Abstract
The Walkerton (Ontario, Canada) outbreak of waterborne Escherichia coli O157:H7 and Campylobacter jejuni was quite limited in both space and time, making it a good model for exploring the utility of different typing and subtyping methods for the characterization of relationships among isolates of these organisms. We have extended previous work with these organisms through analysis by the Oxford multilocus sequence typing (MLST) and the flagellin short variable region (fla-SVR) sequencing methods. Additional isolates not epidemiologically related to the Walkerton outbreak have also been included. Both sequencing methods identified and differentiated between Walkerton outbreak strains 1 and 2. When these strains were compared with isolates that were not part of the outbreak, the information produced by the fla-SVR method more often correlated with epidemiological findings than that produced by MLST, though both methods were required for optimal discrimination. The MLST data were more relevant in terms of the overall population structure of the organisms. Both mutation and recombination appeared to be responsible for generating diversity among the isolates tested.
Bacterial typing, subtyping, and fingerprinting methods must be evaluated to determine how they subdivide specific bacterial populations into smaller groups and at what level of discrimination they operate. This information can be used to target each method under consideration for the most appropriate type of population analysis. Multiple typing methods have been applied to Campylobacter spp. (35). All are useful to some extent, though some are better suited to the analysis of large-scale population structure, while others may be more useful for routine surveillance of bacteria for cluster detection and outbreak analysis. Serotyping of Campylobacter heat-stable (HS) antigens, e.g., lipooligosaccharide or capsule (4), provides a relatively low level of discrimination but measures the production of surface structures that are important for the pathogenesis and severe clinical sequelae (e.g., Guillain-Barré syndrome) of the organism (36). Serological detection of specific heat-labile (HL) surface antigens has fairly good discriminatory power but does not appear to measure characteristics that, on their own, are unambiguously predictive of epidemiologic relationships (5). Phage typing is useful for discriminating among HS types and has been recommended for epidemiological studies (17), especially when the screening of large numbers of isolates is required (11). We found that phage type did not correlate precisely with outbreak strains in the Walkerton (Ontario, Canada) outbreak (5). Thus, although phage typing was somewhat useful in differentiating outbreak from nonoutbreak strains, it is not clear that phage typing is the best method for outbreak detection. Pulsed-field gel electrophoresis (PFGE) is highly discriminatory and can be extraordinarily useful for definition of clones or lineages within Campylobacter populations (10, 12, 33) as well as for analysis of well-defined outbreaks (9, 37). There has been discussion, however, as to whether this method is too discriminatory for routine surveillance for public health purposes (5, 16). Whether other methods, such as amplified fragment length polymorphism protocols, are subject to similar limitations is currently unknown. For Campylobacter spp., amplified fragment length polymorphism methods are at least as discriminatory as multilocus sequence typing (MLST) and may be as discriminatory as PFGE (13, 32); they are certainly less labor-intensive than PFGE (3). Flagellin locus restriction fragment length polymorphism (fla-RFLP) methods have been developed and have proved both epidemiologically relevant and very useful for strain discrimination in an outbreak analysis (14, 21, 22, 23, 25). Analysis of the flagellin short variable region (fla-SVR) yielded results very similar to those obtained by sequencing of the entire gene (21).
Recent work has led to the development of sequence-based subtyping methods for Campylobacter spp. (6, 7, 34). These methods offer the advantages of complete portability among laboratories and ease of interpretation by well-characterized methods and therefore lend themselves to application in large surveillance networks. The Oxford MLST protocol involves sequencing 402 to 507 nucleotide segments of seven housekeeping genes (7). Use of this method has led to the definition of several clonal complexes within the Campylobacter population (6); these clonal complexes correlate well with the results of multilocus enzyme electrophoresis (29). However, fla-SVR typing was found to subtype strains within sequence types (STs), so its discriminatory power was superior to that of MLST and comparable to that of PFGE (30). This latter study assessed a limited number of strains from each of several outbreaks but did not address in detail the question of how well MLST and fla-SVR distinguished outbreak from nonoutbreak strains within a single outbreak.
We have recently reported the results of typing and subtyping Campylobacter sp. isolates from a large outbreak of waterborne organisms in Walkerton in 2000 (5). Of all methods, a combination of HS typing and subtyping using a fla-RFLP method provided the best correlation with epidemiological findings. Since fla-RFLP is a band-based fingerprinting method, it is more difficult to standardize for use by many laboratories. This study was undertaken to investigate alternative, sequence-based methods for Campylobacter subtyping and to determine the relative merits of the Oxford MLST system and fla-SVR sequence typing for outbreak investigations and surveillance for the detection of case clusters. Our results suggest that fla-SVR sequence typing appears to be predictive of epidemiological relationships, while the Oxford MLST scheme may be more appropriate for determining population changes on a broader scale. A comparison of fla-SVR sequencing and an fla-RFLP method suggests that, though the two methods discriminate slightly differently among Campylobacter populations, both are effective for outbreak analysis and large-scale surveillance.
MATERIALS AND METHODS
Strains used in this study.
Most isolates used were collected during the investigation of the Walkerton waterborne Campylobacter jejuni and Escherichia coli O157:H7 outbreak in 2000 and have been described previously (5). Except where otherwise noted, isolates from bovine sources were collected from farms near the town of Walkerton shortly after the outbreak. The Enterics Laboratory, Division of Bacteriology and Enteric Pathogens, National Microbiology Laboratory (NML), Health Canada, received additional strains from the following sources: various provincial public health laboratories; the Enteric Reference Centre, St. John Regional Hospital, New Brunswick; the Ontario Ministry of Agriculture and Food; and research collaborators in Egypt (S. Savarino, Naval Medical Research Center, Silver Spring, Maryland), France (F. Megraud, C. H. U. Pellegrin, Bordeaux), and the United States (R. A. Oberhelman, Tulane School of Public Health and Tropical Medicine, New Orleans, Louisiana) as part of ongoing collaborations. All isolates were subcultured once upon receipt and stored in glycerol peptone water at −70°C until use. Cultures were grown on Mueller-Hinton agar (Oxoid, Ltd, Basingstoke, England) containing 10% sheep erythrocytes at either 37°C or 42°C in a microaerobic atmosphere.
Biotyping, serotyping and phage typing.
Biotyping was performed as described by Lior (18). HL serotyping was performed by the method of Lior and colleagues (19). HS serotyping using passive hemagglutination to detect HS antigens was performed by the method of Penner and Hennessy (24). Phage typing of isolates was performed as described by Frost and colleagues (11).
Typing and subtyping by PFGE and fla-RFLP.
PFGE was done according to the method of Ribot and colleagues (28) with SmaI and KpnI. fla-RFLP typing was performed by the method of Nachamkin and colleagues (22). Gels (4%) for separation of DdeI fragments were electrophoresed for 16 h at 60 V and then for 1 h at 150 V. An abbreviated version of the fla-RFLP method was also used. In this method, 2% agarose gels were run for about 2 h at 120 V. This allowed greater separation of larger fragments, though smaller fragments were not as well resolved, and was similar to methods described by Harrington and colleagues (14). Both 100- and 123-bp size standards were used, and on some gels, both standards were applied to the same lane to give a combined standard profile. Gels were stained in ethidium bromide and visualized by UV transillumination; the results were entered into a database constructed with Bionumerics Version 2.5 software (Applied Maths, Kortrijk, Belgium). The combined 100- and 123-bp lanes were used for gel standardization, and patterns characteristic of HL serotype reference strains as well as clinical and animal strains with unique profiles were included in the database. The Bionumerics software was capable of normalizing banding patterns from both the 2% and 4% agarose gels together in an interpretable manner as long as a few strains were analyzed in both gel systems to indicate where double bands on the 4% gel were compressed to a wider single band on the 2% gels. The numbering system for designating fla-RFLP patterns was changed from that described in the previous report (5). Patterns generated by HL serotype reference strains were assigned the lowest HL type number having that particular pattern. Patterns not represented by the HL serotype strains were given numbers arbitrarily starting with 136.
fla-SVR sequencing and MLST.
fla-SVR sequencing was performed according to previously described methods (21, 30). Allele numbers were determined by querying the existing flaA-SVR databse (http://phoenix.medawar.ox.ac.uk/flaA/). For strains with possible new fla-SVR alleles, DNA trace files from the ABI prism 3100 DNA sequencer (PE Biosystems, Foster City, Calif.) were submitted to the database administrator (K. Dingle) for confirmation.
MLST was done by the methods of Dingle et al. (7, 8) as modified by Dingle and colleagues (6). For some strains, DNA from boiled cell preparations was successfully used (boiled cell preparations were prepared by suspending a half-loop of culture material from blood agar plates in 200 μl distilled water, boiling the water for 5 min, cooling the material on ice, and centrifuging the cooled material to pellet it and remove debris). However, this worked well for only about half the isolates tested, most likely those not producing DNase (31). DNA for the other half was prepared using a Wizard genomic DNA purification kit (Promega, Madison, Wis.). PCR methods were used as described in the references given, except that the annealing temperature was lowered by 2°C in cases where reactions were initially not successful. Alternate primer sets were also used as described previously (6). Amplicons were cleaned up by use of multiscreen PCR plates or Montage PCR centrifugal filter devices (both plates and filter devices were from Millipore, Etobicoke, Ontario, Canada). Sequencing was done by the DNA Core Facility at the National Microbiology Laboratory, using the appropriate primers with a commercial kit (Big Dye Ready Reaction Mix version 2; PE Biosystems) according to the manufacturer's instructions. Reaction products were separated with the ABI prism 3100 DNA sequencer. MLST alleles were determined by querying the Oxford database (http://pubmlst.org/campylobacter/). Sequence files (ABI 3100 trace files) of new alleles not already in the database were checked again to ensure that errors in sequence editing had not been made. Sequence (trace) files of new MLST or fla-SVR alleles were submitted to the appropriate database.
Data analysis and production of dendrograms.
Editing of sequence files was performed using Lasergene software (DNASTAR, Inc., Madison, Wis.). Edited sequence files were entered into a Bionumerics 2.5 software file(s). Dendrograms for all alleles were produced using the unweighted pair group method with arithmetic averages (UPGMA) method using default settings, followed by implementation of the neighbor-joining method with the Kimura 2 parameter and bootstrapping using 1,000 simulations. Simultaneous analysis of all seven MLST was performed with Bionumerics software by both the UPGMA and neighbor-joining (without bootstrapping) methods. To assess the robustness of this analysis, data were also analyzed without randomized inputs or bootstrapping by both the maximum likelihood (DNAML) and parsimony (DNAPARS) methods contained in the PHYLIP 3.6b analytic package.
Statistical analysis.
Analyses were performed using SPSS version 12.0.1. Where the data were adequate, cross tabulations were followed by the calculation of three measures of association appropriate for use with nominal data, namely, the phi coefficient, Cramer's V, and the contingency coefficient. Measures of significance for the results of these tests are reported. Tests of independence were not performed due to inadequate sample size.
RESULTS
Description of isolate relationships.
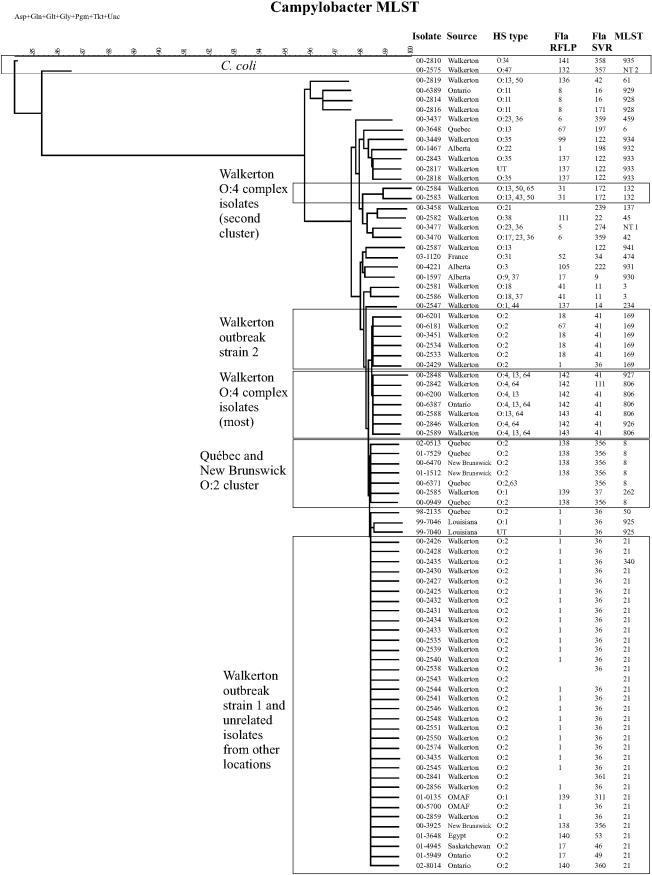
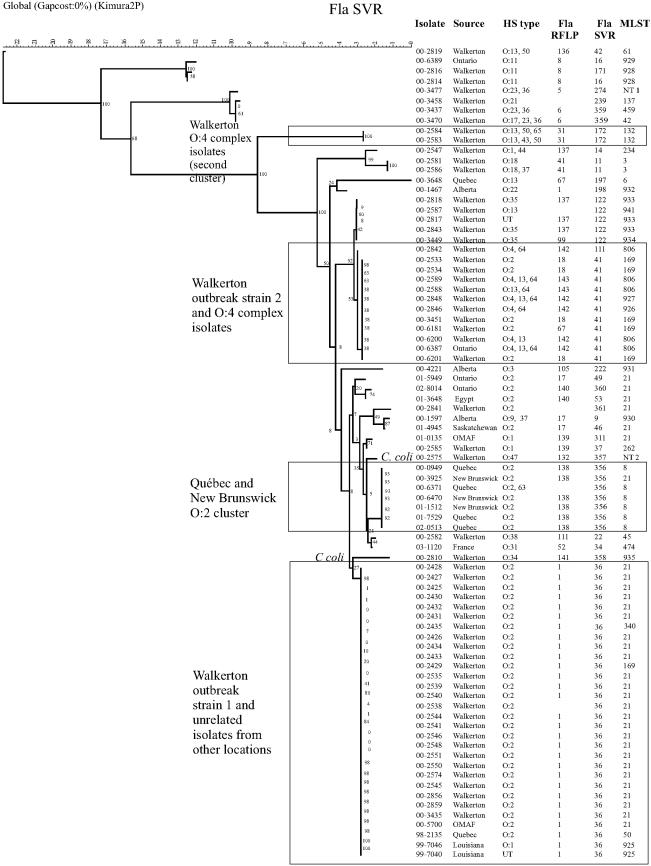
The results of typing and subtyping are shown in Table 1, and the alleles defining the known sequence types are summarized in Table 2. Both Walkerton outbreak types 1 and 2 were quite homogenous with respect to MLST, fla-SVR type, and fla-RFLP (Table 1), and only minor differences were apparent in PFGE patterns, as noted previously (5). Isolate 00-2435 had a single nucleotide change in the aspA allele (Table 2), which was sufficient to classify it into a different ST. One isolate, 00-2429, had an ST characteristic of Walkerton outbreak strain 2 while its fla-SVR type and fla-RFLP pattern were characteristic of Walkerton outbreak strain 1; it clustered with outbreak strain 1 in the fla-SVR dendrogram and with outbreak strain 2 in the MLST dendrogram (Table 1; Fig. 1 and 2). Interestingly, the PFGE patterns obtained with SmaI and KpnI also appeared to indicate a hybrid of Walkerton outbreak strains 1 and 2 (Table 1).
TABLE 1.
Typing comparisons of Campylobacter isolates associated with the Walkerton outbreak and independent of the outbreak
| NML isolate no. | Species | HS type | Bio- typea | ST | PFGE
|
HL sero- type | fla-RFLP type | fla-SVR type | Walkerton outbreak type | Source | Farm | Location | Yr | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | |||||||||||||
| 00-2425 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2428 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2430 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2432 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2435 | C. jejuni | O:2 | II | 340 | 1 | 1 | UT | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2535 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2538 | C. jejuni | O:2 | II | 21 | 11 | 1 | 125 | ND | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2539 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2540 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2541 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2543 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | ND | ND | 1 | Human | Walkerton | 2000 | |
| 00-2544 | C. jejuni | O:2 | II | 21 | 4 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2545 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2546 | C. jejuni | O:2 | II | 21 | 1 | 1 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2550 | C. jejuni | O:2 | II | 21 | 1 | 1 | UT | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2856 | C. jejuni | O:2 | II | 21 | 4 | 1 | 128 | 1 | 36 | 1 | Bovine | 14 | Walkerton | 2000 |
| 00-2859 | C. jejuni | O:2 | II | 21 | 4 | 1 | 112, 125 | 1 | 36 | 1 | Bovine | 2 | Walkerton | 2000 |
| 00-2426 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2427 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2431 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2433 | C. jejuni | O:2 | II | 21 | 2 | 3 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2434 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2548 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2551 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2574 | C. jejuni | O:2 | II | 21 | 2 | 2 | 110 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-3435 | C. jejuni | O:2 | II | 21 | 2 | 2 | 125 | 1 | 36 | 1 | Human | Walkerton | 2000 | |
| 00-2429 | C. jejuni | O:2 | hipp neg | 169 | 2 | 3 | UT | 1 | 36 | 1/2 | Human | Walkerton | 2000 | |
| 00-2533 | C. jejuni | O:2 | hipp neg | 169 | 3 | 3 | 4 | 18 | 41 | 2 | Human | Walkerton | 2000 | |
| 00-2534 | C. jejuni | O:2 | hipp neg | 169 | 3 | 3 | UT | 18 | 41 | 2 | Human | Walkerton | 2000 | |
| 00-3451 | C. jejuni | O:2 | hipp neg | 169 | 3 | 3 | 4 | 18 | 41 | 2 | Human | Walkerton | 2000 | |
| 00-6181 | C. jejuni | O:2 | hipp neg | 169 | 3 | 3 | 128 | 67 | 41 | 2 | Human | Walkerton | 2000 | |
| 00-6201 | C. jejuni | O:2 | hipp neg | 169 | 3 | 3 | 128 | 18 | 41 | 2 | Human | Walkerton | 2000 | |
| 00-2841 | C. jejuni | O:2 | II | 21 | 3 | ND | 4 | ND | 361 | Bovine | 12 | Walkerton | 2000 | |
| 00-0949 | C. jejuni | O:2 | ND | 8 | 9 | 32 | 36 | 138 | 356 | Human | Québec | 2000 | ||
| 00-3925 | C. jejuni | O:2 | ND | 21 | 26 | 31 | 100 | 138 | 356 | Human | New Brunswick | 2000 | ||
| 00-5700 | C. jejuni | O:2 | II | 21 | 4 | 1 | 1 | 1 | 36 | Bovine | OMAF | 2000 | ||
| 00-6371 | C. jejuni | O:2,63 | II | 8 | 9 | 32 | UT | ND | 356 | Human | Québec | 2000 | ||
| 00-6470 | C. jejuni | O:2 | ND | 8 | 9 | 32 | 36 | 138 | 356 | Human | New Brunswick | 2000 | ||
| 01-1512 | C. jejuni | O:2 | II | 8 | 26 | 31 | 90 | 138 | 356 | Human | New Brunswick | 2000 | ||
| 01-3648 | C. jejuni | O:2 | I | 21 | 49 | 37 | 128 | 140 | 53 | Human | Egypt | 2001 | ||
| 01-4945 | C. jejuni | O:2 | II | 21 | 3 | 2 | 128 | 17 | 46 | Human | Saskatchewan | 2001 | ||
| 01-5949 | C. jejuni | O:2 | II | 21 | 50 | 38 | 128 | 17 | 49 | Canine | Ontario | 2001 | ||
| 01-7529 | C. jejuni | O:2 | hipp neg | 8 | 9 | 32 | UT | 138 | 356 | Human | Québec | 2001 | ||
| 02-0513 | C. jejuni | O:2 | hipp neg | 8 | 9 | 32 | 36 | 138 | 356 | Human | Québec | 2001 | ||
| 02-8014 | C. jejuni | O:2 | II | 21 | ND | ND | 4 | 140 | 360 | Human | Ontario | 2002 | ||
| 99-7040 | C. jejuni | UT | I | 925 | ND | ND | 79 | 1 | 36 | Chicken | Louisiana | 1999 | ||
| 99-7046 | C. jejuni | O:1 | II | 925 | ND | ND | UT | 1 | 36 | Chicken | Louisiana | 1999 | ||
| 00-2547 | C. jejuni | O:1,44 | II | 234 | 12 | 10 | 2 | 137 | 14 | Human | Walkerton | 2000 | ||
| 00-2585 | C. jejuni | O:1 | II | 262 | 8 | 7 | 2 | 139 | 37 | Bovine | 5 | Walkerton | 2000 | |
| 01-0135 | C. jejuni | O:1 | II | 21 | ND | ND | 2 | 139 | 311 | Bovine | OMAF | 2001 | ||
| 00-4221 | C. jejuni | O:3 | I | 931 | ND | ND | 94 | 105 | 222 | Human | Alberta | 2000 | ||
| 00-2589 | C. jejuni | O:4,13,64 | II | 806 | 16 | ND | UT | 143 | 41 | Bovine | 7 | Walkerton | 2000 | |
| 00-2842 | C. jejuni | O:4,64 | II | 806 | 26 | ND | UT | 143 | 111 | Bovine | 6 | Walkerton | 2000 | |
| 00-2846 | C. jejuni | O:4,64 | II | 926 | ND | ND | 91 | 142 | 41 | Bovine | 6 | Walkerton | 2000 | |
| 00-2848 | C. jejuni | O:4,13,64 | II | 927 | 30 | ND | 7 | 142 | 41 | Bovine | 6 | Walkerton | 2000 | |
| 00-6200 | C. jejuni | O:4,13 | II | 806 | 30 | ND | 7 | 142 | 41 | Human | Walkerton | 2000 | ||
| 00-6387 | C. jejuni | O:4,13,64 | II | 806 | 30 | ND | 7 | 142 | 41 | Human | Walkerton | 2000 | ||
| 00-2583 | C. jejuni | O:13,43,50 | III | 132 | 6 | ND | 7 | 31 | 172 | Bovine | 3 | Walkerton | 2000 | |
| 00-2584 | C. jejuni | O:13,50,65 | III | 132 | 6 | 7 | 7 | 31 | 172 | Bovine | 3 | Walkerton | 2000 | |
| 00-2587 | C. jejuni | O:13 | II | 941 | 7 | 9 | UT | ND | 122 | Bovine | 7 | Walkerton | 2000 | |
| 00-2588 | C. jejuni | O:13,64 | II | 806 | 15 | ND | 7 | 143 | 41 | Bovine | 7 | Walkerton | 2000 | |
| 00-2819 | C. jejuni | O:13,50 | II | 61 | 22 | ND | 7 | 136 | 42 | Bovine | 10 | Walkerton | 2000 | |
| 00-3648 | C. jejuni | O:13 | I | 6 | ND | ND | UT | 67 | 197 | Human | Québec | 2000 | ||
| 98-2135 | C. jejuni | O:8 | II | 50 | ND | ND | 125 | 1 | 36 | Human | Québec | 1998 | ||
| 00-1597 | C. jejuni | O:9,37 | I | 930 | ND | ND | UT | 17 | 9 | Human | Alberta | 2000 | ||
| 00-2814 | C. jejuni | O:11 | II | 928 | ND | ND | 82 | 8 | 16 | Bovine | 9 | Walkerton | 2000 | |
| 00-2816 | C. jejuni | O:11 | II | 928 | 31 | ND | 82 | 8 | 171 | Bovine | 10 | Walkerton | 2000 | |
| 00-6389 | C. jejuni | O:11 | II | 929 | 21 | ND | 82 | 8 | 16 | Human | Walkerton | 2000 | ||
| 00-2581 | C. jejuni | O:18 | II | 3 | 14 | ND | 41 | 41 | 11 | Bovine | 6 | Walkerton | 2000 | |
| 00-2586 | C. jejuni | O:18,37 | I | 3 | 13 | ND | 41 | 41 | 11 | Bovine | 6 | Walkerton | 2000 | |
| 00-3458 | C. jejuni | O:21 | IV | 137 | ND | ND | 40 | ND | 239 | Human | Walkerton | 2000 | ||
| 00-1467 | C. jejuni | O:22 | I | 932 | ND | ND | UT | 1 | 198 | Human | Alberta | 2000 | ||
| 00-3437 | C. jejuni | O:23,36 | III | 459 | ND | ND | 5 | 6 | 359 | Human | Walkerton | 2000 | ||
| 00-3470 | C. jejuni | O:17,23 | I | 42 | ND | ND | 99 | 6 | 359 | Human | Walkerton | 2000 | ||
| 00-3477 | C. jejuni | O:23,36 | II | NT 1 | ND | ND | 5 | 5 | 274 | Human | Walkerton | 2000 | ||
| 03-1120 | C. jejuni | O:31 | II | 474 | ND | ND | UT | 52 | 34 | Human | France | 2003 | ||
| 00-2817 | C. jejuni | UT | II | 933 | 21 | ND | 51 | 137 | 122 | Bovine | 10 | Walkerton | 2000 | |
| 00-2818 | C. jejuni | O:35 | II | 933 | 21 | ND | 51 | 137 | 122 | Bovine | 10 | Walkerton | 2000 | |
| 00-2843 | C. jejuni | O:35 | II | 933 | 33 | ND | 51 | 137 | 122 | Bovine | 6 | Walkerton | 2000 | |
| 00-3449 | C. jejuni | O:35 | II | 934 | 32 | ND | 51 | 99 | 122 | Human | Walkerton | 2000 | ||
| 00-2582 | C. jejuni | O:38 | III | 45 | 5 | 6 | 2 | 111 | 22 | Bovine | 3 | Walkerton | 2000 | |
| 00-2575 | C. coli | O:47 | I | NT 2 | 10 | 4 | 34 | 132 | 357 | Human | Walkerton | 2000 | ||
| 00-2810 | C. coli | O:34 | I | 935 | 18 | ND | UT | 141 | 358 | Bovine | 8 | Walkerton | 2000 | |
hipp neg, hippurate hydrolysis negative phenotype; ND, not determined; NT, new type; UT, untypeable.
TABLE 2.
MLST allele-specific sequence types for strains used in this study
| ST | ST complex | No. of strains | Allele
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | |||
| 3 | 49 | 2 | 3 | 2 | 5 | 10 | 11 | 11 | 6 |
| 6 | 1 | 63 | 34 | 37 | 33 | 45 | 5 | 7 | |
| 8 | 21 | 7 | 2 | 1 | 1 | 3 | 2 | 1 | 6 |
| 21 | 21 | 31 | 2 | 1 | 1 | 3 | 2 | 1 | 5 |
| 42 | 42 | 1 | 1 | 2 | 3 | 4 | 5 | 9 | 3 |
| 45 | 45 | 1 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 50 | 21 | 1 | 2 | 1 | 12 | 3 | 2 | 1 | 5 |
| 61 | 61 | 1 | 1 | 4 | 2 | 2 | 6 | 3 | 17 |
| 132 | 508 | 2 | 1 | 6 | 22 | 24 | 12 | 28 | 1 |
| 137 | 45 | 1 | 4 | 7 | 10 | 4 | 42 | 7 | 1 |
| 169 | 21 | 6 | 2 | 1 | 1 | 3 | 6 | 1 | 5 |
| 234 | 1 | 8 | 2 | 5 | 53 | 17 | 3 | 1 | |
| 262 | 21 | 1 | 2 | 1 | 1 | 3 | 2 | 1 | 3 |
| 340 | 21 | 1 | 22 | 1 | 1 | 3 | 2 | 1 | 5 |
| 459 | 42 | 1 | 1 | 2 | 3 | 3 | 5 | 9 | 3 |
| 474 | 48 | 1 | 2 | 4 | 1 | 2 | 2 | 1 | 5 |
| 806 | 21 | 4 | 2 | 1 | 1 | 3 | 140 | 3 | 5 |
FIG. 1.
C. jejuni and C. coli isolates clustered by using the neighbor-joining method for comparison of MLST profiles. All seven alleles are included in the analysis. Images from Bionumerics were adjusted in Corel Draw 11.0 for the addition of headings and to make text easier to read.
FIG. 2.
C. jejuni and C. coli isolates clustered by using the neighbor-joining method with bootstrapping for comparison of fla-SVR sequence profiles. Bootstrap values are included adjacent to each branch. Images from Bionumerics were adjusted in Corel Draw 11.0 for the addition of headings and to make text easier to read.
When only Walkerton outbreak strains 1 and 2 were analyzed, the fla-SVR and fla-RFLP subtyping methods were highly associated (P < 0.001). The fla-SVR and MLST test results were also highly associated, but less so than the fla-SVR and fla-RFLP results (P = 0.03, exact test). Finally, the HL serotype was not significantly associated with the fla-SVR type (P = 0.1, exact test).
Several strains had MLST types indistinguishable from the Walkerton outbreak types despite the fact that they appeared to be epidemiologically and either temporally or geographically unrelated to the outbreak. HS serotype O:2 strain 00-5700 was not epidemiologically related to the Walkerton outbreak type 1 strains but had an identical PFGE pattern, MLST, fla-SVR type, and fla-RFLP pattern. This isolate was obtained from a steer at an abattoir east of Toronto, distant from the town of Walkerton. Five other HS serotype O:2 isolates (00-3925, 01-3648, 01-4945, 01-5949, and 02-8014) had ST-21 in common with Walkerton outbreak strain 1 but were from different geographic areas and, in the case of 01-4945, 01-3648, and 02-8014, were obtained at least 1 year later than Walkerton outbreak strain 1. Isolate 01-4945 had PFGE patterns indistinguishable from those of the Walkerton outbreak strains, while isolates 00-3925, 01-3648, and 01-5949 had PFGE patterns different from those of the Walkerton outbreak strains.
Most HS O:2, ST-21 strains not epidemiologically associated with the Walkerton outbreak had fla-SVR sequences distinct from those of the outbreak strains. Isolates 01-3648 and 02-8014 shared the fla-RFLP 140 pattern but had fla-SVR alleles 53 and 360, respectively. fla-SVR allele 360 differed from allele 53 by a single nucleotide (Table 3). Isolate 00-3925 was obtained in New Brunswick in 2000 and carried the ST-21 genotype characteristic of the Walkerton strains but had the fla-RFLP 138 pattern and fla-SVR allele 356.
TABLE 3.
Characterization of new fla-SVR alleles
| Strain no. | Best matching allele | New Oxford allele no. | No. of nt different | Nucleotide change(s) |
|---|---|---|---|---|
| 00-0949 | 45 | 356 | 1 | 270 C > T |
| 00-3925 | 45 | 356 | 1 | 270 C > T |
| 00-6371 | 45 | 356 | 1 | 270 C > T |
| 00-6470 | 45 | 356 | 1 | 270 C > T |
| 01-1512 | 45 | 356 | 1 | 270 C > T |
| 01-7529 | 45 | 356 | 1 | 270 C > T |
| 02-0513 | 45 | 356 | 1 | 270 C > T |
| 00-2841 | 78 | 361 | 1 | 270 T > C |
| 00-3437 | 189 or 239 | 359 | 1 | 105 A > T or 99 T > A |
| 00-3470 | 189 or 239 | 359 | 1 | 105 A > T or 99 T > A |
| 02-8014 | 53 | 360 | 1 | 34 T > C |
| 00-2575 | 23 or 243 | 357 | 1 | 34 C > T or 108 C > T |
| 00-2810 | 320 | 358 | 2 | 117 C > T and 316 A > C |
There was a group of C. jejuni strains in New Brunswick and Québec between early 2000 and 2002 that carried the ST-8 MLST genotype, fla-RFLP 138 pattern, and fla-SVR allele 356 (Tables 1 and 3; Fig. 1 and 2). Four isolates from Québec obtained in three different years and one isolate from New Brunswick (00-6470) also had indistinguishable PFGE patterns (Table 1), suggesting the possibility of clonal derivation. Isolate 00-3925 appeared to be part of this group on the basis of geographical location, fla-SVR type, and fla-RFLP pattern but had different PFGE patterns and carried the ST-21 genotype. This ST differed from ST-8 only at the uncA allele (Table 2). Two isolates from New Brunswick with PFGE patterns 26 (SmaI) and 31 (KpnI) had indistinguishable fla-RFLP patterns and fla-SVR sequences but differed in STs; isolate 00-3925 carried the ST-21 genotype, while 01-1512 carried the ST-8 genotype.
Some isolates epidemiologically unrelated to the Walkerton outbreak strains had fla-SVR types indistinguishable from that of a Walkerton outbreak strain but had different STs. An isolate obtained from Québec in 1998 (98-2135) carried ST-50 (ST-21 complex) and had fla-SVR types and fla-RFLP patterns identical to those of Walkerton outbreak strain 1. Two isolates from chickens in Louisiana (99-7040 and 99-7046) also carried the same fla-SVR allele and fla-RFLP pattern as Walkerton outbreak strain 1 but differed at the glnA allele from ST-50 and three other possible STs; these isolates could be easily distinguished from the Walkerton outbreak strain on the MLST dendrogram (Table 4; Fig. 1). Walkerton outbreak strain 1 clustered separately from outbreak strain 2 on the MLST dendrogram (Fig. 1) on the basis of differences at the pgm allele (Table 2); these isolates also clustered separately on the fla-SVR dendrogram (Fig. 2). Walkerton isolates could be differentiated from non-Walkerton isolate equally well by fla-SVR typing and ST determination (P < 0.001).
TABLE 4.
Characterization of strains with newly described MLST alleles or sequence types
| New ST | No. of strains | Strain no. | Allele
|
Closest ST(s) | Closest ST clonal complex | Possible different allele(s) (close to allele no.) | Nucleotides different | New Oxford database allele number | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||||||
| 925 | 2 | 99-7040 | 2 | 25 | 12 | 3 | 2 | 1 | 5 | 31, 50, 387, 536 | 21 | gln | ||
| 99-7046 | ||||||||||||||
| 926 | 1 | 00-2846 | 2 | 20 | 1 | 3 | 140 | 3 | 5 | 806 | 21 | gln | ||
| 927 | 1 | 00-2848 | 2 | 1 | 1 | 19 | 140 | 3 | 5 | 806 | 21 | gly | ||
| NT 1 | 1 | 00-3477 | 1 | 2 | 3 | 4 | 5 | 9 | NT | 42 | 42 | unc (3) | nt 234 G > A | unc-71 |
| 941 | 1 | 00-2587 | 42 | 63 | 2 | 66 | 6 | 1 | 5 | 35, 38, 476, 804 | 48 | gln | ||
| 928 | 2 | 00-2814 | 9 | 2 | 4 | 62 | 4 | 5 | 38 | 257, 560 | 257 | unc | ||
| 00-2816 | ||||||||||||||
| 929 | 1 | 00-6389 | 9 | 2 | 4 | 62 | 4 | 5 | 17 | 257, 560 | 257 | unc | ||
| 930 | 1 | 00-1597 | 7 | 2 | 5 | 2 | 2 | 3 | 6 | 5, 404 | 353 | gln, pgm | ||
| 931 | 1 | 00-4221 | 8 | 17 | 5 | 2 | 11 | 59 | 6 | 400 | 353 | pgm | ||
| 932 | 1 | 00-1467 | 10 | 27 | 59 | 19 | 10 | 79 | 7 | 403, 435, 552, 556, 557, 605 | 403 | glt, gly, tkt | ||
| 933 | 3 | 00-2817 | 10 | 1 | 59 | 19 | 10 | 5 | 7 | 285, 403, 435, 493, 552 | 403 | gln, glt, gly, tkt, unc | ||
| 00-2818 | ||||||||||||||
| 00-2843 | ||||||||||||||
| 934 | 1 | 00-3449 | 1 | 1 | 59 | 2 | 10 | 5 | 7 | 552 | 403 | asp, gln, gly | ||
| NT 2 | 1 | 00-2575 | 33 | NT | 30 | 82 | 104 | 43 | 17 | 828 | gln (39) | nt 39 C > T | ||
| 935 | 1 | 00-2810 | 33 | 39 | 30 | 115 | 104 | 35 | 42 | 826, 855, 893 | gly, unc | |||
The HS O:4 complex of strains is composed of a group of cross-reacting serotypes, including O:4, O:13, O:16, O:43, and O:50 (27), that may differentially express any single serotype or a combination of serotypes. Members of the HS O:4 complex were distributed through both the MLST and fla-SVR dendrograms (Fig. 1 and 2). Several of these isolates clustered with outbreak strain 2 (HS O:2) as shown in both dendrograms, and many of the isolates from both the HS O:4 complex and the HS O:2 cluster carried the same fla-SVR allele. Isolates from human and bovine sources were indistinguishable. STs (806, 926, and 927) of seven of twelve HS O:4 complex strains belonged to the same clonal complex (ST-21) as both Walkerton outbreak strains (Table 4). ST-927 isolate 00-2848 differed from ST-21 at glyA, pgm, and tkt but differed from ST-806 only at the glyA allele. ST-806 differed from Walkerton outbreak strains 1 and 2 at the pgm and tkt alleles. All these isolates therefore appear to have been derived from ST-21 strains by horizontal transfer and recombination of alleles from outside sources. These HS O:4 isolates also had fla-RFLP patterns that were almost identical, differing only in the width of a single band (Table 1; Fig. 3), as well as fla-SVR sequences that were identical or almost identical (Fig. 2); the isolates were found on farms 6 and 7. The fla-SVR dendrogram (Fig. 2) clustered HS O:2 and HS O:4 complex isolates with fla-SVR type 41 or the closely related type 122 sequence; in this case, MLST results exhibited a variability not seen with fla-SVR.
FIG. 3.
Comparison of fla-RFLP profiles for selected Campylobacter isolates. All profiles associated with isolates analyzed in this study have been included. Profiles were clustered for visualizing of differences in banding patterns using the UPGMA method (Dice coefficient) with 1% optimization and 1.5% tolerance. Images from Bionumerics were adjusted in Corel Draw 11.0 for the addition of headings and to make text easier to read.
The remaining HS O:4 complex isolates comprised a more heterogeneous group. This group included two indistinguishable isolates with ST-132, the fla-RFLP 31 pattern, and fla-SVR allele 172 that were obtained from bovine sources on farm 3. Two other HS O:4 complex isolates had different STs, fla-RFLP patterns, and fla-SVR types; 00-2819 was isolated from bovine sources on farm 10 (Table 1). These latter four isolates were widely separated in the MLST dendrogram (Fig. 1). Within the HS O:4 complex group of isolates, the fla-SVR sequence type and ST were significantly associated (P = 0.02, exact test) while the ST and HL type were not significantly associated (P = 0.22, exact test).
HS O:1 isolates were located close to HS O:2 strains in the MLST dendrogram but were distributed more widely in the fla-SVR dendrogram. Isolate 01-0135 from the Ontario Ministry of Agriculture and Food had ST-21 in common with the Walkerton outbreak strains but clustered separately from the Walkerton outbreak strains on the fla-SVR dendrogram (Fig. 2). An HS O:1 chicken isolate from Louisiana, 99-7046, was ST-925 (ST-21 clonal complex), clustered closely to Walkerton strains in MLST (Fig. 1), and had a fla-SVR genotype identical to that of the Walkerton strains (Table 1). The ST-262 of isolate 00-2585 belonged to ST-21 and differed from ST-8 and ST-21 only in the uncA allele (Table 2); it therefore clustered with ST-8 isolates on the MLST dendrogram (Fig. 1). However, this strain was grouped with HS O:1 isolate 01-0135 on the fla-SVR dendrogram, suggesting recombination may have occurred at either the uncA or the flaA allele. Finally, HS O:1 isolate 00-2547 was separated from HS O:2 isolates on both the MLST and fla-SVR dendrograms.
Most of the remaining isolates had STs and fla-SVR types quite distinct from the Walkerton outbreak strain types (Tables 1, 2, and 4; Fig. 1). All HS O:11 strains clustered together, as did the two HS O:18 strains from farm 6 and three HS O:35 isolates (plus one untypeable isolate) from a Walkerton patient and from farms 6 and 10.
Dendrograms created from composite MLST data of all seven genes indicated that C. jejuni clustered separately from C. coli (Fig. 1). Similar dendrograms were produced for six of the seven individual alleles, namely aspA, glnA, gltA, glyA, pgm, and tkt, confirming that the sequences of these six genes were quite different in C. coli strains (data not shown). Bootstrapping values of 100% were obtained for the branch separating C. jejuni from C. coli for each allele in these dendrograms (data not shown). However, the uncA dendrogram included the C. coli isolates 00-2575 and 00-2810 in a cluster that also contained HS O:11 isolates 00-2814, 00-2816, and 00-6389 (Fig. 4). Also included in this cluster was HS O:4 complex isolate 00-2757. The two C. coli isolates were not clustered separately in the fla-SVR dendrogram (Fig. 2), though they did have unique alleles compared with all other isolates studied. Dendrograms were identical or very similar whether they were constructed using UPGMA, neighbor-joining, or maximum likelihood methods (data not shown).
FIG. 4.
C. jejuni and C. coli isolates clustered by using the neighbor-joining method with bootstrapping for comparison of uncA profiles. Bootstrap values are included adjacent to each branch. Images from Bionumerics were adjusted in Corel Draw 11.0 for the addition of headings and to make text easier to read.
Comparison of HL serotyping, fla-RFLP, and fla-SVR sequencing.
The fla-SVR sequence types were compared with patterns obtained by fla-RFLP, as well as with HL serotypes. For the most part fla-SVR sequencing and fla-RFLP results correlated well. However, some differences were noted. While three isolates (00-2817, 00-2818, 00-2843) were fla-SVR 122 and the fla-RFLP 137 pattern, isolate 00-3449 was fla-SVR 122 and the fla-RFLP 99 pattern (Table 1). The fla-RFLP 137 and 99 patterns appeared quite different (Fig. 3). Of five isolates with fla-SVR allele 41, four had the fla-RFLP 18 pattern and one had the fla-RFLP 67 pattern. In these cases fla-RFLP testing was capable of indexing variation within the flaA allele but outside the fla-SVR region. Two fla-SVR alleles, 14 and 122, were distinguished among strains with the fla-RFLP 137 pattern, indicating that fla-SVR sequencing detected variation not seen with fla-RFLP analysis.
HL serotype 125 predominated in the Walkerton outbreak strain 1 isolates analyzed in this study, correlating well with the fla-RFLP 1 pattern and fla-SVR type 36. However, two of these Walkerton outbreak strain 1 isolates were HL untypeable, one was HL 128, one was HL 110, and one expressed the HL 112, 125 phenotype. Walkerton outbreak strain 2 isolates with fla-SVR sequence type 41 were HL untypeable (one isolate), HL 128 (two isolates), and HL 4 (two isolates). HS O:2 isolates with the fla-RFLP 138 pattern and the new fla-SVR type 356 expressed four different HL types, including the untypeable phenotype. A similar increased discrimination was found when comparing HL serotype with fla genotypes for the HS O:4 complex strains (Table 1). There was a lack of association between fla-SVR sequence type and HL serotype for the HS O:4 complex isolates (P = 0.58). Fewer isolates from HS serogroups other than the HS O:2 and HS O:4 complexes were available for study; however, in the serogroups that were included (Table 1) the association of HL serotype and fla genotype was relatively good. This could not be validated statistically, however, due to inadequate sample size.
DISCUSSION
This investigation has probed a Campylobacter population (strains associated with the Walkerton outbreak during the spring of 2000) that is very well defined geographically and temporally and has compared this population with a small number of isolates from different times and geographic locations. Results from this investigation have provided insights into the population structure and dynamics of Campylobacter at this scale, and into the utility of various typing and subtyping methods for describing Campylobacter populations.
MLST, fla-SVR, and fla-RFLP methods all identified Walkerton outbreak strains 1 and 2 and were capable of distinguishing between these strains. The ST of isolate 00-2435 differed from Walkerton outbreak ST-21 only by a single nucleotide in the aspA allele. Such a change could arise from either recombination or from mutation. It should be noted that, while either mechanism is possible, recombination would require the presence of coexisting strains carrying the variant aspA allele; none were found in isolates analyzed as part of this study. Such isolates may have been present in numbers too low to be included in the small sample analyzed; alternatively, the allele change could have resulted from mutation. Similar changes were not detected within the outbreak and sporadic strain panels used by Sails et al. (30), though this may be because only a small number of isolates from each outbreak were chosen for analysis. However, when the same group assessed the correspondence of MLST with multilocus enzyme electrophoresis, there were several pairs of alleles that differed by only a single nucleotide (29). The differences from at least three of these alleles were attributed to mutations partly on the basis that they resulted in alleles unique to the database. Regardless of the mechanism, small sequence changes in MLST and fla-SVR alleles may therefore occur frequently enough to be detected in the course of larger, or more prolonged, outbreaks. Minor changes in MLST sequence do not appear to cause difficulties in identification of epidemiologically significant relationships among isolates as long as the data are interpreted with care.
ST-21 was characteristic of Walkerton outbreak strain 1 but was also found in several isolates not epidemiologically related to the Walkerton outbreak. Isolate 00-5700 was indistinguishable from outbreak strain 1 in HS serotype, SmaI PFGE pattern, KpnI PFGE pattern, fla-RFLP pattern, fla-SVR type, and MLST. This isolate was obtained from a healthy steer processed at an abattoir east of Toronto, and was obtained as the result of routine testing according to a random sampling plan. Though the place of origin of this steer was not recorded, it is unlikely that an animal from Walkerton would be shipped so far and there is no evidence that there was any contact of this steer with cattle or humans from Walkerton. Nevertheless, there is a remote possibility that a strain of Campylobacter was transferred directly from Walkerton to the steer. At the same time, it is also possible that Walkerton outbreak strain 1 represents a common type found in cattle. This question can be answered only through more extensive testing of Campylobacter sp. isolates from unrelated cattle herds by MLST and fla-SVR. Isolates obtained from Saskatchewan (01-4945) and Ontario (01-5949) in 2001 were both ST-21 and had identical fla-RFLP patterns. However, these isolates had fla-SVR sequence types different from each other and from that of the Walkerton outbreak type 1 strain. Another isolate, 01-0135, was isolated from a cow processed at abattoir specializing in high-risk livestock near Woodstock, Ontario, and may have originated from anywhere in Ontario, including the Walkerton area. This isolate was ST-21 but had a fla-SVR type and fla-RFLP pattern that were different from those of Walkerton outbreak strain 1. Several other isolates from locations outside Ontario were also ST-21 but had fla-RFLP and fla-SVR types different from those of the Walkerton outbreak strain 1. None of these strains were epidemiologically related to the Walkerton outbreak or, to our knowledge, to each other. In these instances, fla-SVR appeared to provide results that were more discriminatory and in closer agreement with epidemiological findings.
The ST-21 complex was the predominant clonal complex overall. In previous studies, strains belonging to the ST-21 complex comprised 32% of all C. jejuni isolates from humans (6). ST-21 isolates may be fairly common in both human and animal populations (20). These observations suggest it may be somewhat difficult to determine whether isolates with this ST are clonally related, limiting the usefulness of MLST for surveillance to detect outbreaks. Dingle and colleagues (6) suggested that the clonal complex defined by MLST is epidemiologically relevant for both long and short-term investigations of C. jejuni epidemiology. Support for this conclusion can be found in the fact that MLST differentiated HS O:2 and HS O:4 complex isolates that had the fla-SVR 41 allele. The results summarized here further suggest that MLST may generally group C. jejuni isolates more broadly, or with a slower rate of change, than methods interrogating flagellar sequences, and that either the fla-RFLP or the fla-SVR sequencing methods may produce information that correlates more closely with epidemiological findings.
Six clonal complexes (ST-21, ST-45, ST-48, ST-61, ST-206, and ST-257) were previously found to contain the majority of isolates from human disease (6). Four of the six were identified in this study (Tables 2 and 4), suggesting that this may be a general phenomenon in different geographic areas.
Several strains of C. jejuni clustered with C. coli on the uncA dendrogram. The simplest explanation is transfer of uncA alleles from some unidentified source and recombination into this group of isolates. Since the uncA alleles of 00-2814, 00-2816, 00-2819, and 00-6389 appear to be at least as distant from all other C. jejuni isolates on the uncA dendrogram as C. coli isolates are from C. jejuni isolates on all other allele dendrograms, it appears likely that these anomalous C. jejuni uncA sequences were derived from C. coli. Recent investigations have found that uncA from C. jejuni ST-61 was very divergent compared with uncA from other STs and may have come from C. coli (6). The fact that isolate 00-2819 was also ST-61 confirms earlier published results that at least some ST-61 isolates appeared to have acquired C. coli-like uncA sequences. It further suggests that ST-61 strains carrying the C. coli-like uncA allele(s) may have a geographically widespread distribution.
Several strains appeared to belong to newly described STs on the basis of a change in a single allele (Table 4), a situation that could have arisen either from mutation or from recombination of the allele in question after horizontal transfer from another source. Isolates belonging to the new type (NT) 1 or new ST-928 differ by only one or two alleles from the closest matching allele. On the other hand, recombination may have been responsible for changes resulting in the apparent exchange of alleles, especially STs 930 to 934, which all differ by two alleles or more from the closest match. Similar observations have been made by others using the Oxford MLST system (29, 30).
Changes in the flaA SVR appeared to be due to mutations rather than recombination, as only one or two nucleotide substitutions were seen and since changes quite often resulted in alleles new to the database (Table 3). This contrasts with other work cataloguing recombination in Campylobacter flaA genes (15, 29) and suggests that both mutation and recombination were operating to create diverse DNA and protein sequences. Stable fla-RFLP patterns exist (10) and outbreak strains often share the same flaA sequence (15), suggesting that both fla-SVR sequencing and fla-RFLP may be appropriate for surveillance to detect clusters. Neither should serve as the only typing or subtyping method used due to instability of the flaA gene; fla-RFLP patterns have been shown to change upon serial subculture (26).
Intraspecific recombination is frequent in C. jejuni and has created extensive diversity in the allelic profiles of this organism (34). The suggestion that most changes in MLST types arise from recombination rather than mutation (7, 32) is only partly supported by the results presented here, as many of the changes found may have arisen from mutations. Some isolates or strains appear to be more susceptible than others to genetic change, especially that arising from horizontal transfer of genes (10, 26). We have also noted that some C. jejuni strains appear to be resistant to natural transformation or transformation mediated by electroporation (data not shown). It is possible that nontransformable isolates comprise a significant proportion of the subset of strains chosen for analysis here.
Few stable C. jejuni populations have been described to date. The demonstration in this study of an apparently geographically delimited population of C. jejuni in Québec and New Brunswick that appeared to be fairly stable during 2000 to 2001 was an unexpected finding. Both MLST and fla-SVR results were in agreement as to the existence of this population, suggesting that the use of both methods may be a powerful tool for describing biological, epidemiological, and evolutionary relationships among C. jejuni.
Isolates belonging to the ST-257 clonal complex have previously been found only in isolates from human disease or chickens (6). Though bovine isolates 00-2814, 00-2816, and 00-6389 all carry newly described STs, they appear to belong to the ST-257 complex as well, suggesting a lack of host restriction of this group of organisms.
HL serotypes either correlated loosely or did not correlate at all with results obtained by using either the fla-RFLP or the fla-SVR subtyping method but tended to provide further discrimination of these types (Table 1). This is consistent with reports that indicate flagellin may not be the HL serotype determinant in many cases (1, 2). Alternately, information responsible for HL serotype may be carried in the long variable region of the fla locus.
Bacterial typing and subtyping systems used for public health surveillance and outbreak identification must have several key features. They must allow high throughput and be relatively inexpensive, both to allow use of the methods with as many isolates as possible and to facilitate the creation of large electronic databases. Turnaround time should be rapid, and both raw and interpreted data must be amenable to storage and transfer in computerized databases via high-speed Internet links. Data should provide information that correlates well with epidemiological findings. Both the fla-RFLP method (31) and fla-SVR sequencing appear to fit these criteria well, though fla-SVR sequencing did group epidemiologically unrelated isolates in a some instances. Additionally, fla-RFLP is more difficult to standardize for portable information sharing among laboratories. Sequencing of the fla-SVR region may be the optimal routine laboratory surveillance method for laboratories with access to medium-throughput sequencing facilities, while fla-RFLP may be adequate for laboratories that do not have access to these facilities but have PCR capability. HS serotyping would be an important accessory technique for routine surveillance, while other methods, such as PFGE and MLST, could be very important for outbreak characterization and traceback investigations. While the Oxford MLST was as effective as fla-SVR at differentiating Walkerton from non-Walkerton isolates, it relied on sequencing of more genes, took longer, and was more costly. Furthermore, in our hands it was sometimes difficult to obtain either PCR amplicon or high-quality sequence data on the first attempt. It appears that, while the Oxford MLST will continue to be the method of choice for investigations into the population structure of Campylobacter, it is not the preferred method for routine surveillance of this organism.
Acknowledgments
This work was supported by Health Canada through the Food Memorandum to Cabinet funding initiative.
We gratefully acknowledge the provision of isolates and accompanying information by the following provincial laboratory partners: Enteric Reference Centre, St. John Regional Hospital, New Brunswick; Québec Public Health Laboratory/Laboratoire de santé publique du Québec (PHL/LSPQ) (Louise Ringuette), Alberta PHL (Linda Chui), and Saskatchewan PHL. We also gratefully acknowledge the ongoing support of the Canadian PHL Forum. Other collaborators who provided valuable information or isolates include Joseph Odumeru (University of Guelph, Guelph, Ontario, Canada), R. A. Oberhelman (Tulane School of Public Health and Tropical Medicine, New Orleans, Louisiana), S. Savarino (Naval Medical Research Center, Silver Spring, Maryland), and F. Megraud (C.H.U. Pellegrin, Bordeaux, France). The staff members of the Molecular Typing Laboratory in the Bacteriology and Enteric Pathogens section of the NML who contributed to this project by performing PFGE include Shelley Johnson, Jennifer Campbell, Russell Easy, Jason Allen, and Jamie Munro. All sequencing was done within the DNA Core Facility, NML, by Brynn Watson and Shari Tyson. Many thanks to these staff members and to Shaun Tyler for troubleshooting sequencing problems. David Woodward, Lawrence Price, and Ali Moterassed are acknowledged for their contributions in isolate serotyping and identification, and for providing training in HS serotyping to C.G.C. Pasquale Melito did the initial work in implementation of fla-RFLP typing.
REFERENCES
- 1.Alm, R. A., P. Guerry, M. E. Power, H. Lior, and T. J. Trust. 1991. Analysis of the role of flagella in the heat-labile Lior serotyping scheme of thermophilic campylobacters by mutant allele exchange. J. Clin. Microbiol. 29:2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnens, A. P., J. Wagner, H. Lior, J. Nicolet, and J. Frey. 1995. Restriction fragment length polymorphisms among the flagellar genes of the Lior heat-labile serogroup reference strains and field strains of Campylobacter jejuni and C. coli. Epidemiol. Infect. 114:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Champion, O. L., E. L. Best, and J. A. Frost. 2002. Comparison of pulsed-field gel electrophoresis and amplified fragment length polymorphism techniques for investigating outbreaks of enteritis due to campylobacters. J. Clin. Microbiol. 40:2263-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chart, H., J. A. Frost, A. Oza, R. Thwaites, S. Gillanders, and B. Rowe. 1996. Heat-stable serotyping antigens expressed by strains of Campylobacter jejuni are probably capsular and not long-chain lipopolysaccharide. J. Appl. Bacteriol. 81:635-640. [DOI] [PubMed] [Google Scholar]
- 5.Clark, C. G., L. Price, R. Ahmed, D. L. Woodward, P. L. Melito, F. G. Rodgers, F. Jamieson, B. Ciebin, A. Li, and A. Ellis. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., N. Van Den Braak, F. M. Colles, L. J. Price, D. L. Woodward, F. G. Rodgers, H. P. Endtz, A. Van Belkum, and M. C. J. Maiden. 2001. Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barré and Miller-Fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J. Clin. Microbiol. 39:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, C., K. Stanley, S. Andrew, and K. Jones. 2001. Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl. Environ. Microbiol. 67:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost, J. A., J. M. Kramer, and S. A. Gillanders. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson, J., E. Lorenz, and R. J. Owen. 1997. Lineages within Campylobacter jejuni defined by numerical analysis of pulsed-field gel electrophoretic DNA profiles. J. Med. Microbiol. 46:157-163. [DOI] [PubMed] [Google Scholar]
- 13.Hänninen, M.-L., P. Perko-Mäkelä, H. Rautelin, B. Duim, and J. A. Wagenaar. 2001. Genomic relatedness within five common Finnish Campylobacter jejuni pulsed-field gel electrophoresis genotypes studied by amplified fragment length polymorphism analysis, ribotyping, and serotyping. Appl. Environ. Microbiol. 67:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington, C. S., L. Moran, A. M. Ridley, D. G. Newell, and R. H. Madden. 2003. Inter-laboratory evaluation of three flagellin PCR/RFLP methods for typing Campylobacter jejuni and C. coli: the CAMPYNET experience. J. Appl. Microbiol. 95:1321-1333. [DOI] [PubMed] [Google Scholar]
- 15.Harrington, C. S., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing system. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedberg, C. W., K. E. Smith, J. M. Besser, D. J. Boxrud, T. W. Hennessy, J. B. Bender, F. A. Anderson, and M. T. Osterholm. 2001. Limitations of pulsed-field gel electrophoresis for the routine surveillance of Campylobacter infections. J. Infect. Dis. 184:242-243. (Letter.) [DOI] [PubMed] [Google Scholar]
- 17.Jackson, C. J., A. J. Fox, D. M. Jones, D. R. A. Wareing, and D. N. Hutchinson. 1998. Associations between heat-stable (O) and heat-labile (HL) serogroup antigens of Campylobacter jejuni: evidence for interstrain relationships within three O/HL serovars. J. Clin. Microbiol. 36:2223-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lior, H. 1984. New, extended biotyping scheme for Campylobacter jejuni, Campylobacter coli, and “Campylobacter laridis.” J. Clin. Microbiol. 20:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lior, H., D. L. Woodward, J. A. Edgar, L. J. Laroche, and P. Gill. 1982. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J. Clin. Microbiol. 15:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nachamkin, I., H. Ung, and C. M. Patton. 1996. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J. Clin. Microbiol. 34:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen, L., and D. G. Newell. 2001. The ability of fla-typing schemes to discriminate between strains of Campylobacter jejuni. J. Appl. Microbiol. 91:217-224. [DOI] [PubMed] [Google Scholar]
- 26.Petersen, L., and S. L. W. On. 2000. Efficacy of flagellin gene typing for epidemiological studies of Campylobacter jejuni in poultry estimated by comparison with macrorestriction profiling. Lett. Appl. Microbiol. 31:14-19. [DOI] [PubMed] [Google Scholar]
- 27.Preston, M. A., and J. L. Penner. 1989. Characterization of cross-reaction serotypes of Campylobacter jejuni. Can. J. Microbiol. 35:265-273. [DOI] [PubMed] [Google Scholar]
- 28.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing correlate with strain associations identified by multilocus enzyme electrophoresis. J. Clin. Microbiol. 41:4058-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santesteban, E., J. Gibson, and R. J. Owen. 1996. Flagellin gene profiling of Campylobacter jejuni heat-stable serotype 1 and 4 complex. Res. Microbiol. 147:641-649. [DOI] [PubMed] [Google Scholar]
- 32.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. L. Willems, K. E. Dingle, F. M. Colles, and J. D. A. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley, J., D. Linton, K. Sutherland, C. Jones, and R. J. Owen. 1995. High-resolution genotyping of Campylobacter coli identifies clones of epidemiologic and evolutionary significance. J. Infect. Dis. 172:1130-1134. [DOI] [PubMed] [Google Scholar]
- 34.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodward, D. L., and F. G. Rodgers. 2002. Identification of Campylobacter heat-stable and heat-labile antigens by combining the Penner and Lior serotyping schemes. J. Clin. Microbiol. 40:741-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]