (Middle East Respiratory Syndrome) outbreaks that took place in 2002 and 2012, respectively.99,153 This faster response is primarily associated with technological advancements in DNA sequencing and the enhanced functionality of molecular kits based on nucleic acid detection, including RT-PCR, which have reduced the time needed to identify clinically significant biomarkers. To minimize the harm caused by upcoming pandemics, however, significant work still needs to be done, since nucleic acid detection techniques are slow, cumbersome, and expensive for decentralized POC applications, particularly in developing countries.99 Rapid tests involving biosensor technology are emerging as excellent candidates to help control future outbreaks through frequent testing. However, the ethical, regulatory, and commercial concerns that emerged during the current COVID-19 pandemic must be addressed prior to implementing biosensor technologies more broadly in our society. Therefore, there is an urgent need to consider the regulatory and practical requirements for each biosensor prototype with an end goal of developing generalizable testing platforms that are readily adaptable for upcoming infectious outbreaks.
Existing biosensor technologies incorporate low-cost materials, miniaturized, portable instrumentation, and reduced testing time, features that are especially valuable for frequent testing in POC settings. In addition to serving as home diagnostic tools, POC biosensor technologies may be used to monitor high-density areas, such as airports, train stations, and hospitals, providing rapid results for decision-making. Furthermore, biosensors can also be used for wastewater and sewage analyses following outbreaks to facilitate tracking the spread of the infectious agent.
Existing strategies and protocols commonly used to manufacture, synthesize, and (bio)functionalize the nanomaterials and transducers constituting currently available biosensors can also be expanded to detect additional infectious agents as well as other biomolecules such as specific enzymes, nucleic acids, antibodies, and cells. The translatability of the biosensor technologies to other sensing targets can be made by choosing adequate bioreceptors (natural or synthetic) and strategies to correctly anchor them onto the transducer surface. We predict that advances made in the biosensing field over the past decade, combined with continued funding for the optimization and diversification of sensing strategies, will enable the design of biosensor tests for screening and surveillance for pathogens and diseases of interest, in addition to COVID-19. Multiplexed biosensor technologies similar to those reported by Torrente-Rodriguez and coauthors,31 where the authors described a multiplexed test that enables the detection of antigen, antibodies, and inflammatory biomarker C-reactive protein related to COVID-19 infection, represent a very attractive approach and should be prioritized for future endemics/pandemics because, in single testing, it provides quantitative and clinically relevant information about the infection status, immune response, and severity of the particular infection. Improved diagnostic testing is a pillar for pandemic prevention, control, and containment.
8. CONCLUSIONS
High-throughput, sensitive, and cost-effective diagnostic devices can be used for accurate and frequent testing of SARS-CoV-2 infection and can contribute to the prevention of viral transmission. Functionalized nanomaterial-based colorimetric and electrochemical biosensors present attractive features for accessible POC testing, such as simple instrumentation (sometimes coupled with smart devices), fast response, and adequate sensitivity. Mass manufacturing and functionalization methods can be used for low-cost production, but it is necessary to ensure that the sensor performance is stable and reproducible. POC and at-home sensing rely on the noninvasive collection of saliva, nasal swabs, or other samples. Recently, the application of antifouling coatings and membranes to biosensor transducers has improved the sensor performance with these complex biofluids, and POC sensing is becoming more accurate. As SARS-CoV-2 persists in the community, it is evident that more tools are needed to monitor mutations and identify variants. Multiplexed sensors that can measure the number and kinds of mutations in the SARS-CoV-2 sequence may act as important screening tools for variant identification and tracking to inform public health measures and future vaccine development. Since SARS-CoV-2 is primarily transmitted via respiratory droplets and aerosols, the capability to monitor the virus directly in exhaled breath would be invaluable in controlling the spread of infections. Future research may lead to the direct detection of viral particles from the breath and the incorporation of flexible printed electronics in face masks for continuous viral sensing.
The methods highlighted here offer sensing capabilities that may easily be modified for monitoring other biomarkers of interest in multiplexed platforms. SARS-CoV-2 symptoms vary greatly and are nonspecific; therefore, it is important to distinguish SARS-CoV-2 from other viral infections for proper treatment. This can be done by using multiplexed platforms that detect a variety of circulating viruses. Multiplexed sensing of general health biomarkers, in addition to SARS-CoV-2 viral products, could present a more complete portrait of the patient’s health for telemedicine monitoring. We envision a near future when accessible biosensor technologies will be widely used for diagnosis. We anticipate that the lessons learned during the current COVID-19 pandemic, in addition to improving technologies for detection and diagnosis, will enable the transition of biosensors from prototypes to diagnostic tests for future pandemics.
VOCABULARY.
| Biosensor | A biosensor is a device that uses biological materials, biologically derived materials, or biomimetics as recognition elements closely coupled to a physicochemical transducer to produce a readable electrical signal that is proportional to the concentration of a specific target analyte or group of analytes. |
| Biomarker | A biomarker is a measurable indicator (biological molecule or process) of some biological state or clinical condition. |
| Functionalization | Introduction of functional groups, materials, or properties onto the surface of a (nano)-material. |
| Diagnostic test | A test used to identify or confirm the presence of a disease or medical condition. |
| Sensitivity | From an analytical perspective: the slope of an analytical or calibration curve. From a clinical perspective: the success rate of a particular test to correctly identify those patients with a particular disease or condition. |
ACKNOWLEDGMENTS
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the Procter & Gamble Company, United Therapeutics, a BBRF Young Investigator Grant, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1–21–1–0014). Research reported in this publication was supported by the Nemirovsky Prize, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, Penn Health-Tech Accelerator Award, the IADR Innovation in Oral Care Award, and by funds provided by the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. Heather Lukas and Wei Gao acknowledge the funding support from National Science Foundation Grant 2145802, Sloan Research Fellowship, and Merkin Institute for Translational Research at California Institute of Technology. We thank Dr. Karen Pepper for editing the manuscript and de la Fuente Lab members for insightful discussions. William Reis de Araujo acknowledges funding from Brazilian funding agencies CAPES, FAPESP (2022/03250–7, 2018/08782–1), and CNPq (438828/2018–6, 310282/2022–5) for supporting the research.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.3c01629
The authors declare no competing financial interest.
Contributor Information
William Reis de Araujo, Portable Chemical Sensors Lab, Department of Analytical Chemistry, Institute of Chemistry, State University of Campinas – UNICAMP, Campinas, SP 13083-970, Brazil.
Heather Lukas, Andrew and Peggy Cherng Department of Medical Engineering, Division of Engineering and Applied Science, California Institute of Technology, Pasadena, California 91125, United States.
Marcelo D. T. Torres, Machine Biology Group, Departments of Psychiatry and Microbiology, Institute for Biomedical Informatics, Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States; Departments of Bioengineering and Chemical and Biomolecular Engineering, School of Engineering and Applied Science and Penn Institute for Computational Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
Wei Gao, Andrew and Peggy Cherng Department of Medical Engineering, Division of Engineering and Applied Science, California Institute of Technology, Pasadena, California 91125, United States.
Cesar de la Fuente-Nunez, Machine Biology Group, Departments of Psychiatry and Microbiology, Institute for Biomedical Informatics, Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States; Departments of Bioengineering and Chemical and Biomolecular Engineering, School of Engineering and Applied Science and Penn Institute for Computational Science, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
REFERENCES
- (1).Koteswara Rao V. Point of Care Diagnostic Devices for Rapid Detection of Novel Coronavirus (SARS-NCoV19) Pandemic: A Review. Front. Nanotechnol. 2021, 2, No. 593619. [Google Scholar]
- (2).Kevadiya BD; Machhi J; Herskovitz J; Oleynikov MD; Blomberg WR; Bajwa N; Soni D; Das S; Hasan M; Patel M; Senan AM; Gorantla S; McMillan J; Edagwa B; Eisenberg R; Gurumurthy CB; Reid SPM; Punyadeera C; Chang L; Gendelman HE. Diagnostics for SARS-CoV-2 Infections. Nat. Mater. 2021, 20 (5), 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Larremore DB; Wilder B; Lester E; Shehata S; Burke JM; Hay JA; Tambe M; Mina MJ; Parker R. Test Sensitivity Is Secondary to Frequency and Turnaround Time for COVID-19 Screening. Sci. Adv. 2021, 7 (1), No. eabd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mina MJ; Parker R; Larremore DB. Rethinking Covid-19 Test Sensitivity – A Strategy for Containment. N. Engl. J. Med. 2020, 383 (22), No. e120. [DOI] [PubMed] [Google Scholar]
- (5).Nasrollahi F; Haghniaz R; Hosseini V; Davoodi E; Mahmoodi M; Karamikamkar S; Darabi MA; Zhu Y; Lee J; Diltemiz SE; Montazerian H; Sangabathuni S; Tavafoghi M; Jucaud V; Sun W; Kim H; Ahadian S; Khademhosseini A. Micro and Nanoscale Technologies for Diagnosis of Viral Infections. Small 2021, 17 (45), No. 2100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yu Y; Bu F; Zhou H; Wang Y; Cui J; Wang X; Nie G; Xiao H. Biosafety Materials: An Emerging New Research Direction of Materials Science from the COVID-19 Outbreak. Mater. Chem. Front. 2020, 4 (7), 1930–1953. [Google Scholar]
- (7).Tang Z; Kong N; Zhang X; Liu Y; Hu P; Mou S; Liljeström P; Shi J; Tan W; Kim JS; Cao Y; Langer R; Leong KW; Farokhzad OC; Tao W. A Materials-Science Perspective on Tackling COVID-19. Nat. Rev. Mater. 2020, 5 (11), 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Weiss C; Carriere M; Fusco L; Capua I; Regla-Nava JA; Pasquali M; Scott JA; Vitale F; Unal MA; Mattevi C; Bedognetti D; Merkoç A; Tasciotti E; Yilmazer A; Gogotsi Y; Stellacci F; Delogu LG. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14 (6), 6383–6406. [DOI] [PubMed] [Google Scholar]
- (9).Ataide VN; Mendes LF; Gama LILM; de Araujo WR; Paixão TRLC. Electrochemical Paper-Based Analytical Devices: Ten Years of Development. Anal. Methods 2020, 12 (8), 1030–1054. [Google Scholar]
- (10).Ozer T; McMahon C; Henry CS. Advances in Paper-Based Analytical Devices. Annu. Rev. Anal. Chem. 2020, 13 (1), 85–109. [DOI] [PubMed] [Google Scholar]
- (11).Ferreira PC; Ataíde VN; Silva Chagas CL; Angnes L; Tomazelli Coltro WK; Longo Cesar Paixão TR; Reis de Araujo W. Wearable Electrochemical Sensors for Forensic and Clinical Applications. TrAC Trends Anal. Chem. 2019, 119, No. 115622. [Google Scholar]
- (12).Madhurantakam S; Muthukumar S; Prasad S. Emerging Electrochemical Biosensing Trends for Rapid Diagnosis of COVID-19 Biomarkers as Point-of-Care Platforms: A Critical Review. ACS Omega 2022, 7 (15), 12467–12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).de Lima LF; de S Moraes A; de T Garcia P,; Araujo WR. Lab on a Paper-Based Device for Coronavirus Biosensing. In Detection and Analysis of SARS Coronavirus; Wiley, 2021; pp 137–161. DOI: 10.1002/9783527832521.ch9. [DOI] [Google Scholar]
- (14).Baldo TA; de Lima LF; Mendes LF; de Araujo WR; Paixão TRLC; Coltro WKT. Wearable and Biodegradable Sensors for Clinical and Environmental Applications. ACS Appl. Electron. Mater. 2021, 3 (1), 68–100. [Google Scholar]
- (15).Alafeef M; Pan D. Diagnostic Approaches For COVID-19: Lessons Learned and the Path Forward. ACS Nano 2022, 16 (8), 11545–11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yari P; Liang S; Chugh VK; Rezaei B; Mostufa S; Krishna VD; Saha R; Cheeran MC-J; Wang J-P; Gómez-Pastora J; Wu K. Nanomaterial-Based Biosensors for SARS-CoV-2 and Future Epidemics. Anal. Chem. 2023, 95 (42), 15419–15449. [DOI] [PubMed] [Google Scholar]
- (17).Mabey D; Peeling RW; Ustianowski A; Perkins MD. Diagnostics for the Developing World. Nat. Rev. Microbiol. 2004, 2 (3), 231–240. [DOI] [PubMed] [Google Scholar]
- (18).Land KJ; Boeras DI; Chen X-S; Ramsay AR; Peeling RW. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2019, 4 (1), 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Global partnership to make available 120 million affordable, quality COVID-19 rapid tests for low- and middle-income countries. https://www.who.int/news/item/28-09-2020-global-partnership-to-make-available-120-million-affordable-quality-covid-19-rapid-tests-for-low--and-middle-income-countries (accessed on October 14, 2023).
- (20).Du Z; Pandey A; Bai Y; Fitzpatrick MC; Chinazzi M; Pastore y Piontti A; Lachmann M; Vespignani A; Cowling BJ; Galvani AP; Meyers LA. Comparative Cost-Effectiveness of SARS-CoV-2 Testing Strategies in the USA: A Modelling Study. Lancet Public Heal. 2021, 6 (3), e184–e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kim D; Lee J-Y; Yang J-S; Kim JW; Kim VN; Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181 (4), 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yao H; Song Y; Chen Y; Wu N; Xu J; Sun C; Zhang J; Weng T; Zhang Z; Wu Z; Cheng L; Shi D; Lu X; Lei J; Crispin M; Shi Y; Li L; Li S. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183 (3), 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kermali M; Khalsa RK; Pillai K; Ismail Z; Harky A. The Role of Biomarkers in Diagnosis of COVID-19 – A Systematic Review. Life Sci. 2020, 254, No. 117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Moitra P; Alafeef M; Dighe K; Frieman MB; Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14 (6), 7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chaibun T; Puenpa J; Ngamdee T; Boonapatcharoen N; Athamanolap P; O’Mullane AP; Vongpunsawad S; Poovorawan Y; Lee SY; Lertanantawong B. Rapid Electrochemical Detection of Coronavirus SARS-CoV-2. Nat. Commun. 2021, 12 (1), 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Qiu G; Gai Z; Tao Y; Schmitt J; Kullak-Ublick GA; Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14 (5), 5268–5277. [DOI] [PubMed] [Google Scholar]
- (27).Torres MDT; de Araujo WR; de Lima LF; Ferreira AL; de la Fuente-Nunez C. Low-Cost Biosensor for Rapid Detection of SARS-CoV-2 at the Point of Care. Matter 2021, 4 (7), 2403–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Torres MDT; de Lima LF; Ferreira AL; de Araujo WR; Callahan P; Dávila A; Abella BS; de la Fuente-Nunez C. Detection of SARS-CoV-2 with RAPID: A Prospective Cohort Study. iScience 2022, 25 (4), No. 104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).de Lima LF; Ferreira AL; Torres MDT; de Araujo WR; de la Fuente-Nunez C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (30), No. e2106724118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ferreira AL; de Lima LF; Torres MDT; de Araujo WR; de la Fuente-Nunez C. Low-Cost Optodiagnostic for Minute-Time Scale Detection of SARS-CoV-2. ACS Nano 2021, 15 (11), 17453–17462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Torrente-Rodríguez RM; Lukas H; Tu J; Min J; Yang Y; Xu C; Rossiter HB; Gao W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3 (6), 1981–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li Z; Yi Y; Luo X; Xiong N; Liu Y; Li S; Sun R; Wang Y; Hu B; Chen W; Zhang Y; Wang J; Huang B; Lin Y; Yang J; Cai W; Wang X; Cheng J; Chen Z; Sun K; Pan W; Zhan Z; Chen L; Ye F. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92 (9), 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhang C; Zhou L; Liu H; Zhang S; Tian Y; Huo J; Li F; Zhang Y; Wei B; Xu D; Hu J; Wang J; Cheng Y; Shi W; Xu X; Zhou J; Sang P; Tan X; Wang W; Zhang M; Wang B; Zhou Y; Zhang K; He K. Establishing a High Sensitivity Detection Method for SARS-CoV-2 IgM/IgG and Developing a Clinical Application of This Method. Emerg. Microbes Infect. 2020, 9 (1), 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yakoh A; Pimpitak U; Rengpipat S; Hirankarn N; Chailapakul O; Chaiyo S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, No. 112912. [DOI] [PMC free article] [PubMed]
- (35).Srivastava M; Srivastava N; Mishra PK; Malhotra BD. Prospects of Nanomaterials-Enabled Biosensors for COVID-19 Detection. Sci. Total Environ. 2021, 754, No. 142363. [DOI] [PMC free article] [PubMed]
- (36).Naikoo GA; Arshad F; Hassan IU; Awan T; Salim H; Pedram MZ; Ahmed W; Patel V; Karakoti AS; Vinu A. Nanomaterials-based Sensors for the Detection of COVID-19: A Review. Bioeng. Transl. Med. 2022, DOI: 10.1002/btm2.10305. [DOI] [PMC free article] [PubMed]
- (37).Walls AC; Park Y-J; Tortorici MA; Wall A; McGuire AT; Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181 (2), 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wrapp D; Wang N; Corbett KS; Goldsmith JA; Hsieh C-L; Abiona O; Graham BS; McLellan JS. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science (80-.) 2020, 367 (6483), 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Aydın EB; Aydın M; Sezgintürk MK. Label-Free and Reagent-Less Electrochemical Detection of Nucleocapsid Protein of SARS-CoV-2: An Ultrasensitive and Disposable Biosensor. New J. Chem. 2022, 46, 9172. [Google Scholar]
- (40).Lan J; Ge J; Yu J; Shan S; Zhou H; Fan S; Zhang Q; Shi X; Wang Q; Zhang L; Wang X. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581 (7807), 215–220. [DOI] [PubMed] [Google Scholar]
- (41).Barton MI; MacGowan SA; Kutuzov MA; Dushek O; Barton GJ; van der Merwe PA. Effects of Common Mutations in the SARS-CoV-2 Spike RBD and Its Ligand, the Human ACE2 Receptor on Binding Affinity and Kinetics. eLife 2021, DOI: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed]
- (42).Ramanathan M; Ferguson ID; Miao W; Khavari PA. SARS-CoV-2 B.1.1.7 and B.1.351 Spike Variants Bind Human ACE2 with Increased Affinity. Lancet Infect. Dis. 2021, 21 (8), 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Sorokina M; Teixeira MC, J.; Barrera-Vilarmau S; Paschke R; Papasotiriou I; Rodrigues JPGLM; Kastritis PL. Structural Models of Human ACE2 Variants with SARS-CoV-2 Spike Protein for Structure-Based Drug Design. Sci. Data 2020, 7 (1), 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Glasgow A; Glasgow J; Limonta D; Solomon P; Lui I; Zhang Y; Nix MA; Rettko NJ; Zha S; Yamin R; Kao K; Rosenberg OS; Ravetch JV; Wiita AP; Leung KK; Lim SA; Zhou XX; Hobman TC; Kortemme T; Wells JA. Engineered ACE2 Receptor Traps Potently Neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (45), 28046–28055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chan KK; Dorosky D; Sharma P; Abbasi SA; Dye JM; Kranz DM; Herbert AS; Procko E. Engineering Human ACE2 to Optimize Binding to the Spike Protein of SARS Coronavirus 2. Science (80-.) 2020, 369 (6508), 1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Daniell H; Nair SK; Guan H; Guo Y; Kulchar RJ; Torres MDT; Shahed-Al-Mahmud M; Wakade G; Liu Y-M; Marques AD; Graham-Wooten J; Zhou W; Wang P; Molugu SK; de Araujo WR; de la Fuente-Nunez C; Ma C; Short WR; Tebas P; Margulies KB; Bushman FD; Mante FK; Ricciardi RP; Collman RG; Wolff MS. Debulking Different Corona (SARS-CoV-2 Delta, Omicron, OC43) and Influenza (H1N1, H3N2) Virus Strains by Plant Viral Trap Proteins in Chewing Gums to Decrease Infection and Transmission. Biomaterials 2022, 288, No. 121671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Vezza VJ; Butterworth A; Lasserre P; Blair EO; MacDonald A; Hannah S; Rinaldi C; Hoskisson PA; Ward AC; Longmuir A; Setford S; Farmer ECW; Murphy ME; Corrigan DK. An Electrochemical SARS-CoV-2 Biosensor Inspired by Glucose Test Strip Manufacturing Processes. Chem. Commun. 2021, 57 (30), 3704–3707. [DOI] [PubMed] [Google Scholar]
- (48).Georgas A; Lampas E; Houhoula DP; Skoufias A; Patsilinakos S; Tsafaridis I; Patrinos GP; Adamopoulos N; Ferraro A; Hristoforou E. ACE2-Based Capacitance Sensor for Rapid Native SARS-CoV-2 Detection in Biological Fluids and Its Correlation with Real-Time PCR. Biosens. Bioelectron. 2022, 202, No. 114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Suh J-S; Kim H-S; Kim T-J. Development of a SARS-CoV-2-Derived Receptor-Binding Domain-Based ACE2 Biosensor. Sensors Actuators B Chem. 2021, 334, No. 129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Raziq A; Kidakova A; Boroznjak R; Reut J; Öpik A; Syritski V. Development of a Portable MIP-Based Electrochemical Sensor for Detection of SARS-CoV-2 Antigen. Biosens. Bioelectron. 2021, 178, No. 113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Vindeirinho JM; Pinho E; Azevedo NF; Almeida C. SARS-CoV-2 Diagnostics Based on Nucleic Acids Amplification: From Fundamental Concepts to Applications and Beyond. Front. Cell. Infect. Microbiol. 2022, 12, No. 799678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Liu D; Shen H; Zhang Y; Shen D; Zhu M; Song Y; Zhu Z; Yang C. A Microfluidic-Integrated Lateral Flow Recombinase Polymerase Amplification (MI-IF-RPA) Assay for Rapid COVID-19 Detection. Lab Chip 2021, 21 (10), 2019–2026. [DOI] [PubMed] [Google Scholar]
- (53).Fassy J; Lacoux C; Leroy S; Noussair L; Hubac S; Degoutte A; Vassaux G; Leclercq V; Rouquié D; Marquette C-H; Rottman M; Touron P; Lemoine A; Herrmann J-L; Barbry P; Nahon J-L; Zaragosi L-E; Mari B. Versatile and Flexible Microfluidic QPCR Test for High-Throughput SARS-CoV-2 and Cellular Response Detection in Nasopharyngeal Swab Samples. PLoS One 2021, 16 (4), No. e0243333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Premraj A; Aleyas AG; Nautiyal B; Rasool TJ. Nucleic Acid and Immunological Diagnostics for SARS-CoV-2: Processes, Platforms and Pitfalls. Diagnostics 2020, 10 (11), 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Vogels CBF; Brito AF; Wyllie AL; Fauver JR; Ott IM; Kalinich CC; Petrone ME; Casanovas-Massana A; Catherine Muenker M; Moore AJ; Klein J; Lu P; Lu-Culligan A; Jiang X; Kim DJ; Kudo E; Mao T; Moriyama M; Oh JE; Park A; Silva J; Song E; Takahashi T; Taura M; Tokuyama M; Venkataraman A; Weizman O-E; Wong P; Yang Y; Cheemarla NR; White EB; Lapidus S; Earnest R; Geng B; Vijayakumar P; Odio C; Fournier J; Bermejo S; Farhadian S; Dela Cruz CS; Iwasaki A; Ko AI; Landry ML; Foxman EF; Grubaugh ND. Analytical Sensitivity and Efficiency Comparisons of SARS-CoV-2 RT–QPCR Primer–Probe Sets. Nat. Microbiol. 2020, 5 (10), 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Artesi M; Bontems S; Göbbels P; Franckh M; Maes P; Boreux R; Meex C; Melin P; Hayette M-P; Bours V; Durkin K. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, DOI: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed]
- (57).Soh JH; Balleza E; Abdul Rahim MN; Chan H-M; Mohd Ali S; Chuah JKC; Edris S; Atef A; Bahieldin A; Ying JY; Sabir JSM. CRISPR-Based Systems for Sensitive and Rapid onSite COVID-19 Diagnostics. Trends Biotechnol. 2022, 40 (11), 1346–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Albright J; Mick E; Sanchez-Guerrero E; Kamm J; Mitchell A; Detweiler AM; Neff N; Tsitsiklis A; Hayakawa Serpa P; Ratnasiri K; Havlir D; Kistler A; DeRisi JL; Pisco AO; Langelier CRA 2-Gene Host Signature for Improved Accuracy of COVID-19 Diagnosis Agnostic to Viral Variants. mSystems 2023, DOI: 10.1128/msystems.00671-22. [DOI] [PMC free article] [PubMed]
- (59).Torul D; Omezli M. Is Saliva a Reliable Biofluid for the Detection of COVID-19? Dent. Med. Probl. 2021, 58 (2), 229–235. [DOI] [PubMed] [Google Scholar]
- (60).Pasomsub E; Watcharananan SP; Boonyawat K; Janchompoo P; Wongtabtim G; Suksuwan W; Sungkanuparph S; Phuphuakrat A. Saliva Sample as a Non-Invasive Specimen for the Diagnosis of Coronavirus Disease 2019: A Cross-Sectional Study. Clin. Microbiol. Infect. 2021, 27 (2), 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).To KK-W; Tsang OT-Y; Yip CC-Y; Chan K-H; Wu T-C; Chan JM-C; Leung W-S; Chik TS-H; Choi CY-C; Kandamby DH; Lung DC; Tam AR; Poon RW-S; Fung AY-F; Hung IF-N; Cheng VC-C; Chan JF-W; Yuen K-Y. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71 (15), 841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Huang C; Wang Y; Li X; Ren L; Zhao J; Hu Y; Zhang L; Fan G; Xu J; Gu X; Cheng Z; Yu T; Xia J; Wei Y; Wu W; Xie X; Yin W; Li H; Liu M; Xiao Y; Gao H; Guo L; Xie J; Wang G; Jiang R; Gao Z; Jin Q; Wang J; Cao B. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Lisboa Bastos M; Tavaziva G; Abidi SK; Campbell JR; Haraoui L-P; Johnston JC; Lan Z; Law S; MacLean E; Trajman A; Menzies D; Benedetti A; Ahmad Khan F. Diagnostic Accuracy of Serological Tests for Covid-19: Systematic Review and Meta-Analysis. BMJ. 2020, 370, m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wyllie AL; Fournier J; Casanovas-Massana A; Campbell M; Tokuyama M; Vijayakumar P; Warren JL; Geng B; Muenker MC; Moore AJ; Vogels CBF; Petrone ME; Ott IM; Lu P; Venkataraman A; Lu-Culligan A; Klein J; Earnest R; Simonov M; Datta R; Handoko R; Naushad N; Sewanan LR; Valdez J; White EB; Lapidus S; Kalinich CC; Jiang X; Kim DJ; Kudo E; Linehan M; Mao T; Moriyama M; Oh JE; Park A; Silva J; Song E; Takahashi T; Taura M; Weizman O-E; Wong P; Yang Y; Bermejo S; Odio CD; Omer SB; Dela Cruz CS; Farhadian S; Martinello RA; Iwasaki A; Grubaugh ND; Ko AI. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383 (13), 1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Russo MJ; Han M; Desroches PE; Manasa CS; Dennaoui J; Quigley AF; Kapsa RMI; Moulton SE; Guijt RM; Greene GW; Silva SM. Antifouling Strategies for Electrochemical Biosensing: Mechanisms and Performance toward Point of Care Based Diagnostic Applications. ACS Sens. 2021, 6 (4), 1482–1507. [DOI] [PubMed] [Google Scholar]
- (66).Fu K; Seo J; Kesler V; Maganzini N; Wilson BD; Eisenstein M; Murmann B; Soh HT. Accelerated Electron Transfer in Nanostructured Electrodes Improves the Sensitivity of Electrochemical Biosensors. Adv. Sci. 2021, 8 (23), No. 2102495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Yoon J; Conley BM; Shin M; Choi J-H; Bektas CK; Choi J-W; Lee K-B. Ultrasensitive Electrochemical Detection of Mutated Viral RNAs with Single-Nucleotide Resolution Using a Nanoporous Electrode Array (NPEA). ACS Nano 2022, 16 (4), 5764–5777. [DOI] [PubMed] [Google Scholar]
- (68).Loh KP; Bao Q; Ang PK; Yang J. The Chemistry of Graphene. J. Mater. Chem. 2010, 20 (12), 2277. [Google Scholar]
- (69).Seo G; Lee G; Kim MJ; Baek S-H; Choi M; Ku KB; Lee C-S; Jun S; Park D; Kim HG; Kim S-J; Lee J-O; Kim BT; Park EC; Kim S, Il Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14 (4), 5135–5142. [DOI] [PubMed] [Google Scholar]
- (70).Li Y; Peng Z; Holl NJ; Hassan MR; Pappas JM; Wei C; Izadi OH; Wang Y; Dong X; Wang C; Huang Y-W; Kim D; Wu C. MXene–Graphene Field-Effect Transistor Sensing of Influenza Virus and SARS-CoV-2. ACS Omega 2021, 6 (10), 6643–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Banga I; Paul A; France K; Micklich B; Cardwell B; Micklich C; Prasad SE Co.Tech-Electrochemical Handheld Breathalyzer COVID Sensing Technology. Sci. Rep. 2022, 12 (1), 4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Shan B; Broza YY; Li W; Wang Y; Wu S; Liu Z; Wang J; Gui S; Wang L; Zhang Z; Liu W; Zhou S; Jin W; Zhang Q; Hu D; Lin L; Zhang Q; Li W; Wang J; Liu H; Pan Y; Haick H. Multiplexed Nanomaterial-Based Sensor Array for Detection of COVID-19 in Exhaled Breath. ACS Nano 2020, 14 (9), 12125–12132. [DOI] [PubMed] [Google Scholar]
- (73).Samper IC; Sánchez-Cano A; Khamcharoen W; Jang I; Siangproh W; Baldrich E; Geiss BJ; Dandy DS; Henry CS. Electrochemical Capillary-Flow Immunoassay for Detecting Anti-SARS-CoV-2 Nucleocapsid Protein Antibodies at the Point of Care. ACS Sens. 2021, 6 (11), 4067–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Singh NK; Ray P; Carlin AF; Magallanes C; Morgan SC; Laurent LC; Aronoff-Spencer ES; Hall DA. Hitting the Diagnostic Sweet Spot: Point-of-Care SARS-CoV-2 Salivary Antigen Testing with an off-the-Shelf Glucometer. Biosens. Bioelectron. 2021, 180, No. 113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Amalfitano E; Karlikow M; Norouzi M; Jaenes K; Cicek S; Masum F; Sadat Mousavi P; Guo Y; Tang L; Sydor A; Ma D; Pearson JD; Trcka D; Pinette M; Ambagala A; Babiuk S; Pickering B; Wrana J; Bremner R; Mazzulli T; Sinton D; Brumell JH; Green AA; Pardee K. A Glucose Meter Interface for Point-of-Care Gene Circuit-Based Diagnostics. Nat. Commun. 2021, 12 (1), 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Beduk T; Beduk D; de Oliveira Filho JI; Zihnioglu F; Cicek C; Sertoz R; Arda B; Goksel T; Turhan K; Salama KN; Timur S. Rapid Point-of-Care COVID-19 Diagnosis with a Gold-Nanoarchitecture-Assisted Laser-Scribed Graphene Biosensor. Anal. Chem. 2021, 93 (24), 8585–8594. [DOI] [PubMed] [Google Scholar]
- (77).Eissa S; Zourob M. Development of a Low-Cost Cotton-Tipped Electrochemical Immunosensor for the Detection of SARS-CoV-2. Anal. Chem. 2021, 93, 1826. [DOI] [PubMed] [Google Scholar]
- (78).Kim HE; Schuck A; Lee SH; Lee Y; Kang M; Kim Y-S. Sensitive Electrochemical Biosensor Combined with Isothermal Amplification for Point-of-Care COVID-19 Tests. Biosens. Bioelectron. 2021, 182, No. 113168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ayankojo AG; Boroznjak R; Reut J; Öpik A; Syritski V. Molecularly Imprinted Polymer Based Electrochemical Sensor for Quantitative Detection of SARS-CoV-2 Spike Protein. Sensors Actuators B Chem. 2022, 353, No. 131160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Lomae A; Preechakasedkit P; Hanpanich O; Ozer T; Henry CS; Maruyama A; Pasomsub E; Phuphuakrat A; Rengpipat S; Vilaivan T; Chailapakul O; Ruecha N; Ngamrojanavanich N. Label Free Electrochemical DNA Biosensor for COVID-19 Diagnosis. Talanta 2023, 253, No. 123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Rocha DS; Baldo TA; Silva-Neto HA; Duarte-Junior GF; Bazílio GS; Borges CL; Parente-Rocha JA; de Araujo WR; de Siervo A; Paixão TLRC; Coltro WKT. Disposable and Eco-Friendly Electrochemical Immunosensor for Rapid Detection of SARS-CoV-2. Talanta 2024, 268, No. 125337. [DOI] [PubMed] [Google Scholar]
- (82).Idili A; Parolo C; Alvarez-Diduk R; Merkoç A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sens. 2021, 6 (8), 3093–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Zhou L; Hao P; Li H; Zhang Z. Electrochemical Resonance of Molecular Motion Enabling Label-, Antibody-, and Enzyme-Free Detection of SARS-CoV-2. ACS Sens. 2021, 6 (4), 1613–1620. [DOI] [PubMed] [Google Scholar]
- (84).Yousefi H; Mahmud A; Chang D; Das J; Gomis S; Chen JB; Wang H; Been T; Yip L; Coomes E; Li Z; Mubareka S; McGeer A; Christie N; Gray-Owen S; Cochrane A; Rini JM; Sargent EH; Kelley SO. Detection of SARS-CoV-2 Viral Particles Using Direct, Reagent-Free Electrochemical Sensing. J. Am. Chem. Soc. 2021, 143 (4), 1722–1727. [DOI] [PubMed] [Google Scholar]
- (85).Wang L; Wang X; Wu Y; Guo M; Gu C; Dai C; Kong D; Wang Y; Zhang C; Qu D; Fan C; Xie Y; Zhu Z; Liu Y; Wei D. Rapid and Ultrasensitive Electromechanical Detection of Ions, Biomolecules and SARS-CoV-2 RNA in Unamplified Samples. Nat. Biomed. Eng. 2022, 6 (3), 276–285. [DOI] [PubMed] [Google Scholar]
- (86).Liu H; Yang A; Song J; Wang N; Lam P; Li Y; Law HK; Yan F. Ultrafast, Sensitive, and Portable Detection of COVID-19 IgG Using Flexible Organic Electrochemical Transistors. Sci. Adv. 2021, DOI: 10.1126/sciadv.abg8387. [DOI] [PMC free article] [PubMed]
- (87).Guo K; Wustoni S; Koklu A; Díaz-Galicia E; Moser M; Hama A; Alqahtani AA; Ahmad AN; Alhamlan FS; Shuaib M; Pain A; McCulloch I; Arold ST; Grünberg R; Inal S. Rapid Single-Molecule Detection of COVID-19 and MERS Antigens via Nanobody-Functionalized Organic Electrochemical Transistors. Nat. Biomed. Eng. 2021, 5 (7), 666–677. [DOI] [PubMed] [Google Scholar]
- (88).Goh B; Ching K; Soares Magalhães RJ; Ciocchetta S; Edstein MD; Maciel-de-Freitas R; Sikulu-Lord MT. The Application of Spectroscopy Techniques for Diagnosis of Malaria Parasites and Arboviruses and Surveillance of Mosquito Vectors: A Systematic Review and Critical Appraisal of Evidence. PLoS Negl. Trop. Dis. 2021, 15 (4), No. e0009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Sahu RK; Mordechai S. Spectroscopic Techniques in Medicine: The Future of Diagnostics. Appl. Spectrosc. Rev. 2016, 51 (6), 484–499. [Google Scholar]
- (90).Shrivastav AM; Cvelbar U; Abdulhalim I. A Comprehensive Review on Plasmonic-Based Biosensors Used in Viral Diagnostics. Commun. Biol. 2021, 4 (1), No. 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Chen Y; Duan W; Xu L; Li G; Wan Y; Li H. Nanobody-Based Label-Free Photoelectrochemical Immunoassay for Highly Sensitive Detection of SARS-CoV-2 Spike Protein. Anal. Chim. Acta 2022, 1211, No. 339904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Liu B; Zhuang J; Wei G. Recent Advances in the Design of Colorimetric Sensors for Environmental Monitoring. Environ. Sci. Nano 2020, 7 (8), 2195–2213. [Google Scholar]
- (93).Maddali H; Miles CE; Kohn J; O’Carroll DM. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem. 2021, 22 (7), 1176–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Ince B; Sezgintürk MK. Lateral Flow Assays for Viruses Diagnosis: Up-to-Date Technology and Future Prospects. TrAC Trends Anal. Chem. 2022, 157, No. 116725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Lou T; Chen L; Chen Z; Wang Y; Chen L; Li J. Colorimetric Detection of Trace Copper Ions Based on Catalytic Leaching of Silver-Coated Gold Nanoparticles. ACS Appl. Mater. Interfaces 2011, 3 (11), 4215–4220. [DOI] [PubMed] [Google Scholar]
- (96).Xie Q-Y; Wu Y-H; Xiong Q-R; Xu H-Y; Xiong Y-H; Liu K; Jin Y; Lai W-H. Advantages of Fluorescent Microspheres Compared with Colloidal Gold as a Label in Immunochromatographic Lateral Flow Assays. Biosens. Bioelectron. 2014, 54, 262–265. [DOI] [PubMed] [Google Scholar]
- (97).Abdellatif AA; Younis MA; Alsharidah M; Al Rugaie O; Tawfeek HM. Biomedical Applications of Quantum Dots: Overview, Challenges, and Clinical Potential. Int. J. Nanomedicine 2022, 17, 1951–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Koczula KM; Gallotta A. Lateral Flow Assays. Essays Biochem. 2016, 60 (1), 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Rosati G; Idili A; Parolo C; Fuentes-Chust C; Calucho E; Hu L; Castro e Silva C de C; Rivas L; Nguyen EP; Bergua JF; Alvárez-Diduk R; Muñoz J; Junot C; Penon O; Monferrer D; Delamarche E; Merkoç A. Nanodiagnostics to Face SARS-CoV-2 and Future Pandemics: From an Idea to the Market and Beyond. ACS Nano 2021, 15 (11), 17137–17149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Zhou Y; Wu Y; Ding L; Huang X; Xiong Y. Point-of-Care COVID-19 Diagnostics Powered by Lateral Flow Assay. TrAC Trends Anal. Chem. 2021, 145, No. 116452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Carrell C; Kava A; Nguyen M; Menger R; Munshi Z; Call Z; Nussbaum M; Henry C. Beyond the Lateral Flow Assay: A Review of Paper-Based Microfluidics. Microelectron. Eng. 2019, 206, 45–54. [Google Scholar]
- (102).Patchsung M; Jantarug K; Pattama A; Aphicho K; Suraritdechachai S; Meesawat P; Sappakhaw K; Leelahakorn N; Ruenkam T; Wongsatit T; Athipanyasilp N; Eiamthong B; Lakkanasirorat B; Phoodokmai T; Niljianskul N; Pakotiprapha D; Chanarat S; Homchan A; Tinikul R; Kamutira P; Phiwkaow K; Soithongcharoen S; Kantiwiriyawanitch C; Pongsupasa V; Trisrivirat D; Jaroensuk J; Wongnate T; Maenpuen S; Chaiyen P; Kamnerdnakta S; Swangsri J; Chuthapisith S; Sirivatanauksorn Y; Chaimayo C; Sutthent R; Kantakamalakul W; Joung J; Ladha A; Jin X; Gootenberg JS; Abudayyeh OO; Zhang F; Horthongkham N; Uttamapinant C. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4 (12), 1140–1149. [DOI] [PubMed] [Google Scholar]
- (103).Broughton JP; Deng X; Yu G; Fasching CL; Servellita V; Singh J; Miao X; Streithorst JA; Granados A; Sotomayor-Gonzalez A; Zorn K; Gopez A; Hsu E; Gu W; Miller S; Pan C-Y; Guevara H; Wadford DA; Chen JS; Chiu CY. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38 (7), 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Budd J; Miller BS; Weckman NE; Cherkaoui D; Huang D; Decruz AT; Fongwen N; Han G-R; Broto M; Estcourt CS; Gibbs J; Pillay D; Sonnenberg P; Meurant R; Thomas MR; Keegan N; Stevens MM; Nastouli E; Topol EJ; Johnson AM; Shahmanesh M; Ozcan A; Collins JJ; Fernandez Suarez M; Rodriguez B; Peeling RW; McKendry RA. Lateral Flow Test Engineering and Lessons Learned from COVID-19. Nat. Rev. Bioeng. 2023, 1 (1), 13–31. [Google Scholar]
- (105).Zheng C; Wang K; Zheng W; Cheng Y; Li T; Cao B; Jin Q; Cui D. Rapid Developments in Lateral Flow Immunoassay for Nucleic Acid Detection. Analyst 2021, 146 (5), 1514–1528. [DOI] [PubMed] [Google Scholar]
- (106).Zhang Y; Chai Y; Hu Z; Xu Z; Li M; Chen X; Yang C; Liu J. Recent Progress on Rapid Lateral Flow Assay-Based Early Diagnosis of COVID-19. Front. Bioeng. Biotechnol. 2022, DOI: 10.3389/fbioe.2022.866368. [DOI] [PMC free article] [PubMed]
- (107).Hsiao WW-W; Le T-N; Pham DM; Ko H-H; Chang H-C; Lee C-C; Sharma N; Lee C-K; Chiang W-H. Recent Advances in Novel Lateral Flow Technologies for Detection of COVID-19. Biosensors 2021, 11 (9), 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Liu Y; Tan Y; Fu Q; Lin M; He J; He S; Yang M; Chen S; Zhou J. Reciprocating-Flowing on-a-Chip Enables Ultra-Fast Immunobinding for Multiplexed Rapid ELISA Detection of SARS-CoV-2 Antibody. Biosens. Bioelectron. 2021, 176, No. 112920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Gong F; Wei H; Qi J; Ma H; Liu L; Weng J; Zheng X; Li Q; Zhao D; Fang H; Liu L; He H; Ma C; Han J; Sun A; Wang B; Jin T; Li B; Li B. Pulling-Force Spinning Top for Serum Separation Combined with Paper-Based Microfluidic Devices in COVID-19 ELISA Diagnosis. ACS Sens. 2021, 6 (7), 2709–2719. [DOI] [PubMed] [Google Scholar]
- (110).Tomita N; Mori Y; Kanda H; Notomi T. Loop-Mediated Isothermal Amplification (LAMP) of Gene Sequences and Simple Visual Detection of Products. Nat. Protoc. 2008, 3 (5), 877–882. [DOI] [PubMed] [Google Scholar]
- (111).Amaral C; Antunes W; Moe E; Duarte AG; Lima LMP; Santos C; Gomes IL; Afonso GS; Vieira R; Teles HSS; Reis MS; da Silva MAR; Henriques AM; Fevereiro M; Ventura MR; Serrano M; Pimentel C. A Molecular Test Based on RT-LAMP for Rapid, Sensitive and Inexpensive Colorimetric Detection of SARS-CoV-2 in Clinical Samples. Sci. Rep. 2021, 11 (1), No. 16430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Dao Thi VL; Herbst K; Boerner K; Meurer M; Kremer LP; Kirrmaier D; Freistaedter A; Papagiannidis D; Galmozzi C; Stanifer ML; Boulant S; Klein S; Chlanda P; Khalid D; Barreto Miranda I; Schnitzler P; Kräusslich H-G; Knop M; Anders S. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, DOI: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed]
- (113).Garneret P; Coz E; Martin E; Manuguerra J-C; Brient-Litzler E; Enouf V; González Obando DF; Olivo-Marin J-C; Monti F; van der Werf S; Vanhomwegen J; Tabeling P. Performing Point-of-Care Molecular Testing for SARS-CoV-2 with RNA Extraction and Isothermal Amplification. PLoS One 2021, 16 (1), No. e0243712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Ganguli A; Mostafa A; Berger J; Aydin MY; Sun F; Ramirez SAS de Valera E; Cunningham BT; King WP; Bashir R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (37), 22727–22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Jalali Moghaddam M; Ghavipour M. Towards Smart Diagnostic Methods for COVID-19: Review of Deep Learning for Medical Imaging. IPEM-Translation 2022, 3–4, No. 100008. [DOI] [PMC free article] [PubMed]
- (116).Alafeef M; Moitra P; Dighe K; Pan D. Hyperspectral Mapping for the Detection of SARS-CoV-2 Using Nanomolecular Probes with Yoctomole Sensitivity. ACS Nano 2021, 15 (8), 13742–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Tang L; Li J. Plasmon-Based Colorimetric Nanosensors for Ultrasensitive Molecular Diagnostics. ACS Sens. 2017, 2 (7), 857–875. [DOI] [PubMed] [Google Scholar]
- (118).Ventura BD; Cennamo M; Minopoli A; Campanile R; Censi SB; Terracciano D; Portella G; Velotta R. Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. ACS Sens. 2020, 5 (10), 3043–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Nguyen PQ; Soenksen LR; Donghia NM; Angenent-Mari NM; de Puig H; Huang A; Lee R; Slomovic S; Galbersanini T; Lansberry G; Sallum HM; Zhao EM; Niemi JB; Collins JJ. Wearable Materials with Embedded Synthetic Biology Sensors for Biomolecule Detection. Nat. Biotechnol. 2021, 39 (11), 1366–1374. [DOI] [PubMed] [Google Scholar]
- (120).Leung NHL; Chu DKW; Shiu EYC; Chan K-H; McDevitt JJ; Hau BJP; Yen H-L; Li Y; Ip DKM; Peiris JSM; Seto W-H; Leung GM; Milton DK; Cowling BJ. Respiratory Virus Shedding in Exhaled Breath and Efficacy of Face Masks. Nat. Med. 2020, 26 (5), 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Celiker T; Ghorbanizamani F; Moulahoum H; Guler Celik E; Tok K; Zihnioglu F; Cicek C; Sertoz R; Arda B; Goksel T; Turhan K; Timur S; Yagci Y. Fluorescent Bioassay for SARS-CoV-2 Detection Using Polypyrene-g-Poly(ε-Caprolactone) Prepared by Simultaneous Photoinduced Step-Growth and Ring-Opening Polymerizations. Microchim. Acta 2022, 189 (5), No. 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Zheng Y; Song K; Cai K; Liu L; Tang D; Long W; Zhai B; Chen J; Tao Y; Zhao Y; Liang S; Huang Q; Liu Q; Zhang Q; Chen Y; Liu Y; Li H; Wang P; Lan K; Liu H; Xu K. B-Cell-Epitope-Based Fluorescent Quantum Dot Biosensors for SARS-CoV-2 Enable Highly Sensitive COVID-19 Antibody Detection. Viruses 2022, 14 (5), 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Lin P-Y; Chung Y-W; Chuang E-Y; Wang Y-C; Lai M-C; Hsu Y-C; Lin C-C; Hsiao Y-C. Cholesteric Liquid Crystal Biosensor Platform with Image Analysis for Rapid Detection of COVID-19. Front. Bioeng. Biotechnol. 2023, DOI: 10.3389/fbioe.2023.1148446. [DOI] [PMC free article] [PubMed]
- (124).Zhang C; Zhou L; Du K; Zhang Y; Wang J; Chen L; Lyu Y; Li J; Liu H; Huo J; Li F; Wang J; Sang P; Lin S; Xiao Y; Zhang K; He K. Foundation and Clinical Evaluation of a New Method for Detecting SARS-CoV-2 Antigen by Fluorescent Microsphere Immunochromatography. Front. Cell. Infect. Microbiol. 2020, DOI: 10.3389/fcimb.2020.553837. [DOI] [PMC free article] [PubMed]
- (125).Grant BD; Anderson CE; Williford JR; Alonzo LF; Glukhova VA; Boyle DS; Weigl BH; Nichols KP. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92 (16), 11305–11309. [DOI] [PubMed] [Google Scholar]
- (126).Mertens P; De Vos N; Martiny D; Jassoy C; Mirazimi A; Cuypers L; Van den Wijngaert S; Monteil V; Melin P; Stoffels K; Yin N; Mileto D; Delaunoy S; Magein H; Lagrou K; Bouzet J; Serrano G; Wautier M; Leclipteux T; Van Ranst M; Vandenberg O. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020, DOI: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed]
- (127).Wang C; Shi D; Wan N; Yang X; Liu H; Gao H; Zhang M; Bai Z; Li D; Dai E; Rong Z; Wang S. Development of Spike Protein-Based Fluorescence Lateral Flow Assay for the Simultaneous Detection of SARS-CoV-2 Specific IgM and IgG. Analyst 2021, 146 (12), 3908–3917. [DOI] [PubMed] [Google Scholar]
- (128).Huang C; Wen T; Shi F-J; Zeng X-Y; Jiao Y-J. Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega 2020, 5 (21), 12550–12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Wang Z; Zheng Z; Hu H; Zhou Q; Liu W; Li X; Liu Z; Wang Y; Ma Y. A Point-of-Care Selenium Nanoparticle-Based Test for the Combined Detection of Anti-SARS-CoV-2 IgM and IgG in Human Serum and Blood. Lab Chip 2020, 20 (22), 4255–4261. [DOI] [PubMed] [Google Scholar]
- (130).Chen Z; Zhang Z; Zhai X; Li Y; Lin L; Zhao H; Bian L; Li P; Yu L; Wu Y; Lin G. Rapid and Sensitive Detection of Anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92 (10), 7226–7231. [DOI] [PubMed] [Google Scholar]
- (131).Davidson JL; Wang J; Maruthamuthu MK; Dextre A; Pascual-Garrigos A; Mohan S; Putikam SVS; Osman FOI; McChesney D; Seville J; Verma MS. A Paper-Based Colorimetric Molecular Test for SARS-CoV-2 in Saliva. Biosens. Bioelectron. X 2021, 9, No. 100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Harun-Ur-Rashid M; Foyez T; Jahan I; Pal K; Imran AB. Rapid Diagnosis of COVID-19 via Nano-Biosensor-Implemented Biomedical Utilization: A Systematic Review. RSC Adv. 2022, 12 (15), 9445–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Parihar A; Ranjan P; Sanghi SK; Srivastava AK; Khan R. Point-of-Care Biosensor-Based Diagnosis of COVID-19 Holds Promise to Combat Current and Future Pandemics. ACS Appl. Bio Mater. 2020, 3 (11), 7326–7343. [DOI] [PubMed] [Google Scholar]
- (134).Bhalla N; Pan Y; Yang Z; Payam AF. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14 (7), 7783–7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).At-Home OTC COVID-19 Diagnostic Tests | FDA. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/home-otc-covid-19-diagnostic-tests (accessed on October 14, 2023).
- (136).Jiang C; Mu X; Du B; Tong Z. A Review of Electrochemical Biosensor Application in the Detection of the SARS-COV-2. Micro Nano Lett. 2022, 17 (3), 49–58. [Google Scholar]
- (137).Cao E; Chen Y; Cui Z; Foster PR. Effect of Freezing and Thawing Rates on Denaturation of Proteins in Aqueous Solutions. Biotechnol. Bioeng. 2003, 82 (6), 684–690. [DOI] [PubMed] [Google Scholar]
- (138).Yüce M; Kurt H. How to Make Nanobiosensors: Surface Modification and Characterisation of Nanomaterials for Biosensing Applications. RSC Adv. 2017, 7 (78), 49386–49403. [Google Scholar]
- (139).Xu T; Geng Z. Strategies to Improve Performances of LSPR Biosensing: Structure, Materials, and Interface Modification. Biosens. Bioelectron. 2021, 174, No. 112850. [DOI] [PubMed] [Google Scholar]
- (140).Lin P-H; Li B-R. Antifouling Strategies in Advanced Electrochemical Sensors and Biosensors. Analyst 2020, 145 (4), 1110–1120. [DOI] [PubMed] [Google Scholar]
- (141).Chen X; Noy A. Antifouling Strategies for Protecting Bioelectronic Devices. APL Mater. 2021, 9 (2), No. 020701. [Google Scholar]
- (142).Wei H; Zhang C; Du X; Zhang Z. Research Progress of Biosensors for Detection of SARS-CoV-2 Variants Based on ACE2. Talanta 2023, 251, No. 123813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Jackson CB; Farzan M; Chen B; Choe H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23 (1), 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Song M; Lin X; Peng Z; Xu S; Jin L; Zheng X; Luo H. Materials and Methods of Biosensor Interfaces With Stability. Front. Mater. 2021, DOI: 10.3389/fmats.2020.583739. [DOI]
- (145).Shaver A; Arroyo-Currás N. The Challenge of Long-Term Stability for Nucleic Acid-Based Electrochemical Sensors. Curr. Opin. Electrochem. 2022, 32, No. 100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).de Lima LF; Ferreira AL; Awasthi S; Torres MDT; Friedman HM; Cohen GH; de Araujo WR; de la Fuente-Nunez C. Rapid and Accurate Detection of Herpes Simplex Virus Type 2 Using a Low-Cost Electrochemical Biosensor. Cell Reports Phys. Sci. 2023, 4, No. 101513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Yabuki S. How to Lengthen the Long-Term Stability of Enzyme Membranes: Trends and Strategies. Catalysts 2017, 7 (12), 36. [Google Scholar]
- (148).McClements J; Bar L; Singla P; Canfarotta F; Thomson A; Czulak J; Johnson RE; Crapnell RD; Banks CE; Payne B; Seyedin S; Losada-Pérez P; Peeters M. Molecularly Imprinted Polymer Nanoparticles Enable Rapid, Reliable, and Robust Point-of-Care Thermal Detection of SARS-CoV-2. ACS Sens. 2022, 7 (4), 1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Yarman A; Kurbanoglu S. Molecularly Imprinted Polymer-Based Sensors for SARS-CoV-2: Where Are We Now? Biomimetics 2022, 7 (2), 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Larkins MC; Thombare A. Point-of-Care Testing; StatPearls Publishing, 2023. [PubMed] [Google Scholar]
- (151).de Lima LF; Ferreira AL; Ranjan I; Collman RG; de Araujo WR; de la Fuente-Nunez C. A Bacterial Cellulose-Based and Low-Cost Electrochemical Biosensor for Ultrasensitive Detection of SARS-CoV-2. Cell Reports Phys. Sci. 2023, 4 (8), No. 101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Evans R; McNamee M; Guy O. Ethics, Nanobiosensors and Elite Sport: The Need for a New Governance Framework. Sci. Eng. Ethics 2017, 23 (6), 1487–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Abdelghany TM; Ganash M; Bakri MM; Qanash H; Al-Rajhi AMH; Elhussieny NI. SARS-CoV-2, the Other Face to SARS-CoV and MERS-CoV: Future Predictions. Biomed. J. 2021, 44 (1), 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]