Abstract
Classical plant breeding methods are limited in their ability to confer disease resistance on plants. However, in recent years, advancements in molecular breeding and biotechnological have provided new approaches to overcome these limitations and protect plants from disease. Antimicrobial peptides (AMPs) constitute promising agents that may be able to protect against infectious agents. Recently, peptides have been recombinantly produced in plants at scale and low cost. Because AMPs are less likely than conventional antimicrobials to elicit resistance of pathogenic bacteria, they open up exciting new avenues for agricultural applications. Here, we review recent advances in the design and production of bioactive recombinant AMPs that can effectively protect crop plants from diseases.
Keywords: Antimicrobial peptides, Recombinant technology, Genetic engineering, Transgenic plants
1. Introduction
In the universe of extant antimicrobial molecules, antimicrobial peptides (AMPs) hold great promise to protect crop plants, farm animals, and even humans against pathogens and insects (Lei et al., 2019). Unlike the existing armamentarium of antimicrobial drugs, one interesting feature of AMPs is their nonspecific mode of action, which is to destabilize the membrane of microbial pathogens. This results in considerably less development of bacterial resistance than seen with conventional antibiotics (Lei et al., 2019; Mwangi et al., 2019).
AMPs are peptides chains composed of 6 to 50 amino acid residues (Srivastava et al., 2021) expressed by almost all living organisms as part of their innate immune system (Nguyen et al., 2011; Zasloff, 2002, 2006). In eukaryotic cells, they are either constitutively expressed or induced upon abiotic and abiotic stresses (Nawrot et al., 2014). The prevalence of cationic amino acid residues such as lysine and arginine over the acidic ones make AMPs have positive net charged (+2 to +12), enabling more effective electrostatic interactions with negatively charged membrane components (Da Costa et al., 2015; Sani and Separovic, 2016). As of December 2023, around 21,367 AMPs have been characterized, including 1923 AMPs from plants (https://dbaasp.org/home).
Purothionin, which is found in the endosperm of wheat kernels, was the first plant-derived AMP isolated and characterized (Balls et al., 1942). Since then, hundreds of AMPs belonging to different classes have been isolated from plant organs and species (Table 1). Similar to other AMPs classes, plant peptides have been also classified based on their amino acids sequence, number of disulfide bridges, mechanism of action (Lay and Anderson, 2005), and net charge (Barbosa Pelegrini et al., 2011). In addition to their antimicrobial activities, some plant AMPs have been shown to regulate plant growth and development (Li et al., 2021). Although a number of plant-derived AMPs have been proposed as potential alternatives to conventional antibiotics, none are currently used in clinical practice to treat fungal or bacterial infections (Porto et al., 2018; Divyashree et al., 2020), partly because most plant encoding AMPs undergo post-translational modifications, so often to achieve a therapeutic effect, a high dose is needed.
Table 1.
Overview of the main groups of plant-derived AMPs isolated and cloned from different plant species.
| Peptide name | Length (number of amino acids), Mass (kDa) |
Localization | Target organisms | Host organ target | Activity |
|---|---|---|---|---|---|
| Thionins (α/β/γ) | 45–48, ~5 | L/F/S | Bacteria/Fungi | Membrane lipids | MPA and lysis |
| Defensins | 45–54, ~5–7 | Fl/R/F/S | Bacteria/Fungi/Insects | Membrane and intracellular signaling cascades | Hinders pathogen growth/inhibits insect digestive proteins |
| Heveins | 30–45, ~5 | L/B | Bacterial/Fungi/Mammals | Microbial cell wall | Cell wall biosynthesis inhibition/Allergens |
| Knottins | 28–37, ~4 | Fl/R/F/S | Fungi | Microbial membrane and intracellular components. | α-amylase and protease inhibitors |
| Cyclotides | 28–37, ~4 | Fl/R/F/S | Bacteria/Fungi/Arthropod | Membrane lipids | MPA/Disruption of arthropod digestive systems |
| Lipid transfer proteins | 70–90, ~9–10 | S/L | Bacteria/Fungi | Microbial membranes | MPA/Increasing membrane permeabilization |
| Snakins | 60–70, ~7 | L/F/ Bulbs/ Tubers | Bacteria/Fungi | Unknown | Unknown |
| A | 31–50, ~4–5 | S/L | Bacteria/Fungi | Unknown | Antimicrobial and trypsin-inhibitory activity, Ribosome-iAnactivating activity |
L: Leaves; F: Fruits; S: Seeds; B: Bark; R; Roots; MPA: Membrane Permeability Activity.
2. Mechanisms of action of antimicrobial peptides
Unlike antibiotics, which usually have specific intracellular targets, AMPs have diverse, and sometimes multiple, mechanisms of action (Sumi et al., 2015; Torres et al., 2019). According to their mechanism of action, AMPs can be grouped into membrane-targeting or intracellular-targeting agents. Most known AMPs actively engage with the bacterial cell membrane via electrostatic interactions (Hollmann et al., 2018). When the peptide is sufficiently close to the phospholipids of bacterial membranes, van der Waals and hydrogen bonds come into play, and the amphiphilic sequence of the AMP effectively interacts with the lipid bilayer of the membrane via non-specific interactions. As a result, pathogens rarely develop resistance to membrane-targeting peptides (Pfalzgraff et al., 2018), so these are excellent candidates as ways to help plants resist pathogenic invaders. Membrane-targeting AMPs are mainly amphipathic molecules, i.e., they present a balance between cationic and non-polar residues, leading to pathogen membrane disruption mechanisms, such as destabilization, changes in fluidity, and depolarization; whereas AMPs such as dermaseptin act through either cytolysis or cell membrane disruption, killing the cell (Belmadani et al., 2018).
Despite their ability to disrupt the bacterial cell membrane, dermaseptins (except dermaseptin S4) exhibit no toxicity against erythrocytes or other mammalian cells, a desirable property for antimicrobial agents (Feder et al., 2000). Some AMPs, such as defensins (Shafee et al., 2017) and melittin (Sun et al., 2017), act by forming pores on the bacterial membrane, resulting in its depolarization. Most AMPs exhibit various structures upon interactions with cell membranes (Haney et al., 2017; Mingeot-Leclercq and Décout, 2016). It has also been reported that AMPs suppress the synthesis of proteins, nucleic acids, and cell walls, as well as host enzymatic activities (Cudic and Otvos Jr, 2002; Krizsan et al., 2014; Mansour et al., 2014). Although most AMPs when interacting with the membrane present amphipathic conformations (Matsuzaki, 1999), additional evidence suggests that some AMPs can penetrate the membrane lipid layer, subsequently targeting intracellular processes (Savini et al., 2018). Various models have been devised to illustrate the mode of action of AMPs on the membrane, including the barrel-stave model (Shabir et al., 2018), the carpet model (Han et al., 2017),and the toroidal pore model (Wimley, 2010). However, we still do not fully understand how peptides act on the membrane because nonspecific interactions make it challenging to predict the optimal physicochemical balance needed for each of the membrane-targeting mechanisms to take place. Indeed, AMPs are often found to operate through multiple mechanisms of action.
3. Role of AMPs in plant defense responses
As they evolve, plants are continuously exposed to diverse biotic and abiotic stresses. Abiotic stresses such as drought, heat, salinity, and cold stresses could reduce the yield of economically important crops worldwide by more than 50%. Also, biotic stresses such as living organisms (including infectious agents) can decrease yields by as much as 35% (Flood, 2010; Spence et al., 2015).
To counteract pathogenic infections, plants have developed various mechanisms of defense, including the generation of antimicrobial compounds (Tam et al., 2015; Campos et al., 2018). Plants can respond to diverse biotic stresses via the recognition of pathogen/microbe-associated molecular patterns (PAMPs/MAMPs). PAMPs, once sensed by plant membrane receptors, trigger an immediate immune response that can prevent the spread of infection to other plant organs (Bigeard et al., 2015). Even though plant immunity is robust, many pathogens have evolved mechanisms that suppress plant immune responses by synthesizing effector molecules, which can facilitate plant cell infection. According to the zigzag model, plants have evolved specific receptors to sense PAMPs (Thomma et al., 2011). Upon pathogen recognition, plant cells initiate the expression of the conserved mitogen-activated protein kinase (MAPK) cascade, involving the activation of MYB, WRKY, AP2/ERF, and bZIP genes belonging to the specific transcription factor (TF) families. Consequently, MAPK signal transduction pathways activate the biosynthesis of AMPs, as well as secondary metabolites such as phytoanticipins, phytoalexins, and pathogenesis-associated proteins (Karpun et al., 2015; Miller et al., 2017; Onaga and Wydra, 2016; Piasecka et al., 2015).
The expression of AMPs seems to share the same signaling module associated with the control of several other plant defense-related responses; therefore, it has been proposed that AMP-encoding genes are part of a general immunity-signaling network. For instance, plant growth regulators such as jasmonic acid, ethylene, and salicylic acid induce the expression of AMP genes in plant species. The correlation between AMP expression and the involvement of various families of plant TFs in plant innate immunity indicates that AMPs are a vital component of the plant’s resistance to pathogens.
AMPs play bifunctional roles as either positive or negative regulators of abiotic and biotic stress responses related to reactive oxygen species (ROS), hormone production, heat shock protein synthesis, and MAPK cascade initiation steps. For instance, a pea defensin that is constitutively expressed in seeds and leaves crosses fungal membranes to interact with nuclear-localized proteins involved with the regulation of fungal cell wall synthesis as well as cell division cycling, thereby hampering pathogen cell growth (Almeida et al., 2000; Lobo et al., 2007). Furthermore, AMPs directly regulate cellular redox status. For instance, a defensin peptide from sweet potato (SPD1) was shown to regulate the redox status of ascorbate (Huang et al., 2008). Similarly, the expression of a chickpea defensin gene (Ca-AFP) led to increased drought tolerance through the regulation of catalase, ascorbate peroxidase, superoxide dismutase, and the amount of proline in Arabidopsis transgenic plants (Kumar et al., 2019).
Thus, AMPs are part of complex immune defense responses in plant systems. Some of these responses may be redundant, as it was found that knocking out the expression of certain AMPs and analyzing the loss of function of mutated AMPs from plants may not necessarily result in a detectable imbalance in the response of plants to pathogenic infection (Campos et al., 2018); this study suggested that identifying AMPs via traditional molecular genetic approaches (e.g., gene knockouts or gene silencing) may not yield effective results.
4. Important features for the antimicrobial activity of AMPs
The physicochemical features of AMPs are essential for their anti-microbial activity. By understanding these features, AMPs can be identified and designed with improved biological activities. The most studied and accepted physicochemical features of AMPs from plants include: (i) positive net charge, containing cationic (+2 to +9) amino acid residues such as lysine and arginine; (ii) hydrophobicity, AMPs from plants are usually slightly more hydrophobic than other AMPs; (iii) amphiphilicity, i.e., they present hydrophilic and hydrophobic portions; (iv) length, AMPs from plants are short peptides, usually containing 10 to 50 amino acid residues in the sequence; and (v) secondary structure, AMPs from plants present varied structure ranging from α-helical to constrained while also and adopting β-like structures.
In addition to the physicochemical features listed above, two additional factors influence the antimicrobial activity of AMPs from plants. One is the presence of disulfide bonds stabilizing their secondary structure and favoring insertion and destabilization of the microorganisms’ membrane (Nawrot et al., 2014; de Oliveira and Gomes, 2009). The second is the presence of post-translational modifications made by plant cells such as glycosylation that will directly impact how the peptide will interact with the membrane and how the AMP will get around resistance mechanisms of bacterial cells (Grimsey et al., 2020; Bednarska et al., 2017).
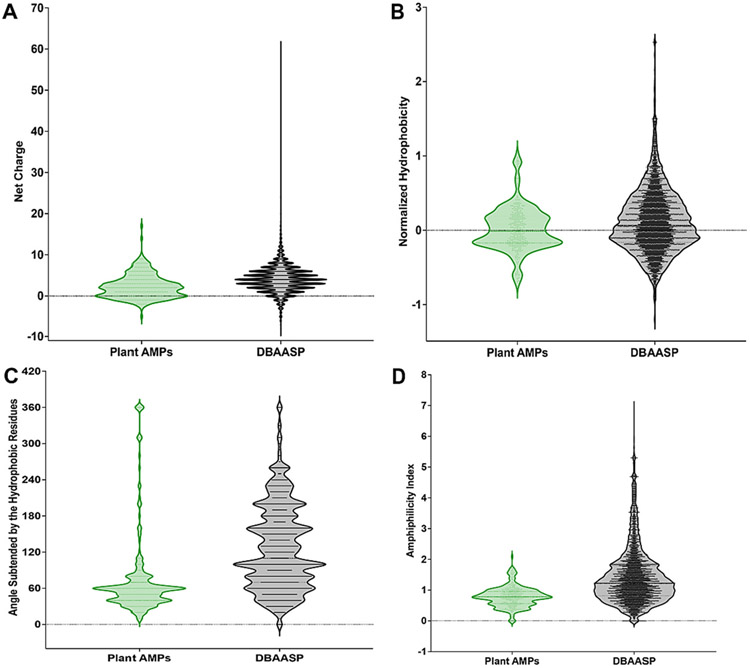
To evaluate the importance of these physicochemical features in the context of plant AMPs, we calculated their main physicochemical features compared to all other AMPs not from plants, using the Database of Antimicrobial Activity and Structure of Peptides (DBAASP) server (Fig. 1).
Fig. 1. Physicochemical features of plant-derived AMPs.
The AMPs were obtained from the Database of Antimicrobial Activity and Structure of Peptides (DBAASP) server, as well as the physicochemical features that are most relevant to their antimicrobial activity: (A) net charge, (B) normalized hydrophobicity, (C) angle subtended by the hydrophobic residues, and (D) amphiphilicity index.
5. Net charge
Most reported active AMPs are cationic, i.e., the frequency of positively charged amino acid residues (arginine and lysine) present in AMPs is higher compared to those negatively charged (aspartic and glutamic acids). Plant AMPs, on average, are slightly less cationic compared to other AMPs (Fig. 1A). Despite this, plant AMPs are as active as other AMPs, likely because of the balance between cationic and hydrophobic (Fig. 1B) amino acid residues and other structural descriptors, such as angle subtended by polar residues (Fig. 1C), and amphipathicity (Fig. 1D).
A positive net charge is important for a peptide because the bacterial outer and cytoplasmic membranes have an overall negative net charge, and plant cytoplasmic membranes mostly display zwitterionic and neutral membrane lipids. This suggests that net charge might not be a crucial determinant for toxicity toward plant cells, which may have implications to improve the recombinant expression of peptides using transgenic plants (Shagaghi et al., 2018). Since fusion partners are translationally linked, an amino acid substitution in one fusion partner may not affect the activity of the other. Hence, increasing or decreasing the net charge of the fusion proteins will not necessarily cause a net charge shift in the AMP molecule. This is particularly important because sometimes it is necessary to replace certain amino acids within AMP fusion proteins for purposes other than to improve antimicrobial activity. For example, changes might be needed to modulate adhesiveness to host membrane components. Since hybrid peptides are typically fused with a stable and flexible linker, the chimeric peptide partners of the new hybrid AMP maintain their original functions.
6. Hydrophobicity
The hydrophobicity of plant AMPs is generally higher than that of other antimicrobial peptides (Fig. 1B), such as those produced by animals or bacteria (Edwards et al., 2016). This is likely because plant cells are surrounded by a thick cell wall that makes it difficult for hydrophilic peptides to penetrate. As a result, plant AMPs have evolved to be more hydrophobic to be able to reach their target (Li et al., 2021).
The hydrophobicity of plant AMPs is also thought to play a role in their selectivity. More hydrophobic AMPs are more likely to interact with and disrupt the membranes of Gram-negative bacteria, which have a more hydrophobic outer membrane than Gram-positive bacteria (Li et al., 2021), and fungi that have a more rigid cellular membrane because of the ergosterol molecules embedded on them and with a higher proportion of unsaturated fatty acids to confer them flexibility.
7. Amphiphilicity
According to the predictions obtained from DBAASP, we observed that AMPs from plants present a smaller angle subtended by the hydrophobic portion of the peptide (60° on average) compared to AMPs from other sources (110° on average) (Fig. 1C). This impacts directly the amphiphilicity of these peptides. Plant AMPs show lower amphiphilicity than other reported AMPs, i.e., plant AMPs present a more even distribution of hydrophobic and hydrophilic residues than other AMPs, which have a more amphipathic sequence (Fig. 1D). The theoretical implications of this are that the interactions of plant AMPs would not be as effective with the amphipathic phospholipids of membrane bilayers such as the bacterial membranes, compared to AMPs from other sources. However, the activity range and efficacy of plant-derived AMPs has been shown to be broad and considerably high, indicating that amphiphilicity is not a determinant of activity for this class of peptides.
8. Length
Most AMPs are composed of approximately 4 to 70 amino acids. The effect of peptide length on antimicrobial activity is related to secondary structure and how much the peptide can diffuse in biological membranes. The minimal structural motifs present in α-helical and β-like structures are heptads and octets (seven and eight amino acid residues, respectively), but longer motifs leading to amphiphilic structures have also been reported (Bahar and Ren, 2013; Shagaghi et al., 2018). While there are some reports indicating that variations in length do not directly correlate with the antimicrobial activity of AMPs (Scott et al., 1999), other studies show that longer peptides tend to be more active and with a higher net charge (Wang, 2020). For instance, a short derivative of melittin composed of 15 residues demonstrated almost 5- to 7-fold lower antimicrobial activity and 300-fold lower toxicity toward rat red blood cells as compared with the native peptide. It has also been documented that the antibacterial activity of cationic peptides decreases with increased peptide length (Niidome et al., 2005). Thus, length by itself is not a determinant predictive of the antimicrobial activity of the peptide; rather, the amino acid composition and distribution should be taken into account as these play a crucial role in biological activities.
9. The impact of structure on the antimicrobial activity of AMPs
Secondary structures play a crucial role in the activity of AMPs. The well-defined conformations adopted by AMPs facilitate their interaction with the lipid bilayer of microbial membranes. These structures allow AMPs to insert into the membrane, disrupt its integrity, and form pores, channels, or destabilize the membrane lipid bilayer by changing its transmembrane potential or fluidity. There are many examples of AMPs that are highly dependent on their secondary structure to be active (Ageitos et al., 2022; Torres et al., 2017; Pedron et al., 2017; Silva et al., 2020; Torres et al., 2018).
The most common secondary structures adopted by AMPs are α-helical, β-like structures, and turns, but there are examples of families of AMPs that even upon contact with the membrane are still coiled or unstructured (Torres et al., 2022). Although, the latter are exceptions. Secondary structures confer structural integrity when those molecules are in the presence of proteases besides of enabling more effective interaction with the membrane when the AMPs are in the interface of the lipid bilayer and the extracellular environment.
The predominant secondary structure observed in AMPs is the α-helical since most are amphipathic and cationic, and in a peptide design perspective, any significant alterations in the overall conformation of the α-helix caused by amino acid residues substitutions can greatly affect the antimicrobial activity (Lee et al., 2016). The β-sheet structures of AMPs has also been connected to highly stable and active peptides (Akishiba et al., 2017; Fan et al., 2020). AMPs with β-sheet structures tend to form supramolecular structures, creating pores that allow the peptide to traverse the host membrane. This enables AMPs with β-sheet structures to interact with intracellular targets, including promoters, encoding regions, mRNA-binding motifs, enzyme regulatory regions, and protein folding domains, ultimately resulting in inhibitory effects and cell death of pathogenic microorganisms (Sharma et al., 2016). Overall, despite recent successful attempts (Silva et al., 2020; Torres et al., 2018; Pedron et al., 2023; Boaro et al., 2023; Pirtskhalava et al., 2021; Liu et al., 2019), it is challenging to elucidate the structure-activity relationship (SAR) of AMPs. A general SAR behavior to explain AMP function is still missing, and most likely, computational methods (Wan et al., 2022; Torres et al., 2022; Wan and de la Fuente-Nunez, 2023; de la Fuente-Nunez, 2022; Maasch et al., 2023; Cesaro et al., 2023; Liu et al., 2019) will serve as useful tools in future studies as they can account for complex physicochemical and structural features.
10. Design of recombinant antimicrobial peptides
Although AMPs are promising antimicrobials, their commercial application to plants is not widespread. The most apparent obstacles are: (1) damage to plant cell membranes; (2) production costs and technical problems limiting scale-up manufacturing; (3) instability (breakdown by proteases or under non-standard storage conditions); (4) diminishing activity in the presence of cations and serum with proteins, lipids, and proteases. Therefore, it has become a priority to design new AMPs that are safe for plant cells while effective at killing pathogens, easily expressed, and stable and active in complex environments (Li et al., 2017; Wan et al., 2022; Torres et al., 2021).
To protect AMPs from degradation by host proteases, various approaches have been developed. For example, chemical alteration of the AMP backbone, including N-terminal acetylation, C-terminal amidation, and the substitution of certain amino acids with amino acid analogs are strategies that have been devised to protect AMPs from degradation in vivo (Shao et al., 2019; Rozek et al., 2003). However, modification, such as the substitution of amino acids (between L- and D-amino acids) in a peptide, may be an ineffective strategy because changes in the composition of the peptides and the side chain direction can alter the geometry needed for target binding (Durani, 2008; Gentilucci et al., 2007; Li et al., 2013; Pallerla et al., 2018). Furthermore, the high cost of such modifications limits their widespread use (Durani, 2008; Gentilucci et al., 2007; Li et al., 2013; Pallerla et al., 2018).
Currently, the main approaches to obtaining AMPs from plants involve direct isolation, chemical synthesis, and heterologous expression. However, both extraction from natural resources and chemical synthesis of AMPs are costly, complex, and unpredictable (Da Cunha et al., 2017). Inexpensive genetic engineering strategies have been proposed; however, they cannot produce peptides containing certain chemical modifications (Holaskova et al., 2015).
Recently, computational approaches have been efficiently used for predicting recombinant peptide behavior, accelerating the identification, design, and synthesis of bioactive peptides (D’Annessa et al., 2020; Torres et al., 2019). Such approaches can be used to predict the anti-microbial activity of recombinant peptide sequences, thus reducing the number of cloning steps and in vitro bioassays performed in the lab.
The in silico analysis of peptides and recombinant derivatives has enabled researchers and pharmaceutical companies to rationally design new bioactive peptides (Loose et al., 2006; Marcos et al., 2008; Soltani et al., 2007; Wang, 2020). The effects of the most accessible physicochemical features of AMPs, such as length, net charge, hydrophobicity, and amphipathicity have been extensively evaluated.
A few approaches are being taken for the heterologous expression of AMPs in plants, including nuclear and plastidial transformation, expression in cell suspensions, and the use of hairy root (HR) systems (Desai et al., 2010). In all instances, the heterologous transgene is integrated into the host plant genome (nuclear or plastidial), where it is passed on to future generations (Abiri et al., 2016; Sampaio de Oliveira et al., 2020).
Although several research groups have developed biological expression systems for AMPs, especially in the bacterium Escherichia coli and in yeast (Lee et al., 2000; Kaur et al., 2018), so far only a few attempts have been made to fuse AMPs to other fusion partners to express them in plants for molecular farming or to generate infection-resistant transgenic plants (Badrhadad et al., 2018; Khademi et al., 2019; Khademi et al., 2020; Lee et al., 2011; Varasteh Shams et al., 2019). To enhance the expression efficiency and increase the amount of AMP produced, Okamoto et al. (1998) fused sarcotoxin IA, an antimicrobial peptide from an insect, with GUS protein and expressed the fused protein in tobacco plants by using Agrobacterium-mediated transformation. This strategy yielded a higher level of recombinant protein production as compared to the transgenic plants expressing sarcotoxin IA alone.
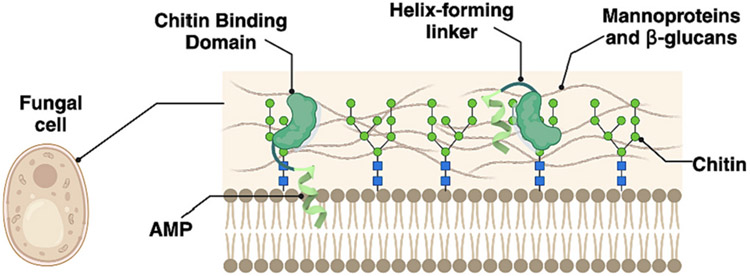
So far, new recombinant peptides have been constructed by fusing (1) two different AMPs, (2) an AMP to an antimicrobial active enzyme, or (3) an AMP to a truncated and functionally active domain from an antimicrobial enzyme (Chahardoli et al., 2018; Khademi et al., 2020; Osusky et al., 2000; Varasteh Shams et al., 2019). In most cases, the new recombinant peptides had increased antimicrobial activity compared to the native AMPs (Table 2), suggesting that AMP length might not be the sole determinant of antimicrobial activity. However, one can conclude that AMPs in the fusion forms are functionally independent because the linker sequences keep the structure of AMPs intact. Recently, a fusion peptide comprising dermaseptin B1 (DrsB1) peptide fused to tandem repeats of chitin-binding domain (CBD) was recombinantly produced in HRs. The recombinant fusion proteins demonstrated enhanced antimicrobial activity against plant phytopathogens, especially Alternaria alternata, with a minimal inhibitory concentration (MIC) of 11.25 μg L−1 in comparison with DrsB1-expressing transgenic plants (Varasteh Shams et al., 2019). Because the CBD has an intrinsic affinity for fungal cell wall chitin and glucans, the fused peptide aggregates on the fungal cell wall surface, bringing the AMP in proximity with it, such that the AMP eventually permeabilizes the plasma membrane by generating pores that eventually result in cell leakage (Fig. 2).
Table 2.
Recombinant AMPs expressed in plants.
| As | Transgenic Plant |
Pathogens targeted | Reference | |
|---|---|---|---|---|
| Name | Origin | |||
| Magainin II | Frog | Potato | E. carotovora | (Barrell and Conner, 2009) |
| Myp30, analog of magainin-2 | Frog | Tobacco | P. tabacina, E. carotovora | (Li et al., 2001) |
| MSI-99, analog of magainin-2 | Frog | Tobacco | P. syringae pv tabaci, A. flavus, F. moniliforme, V. dahlia | (DeGray et al., 2001) |
| MsrA1(Cec A-Mel) | Silk moth/Frog | Potato, Tobacco | F. solani, F. oxysporum, P. cactorum, P. infestans, P. eryroseptica, Alternaria alternata, C. beticola, B. cinerea, P. irregulare, irregular, Rhizoctonia and Verticillium, Erwinia carotovora | (Osusky et al., 2004; Osusky et al., 2000) |
| MsrA2 (N-methionine dermaseptin B1) | Silk moth/Frog | Potato | Alternaria, Cercospora, Fusarium, Phytophthora, Pythium, Rhizoctonia, Verticillium Erwinia carotovora | (Osusky et al., 2005) |
| MsrA2 (N-methionine dermaseptin B1) | Frog | Tobacco | F. solani, F. oxysporum, A. alternata, B. cinerea, S. sclerotiorum, P.aphanidermatum, P. carotovorum | (Yevtushenko and Misra, 2007) |
| AP24 osmotine | Tobacco | Potato | E.carotovora, S. scabies, P. infestans, R. solani, F. solani | (Rivero et al., 2012) |
| Cecropin B | Giant silk moth | Tobacco | P. syringae, P. solanacearum | (Florack et al., 1995) |
| MsrA1, CEMA | Giant silk mothmelittin | Potato | P. infestans, A. solani, E. carotovora | (Vutto et al., 2010) |
| MsrA1, CEMA | Silk moth | Tobacco | F. solani | (Yevtushenko et al., 2005) |
| Heliomicin | Twirler moths | Tobacco | C. nicotianae | (Banzet et al., 2002) |
| Defensin-NP1 | Rabbit | Tobacco | R. solanacearum | (Fu et al., 1998) |
| DrsB1-CBDAvr4 | Frog/ C. fulvum | Tobacco | F. solani, F. oxysporum, Alternaria, alternata, | (Khademi et al., 2019) |
| CBDAvr4-DrsB1 | C. fulvum /Frog | Tobacco | F. solani, F. oxysporum, Alternaria, alternata, | (Khademi et al., 2019) |
| CBDrice-alfAFP | Rice/Alfalfa | Tobacco | F. solani, F. oxysporum, Alternaria, alternata, | (Badrhadad et al., 2018) |
| LFcin-LFampin | Bovine | Tobacco | E. coli, S. aureus, E. amylovira and R. solanacearum | (Chahardoli et al., 2018) |
| colicin M | E. coli | Tobacco | E. coli, K. pneumoniae | (Łojewska et al., 2020) |
Cec: Cecropin; Mag: magainin; Mel: melittin; DrsB1: Dermaseptin B1: CBD: Carbohydrate-Binding Domain; Avr; Avirulence gene CBD from C. fulvum.
Fig. 2. The mode of action proposed for the activity of a recombinant AMP against a fungal pathogen.
First, the chitin-binding domain (CBD) aids and guides the AMP to aggregate on the pathogen cell wall surface. Second, the CBD anchors the AMP in the cell wall by covering the surface. Third, AMP permeabilizes the pathogen plasma membrane, resulting in pore formation, cell leakage, and eventually pathogen cell death (Badrhadad et al., 2018).
11. In planta expression of AMPs
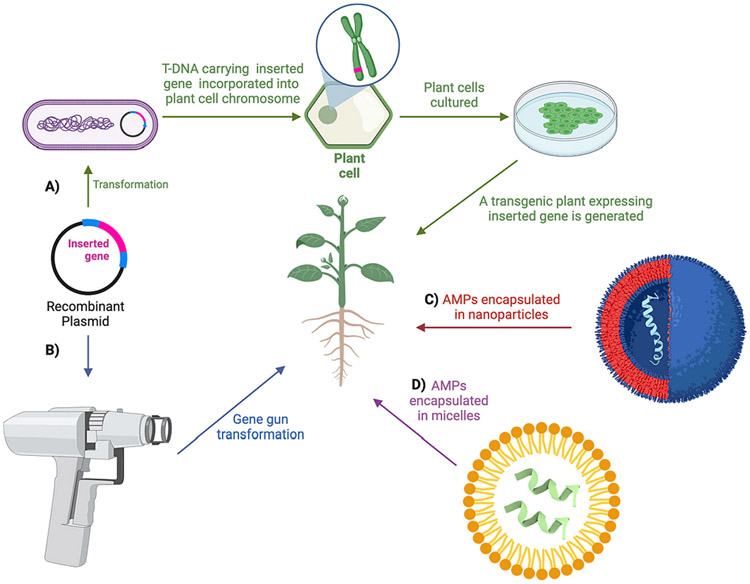
To date, a wide range of organisms, including bacteria and animals, have been the source of AMPs that have been isolated, cloned, and incorporated into plants, resulting in the generation of transgenic plants with resistance against various plant pathogens and insects (Table 2). In molecular plant breeding programs, diverse strategies have been employed to deliver/introduce AMPs into plant cells (Fig. 3) to enhance their expression in transgenic plants, including the use of different promoters, N- and C-terminal peptide modifications, and various cloning techniques (Soleymani-Goloujeh et al., 2018). For example, cecropin, an AMP with significant antibacterial activity against plant pathogenic bacteria in vitro, has been extracted from silk moths and has been successfully used to confer protection against invading pathogens when introduced into plants. Numerous AMPs have been transgenically produced in plants, leading to resistance against different pathogens. For instance, transgenic tobacco plants expressing an AMP from radish seeds (RS-AFP2) demonstrated resistance to Alternaria longipes, a fungus responsible for brown spot in tobacco plants (Terras et al., 1992). Similarly, the expression of a pea defensin in oilseed rape resulted in transgenic lines resistant to Leptosphaeria maculans, the causal agent of blackleg disease (Wang et al., 1999). Gao et al. (2000) showed that transgenic potatoes expressing an AMP from alfalfa seeds (alfAFP) exhibited significant resistance to Verticillium wilt caused by V. dahliae. Furthermore, the introduction of genes encoding plant defensins into crop plants has led to enhanced resistance against fungal pathogens (Kazan et al., 2002; Lee et al., 2008). In a recent study, Lee et al. (2018) introduced a defensin-encoding gene from pepper into tobacco and demonstrated their resistance to Phytophthora parasitica and Pythium aphanidermatum.
Fig. 3. AMP-delivery methods to plant cells.
Gene cloning approach for AMP and AMP fusion introduction either by: A) Agrobacterium-mediated transformation; or B) through biolistic method. Gene elements are colored and named flanked by T-DNA borders. C) Nanoparticle-mediated and D) micelle-mediated AMP delivery. Micelles and nanoparticles are loaded with cationic AMPs prior to plant transformation procedure.
Despite these successes, it is common for pathogenic bacteria exposed to recombinantly expressed intact AMPs to develop partial resistance over the course of pathogen-host evolution. Therefore, additional modifications are often necessary to enhance their effectiveness. To this end, Tugyi et al. (2005) structurally modified peptides to increase their bioavailability. Specifically, they replaced three L-amino acids (TPT) with their corresponding D-amino acids at both the N- and C-termini of the MUC2 mucin glycoprotein. The resulting alternative peptide displayed remarkable resistance to proteolytic degradation in vitro, as observed in both in human serum and lysosomal fractions (Tugyi et al., 2005). The simultaneous modification of the N-terminus and C-terminus residues with D-amino acids proved to be the most effective approach in enhancing the stability of mucin in the presence of proteolytic enzymes (Tugyi et al., 2005). In another study, (Arias et al., 2018) demonstrated that substitutions of arginine with lysine, coupled with side chain modifications, led to increased antimicrobial activity and stability of certain AMPs. Altogether, existing evidence indicates that the antimicrobial activity and increased resistance to degradation of AMPs in host cells can be optimized.
12. Heterologous expression of AMPs in planta
The expression of intact AMPs in plants encounters various challenges. As previously mentioned, both endogenous proteolytic enzymes in the host and those expressed in transgenic plants can degrade AMPs (Flavia Cancado Viana et al., 2013; Li et al., 2011). The small size and low molecular weight of AMPs make them susceptible to protease digestion within eukaryotic cells. To overcome these obstacles, fusion partners have been developed. These partners not only enhance the stability of AMPs but also influence the expression and accumulation of peptides within cells. Moreover, heterologous expression of AMPs offers advantages such as facilitating purification, increasing peptide efficiency through synergistic interactions with fusion partners, and reducing potential toxic effects on host cells. For example, the fusion of two AMPs has resulted in high-yield expression of AMPs with increased antimicrobial activity and low cytotoxicity toward host cells (Ferre et al., 2009).
Much effort has been devoted to designing chimeric recombinant peptides, particularly highly active AMPs sourced from non-plant origins, with the aim of generating transgenic plants that are resistant to specific target pathogens (Table 2). For instance, Osusky et al. (2000) introduced an N-terminus-modified cecropin-melittin cationic peptide chimera (MsrA1) into potato plants to enhance their resistance against bacterial and fungal pathogens. While the expression of MsrA1 appeared to be influenced by the cultivar, transgenic potato tubers exhibited improved resistance to phytopathogens.
To control Xylella fastidiosa, the causal agent of Pierce’s disease, which affects numerous woody plant species, a chimera was created by fusing a human elastase with cecropin from the silk moth Bombyx mori. Analysis of transgenic grapevine lines expressing the chimeric protein demonstrated enhanced resistance to Xylella fastidiosa by suppressing its growth and significantly reducing leaf scorching and xylem clogging (Dandekar et al., 2012). Lactoferricin (LFcin) and lactoferrampin (LFampin), derived from bovine lactoferrin, possess a wide range of activities, including antibacterial, antifungal, antiviral, and antitumor properties (García-Montoya et al., 2012; Gifford et al., 2005). Expression of a chimeric peptide consisting of LFcin and LFampin in tobacco plants resulted in the production of an active recombinant peptide with anti-microbial activity (Chahardoli et al., 2018). Although the antimicrobial activity of recombinant LFampin-Lfcin was demonstrated, transgenic plants were not challenged with plant pathogens in tissue culture or in the greenhouse.
A chimeric protein, known as SlP14a-PPC20, was created by combining a linear alpha-helical peptide derived from sunflower phosphoenolpyruvate carboxylase (PPC20) with a segment of the pathogenesis-related protein SlP14a. This chimeric protein demonstrated potent antibacterial activity in vitro against Ralstonia solanacearum, the causal agent of bacterial wilt in tobacco and tomato plants. In transgenic tomato plants expressing SlP14a-PPC20, the stems exhibited significantly lower microbial populations compared to wild-type control plants when inoculated with R. solanacearum, indicating enhanced resistance to the pathogen (Morais et al., 2019).
DrsB1, a 31-amino acid cationic peptide with an α-helix conformation, has been isolated from Phyllomedusa frogs (Mor et al., 1994; Mor and Nicolas, 1994). Unlike other dermaseptins, DrsB1 exhibits potent antimicrobial activity, effectively inhibiting the growth of various bacterial and fungal pathogens (Yevtushenko and Misra, 2007; Osusky et al., 2004). To enhance the interaction between DrsB1 and pathogen membranes and increase its efficacy against them, the coding sequence of DrsB1 was fused with different CBDs (chitin-binding domains) from Cladosporium fulvum Avr4 effector protein and rice chitinases (Alibakhshi et al., 2018; Khademi et al., 2019; Khademi et al., 2020; Varasteh Shams et al., 2019; Badrhadad et al., 2018; Nazari et al., 2017). The fusion of DrsB1 or alfAFP (an AMP from alfalfa seeds) with CBDs significantly enhanced resistance against a wide range of pathogens compared to transgenic lines expressing DrsB1 alone. Scanning electron microscopy imaging of pathogens treated with recombinant peptides revealed significant membrane damage. In vitro and greenhouse studies involving plants expressing DrsB1 or alfAFP demonstrated elevated resistance levels against fungal and bacterial pathogens, suggesting that targeting these peptides to the pathogen surface led to higher peptide accumulation (Morais et al., 2019). The heterologous expression of AMPs in host plants not only hinders plant pathogens from causing severe damage but also helps AMPs reach target cells (Neundorf et al., 2009) and cellular components such as the cell membrane (Khademi et al., 2020), the ribosome (Graf et al., 2017), and prevents their degradation by cell proteases.
13. AMP construct elements
The successful engineering of plants to express a transgene largely depends on the appropriate choice of a promoter and a terminator to ensure successful gene transcription and RNA transcript processing. To date, a diverse range of promoters and terminators has been employed for expressing AMPs in various plants. The widely utilized CaMV 35S promoter, derived from the cauliflower mosaic virus CaMV, has been extensively used in vectors to introduce AMPs into plants. While this promoter typically results in high AMP expression in plants, it can lead to gene silencing (Al-Kaff et al., 2000) and, due to homologous recombination, may deactivate the expression of heterologous genes (Stam et al., 1997). In addition to the CaMV 35S promoter, several other viral promoters have also been used for AMP expression in plants (Porto et al., 2014).
Apart from viral promoters, strong and constitutive promoters native to plants have been engineered into vectors for expressing AMPs during plant transformation. For example, actin promoters (Act1, Act2) and ubiquitin promoters (Ubi1, Ubi2) from different monocotyledon plants have been characterized and employed for the expression of various AMPs in plants (Coca et al., 2006; Li et al., 2011; Madzharova et al., 2018). In some instances, plant promoters have shown higher activity and recombinant protein expression compared to CaMV 35S (Coussens et al., 2012; Wang and Oard, 2003).
Due to the nature of AMPs in combating plant pathogens, a variety of inducible promoters responsive to factors such as heat shock proteins (Hsp18, Hsp19) (Company et al., 2014), auxin, and wounding (Langen et al., 2006; Yevtushenko et al., 2005) have also been employed to confer resistance to transgenic plants upon pathogen contact. Additionally, tissue-specific promoters, like oleosin (Montesinos et al., 2016) and glutenin (Holásková et al., 2018), have been used to drive AMP expression in specific plant tissues. In summary, there are numerous promoter options to choose from based on the intended purpose of expression, whether for managing biotic and abiotic stresses or for molecular farming purposes.
When designing gene constructs for expression, transcription terminators play two crucial roles: terminating gene transcription and enhancing the stability of mRNA transcripts during translation (He et al., 2020). The Nopaline synthase terminator (T-nos) and the octopine synthase terminator (T-ocs), both from Agrobacterium tumefaciens, are frequently employed to terminate the expression of AMPs in designed gene circuits (de Felippes, 2020). Both terminators appear to lead to the production of small interfering RNAs (siRNAs), which are the primary factors in transgene silencing in transgenic plants. Although some other terminators, such as the HEAT SHOCK PROTEIN 18.2 gene from Arabidopsis thaliana, have been used for gene expression termination, the majority of transformation binary vectors are equipped with either T-nos or T-ocs terminators (Pérez-González and Caro, 2018).
14. Effects of AMP chimeras on pathogens
Plant-pathogen interactions have undergone coevolution over millions of years, leading to the development of a coevolutionary model known as the zigzag model. This model depicts a continuous arms race between plants and pathogens, with hosts evolving proteins to evade pathogens, and pathogens evolving strategies to evade or suppress host resistance genes (Woolhouse et al., 2002). In this dynamic relationship, both plants and pathogens constantly adapt, and counter adapt to gain the upper hand. To counteract the detrimental effects of pathogens and enhance crop productivity, interventions such as plant breeding or the introduction of new resistance genes can be employed. These approaches aim to bolster plant defenses and provide protection against pathogens. Additionally, plants need to express proteins that not only recognize pathogen effectors during the early stages of invasion but also trigger immune signaling to prevent pathogens from suppressing the host’s immune system. This intricate interplay between plants and pathogens highlights the importance of understanding their coevolutionary dynamics for effective disease management and crop improvement strategies.
The design and expression of new recombinant resistance genes (R-genes) offer a promising approach to generate transgenic plant lines that are resistant to pathogens. By introducing recombinant proteins that pathogens have not yet developed ways to counteract, crop plants can be engineered to enhance their resistance. The expression of these recombinant proteins disrupts the growth and development of the pathogens, effectively reducing their pathogenicity. Several recombinant proteins have been identified that exhibit enhanced antimicrobial activity compared to their individual counterparts (Zhou et al., 2021; Wang et al., 2017; Khademi et al., 2019). This suggests that the combination of different protein partners in chimeric constructs may have a synergistic effect, further bolstering their effectiveness in combating pathogens. This research direction holds promise for developing novel strategies to enhance plant resistance and improve crop protection against pathogens.
As noted above, most cationic peptides interfere with the microbial cell membrane, leading to pore formation and eventually causing cell death. Indeed, when the surface of microbial cells becomes saturated with a high concentration of recombinant peptides, it appears that their overall integrity is compromised, resulting in damage to the cell membrane (Fig. 2). However, microorganisms have the ability to evolve and counteract the negative charge of their phospholipid layers, thereby reducing the susceptibility to AMPs (Maria-Neto et al., 2015). For example, Pseudomonas aeruginosa and Staphylococcus epidermidis present the PhoQ/PhoP and ApsR/ApsS systems, respectively, which increase resistance to cationic AMPs. Bacteria can also develop resistance to AMPs by D-alanylation of teichoic acids in the cell wall. As a result, an increase in the net positive charge of lipids decreases the affinity of bacterial cell wall lipopolysaccharides (Da Cunha et al., 2017; Henderson et al., 2014). These adaptations highlight the dynamic nature of the microbial defense mechanisms against AMPs and their potential to overcome the antimicrobial effects of recombinant peptides.
Engineering AMPs with non-catalytic domains, especially domains with affinity for bacterial cell membranes, offers a promising avenue for enhancing the interaction between these peptides and the microbial membrane (Table 2). Fusing domains with high affinity for cell membranes rather than tuning their net charge may be an interesting alternative to pursue in the future. For instance, fusing AMPs to CBDs can increase the number of AMP molecules that accumulate on the pathogen surface. This approach holds potential for optimizing the effectiveness of AMPs in combatting microbial pathogens.
15. Plant expression systems for AMPs
Despite the need to produce AMPs at industrial large scale, their purification from the native host is usually not feasible. Furthermore, except for linear AMPs which require no disulfide bridges formation and/or other post-translational modifications, direct chemical synthesis is too costly. To circumvent this drawback, recombinant expression of AMPs using transgenic methods holds promise for commercial-level AMP production and purification. Among different expression systems, plants exhibit great advantages over yeast and bacterial expression systems. First, large quantities of AMPs can be produced at low cost, scaling up production simply by the cultivation of transgenic plants in a high cultivation area (Tusé et al., 2014). Second, plant systems offer two types of expression of a transgene, namely transient and stable expression. Stable expression of AMPs can be achieved by introducing them in either nuclear or plastid genomes (Hoelscher et al., 2022). Up to now, expression of a vast number of AMPs in different plants has been attempted by transient, nuclear and plastid transformation (Lee et al., 2011; Shanmugaraj et al., 2021). Both nuclear and plastid transformation implement the stable transgene expression, however, plastids offer some additional benefits, including exclusion from pollen transmission as well as preventing epigenetic transgene silencing (Bock, 2015). Although plastid expression shows a number of significant advantages over nuclear and transient expression for AMPs production, toxicity of AMPs toward plastid membrane, instability due to small size and presence of hydrophobic domains are still major concerns (Giomarelli et al., 2006; Li, 2011; Scotti et al., 2015).
16. Limitations and future perspectives
Similar to other expression platforms, the expression of natural AMPs in plant systems is associated with certain drawbacks, including variable efficacy, stability in successive generations, and potential toxicity to plant cells and tissues. Recently, machine learning approaches have been described to predict the molecular modifications necessary to overcome the drawbacks of AMP expression in plants (Jaiswal et al., 2023). Although transgenic plants expressing AMPs confer varying degrees of disease protection, the expression of AMPs has resulted in different levels of protection against plant pathogens (Montesinos, 2007). To mitigate potential drawbacks associated with AMP-expressing plants, engineering the chloroplast genome has been proposed as a potential avenue to create crop plants with disease resistance. This approach may also prevent pollen-mediated transfer of AMPs to weed and wild relatives of transgenic crop plants (DeGray et al., 2001). Infertility is another issue associated with AMP expression. To address this drawback, targeting AMPs into the endoplasmic reticulum may alleviate transgenic infertility in the next generation (Coca et al., 2006). Altogether, addressing issues related to AMP expression in plant systems requires further research to assess the stability, economic feasibility, and biosafety of AMPs in transgenic plants.
AMPs are an essential part of the innate immunity of almost all living organisms, suggesting that, evolutionarily, they must be safe for eukaryotic cells. However, considering ecological concerns, the expression of foreign AMPs in host plants may pose a threat to the mutualistic interactions between crop plants and beneficial microbes and insects. Despite the cultivation of genetically modified (GM) plants for three decades, no clear and significant trend has been observed regarding the negative effects of transgenes, including AMPs, on the safety of beneficial microorganisms (Stefani and Hamelin, 2010). Rahnamaeian et al. (2009) investigated the adverse effect of metchnikowin (Mtk), a 26-amino acid residue AMP found in the fat body of Drosophila melanogaster, on the beneficial root endophyte Piriformospora indica. This study revealed that Mtk did not inhibit the growth of P. indica in the roots of transgenic plants, whereas it impaired the growth and development of devastating plant fungal pathogens. The toxicological investigation of an antifungal defensin from chickpea (Ca-AFP) led to the conclusion that Ca-AFP was safe for humans, insects, and beneficial bacteria (Islam, 2008). In another study, two cultivars of Nicotiana tabacum L. transgenic seeds expressing plastids D4E1 (an analog of the antimicrobial peptide cecropin) and MSI-99 (an analog of Magainin 2) were tested for colonization by two mycorrhizal fungal species, Glomus mosseae and Gigaspora rosea. No significant differences were found in the symbiosis ability of mycorrhizal fungi between transgenic lines and non-transgenic lines, suggesting that transgenic plants expressing AMPs were able to establish mycorrhizal associations without adverse effects on beneficial fungi (Stewart et al., 2007). Taken together, based on the available literature, these findings may be interpreted as suggesting that transgenic lines expressing AMPs can be considered as safe.
One of the most desirable features of AMPs is their ability to quickly kill resistant pathogens even at low concentrations, reducing the likelihood of the emergence of AMP-resistant plant pathogens. For instance, Niu et al. (2020) demonstrated that overexpression of an AMP from Capsicum annuum (CaAMP1) exhibited enhanced and stable pathogen tolerance even after four generations compared to the wild-type. Similarly, the expression of the Snakin-1 (SN1) gene from potatoes in wheat led to the stable expression of the AMP even in the T4 generation (Rong et al., 2013). The expression of a synthetic BP100-magainin derivative AMP (BP178) in rice remained stable in T4 transgenic lines, indicating that BP178 remained functional and stable following Mendelian segregation inheritance (Montesinos et al., 2017).
Although developing resistance against AMPs is difficult and complicated due to the coevolution of host and pathogen interactions, plant pathogens may develop mechanisms to sense and exhibit an adaptive response against AMP transgenes in plants. The emergence of resistance against AMPs represents a significant challenge to human health. However, it seems that the emergence of such resistance mechanisms is relatively nonspecific, negligible, and occurs at a low frequency (Baindara et al., 2020). To the best of our knowledge, there is no report indicating the development of resistance against AMPs as transgenes by plant pathogens. This can be explained by the fact that the coevolutionary changes between transgenic plants harboring new AMPs and plant pathogens occur over an extended period of time.
17. Gene flow of AMP-expressing transgenic plants
Another concern related to AMP-expressing transgenic plants is that of gene flow from transgenic plants to related wild and weedy species. Generally, there is evidence that gene flow has occurred before the development of transgenic crops (Chandler and Dunwell, 2008). Therefore, gene flow from transgenic plants can be expected, and for some species, it is even inevitable (Dlugosch and Whitton, 2008). Many commercial transgenic plants have been extensively cultivated over the past three decades. So far, there has been no convincing evidence that the cultivation of transgenic plants, such as maize, soybean, cotton, canola, etc., has posed a greater environmental and human health risk compared to cultivars released by traditional plant breeding programs. Various techniques have been proposed to reduce the risk of gene flow, including plastid transformation through transplastomic approaches (Chandler and Dunwell, 2008).
When AMPs are expressed in plants as pharmaceutical compounds, control of gene flow becomes even more important than with approved transgenic plants used as food supplies (Murphy, 2007). The risk assessment for AMP-expressing transgenic plants should be based on crop-to-crop evaluation, especially when considering the use of transgenic plants as pharmaceuticals.
18. AMPs and potential health concerns
In animals, AMPs also play roles in cell differentiation, the biosynthesis of chemokines and cytokines, the maturation of germ cells, and the regulation of inflammation (Guryanova and Ovchinnikova, 2022). Therefore, it is possible that the expression of AMPs in crop plants may trigger allergic reactions in humans. Among the diverse group of AMPs from various sources, plant-derived AMPs belonging to pathogenesis-related (PR) proteins, which are naturally expressed in different plant organs and tissues, may cause severe allergic reactions in humans, such as anaphylactic shock (Midoro-Horiuti et al., 2001).
Since the expression of AMPs is inducible by environmental factors, biotic and abiotic stresses may result in varying levels of AMP expression in transgenic plants, potentially leading to acute allergic reactions in humans. Hence, it would be advisable to utilize promoters for driving AMPs in plants whose regulation cannot be influenced by environmental factors. Another approach could involve expressing peptides naturally present in the human or animal innate immune system, which have long been recognized as safe. Despite ongoing debates regarding the allergenic or non-allergenic nature of some AMPs, a thorough examination is essential to ensure that AMPs, when used as transgenes, do not trigger allergic reactions in humans or any disbalances to the surrounding biodiversity.
19. Concluding remarks
AMPs have a crucial role in plant defense against microbial pathogens (Zasloff, 2006; Porto et al., 2018). However, these pathogens can still cause significant damage to crops. To achieve the goal of developing transgenic plants resistant to pathogenic invasion, it is important to expand our search for novel peptides with enhanced antimicrobial activity and diverse structures and mechanisms of action. While plant-derived AMPs are valuable in combating pathogens, modifications to these peptides, including the design of recombinant peptides, can improve their effectiveness and mitigate potential bacterial resistance during pathogen-host evolution. Genetic engineering techniques, such as the use of recombinant fusion peptides and heterologous expression, offer promising approaches for generating pathogen-resistant crop plants. Additionally, synthetic biology and computational methods can expedite the design of peptides with optimized functionality (Torres and de la Fuente-Nunez, 2019; Wan et al., 2022; Wong et al., 2023; Torres et al., 2022; Maasch et al., 2023). Numerous transgenic plants expressing various AMPs from different sources have been created, providing varying degrees of protection against devastating plant pathogens. Thus, AMPs hold significant potential in plant molecular breeding as effective plant protection agents.
Acknowledgments
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania and acknowledges funding from the Procter & Gamble Company, United Therapeutics, a BBRF Young Investigator Grant, the Nemirovsky Prize, Penn Health-Tech Accelerator Award, and the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. Research reported in this publication was supported by the Langer Prize (AIChE Foundation), the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041, HDTRA1-21-1-0014, and HDTRA1-23-1-0001).
Footnotes
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Cesar de la Fuente-Nunez provides consulting services to Invaio Sciences and is a member of the Scientific Advisory Boards of Nowture S.L., Peptidus, and Phare Bio. De la Fuente is also on the Advisory Board of the Peptide Drug Hunting Consortium (PDHC). The de la Fuente Lab has received research funding or in-kind donations from United Therapeutics, Strata Manufacturing PJSC, and Procter & Gamble, none of which were used in support of this work.
References
- Abiri R, Valdiani A, Maziah M, Shaharuddin NA, Sahebi M, Yusof ZNB, Atabaki N, Talei D, 2016. A critical review of the concept of transgenic plants: insights into pharmaceutical biotechnology and molecular farming. Curr. Issues Mol. Biol 18 (1), 21–42. [PubMed] [Google Scholar]
- Ageitos L, Torres MD, de la Fuente-Nunez C, 2022. Biologically active peptides from venoms: applications in antibiotic resistance, cancer, and beyond. Int. J. Mol. Sci 23 (23), 15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akishiba M, Takeuchi T, Kawaguchi Y, Sakamoto K, Yu H-H, Nakase I, Takatani-Nakase T, Madani F, Gräslund A, Futaki S, 2017. Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem 9 (8), 751–761. [DOI] [PubMed] [Google Scholar]
- Alibakhshi A, Nazarian Firouzabadi F, Ismaili A, 2018. Expression and antimicrobial activity analysis of a Dermaseptin B1 antibacterial peptide in tobacco hairy roots. Plant Protection (Scientific Journal of Agriculture) 41 (3), 87–96. [Google Scholar]
- Al-Kaff NS, Kreike MM, Covey SN, Pitcher R, Page AM, Dale PJ, 2000. Plants rendered herbicide-susceptible by cauliflower mosaic virus–elicited suppression of a 35S promoter-regulated transgene. Nat. Biotechnol 18 (9), 995–999. [DOI] [PubMed] [Google Scholar]
- Almeida MS, Cabral KM, Zingali RB, Kurtenbach E, 2000. Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch. Biochem. Biophys 378 (2), 278–286. [DOI] [PubMed] [Google Scholar]
- Arias M, Piga KB, Hyndman ME, Vogel HJ, 2018. Improving the activity of Trp-rich antimicrobial peptides by Arg/Lys substitutions and changing the length of cationic residues. Biomolecules 8 (2), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrhadad A, Nazarian-Firouzabadi F, Ismaili A, 2018. Fusion of a chitin-binding domain to an antibacterial peptide to enhance resistance to fusarium solani in tobacco (Nicotiana tabacum). 3. Biotech 8 (9), 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar AA, Ren D, 2013. Antimicrobial peptides. Pharmaceuticals 6 (12), 1543–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baindara P, Ghosh AK, Mandal SM, 2020. Coevolution of resistance against antimicrobial peptides. Microb. Drug Resist 26 (8), 880–899. [DOI] [PubMed] [Google Scholar]
- Balls A, Hale W, Harris T, 1942. A crystalline protein obtained from a lipoprotein of wheat flour. Cereal Chem. 19 (19), 279–288. [Google Scholar]
- Banzet N, Latorse M-P, Bulet P, Fraois E, Derpierre C, Dubald M, 2002. Expression of insect cystein-rich antifungal peptides in transgenic tobacco enhances resistance to a fungal disease. Plant Sci. 162 (6), 995–1006. [Google Scholar]
- Barbosa Pelegrini P, Del Sarto RP, Silva ON, Franco OL, 2011. Grossi-de-Sa MF (2011) Antibacterial peptides from plants: what they are and how they probably work. Biochem. Res. Int 9, 250349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell PJ, Conner AJ, 2009. Expression of a chimeric magainin gene in potato confers improved resistance to the phytopathogen Erwinia carotovora. The Open Plant Sci. J 3 (1). [Google Scholar]
- Bednarska NG, Wren BW, Willcocks SJ, 2017. The importance of the glycosylation of antimicrobial peptides: natural and synthetic approaches. Drug Discov. Today 22 (6), 919–926. [DOI] [PubMed] [Google Scholar]
- Belmadani A, Semlali A, Rouabhia M, 2018. Dermaseptin-S1 decreases Candida albicans growth, biofilm formation and the expression of hyphal wall protein 1 and aspartic protease genes. J. Appl. Microbiol 125 (1), 72–83. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H, 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8 (4), 521–539. [DOI] [PubMed] [Google Scholar]
- Boaro A, Ageitos L, Torres MDT, Blasco EB, Oztekin S, de la Fuente-Nunez C, 2023. Structure-function-guided design of synthetic peptides with anti-infective activity derived from wasp venom. Cell Rep. Phys. Sci 4 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R., 2015. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol 66, 211–241. [DOI] [PubMed] [Google Scholar]
- Campos ML, de Souza CM, de Oliveira KBS, Dias SC, Franco OL, 2018. The role of antimicrobial peptides in plant immunity. J. Exp. Bot 69 (21), 4997–5011. [DOI] [PubMed] [Google Scholar]
- Cesaro A, Bagheri M, Torres M, Wan F, de la Fuente-Nunez C, 2023. Deep learning tools to accelerate antibiotic discovery. Expert Opinion on Drug Discovery 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahardoli M, Fazeli A, Ghabooli M, 2018. Recombinant production of bovine Lactoferrin-derived antimicrobial peptide in tobacco hairy roots expression system. Plant Physiol. Biochem 123, 414–421. [DOI] [PubMed] [Google Scholar]
- Chandler S, Dunwell JM, 2008. Gene flow, risk assessment and the environmental release of transgenic plants. Crit. Rev. Plant Sci 27 (1), 25–49. [Google Scholar]
- Coca M, Penas G, Gómez J, Campo S, Bortolotti C, Messeguer J, Segundo BS, 2006. Enhanced resistance to the rice blast fungus Magnaporthe grisea conferred by expression of a cecropin a gene in transgenic rice. Planta 223 (3), 392–406. [DOI] [PubMed] [Google Scholar]
- Company N, Nadal A, Ruiz C, Pla M, 2014. Production of phytotoxic cationic α-helical antimicrobial peptides in plant cells using inducible promoters. PLoS One 9 (11), e109990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens G, Aesaert S, Verelst W, Demeulenaere M, De Buck S, Njuguna E, Inze D, Van Lijsebettens M, 2012. Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J. Exp. Bot 63 (11), 4263–4273. [DOI] [PubMed] [Google Scholar]
- Cudic M, Otvos L Jr., 2002. Intracellular targets of antibacterial peptides. Curr. Drug Targets 3 (2), 101–106. [DOI] [PubMed] [Google Scholar]
- Da Costa JP, Cova M, Ferreira R, Vitorino R, 2015. Antimicrobial peptides: an alternative for innovative medicines? Appl. Microbiol. Biotechnol 99 (5), 2023–2040. [DOI] [PubMed] [Google Scholar]
- Da Cunha NB, Cobacho NB, Viana JF, Lima LA, Sampaio KB, Dohms SS, Ferreira AC, de la Fuente-Núñez C, Costa FF, Franco OL, 2017. The next generation of antimicrobial peptides (AMPs) as molecular therapeutic tools for the treatment of diseases with social and economic impacts. Drug Discov. Today 22 (2), 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar AM, Gouran H, Ibáñez AM, Uratsu SL, Agüero CB, McFarland S, Borhani Y, Feldstein PA, Bruening G, Nascimento R, 2012. An engineered innate immune defense protects grapevines from Pierce disease. Proc. Natl. Acad. Sci 109 (10), 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Annessa I, Di Leva FS, La Teana A, Novellino E, Limongelli V, Di Marino D, 2020. Bioinformatics and biosimulations as toolbox for peptides and peptidomimetics design: where are we? Front. Mol. Biosci 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes F, Mchale M, Doran RL, Roden S, Eamens AL, Finnegan EJ, Waterhouse PM, 2020. The key role of terminators on the expression and post-transcriptional gene silencing of transgenes. Plant J. 104 (1), 96–112. [DOI] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., 2022. Antibiotic discovery with machine learning. Nat. Biotechnol 40 (6), 833–834. [DOI] [PubMed] [Google Scholar]
- de Oliveira Carvalho A., Gomes VM, 2009. Plant defensins—prospects for the biological functions and biotechnological properties. Peptides 30 (5), 1007–1020. [DOI] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H, 2001. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 127 (3), 852–862. [PMC free article] [PubMed] [Google Scholar]
- Desai PN, Shrivastava N, Padh H, 2010. Production of heterologous proteins in plants: strategies for optimal expression. Biotechnol. Adv 28 (4), 427–435. [DOI] [PubMed] [Google Scholar]
- Divyashree M, Mani MK, Reddy D, Kumavath R, Ghosh P, Azevedo V, Barh D, 2020. Clinical applications of antimicrobial peptides (AMPs): where do we stand now? Protein Pept. Lett 27 (2), 120–134. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Whitton J, 2008. Can we Stop Transgenes from Taking a Walk on the Wild Side? Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- Durani S., 2008. Protein design with L-and D-α-amino acid structures as the alphabet. Acc. Chem. Res 41 (10), 1301–1308. [DOI] [PubMed] [Google Scholar]
- Edwards IA, Elliott AG, Kavanagh AM, Zuegg J, Blaskovich MA, Cooper MA, 2016. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect. Dis 2 (6), 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Li X-D, He P-P, Hu X-X, Zhang K, Fan J-Q, Yang P-P, Zheng H-Y, Tian W, Chen Z-M, 2020. A biomimetic peptide recognizes and traps bacteria in vivo as human defensin-6. Sci. Adv 6 (19):eaaz4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder R, Dagan A, Mor A, 2000. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem 275 (6), 4230–4238. [DOI] [PubMed] [Google Scholar]
- Ferre R, Melo MN, Correia AD, Feliu L, Bardají E, Planas M, Castanho M, 2009. Synergistic effects of the membrane actions of cecropin-melittin antimicrobial hybrid peptide BP100. Biophys. J 96 (5), 1815–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavia Cancado Viana J, Campos Dias S, Luiz Franco O, Lacorte C, 2013. Heterologous production of peptides in plants: fusion proteins and beyond. Curr. Protein Pept. Sci 14 (7), 568–579. [DOI] [PubMed] [Google Scholar]
- Flood J., 2010. The importance of plant health to food security. Food Sec. 2 (3), 215–231. [Google Scholar]
- Florack D, Allefs S, Bollen R, Bosch D, Visser B, Stiekema W, 1995. Expression of giant silkmoth cecropin B genes in tobacco. Transgenic Res. 4, 132–141. [DOI] [PubMed] [Google Scholar]
- Fu R, Peng Y, Cao G, Ma J, Chen C, Zhang L, Li W, Sun Y, 1998. Expression of rabbit defensin NP-1 gene in transgenic tobacco plants and its activity against bacterial wilt. Chin. Sci. Bull 43 (18), 1544–1550. [Google Scholar]
- Gao A-G, Hakimi SM, Mittanck CA, Wu Y, Woerner BM, Stark DM, Shah DM, Liang J, Rommens CM, 2000. Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol 18 (12), 1307–1310. [DOI] [PubMed] [Google Scholar]
- García-Montoya IA, Cendón TS, Arévalo-Gallegos S, Rascón-Cruz Q, 2012. Lactoferrin a multiple bioactive protein: an overview. Biochimica et Biophysica Acta (BBA)-General Subjects 1820 (3), 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentilucci L, Cardillo G, Squassabia F, Tolomelli A, Spampinato S, Sparta A, Baiula M, 2007. Inhibition of cancer cell adhesion by heterochiral pro-containing RGD mimetics. Bioorg. Med. Chem. Lett 17 (8), 2329–2333. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Hunter HN, Vogel H, 2005. Lactoferricin. Cell. Mol. Life Sci 62 (22), 2588–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giomarelli B, Schumacher KM, Taylor TE, Sowder RC II, Hartley JL, McMahon JB, Mori T, 2006. Recombinant production of anti-HIV protein, griffithsin, by auto-induction in a fermentor culture. Protein Expr. Purif 47 (1), 194–202. [DOI] [PubMed] [Google Scholar]
- Graf M, Mardirossian M, Nguyen F, Seefeldt AC, Guichard G, Scocchi M, Innis CA, Wilson DN, 2017. Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep 34 (7), 702–711. [DOI] [PubMed] [Google Scholar]
- Grimsey E, Collis DW, Mikut R, Hilpert K, 2020. The effect of lipidation and glycosylation on short cationic antimicrobial peptides. Biochimica et Biophysica Acta (BBA)-Biomembranes 1862 (8), 183195. [DOI] [PubMed] [Google Scholar]
- Guryanova SV, Ovchinnikova TV, 2022. Immunomodulatory and allergenic properties of antimicrobial peptides. Int. J. Mol. Sci 23 (5), 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhao S, Ma Z, Gao L, Liu H, Muhammad U, Lu Z, Lv F, Bie X, 2017. The antibacterial activity and modes of LI-F type antimicrobial peptides against Bacillus cereus in vitro. J. Appl. Microbiol 123 (3), 602–614. [DOI] [PubMed] [Google Scholar]
- Haney EF, Mansour SC, Hancock RE, 2017. Antimicrobial peptides: an introduction. Antimicrobial Peptides 3–22. [DOI] [PubMed] [Google Scholar]
- He Z, Duan Y, Zhai W, Zhang X, Shi J, Zhang X, Xu Z, 2020. Evaluating terminator strength based on differentiating effects on transcription and translation. ChemBioChem 21 (14), 2067–2072. [DOI] [PubMed] [Google Scholar]
- Henderson JC, Fage CD, Cannon JR, Brodbelt JS, Keatinge-Clay AT, Trent MS, 2014. Antimicrobial peptide resistance of vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem. Biol 9 (10), 2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelscher MP, Forner J, Calderone S, Krämer C, Taylor Z, Loiacono FV, Agrawal S, Karcher D, Moratti F, Kroop X, 2022. Expression strategies for the efficient synthesis of antimicrobial peptides in plastids. Nat. Commun 13 (1), 5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaskova E, Galuszka P, Frebort I, Oz MT, 2015. Antimicrobial peptide production and plant-based expression systems for medical and agricultural biotechnology. Biotechnol. Adv 33 (6), 1005–1023. [DOI] [PubMed] [Google Scholar]
- Holásková E, Galuszka P, Mičúchová A, Šebela M, Öz MT, Frébort I, 2018. Molecular farming in barley: development of a novel production platform to produce human antimicrobial peptide LL-37. Biotechnol. J 13 (6), 1700628. [DOI] [PubMed] [Google Scholar]
- Hollmann A, Martinez M, Maturana P, Semorile LC, Maffia PC, 2018. Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem 6, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G-J, Lai H-C, Chang Y-S, Sheu M-J, Lu T-L, Huang S-S, Lin Y-H, 2008. Antimicrobial, dehydroascorbate reductase, and monodehydroascorbate reductase activities of defensin from sweet potato [Ipomoea batatas (L.) lam.‘tainong 57’] storage roots. J. Agric. Food Chem 56 (9), 2989–2995. [DOI] [PubMed] [Google Scholar]
- Islam A., 2008. Preliminary risk assessment of a novel antifungal defensin peptide from chickpea (Cicer arietinum L.). Appl. Biosaf 13 (4):222–230. [Google Scholar]
- Jaiswal M, Singh A, Kumar S, 2023. PTPAMP: prediction tool for plant-derived antimicrobial peptides. Amino Acids 55 (1), 1–17. [DOI] [PubMed] [Google Scholar]
- Karpun N, Yanushevskaya E, Mikhailova YV (2015) Formation of plants nonspecific induced immunity at the biogenous stress. Сельскохозяйственная биология (5 (eng)). [Google Scholar]
- Kaur J, Kumar A, Kaur J, 2018. Strategies for optimization of heterologous protein expression in E. Coli: roadblocks and reinforcements. Int. J. Biol. Macromol 106, 803–822. [DOI] [PubMed] [Google Scholar]
- Kazan K, Rusu A, Marcus JP, Goulter KC, Manners JM, 2002. Enhanced quantitative resistance to Leptosphaeria maculans conferred by expression of a novel antimicrobial peptide in canola (Brassica napus L.). Mol. Breed 10 (1):63–70. [Google Scholar]
- Khademi M, Nazarian-Firouzabadi F, Ismaili A, Shirzadian Khorramabad R, 2019. Targeting microbial pathogens by expression of new recombinant dermaseptin peptides in tobacco. MicrobiologyOpen 8 (11), e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi M, Varasteh-Shams M, Nazarian-Firouzabadi F, Ismaili A, 2020. New recombinant antimicrobial peptides confer resistance to fungal pathogens in tobacco plants. Front. Plant Sci 11, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizsan A, Volke D, Weinert S, Sträter N, Knappe D, Hoffmann R, 2014. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew. Chem. Int. Ed 53 (45), 12236–12239. [DOI] [PubMed] [Google Scholar]
- Kumar M, Yusuf MA, Yadav P, Narayan S, Kumar M, 2019. Overexpression of chickpea defensin gene confers tolerance to water-deficit stress in Arabidopsis thaliana. Front. Plant Sci 10, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen G, Imani J, Altincicek B, Kieseritzky G, Kogel K-H, Vilcinskas A, 2006. Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth galleria mellonella, confers resistance to pathogenic fungi in tobacco. [DOI] [PubMed] [Google Scholar]
- Lay F, Anderson M, 2005. Defensins-components of the innate immune system in plants. Curr. Protein Pept. Sci 6 (1), 85–101. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim J, Hwang S, Lee W, Yoon H, Lee H, Hong S, 2000. High-level expression of antimicrobial peptide mediated by a fusion partner reinforcing formation of inclusion bodies. Biochem. Biophys. Res. Commun 277 (3), 575–580. [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang IS, Choi HW, Hwang BK, 2008. Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol. 148 (2), 1004–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Li B, Jin S, Daniell H, 2011. Expression and characterization of antimicrobial peptides Retrocyclin-101 and Protegrin-1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol. J 9 (1), 100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Oh M, Kim HS, Lee H, Im W, Lim H-S, 2016. Converting one-face α-helix mimetics into amphiphilic α-helix mimetics as potent inhibitors of protein–protein interactions. ACS Comb. Sci 18 (1), 36–42. [DOI] [PubMed] [Google Scholar]
- Lee H-H, Kim J-S, Hoang QT, Kim JI, Kim YS, 2018. Root-specific expression of defensin in transgenic tobacco results in enhanced resistance against Phytophthora parasitica var. nicotianae. Eur. J. Plant Pathol 151 (3):811–823. [Google Scholar]
- Lei J, Sun L, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He Q, 2019. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res 11 (7), 3919. [PMC free article] [PubMed] [Google Scholar]
- Li Y., 2011. Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expr. Purif 80 (2), 260–267. [DOI] [PubMed] [Google Scholar]
- Li Q, Lawrence CB, Xing H-Y, Babbitt RA, Bass WT, Maiti IB, Everett NP, 2001. Enhanced disease resistance conferred by expression of an antimicrobial magainin analog in transgenic tobacco. Planta 212 (4), 635–639. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhou M, Zhang Z, Ren L, Du L, Zhang B, Xu H, Xin Z, 2011. Expression of a radish defensin in transgenic wheat confers increased resistance to fusarium graminearum and Rhizoctonia cerealis. Funct. Integr. Genomics 11, 63–70. [DOI] [PubMed] [Google Scholar]
- Li C, Zhan C, Zhao L, Chen X, Lu W-Y, Lu W, 2013. Functional consequences of retro-inverso isomerization of a miniature protein inhibitor of the p53–MDM2 interaction. Bioorg. Med. Chem 21 (14), 4045–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Koh J-J, Liu S, Lakshminarayanan R, Verma CS, Beuerman RW, 2017. Membrane active antimicrobial peptides: translating mechanistic insights to design. Front. Neurosci 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hu S, Jian W, Xie C, Yang X, 2021. Plant antimicrobial peptides: structures, functions, and applications. Bot. Stud 62 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du Q, Ma C, Xi X, Wang L, Zhou M, Burrows JF, Chen T, Wang H, 2019. Structure–Activity Relationship of an Antimicrobial Peptide, Phylloseptin-PHa: Balance of Hydrophobicity and Charge Determines the Selectivity of Bioactivities. Drug Design, Development and Therapy, pp. 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo DS, Pereira IB, Fragel-Madeira L, Medeiros LN, Cabral LM, Faria J, Bellio M, Campos RC, Linden R, Kurtenbach E, 2007. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 46 (4), 987–996. [DOI] [PubMed] [Google Scholar]
- Łojewska E, Sakowicz T, Kowalczyk A, Konieczka M, Grzegorczyk J, Sitarek P, Skała E, Czarny P, Šliwiński T, Kowalczyk T, 2020. Production of recombinant colicin M in Nicotiana tabacum plants and its antimicrobial activity. Plant Biotechnol. Rep 14 (1), 33–43. [Google Scholar]
- Loose C, Jensen K, Rigoutsos I, Stephanopoulos G, 2006. A linguistic model for the rational design of antimicrobial peptides. Nature 443 (7113), 867–869. [DOI] [PubMed] [Google Scholar]
- Maasch JR, Torres MD, Melo MC, de la Fuente-Nunez C, 2023. Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning. Cell Host & Microbe 31 (8), 1260–1274 e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzharova N, Kazakova K, Strelnikova S, Snycheva O, Vetchinkina E, Efremova L, Vysotskii D, Babakov A, Komakhin R, 2018. Promoters pro-SmAMP1 and pro-SmAMP2 from wild plant Stellaria media for the biotechnology of Dicotyledons. Russ. J. Plant Physiol 65, 750–761. [Google Scholar]
- Mansour SC, Pena OM, Hancock RE, 2014. Host defense peptides: front-line immunomodulators. Trends Immunol. 35 (9), 443–450. [DOI] [PubMed] [Google Scholar]
- Marcos JF, Munoz A, Perez-Paya E, Misra S, Lopez-Garcia B, 2008. Identification and rational design of novel antimicrobial peptides for plant protection. Annu. Rev. Phytopathol 46, 273–301. [DOI] [PubMed] [Google Scholar]
- Maria-Neto S, de Almeida KC, Macedo MLR, Franco OL, 2015. Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochimica et Biophysica Acta (BBA)-Biomembranes 1848 (11), 3078–3088. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K., 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochimica et Biophysica Acta (BBA)-Biomembranes 1462 (1–2), 1–10. [DOI] [PubMed] [Google Scholar]
- Midoro-Horiuti T, Brooks EG, Goldblum RM, 2001. Pathogenesis-related proteins of plants as allergens. Ann. Allergy Asthma Immunol 87 (4), 261–271. [DOI] [PubMed] [Google Scholar]
- Miller RNG, Costa Alves GS, Van Sluys M-A, 2017. Plant immunity: unravelling the complexity of plant responses to biotic stresses. Ann. Bot 119 (5), 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingeot-Leclercq M-P, Décout J-L, 2016. Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides. MedChemComm 7 (4), 586–611. [Google Scholar]
- Montesinos E., 2007. Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett 270 (1), 1–11. [DOI] [PubMed] [Google Scholar]
- Montesinos L, Bundó M, Izquierdo E, Campo S, Badosa E, Rossignol M, Montesinos E, San Segundo B, Coca M, 2016. Production of biologically active cecropin a peptide in rice seed oil bodies. PLoS One 11 (1), e0146919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos L, Bundó M, Badosa E, San Segundo B, Coca M, Montesinos E, 2017. Production of BP178, a derivative of the synthetic antibacterial peptide BP100, in the rice seed endosperm. BMC Plant Biol. 17 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Nicolas P, 1994. The NH2-terminal alpha-helical domain 1-18 of dermaseptin is responsible for antimicrobial activity. J. Biol. Chem 269 (3), 1934–1939. [PubMed] [Google Scholar]
- Mor A, Hani K, Nicolas P, 1994. The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J. Biol. Chem 269 (50), 31635–31641. [PubMed] [Google Scholar]
- Morais TP, Zaini PA, Chakraborty S, Gouran H, Carvalho CP, Almeida-Souza HO, Souza JB, Santos PS, Goulart LR, Luz JM, 2019. The plant-based chimeric antimicrobial protein SlP14a-PPC20 protects tomato against bacterial wilt disease caused by Ralstonia solanacearum. Plant Sci. 280, 197–205. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, 2007. Improving containment strategies in biopharming. Plant Biotechnol. J 5 (5), 555–569. [DOI] [PubMed] [Google Scholar]
- Mwangi J, Hao X, Lai R, Zhang Z-Y, 2019. Antimicrobial peptides: new hope in the war against multidrug resistance. Zool. Res 40 (6), 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Goździcka-Józefiak A, 2014. Plant antimicrobial peptides. Folia Microbiologica 59 (3), 181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari Z, Nazarian Firouzabadi F, Ismaili A, Darvishnia M, 2017. Production of a recombinant antimicrobial Dermaseptine B1 peptide in Nicotiana tabacum L. hairy roots with antibacterial activity. Yafte 19 (2), 75–84. [Google Scholar]
- Neundorf I, Rennert R, Hoyer J, Schramm F, Löbner K, Kitanovic I, Wölfl S, 2009. Fusion of a short HA2-derived peptide sequence to cell-penetrating peptides improves cytosolic uptake, but enhances cytotoxic activity. Pharmaceuticals 2 (2), 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Haney EF, Vogel HJ, 2011. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29 (9), 464–472. [DOI] [PubMed] [Google Scholar]
- Niidome T, Matsuyama N, Kunihara M, Hatakeyama T, Aoyagi H, 2005. Effect of chain length of cationic model peptides on antibacterial activity. Bull. Chem. Soc. Jpn 78 (3), 473–476. [Google Scholar]
- Niu L, Zhong X, Zhang Y, Yang J, Xing G, Li H, Liu D, Ma R, Dong Y, Yang X, 2020. Enhanced tolerance to Phytophthora root and stem rot by over-expression of the plant antimicrobial peptide CaAMP1 gene in soybean. BMC Genet. 21 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Mitsuhara I, Ohshima M, Natori S, Ohashi Y, 1998. Enhanced expression of an antimicrobial peptide sarcotoxin IA by GUS fusion in transgenic tobacco plants. Plant Cell Physiol. 39 (1), 57–63. [DOI] [PubMed] [Google Scholar]
- Onaga G, Wydra K, 2016. Advances in plant tolerance to abiotic stresses. Plant Genom. 10, 229–272. [Google Scholar]
- Osusky M, Zhou G, Osuska L, Hancock RE, Kay WW, Misra S, 2000. Transgenic plants expressing cationic peptide chimeras exhibit broad-spectrum resistance to phytopathogens. Nat. Biotechnol 18 (11), 1162–1166. [DOI] [PubMed] [Google Scholar]
- Osusky M, Osuska L, Hancock RE, Kay WW, Misra S, 2004. Transgenic potatoes expressing a novel cationic peptide are resistant to late blight and pink rot. Transgenic Res. 13 (2), 181–190. [DOI] [PubMed] [Google Scholar]
- Osusky M, Osuska L, Kay W, Misra S, 2005. Genetic modification of potato against microbial diseases: in vitro and in planta activity of a dermaseptin B1 derivative, MsrA2. Theor. Appl. Genet 111 (4), 711–722. [DOI] [PubMed] [Google Scholar]
- Pallerla S, Naik H, Singh S, Gauthier T, Sable R, Jois SD, 2018. Design of cyclic and d-amino acids containing peptidomimetics for inhibition of protein-protein interactions of HER2-HER3. J. Pept. Sci 24 (2), e3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedron CN, Torres MDT, da Silva Lima JA, Silva PI, Silva FD, Oliveira VX, 2017. Novel designed VmCT1 analogs with increased antimicrobial activity. Eur. J. Med. Chem 126, 456–463. [DOI] [PubMed] [Google Scholar]
- Pedron CN, Torres MDT, Oliveira CS, Silva AF, Andrade GP, Wang Y, Pinhal MAS, Cerchiaro G, da Silva Junior PI, da Silva FD, 2023. Molecular hybridization strategy for tuning bioactive peptide function. Comm. Biol 6 (1), 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González A, Caro E, 2018. Effect of transcription terminator usage on the establishment of transgene transcriptional gene silencing. BMC. Res. Notes 11, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalzgraff A, Brandenburg K, Weindl G, 2018. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol 9, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka A, Jedrzejczak-Rey N, Bednarek P, 2015. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206 (3), 948–964. [DOI] [PubMed] [Google Scholar]
- Pirtskhalava M, Vishnepolsky B, Grigolava M, Managadze G, 2021. Physicochemical features and peculiarities of interaction of AMP with the membrane. Pharmaceuticals 14 (5), 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto MS, Pinheiro MPN, Batista VGL, dos Santos RC, de Albuquerque Melo Filho P, de Lima LM, 2014. Plant promoters: an approach of structure and function. Mol. Biotechnol 56, 38–49. [DOI] [PubMed] [Google Scholar]
- Porto WF, Irazazabal L, Alves ES, Ribeiro SM, Matos CO, Pires ÁS, Fensterseifer I, Miranda VJ, Haney EF, Humblot V, 2018. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun 9 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamaeian M, Langen G, Imani J, Khalifa W, Altincicek B, Von Wettstein D, Kogel K-H, Vilcinskas A, 2009. Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J. Exp. Bot 60 (14), 4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero M, Furman N, Mencacci N, Picca P, Toum L, Lentz E, Bravo-Almonacid F, Mentaberry A, 2012. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J. Biotechnol 157 (2), 334–343. [DOI] [PubMed] [Google Scholar]
- Rong W, Qi L, Wang J, Du L, Xu H, Wang A, Zhang Z, 2013. Expression of a potato antimicrobial peptide SN1 increases resistance to take-all pathogen Gaeumannomyces graminis var. tritici in transgenic wheat. Funct. Integr. Genomics 13, 403–409. [DOI] [PubMed] [Google Scholar]
- Rozek A, Powers J-PS, Friedrich CL, Hancock RE, 2003. Structure-based design of an indolicidin peptide analogue with increased protease stability. Biochemistry 42 (48), 14130–14138. [DOI] [PubMed] [Google Scholar]
- Sampaio de Oliveira KB, Leite ML, Rodrigues GR, Duque HM, da Costa RA, Cunha VA, de Loiola Costa LS, da Cunha NB, Franco OL, Dias SC, 2020. Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Expert. Rev. Clin. Pharmacol 13 (4), 367–390. [DOI] [PubMed] [Google Scholar]
- Sani M-A, Separovic F, 2016. How membrane-active peptides get into lipid membranes. Acc. Chem. Res 49 (6), 1130–1138. [DOI] [PubMed] [Google Scholar]
- Savini F, Bobone S, Roversi D, Mangoni ML, Stella L, 2018. From liposomes to cells: filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Pept. Sci 110 (5), e24041. [Google Scholar]
- Scott MG, Yan H, Hancock RE, 1999. Biological properties of structurally related α-helical cationic antimicrobial peptides. Infect. Immun 67 (4), 2005–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti N, Sannino L, Idoine A, Hamman P, De Stradis A, Giorio P, Maréchal-Drouard L, Bock R, Cardi T, 2015. The HIV-1 Pr55 gag polyprotein binds to plastidial membranes and leads to severe impairment of chloroplast biogenesis and seedling lethality in transplastomic tobacco plants. Transgenic Res. 24, 319–331. [DOI] [PubMed] [Google Scholar]
- Shabir U, Ali S, Magray AR, Ganai BA, Firdous P, Hassan T, Nazir R, 2018. Fish antimicrobial peptides (AMP’s) as essential and promising molecular therapeutic agents: a review. Microb. Pathog 114, 50–56. [DOI] [PubMed] [Google Scholar]
- Shafee TM, Lay FT, Phan TK, Anderson MA, Hulett MD, 2017. Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci 74 (4), 663–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagaghi N, Palombo EA, Clayton AH, Bhave M, 2018. Antimicrobial peptides: biochemical determinants of activity and biophysical techniques of elucidating their functionality. World J. Microbiol. Biotechnol 34 (4), 1–13. [DOI] [PubMed] [Google Scholar]
- Shanmugaraj B, Bulaon CJI, Malla A, Phoolcharoen W, 2021. Biotechnological insights on the expression and production of antimicrobial peptides in plants. Molecules 26 (13), 4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Zhu Y, Lai Z, Tan P, Shan A, 2019. Antimicrobial Peptides with Protease Stability: Progress and Perspective. Future Science. [DOI] [PubMed] [Google Scholar]
- Sharma S, Sahoo N, Bhunia A, 2016. Antimicrobial peptides and their pore/ion channel properties in neutralization of pathogenic microbes. Curr. Top. Med. Chem 16 (1), 46–53. [DOI] [PubMed] [Google Scholar]
- Silva ON, Torres MD, Cao J, Alves ES, Rodrigues LV, Resende JM, Lião LM, Porto WF, Fensterseifer IC, Lu TK, 2020. Repurposing a peptide toxin from wasp venom into antiinfeetives with dual antimicrobial and immunomodulatory properties. Proc. Natl. Acad. Sci 117 (43), 26936–26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani-Goloujeh M, Nokhodchi A, Niazi M, Najafi-Hajivar S, Shahbazi-Mojarrad J, Zarghami N, Zakeri-Milani P, Mohammadi A, Karimi M, Valizadeh H, 2018. Effects of N-terminal and C-terminal modification on cytotoxicity and cellular uptake of amphiphilic cell penetrating peptides. Artificial cells, nanomedicine, and biotechnology 46 (supl), 91–103. [DOI] [PubMed] [Google Scholar]
- Soltani S, Keymanesh K, Sardari S, 2007. In silico analysis of antifungal peptides: determining the lead template sequence of potent antifungal peptides. Expert Opin. Drug Discovery 2 (6), 837–847. [DOI] [PubMed] [Google Scholar]
- Spence CA, Lakshmanan V, Donoffio N, Bais HP, 2015. Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae. Front. Plant Sci 6, 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Dashora K, Ameta KL, Singh NP, El-Enshasy HA, Pagano MC, Hesham AEL, Sharma GD, Sharma M, Bhargava A, 2021. Cysteine-rich antimicrobial peptides from plants: the future of antimicrobial therapy. Phytother. Res 35 (1), 256–277. [DOI] [PubMed] [Google Scholar]
- Stam M, Mol JN, Kooter JM, 1997. The silence of genes in transgenic plants. Ann. Bot 79 (1), 3–12. [Google Scholar]
- Stefani F, Hamelin R, 2010. Current state of genetically modified plant impact on target and non-target fungi. Environ. Rev 18 (NA):441–475. [Google Scholar]
- Stewart JM, Nader C, Rajasekaran K, 2007. Effect of antimicrobial peptides (AMPS) on Micorrhizal associations. Summaries of Arkansas Cotton Res. 2007. [Google Scholar]
- Sumi CD, Yang BW, Yeo IC, Hahm YT, 2015. Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can. J. Microbiol 61 (2), 93–103. [DOI] [PubMed] [Google Scholar]
- Sun D, Forsman J, Woodward CE, 2017. Molecular simulations of melittin-induced membrane pores. J. Phys. Chem. B 121 (44), 10209–10214. [DOI] [PubMed] [Google Scholar]
- Tam JP, Wang S, Wong KH, Tan WL, 2015. Antimicrobial peptides from plants. Pharmaceuticals 8 (4), 711–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras F, Schoofs H, De Bolle M, Van Leuven F, Rees SB, Vanderleyden J, Cammue B, Broekaert WF, 1992. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem 267 (22):15301–15309. [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH, 2011. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23 (1), 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MDT, de la Fuente-Nunez C, 2019. Toward computer-made artificial antibiotics. Curr. Opin. Microbiol 51, 30–38. [DOI] [PubMed] [Google Scholar]
- Torres MD, Pedron CN, Araújo I, Silva PI Jr., Silva FD, Oliveira VX, 2017. Decoralin analogs with increased resistance to degradation and lower hemolytic activity. ChemistrySelect 2 (1), 18–23. [Google Scholar]
- Torres MD, Pedron CN, Higashikuni Y, Kramer RM, Cardoso MH, Oshiro KG, Franco OL, Silva PI Junior, Silva FD, Oliveira Junior VX, 2018. Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates. Communications biology 1 (1), 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MD, Sothiselvam S, Lu TK, de la Fuente-Nunez C, 2019. Peptide design principles for antimicrobial applications. J. Mol. Biol 431 (18), 3547–3567. [DOI] [PubMed] [Google Scholar]
- Torres MD, Cao J, Franco OL, Lu TK, de la Fuente-Nunez C, 2021. Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 15 (2), 2143–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MD, Melo MC, Flowers L, Crescenzi O, Notomista E, de la Fuente-Nunez C, 2022. Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng 6 (1), 67–75. [DOI] [PubMed] [Google Scholar]
- Tugyi R, Uray K, Iván D, Fellinger E, Perkins A, Hudecz F, 2005. Partial D-amino acid substitution: improved enzymatic stability and preserved ab recognition of a MUC2 epitope peptide. Proc. Natl. Acad. Sci 102 (2), 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varasteh Shams M, Nazarian-Firouzabadi F, Ismaili A, Shirzadian-Khorramabad R (2019) Production of a Recombinant Dermaseptin Peptide in Nicotiana tabacum Hairy Roots with Enhanced Antimicrobial Activity. Molecular biotechnology: 1–12. [DOI] [PubMed] [Google Scholar]
- Tusé D, Tu T, McDonald KA, 2014. Manufacturing economics of plant-made biologics: case studies in therapeutic and industrial enzymes. Biomed. Res. 16, 256135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vutto N, Gapeeva T, Pundik A, Tretyakova T, Volotovski I, 2010. Transgenic Belarussian-bred potato plants expressing the genes for antimicrobial peptides of the cecropin-melittin type. Russ. J. Genet 46 (12), 1433–1439. [PubMed] [Google Scholar]
- Wan F, de la Fuente-Nunez C (2023) Mining for antimicrobial peptides in sequence space. Nature Biomedical Engineering: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Kontogiorgos-Heintz D, de la Fuente-Nunez C, 2022. Deep generative models for peptide design. Dig. Dis 1 (3), 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., 2020. Bioinformatic analysis of 1000 amphibian antimicrobial peptides uncovers multiple length-dependent correlations for peptide design and prediction. Antibiotics 9 (8), 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Oard J, 2003. Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 22, 129–134. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nowak G, Culley D, Hadwiger LA, Fristensky B, 1999. Constitutive expression of pea defense gene DRR206 confers resistance to blackleg (Leptosphaeria maculans) disease in transgenic canola (Brassica napus). Mol. Plant-Microbe Interact 12 (5), 410–418. [Google Scholar]
- Wang Z. p., Deng L. h., Weng L. s., Deng X. y., Fu X. q., Xin Y. y., Xiao G. y., 2017. Transgenic rice expressing a novel phytase-lactoferricin fusion gene to improve phosphorus availability and antibacterial activity. J. Integr. Agric 16 (4), 774–788. [Google Scholar]
- Wimley WC, 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol 5 (10), 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F, de la Fuente-Nunez C, Collins JJ, 2023. Leveraging artificial intelligence in the fight against infectious diseases. Science 381 (6654), 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR, 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet 32 (4), 569–577. [DOI] [PubMed] [Google Scholar]
- Yevtushenko DP, Misra S, 2007. Comparison of pathogen-induced expression and efficacy of two amphibian antimicrobial peptides, MsrA2 and temporin a, for engineering wide-spectrum disease resistance in tobacco. Plant Biotechnol. J 5 (6), 720–734. [DOI] [PubMed] [Google Scholar]
- Yevtushenko DP, Romero R, Forward BS, Hancock RE, Kay WW, Misra S, 2005. Pathogen-induced expression of a cecropin A-melittin antimicrobial peptide gene confers antifungal resistance in transgenic tobacco. J. Exp. Bot 56 (416), 1685–1695. [DOI] [PubMed] [Google Scholar]
- Zasloff M., 2002. Antimicrobial peptides of multicellular organisms. Nature 415 (6870), 389–395. [DOI] [PubMed] [Google Scholar]
- Zasloff M., 2006. Defending the epithelium. Nat. Med 12 (6), 607–608. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang K, Yan Q, Wang X, Cheng M, Si J, Xue X, Shen D, Jing M, Tyler BM, 2021. Targeting of anti-microbial proteins to the hyphal surface amplifies protection of crop plants against Phytophthora pathogens. Mol. Plant 14 (8), 1391–1403. [DOI] [PubMed] [Google Scholar]