Abstract
IncA variation among Dutch Chlamydia trachomatis isolates was investigated. Of 98 strains, two carried an incA with a premature stop codon, lacked IncA, and were nonfusogenic, while 96 contained an intact incA, expressed IncA, and were fusogenic. Among these 96 strains, nine IncA sequence types were found, of which the three most frequently encountered (88% of the strains) were randomly distributed among symptomatic and asymptomatic patients.
Chlamydia trachomatis is an obligate intracellular pathogen, responsible for sexually transmitted disease (STD) (4, 6). The bacteria replicate intracellularly in a membrane-bound vacuole (inclusion) that is modified through the insertion of chlamydial proteins, collectively classified as Inc proteins, including IncA (5, 11). Inc proteins are generally characterized by a typical bi-lobed hydrophobic domain of approximately 60 amino acids but lack overall amino acid homology (1). Strains with mutations in incA, resulting into a premature stop codon, reside in multiple small homotypic inclusions that, in contrast to inclusions containing wild-type incA strains, do not fuse with each other (12). This strongly suggests involvement of IncA in the homotypic fusion of the inclusions. It has been previously shown from the study of 11,440 isolates that only 176 (1.5%) had the nonfusogenic phenotype (16). The observation that the nonfusogenic strains are identified among patients with less-severe disease symptoms suggests that incA polymorphisms may be involved in the clinical course of the infection (3).
We have previously reported the incA sequence polymorphisms and fusogenic properties of 25 C. trachomatis laboratory reference strains comprising most serovars (9). Among these laboratory strains, all of which contained an incA gene encoding a full-length 273-amino-acid-long protein, 12 IncA amino acid sequence types (STs) were identified. In addition, these strains were fusogenic and expressed IncA in the inclusion membrane (IM), irrespective of their IncA ST (9). For the present study, we report the incA sequence polymorphisms, expression patterns of IncA, and the fusogenic properties of 98 C. trachomatis isolates from Dutch female patients. The relationship between IncA ST and the clinical outcome of infection was also evaluated.
Females (age range, 14 to 33 years; median, 22 years), visiting our Public Health Laboratory, participated after signing an informed consent. From each female two cervical swabs were taken. One was assessed for the presence of C. trachomatis DNA as described previously (14). A second cervical swab was placed in 0.4 M sucrose phosphate buffer (4SP) medium and used for C. trachomatis culture as well as DNA extraction. Females with coinfections with other STD-causing microorganisms (Candida albicans, Neisseria gonorrhoeae, Trichomonas vaginalis, and herpes simplex virus type 1 or type 2), detected by using the described techniques, were excluded from the study (2, 13, 17). Selected females were classified as symptomatic based upon at least one positive answer on a questionnaire regarding their complaints, which included increased discharge, having bloody discharge during and/or after coitus, abdominal pain, and/or dysuria. The first 98 consecutively enrolled C. trachomatis-positive females without coinfection were included.
Of these females, 43 showed symptoms of infection (44%). Two-tailed chi-square tests or Fisher's exact test was used to compare groups. A P value of <0.05 was considered statistically significant (SPSS Inc., Chicago, Ill.). The 4SP-collected clinical sample was used to culture C. trachomatis in HeLa 229 cells (ATCC CCL2). Cultures were harvested and stored as described elsewhere (7, 8). DNA for incA amplification was extracted from the 4SP-collected clinical sample (7). incA was amplified by thermocycling and sequenced as described previously (9). Sequence analysis (Staden Package v4.7 [www.mrc-lmb.cam.ac.uk/pubseq/]) revealed that 96 females were infected with a C. trachomatis strain harboring an incA gene encoding a putative full-length protein. Among the 96 IncA sequences, nine STs were observed. Four STs represented 94% of the strains, indicating that incA variation is limited (Fig. 1). These four predominant types are ST1 (identical to the prototypic sequence) (27%), ST2 (I47T) (36%), ST3 (I47T and E116K) (26%), and ST4 (I218T) (5%), respectively (Table 1). The ratio of ST1 to ST3 among the 96 isolates was 88%, supporting previous data of a study of a collection of 25 reference laboratory strains. The ratio of ST1 to ST3 among these strains was 64% (9). ST4 has also been described previously (12), but ST5 to ST9 represent novel incA alleles (Table 1).
FIG. 1.
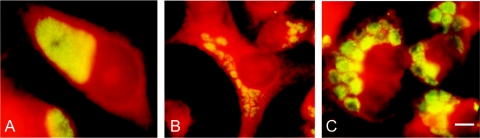
Fluorescence microscopic analyses of HeLa cells infected with clinical isolates (multiplicity of infection, 5 to 10) containing an incA gene encoding full-length IncA (A) or an incA gene with insertions, leading to premature stop codons (B, C). Immunostaining with anti-MOMP revealed a single inclusion in HeLa cells infected with C. trachomatis isolates (n = 86), containing full-length IncA, irrespective of its ST. In contrast, multiple small inclusions were observed in HeLa cells infected with strain 190-163-02 (B) or strain 190-059-03 (C) carrying an incA with a premature stop codon. Bar, 10 μm.
TABLE 1.
C. trachomatis IncA amino acid sequence types and the distribution among symptomatic and asymptomatic females
| STa | Accession no.b | Polymorphic amino acid sitesc | No. of females:
|
Pd | |
|---|---|---|---|---|---|
| Symptomatic (n = 43) (%) | Asymptomatic (n = 53) (%) | ||||
| 1 | AY683465 | MKIPQENT | 11 (26) | 15 (28) | NS |
| 2 | AF683466 | ..T..... | 12 (28) | 23 (43) | NS |
| 3 | AF683467 | ..T..K.. | 14 (33) | 10 (19) | NS |
| 4 | AF683468 | .......I | 2 (5) | 3 (6) | ND |
| 5 | AF683469 | L.T..... | 1 (2) | 1 (2) | ND |
| 6 | AF683470 | ..T...S. | 1 (2) | ND | |
| 7 | AF683471 | ...L.... | 1 (2) | ND | |
| 8 | AF683472 | ....E... | 1 (2) | ND | |
| 9 | AF683473 | .R.....I | 1 (2) | ND | |
The prototype amino acid sequence IncA deduced from the DNA sequence of strain D/UW-3/Cx is indicated as ST1 (15). ST1, ST2, and ST3 were previously annotated as STa, STb, and STc, respectively (9).
The GenBank accession number of the incA sequence, representative for a ST. The sequences of two strains with insertions in incA, resulting in premature stop codons, are not presented in the table but are available at GenBank through accession no. AY683474 (strain 190-163-02) and AY683475 (strain 190-059-03).
Polymorphic amino acid sites of IncA at positions 29 (M), 33 (K), 47 (I), 63 (P), 64 (Q), 116 (E), 161 (N), and 218 (T), respectively. Numbering according to the amino acid numbering of the prototype IncA sequence (15). A dot indicates amino acid identity with ST1.
P values (two-tailed) are based on chi-square analysis. NS, not significant; ND, not determined.
These data, in combination with the incA sequences already deposited in GenBank, bring the total number of reported STs to 19. Silent mutations were only observed in incA of two strains with an ST3 IncA (not shown). Two females (2%) were infected with C. trachomatis harboring an incA gene with a mutation leading to a premature stop codon. The incA gene, amplified using DNA extracted from the cotton swab specimen of subject 46, had a single-nucleotide insertion at position 175, leading to a frame shift and a premature stop codon at position 265. In addition, the incA amplified using DNA extracted from the cotton swab specimen of subject 64 had a 20-bp nucleotide duplication at position 54, leading to a frame shift and a premature stop codon at position 142. We next assessed inclusion morphology and IncA expression by fluorescence microscopy of infected cells using anti-major outer membrane protein (Chlamydia direct IF identification kit; bioMérieux, sa Marcy l'Etoile, France) and anti-IncA (10). HeLa 229 cells cultured on coverslips were infected with each of the isolates at a multiplicity of infection of 5 to 10. Inclusion morphology and IncA expression were independently judged by two persons blinded for the code of the isolates.
Repropagation was successful with 88 of the 98 isolates. Of 88 isolates, 86 resulted in one large single bacterium containing inclusion in infected HeLa cells observed by immunofluorescence using anti-MOMP (Fig. 1A). Of these 86 isolates, expression of IncA and its localization to the IM of the inclusion was revealed by bright fluorescent labeling of the IM of the infected cells, using anti-IncA (not shown). Chlamydia isolates of two females resulted in a completely different inclusion morphology after infection of HeLa cells (Fig. 1B and C). These isolates were phenotypically characterized by the formation and persistence of multiple inclusions per infected cell, indicating nonfusogenicity. Apparently, these multiple inclusions lack IncA, as fluorescence was absent in the infected HeLa cells when anti-IncA was used in fluorescence microscopy (not shown). The nonfusogenic phenotype and absence of IncA expression was observed during several serial passages of both isolates in HeLa cells, indicating these isolates had a stable phenotype. Decoding of the identity of the isolates revealed that the nonfusogenic isolates, lacking IncA expression, corresponded to the two isolates carrying incA genes with an insertion leading to a premature stop codon.
We demonstrated that 2% (2 of 88) of the isolates studied were nonfusogenic, consistent with the results of Suchland and coworkers from the United States (16), indicating that in The Netherlands nonfusogenicity of C. trachomatis isolates as seen in the United States is a rare phenomenon. The two nonfusogenic strains described in our study were isolated from symptomatic female patients. Geisler and colleagues have reported that females infected with nonfusogenic strains had fewer signs and symptoms of infections than females infected with fusogenic strains (3, 16). In our study population, it was impossible to predict whether the presence or absence of IncA may lead to differences in the course of infection, due to the low proportion of nonfusogenic strains. In addition, association between IncA ST and disease was also absent. Chlamydial isolates harboring ST1, ST2, or ST3, the prevalent IncA STs among chlamydial isolates of our study population, were randomly distributed among the symptomatic and asymptomatic patients (Table 1). Moreover, a relationship between ST1, ST2, or ST3 and individual symptoms or a combination of symptoms was not found. We conclude that, in this study population, IncA ST variation is limited and that ST1 to ST3 are not related to clinical manifestations of a C. trachomatis infection.
Nucleotide sequence accession numbers.
The nucleotide sequences of incA, representative of ST1 to ST9, and the sequences of incA with a premature stop codon are deposited in the GenBank under the accession numbers indicated in the Table 1.
Acknowledgments
We gratefully acknowledge Daniel D. Rockey for providing us with anti-IncA serum, Wim van Est for the artwork, and Bill Paxton for critical reading of the manuscript.
REFERENCES
- 1.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 2.Bruisten, S. M., I. Cairo, H. Fennema, A. Pijl, M. Buimer, P. G. Peerbooms, E. Van Dyck, A. Meijer, J. M. Ossewaarde, and G. J. van Doornum. 2001. Diagnosing genital ulcer disease in a clinic for sexually transmitted diseases in Amsterdam, The Netherlands. J. Clin. Microbiol. 39:601-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geisler, W. M., R. J. Suchland, D. D. Rockey, and W. E. Stamm. 2001. Epidemiology and clinical manifestations of unique Chlamydia trachomatis isolates that occupy nonfusogenic inclusions. J. Infect. Dis. 184:879-884. [DOI] [PubMed] [Google Scholar]
- 4.Gerbase, A. C., J. T. Rowley, D. H. Heymann, S. F. Berkley, and P. Piot. 1998. Global prevalence and incidence estimates of selected curable STDs. Sex. Transm. Infect. 74(Suppl. 1):S12-S16. [PubMed] [Google Scholar]
- 5.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5:288-293. [DOI] [PubMed] [Google Scholar]
- 6.Marrazzo, J. M., and W. E. Stamm. 1998. New approaches to the diagnosis, treatment, and prevention of chlamydial infection. Curr. Clin. Top. Infect. Dis. 18:37-59. [PubMed] [Google Scholar]
- 7.Morre, S. A., J. M. Ossewaarde, J. Lan, G. J. van Doornum, J. M. Walboomers, D. M. MacLaren, C. J. Meijer, and A. J. van den Brule. 1998. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J. Clin. Microbiol. 36:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ossewaarde, J. M., M. Rieffe, A. de Vries, R. P. Derksen-Nawrocki, H. J. Hooft, G. J. van Doornum, and A. M. van Loon. 1994. Comparison of two panels of monoclonal antibodies for determination of Chlamydia trachomatis serovars. J. Clin. Microbiol. 32:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pannekoek, Y., A. van der Ende, P. P. Eijk, J. van Marle, M. A. de Witte, J. M. Ossewaarde, A. J. van den Brule, S. A. Morre, and J. Dankert. 2001. Normal IncA expression and fusogenicity of inclusions in Chlamydia trachomatis isolates with the incA I47T mutation. Infect. Immun. 69:4654-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617-626. [DOI] [PubMed] [Google Scholar]
- 11.Rockey, D. D., M. A. Scidmore, J. P. Bannantine, and W. J. Brown. 2002. Proteins in the chlamydial inclusion membrane. Microbes Infect. 4:333-340. [DOI] [PubMed] [Google Scholar]
- 12.Rockey, D. D., W. Viratyosin, J. P. Bannantine, R. J. Suchland, and W. E. Stamm. 2002. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology 148:2497-2505. [DOI] [PubMed] [Google Scholar]
- 13.Spaargaren, J., J. Stoof, H. Fennema, R. Coutinho, and P. Savelkoul. 2001. Amplified fragment length polymorphism fingerprinting for identification of a core group of Neisseria gonorrhoeae transmitters in the population attending a clinic for treatment of sexually transmitted diseases in Amsterdam, The Netherlands. J. Clin. Microbiol. 39:2335-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spaargaren, J., I. Verhaest, S. Mooij, C. Smit, H. S. Fennema, R. A. Coutinho, A. Salvador Pena, and S. A. Morre. 2004. Analysis of Chlamydia trachomatis serovar distribution changes in the Netherlands (1986-2002). Sex. Transm. Infect. 80:151-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 16.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Schee, C., H. J. Sluiters, W. I. van der Meijden, P. van Beek, P. Peerbooms, H. Verbrugh, and A. van Belkum. 2001. Host and pathogen interaction during vaginal infection by Trichomonas vaginalis and Mycoplasma hominis or Ureaplasma urealyticum. J. Microbiol. Methods 45:61-67. [DOI] [PubMed] [Google Scholar]