Abstract
Norway has a low prevalence of antimicrobial resistance, including macrolide-resistant Streptococcus pneumoniae (MRSP). In a nationwide surveillance program, a total of 2,200 S. pneumoniae isolates were collected from blood cultures and respiratory tract specimens. Macrolide resistance was detected in 2.7%. M-type macrolide resistance was found in 60% of resistant isolates, and these were mainly mef(A)-positive, serotype-14 invasive isolates. The erm(B)-encoded macrolide-lincosamide-streptogramin B (MLSB) type dominated among the noninvasive isolates. One strain had an A2058G mutation in the 23S rRNA gene. Coresistance to other antibiotics was seen in 96% of the MLSB-type isolates, whereas 92% of the M-type isolates were susceptible to other commonly used antimicrobial agents. Serotypes 14, 6B, and 19F accounted for 84% of the macrolide-resistant isolates, with serotype 14 alone accounting for 67% of the invasive isolates. A total of 29 different sequence types (STs) were detected by multilocus sequence typing. Twelve STs were previously reported international resistant clones, and 75% of the macrolide-resistant isolates had STs identical or closely related to these clones. Eleven isolates displayed 10 novel STs, and 7/11 of these “Norwegian strains” coexpressed MLSB and tetracycline resistance, indicating the presence of Tn1545. The invasive serotype-14 isolates were all classified as ST9 or single-locus variants of this clone. ST9 is a mef-positive M-type clone, commonly known as England14-9, reported from several European countries. These observations suggest that the import of major international MRSP clones and the local spread of Tn1545 are the major mechanisms involved in the evolution and dissemination of MRSP in Norway.
Antimicrobial resistance is an increasing problem worldwide, and several countries have implemented surveillance systems in recent years. The Norwegian surveillance program for antimicrobial resistance (NORM) was established in 1999 and has reported yearly on the epidemiology of antimicrobial resistance and the usage of antimicrobial agents. Antimicrobial susceptibility data on common pathogens such as Escherichia coli, Klebsiella spp., Haemophilus influenzae, Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes confirm that antimicrobial resistance is still a limited problem in Norway (28). The prevalence of macrolide-resistant S. pneumoniae (MRSP) has increased dramatically in several European countries (16) but is still relatively low in Northern Europe (31, 34). In Norway, macrolide resistance in S. pneumoniae blood culture isolates has increased from 2.4% in 2000 to 4.8% in 2002 and 6.0% in 2003 (28). Macrolides are used as second-line alternatives in the treatment of respiratory tract infections, because the prevalence of non-penicillin-susceptible S. pneumoniae is below 3% in Norway. They are recommended only for patients who are allergic to penicillin or are suspected of having atypical pneumonia. Thus, the increase in MRSP is of clinical concern as well as public health interest.
There are three well-characterized mechanisms of macrolide resistance in S. pneumoniae (23, 36, 46, 53). (i) Target site modification is mediated by methylases encoded by erythromycin ribosome methylation (erm) genes. Methylation of adenine at position 2058 of the peptidyl transferase loop of 23S rRNA prevents binding of 14-, 15-, and 16-member ring macrolides, lincosamides, and streptogramin B, thus leading to resistance to all these compounds (MLSB-type resistance). Expression of MLSB-type resistance can be either constitutive (cMLS) or inducible (iMLS). The erm(B) gene is the most common erm gene in S. pneumoniae, but erm(A) has been reported in rare cases (48). (ii) The active efflux mechanism encoded by macrolide efflux (mef) genes is more specific and causes so-called M-type resistance only to 14-and 15-member ring macrolides. (iii) Ribosomal mutations in the 23S rRNA gene or the ribosomal protein L4 or L22 have been shown to cause macrolide resistance in S. pneumoniae (4, 51, 52).
The aims of this study were to examine the true prevalence of macrolide resistance in Norwegian clinical isolates of S. pneumoniae and to determine their resistance phenotypes and genotypes. We also performed serotyping and multilocus sequence typing (MLST) to investigate the clonal relationship among MRSP strains (10).
MATERIALS AND METHODS
Bacterial strains.
The study included all strains with reduced susceptibility to erythromycin (MIC, ≥1 μg/ml) among 2,200 clinical isolates of S. pneumoniae collected by Norwegian clinical microbiology laboratories during four different periods from 1993 to 2002. The isolates were either invasive isolates from blood cultures (n = 998) or noninvasive isolates from respiratory tract specimens (n = 1,202). Table 1 gives the study periods in relation to national population coverage, number of isolates, and distribution of invasive versus noninvasive isolates. Briefly, the 494 strains collected in 1993 to 1994 and 1997 were from three and five laboratories covering 15.3% and 27.9% of the population in Norway, respectively (22). The remaining 1,706 strains were collected in 2001 and 2002, as part of the NORM surveillance program, and included all 24 Norwegian laboratories. The surveillance strategy is based on local sampling and antimicrobial susceptibility testing by Etest (AB Biodisk, Solna, Sweden).
TABLE 1.
Sample periods, population coverage, number of strains, and prevalence of macrolide resistance among 2,200 invasive and noninvasive S. pneumoniae isolates collected in Norway, 1993 to 2002
| Study period | Norwegian population coverage (%) | No. of strains | No. (%) of macrolide-resistant strains
|
Reference | ||
|---|---|---|---|---|---|---|
| Total | Invasive | Noninvasivea | ||||
| 1993-1994 | 15.3 | 176 | 4 | 4 (2.3) | 22 | |
| 1997 | 27.9 | 318 | 4 | 4 (1.3) | ||
| 2001 | 100 | 460 | 11 | 11 (2.4) | 28 | |
| 2001 | 708 | 19 | 19 (2.7) | |||
| 2002 | 100 | 538 | 22 | 22 (4.1) | ||
| Total | 2,200 | 60 (2.7) | 33 (3.3) | 27 (2.3) | ||
Isolated from sputum, throat, middle ear, sinus, and eye specimens.
Bacterial identification.
Strains were phenotypically identified as S. pneumoniae based on colony morphology, Gram staining, catalase reaction, and α-hemolysis on Mueller-Hinton agar plates supplemented with 5% (vol/vol) sheep blood (Difco Laboratories, Detroit, MI). The strains were further tested for optochin susceptibility (AB Biodisk, Solna, Sweden), agglutination in the Pneumo-Kit slidex test (bioMérieux, Marcy l'Étoile, France), and bile solubility (Sigma-Aldrich Chemie, Gmbh, Steinheim, Germany). A total of 64 strains phenotypically identified as S. pneumoniae and with reduced susceptibility to erythromycin were selected for further investigations (38).
Antimicrobial susceptibility testing.
Strains were examined by Etest on Mueller-Hinton agar supplemented with 5% sheep blood according to the recommendation of the manufacturer. The antimicrobials tested were erythromycin, azithromycin, clindamycin, tetracycline, penicillin G, ciprofloxacin, chloramphenicol, and trimethoprim-sulfamethoxazole. Defibrinated horse blood was used for trimethoprim-sulfamethoxazole. S. pneumoniae ATCC 49619 was included for quality control in each run. Susceptibility results were categorized according to NCCLS breakpoints (27). Interpretive criteria for ciprofloxacin are not given by the NCCLS. Thus, we used the breakpoints for ciprofloxacin obtained from the Swedish Reference Group for Antibiotics (47).
Macrolide resistance phenotypes.
The double-disk method with erythromycin and clindamycin disks (AB Biodisk, Solna, Sweden), as well as MIC data, was used for determination of macrolide resistance phenotypes as previously described (42). Blunting of the clindamycin inhibition zone near the erythromycin disk indicated iMLSB, and resistance to both erythromycin and clindamycin indicated cMLSB. Susceptibility to clindamycin with no blunting indicated the M resistance phenotype (M type) (42).
Detection of macrolide resistance determinants.
The isolates were examined by PCR for the macrolide resistance genes erm(A), erm(B), and mef(A) as previously described (45, 50, 52), except for the forward erm(B) primer, which was redesigned (5′-GTA CTC AAC CAA ATA ATA AAA CAA-3′). S. pyogenes 200A [erm(A)], S. pneumoniae K-96 [erm(B)], and S. pneumoniae NT896 [mef(A)], kindly provided by Jari Jalava, National Institute of Public Health, Turku, Finland, were used as positive controls. A single strain with no detectable acquired macrolide resistance gene was examined by partial pyrosequencing of 23S rRNA and genes encoding ribosomal proteins L4 and L22 at the National Institute of Public Health in Turku (37).
Serotyping.
All the strains were serotyped using Neufeld's Quellung reaction with serum obtained from Statens Serum Institut, Denmark. Nontypeable isolates that were optochin susceptible and bile soluble were considered presumptive pneumococci.
MLST.
MLST was performed according to the work of Enright et al. (10). Briefly, internal fragments of the seven housekeeping genes aroE, gdh, gki, recP, spi, xpt, and ddl were amplified by PCR and sequenced on each strand using the ABI Prism BigDye cycle sequencing kit (Perkin-Elmer Applied Biosystems). Sequencing reaction products were electrophoresed on an ABI Prism 377 automated DNA sequencer (Applied Biosystems). Sequences were analyzed using the Sequence Navigator DNA and Protein Sequence Comparison software (Applied Biosystems). Alleles were assigned by comparing the sequence at each locus to all known alleles at that locus, and the combination of seven alleles (the allelic profile) determined the sequence type (ST). Allele and ST assignments were made through the S. pneumoniae MLST website at http://www.mlst.net. Strains were clustered using the program for tree building provided by the MLST database.
Clone complex designation.
An isolate was assigned to a clone complex if it had the allelic profile of the reference isolate of the clone or differed from the reference strain at one (single-locus variant [SLV]) or two (double-locus variant [DLV]) loci. Thus, to be included in a clone complex, strains must have at least five out of seven alleles identical to the reference isolate and preferably the same serotype (13).
Statistical analysis.
The χ2 test was used for statistical analysis. P < 0.05 was considered statistically significant.
RESULTS
Bacterial identification and prevalence of macrolide resistance.
Sixty-four out of 2,200 isolates phenotypically identified as S. pneumoniae expressed reduced susceptibility to erythromycin. Four nontypeable isolates showed less than 97% sequence homology of their seven MLST loci to those found in the MLST database, and they were all identified as Streptococcus mitis by sodA sequence analysis (33) (data not shown). This observation illustrates the close phylogenetic relatedness between S. pneumoniae and S. mitis and emphasizes the importance of accurate bacterial identification. Thus, the overall prevalence of macrolide resistance in the strain collection was 2.7% (60/2,200) (Table 1). The prevalences among invasive and noninvasive isolates were 3.3% (33/998) and 2.3% (27/1,202), respectively (not statistically significant; P = 0.13). The difference in the prevalence of macrolide resistance between invasive isolates in 2001 (2.4%) and 2002 (4.1%) was not statistically significant (P = 0.14), nor was the difference in prevalence between noninvasive isolates in 1993 to 1997 (1.6%) and 2001 (2.7%) (P = 0.22).
Resistance phenotypes and genotypes.
Results for resistance phenotypes and genotypes are presented in Table 2. The most common macrolide resistance phenotype was the M type, comprising 36/60 (60%) of the resistant isolates. Twenty-two strains (37%) harbored cMLSB-type resistance. A single isolate expressed an iMLSB phenotype. One strain (01-41) displayed a weak cMLSB phenotype with high-level resistance to erythromycin and azithromycin (MIC, ≥256 μg/ml) and intermediate susceptibility to clindamycin (MIC, 2 μg/ml). The M-type S. pneumoniae isolates all carried the mef(A) gene. The difference in the prevalence of mef among invasive strains in 2001 (5/11; 45%) and 2002 (18/22; 82%) (data not shown) was statistically significant (P = 0.034). All MLSB-type strains harbored the erm(B) gene, except for the weak-cMLSB-phenotype strain 01-41, which was negative for all macrolide resistance genes. Pyrosequence analysis of this strain has revealed that two out of four 23S rRNA alleles had an A-to-G mutation at position 2058. This mutation mediates reduced susceptibility to macrolides (M. Haanperä, personal communication). No strains carried the erm(A) gene. No isolates were found to carry more than one macrolide resistance gene.
TABLE 2.
Distribution of macrolide resistance phenotypes, genotypes, and MIC ranges for erythromycin, clindamycin, and azithromycin among invasive and noninvasive S. pneumoniae isolates
| Phenotype | Genotype | MIC range (μg/ml)
|
No. of isolates
|
||||
|---|---|---|---|---|---|---|---|
| Erythromycin | Clindamycin | Azithromycin | Total | Invasive | Noninvasive | ||
| M type | mef(A) | 1-48 | 0.016-0.19 | 1-256 | 36 | 24 | 12 |
| MLSBa | erm(B) | 48->256 | >256 | >256 | 23 | 9 | 14 |
| MLSBb | A2058G in 23S rRNA gene | >256 | 2 | >256 | 1 | 1 | |
| Total | 60 | 33 | 27 | ||||
Including one inducible strain (iMLSB).
One weak cMLSB strain (strain 01-41).
Coresistance.
Table 3 shows the pattern of coresistance to other antimicrobials in relation to serotype and macrolide resistance genotype. Resistance to one or more commonly used antimicrobial agents was detected in 30/60 (50%) of the isolates, and it was mostly confined to erm(B)-positive strains (22/30 [73%]). The predominating mef(A)-containing serotype-14 strain (see below) did not express coresistance to other antimicrobials. The overall rate of nonsusceptibility to penicillin G was 14/60 (23%). Six strains were resistant to penicillin (MIC, ≥2 μg/ml). Tetracycline resistance was detected in 26 (43%) isolates. Resistance to trimethoprim-sulfamethoxazole and chloramphenicol was detected in 16 (27%) and 6 (10%) isolates, respectively. All strains were intermediately susceptible to ciprofloxacin.
TABLE 3.
Distribution of different coresistances within the respective resistance genotypes and serotypes among 60 macrolide-resistant S. pneumoniae isolates
| Antibiotica | No. of strains of the following type resistant to the indicated antibiotic
|
||||||
|---|---|---|---|---|---|---|---|
| Resistance genotype
|
Serotype
|
||||||
| erm(B) | mef(A) | Mutation in 23S rRNAb | 14 | 6B | 19F | Other | |
| No coresistance | 1 | 28 | 1 | 24 | 4 | 2b | |
| CHL | 1 | 1 | |||||
| TET | 4 | 1 | 2 | 3 | |||
| SXT | 2 | 2 | |||||
| SXT, CHL | 1 | 1 | |||||
| TET, CHL | 1 | 1 | |||||
| TET, SXT | 3c | 2 | 1 | 2 | 2c | ||
| PEN, SXT | 1 | 1 | |||||
| PEN, TET | 4 | 2 | 3 | 2 | 1 | ||
| TET, SXT, CHL | 1 | 1 | |||||
| PEN, TET, SXT | 4 | 1 | 2 | 1 | 2 | ||
| PEN, TET, SXT, CHL | 2 | 2 | |||||
| Total (n = 60) | 23 | 36 | 1 | 26 | 16 | 8 | 10 |
PEN, penicillin G; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol.
Includes a single strain with a weak cMLSB phenotype and no detectable macrolide resistance gene but a mutation in the 23S rRNA gene at position 2058 (A2058G).
Including one inducible strain (iMLSB).
Serotypes.
The 60 MRSP strains belonged to 12 different serotypes (Table 3). More than 80% of the strains were confined to three serotypes: serotypes 14 (43%), 6B (27%), and 19F (13%). M-type serotype-14 isolates accounted for 25 (42%) of all macrolide-resistant isolates and 22/33 (67%) of the invasive strains. Only a single serotype-14 isolate was isolated from a patient in North Norway, a region that contains approximately 10% of the Norwegian population. Serotype 14 accounted for 21/24 (88%) of M-type invasive isolates. In contrast, MLSB-type isolates, which accounted for 23 (38%) of the resistant strains, included nine different serotypes, of which 6B (52%) and 19F (17%) were the most prevalent. Penicillin nonsusceptibility was found only among serotype-6B and -19F isolates. The majority of isolates resistant to more than two antibiotics in addition to macrolides belonged to either serogroup 6B (12/30; 40%) or serogroup 19F (6/30; 20%).
MLST results in relation to previously described allelic profiles, pandemic clones, serotypes, and antimicrobial resistance patterns.
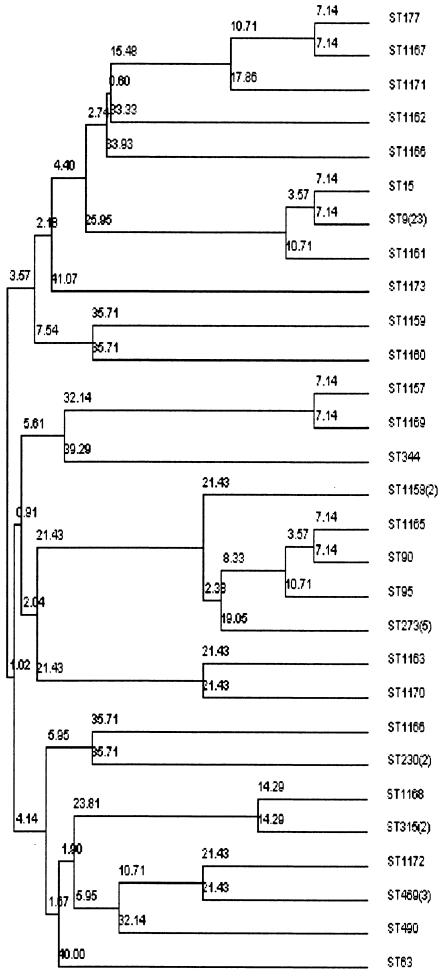
The clonal relationships among the 60 MRSP strains were defined by MLST. A total of 29 different STs were identified (Table 4 and Fig. 1). Forty-two strains (70%) displayed previously described allelic profiles, and 38 of these were identical to (n = 33) or variants of (5 SLVs or DLVs) seven pandemic clones: ST9 (and its SLV ST15; England14-9) (n = 24) (19), ST273 (Greece6B-22) (n = 5) (49), ST315 (Poland6B-20) (n = 2) (29), ST177 (Portugal19F-21) (n = 1) (40), ST90 (and its SLV ST95; Spain6B-2) (n = 2) (26), ST63 (Sweden15A-25) (n = 1) (39), and ST226 (represented by its SLV ST469; Hungary19A-6) (n = 3) (25). The remaining four strains with previously described allelic profiles belonged to ST230 (n = 2) (30), ST344 (n = 1), and ST490 (n = 1). A total of 18 strains revealed new allele combinations and were assigned to STs not previously included in the database. Eleven of these strains displayed 10 unique STs, while seven strains displayed allelic profiles that were SLVs or DLVs of previously reported pandemic clones: ST1161 (SLV of ST9; England14-9), ST1165 (SLV of ST90; Spain6B-2), ST1166 (SLV of ST242; Taiwan23F-15) (44), ST1167 and ST1171 (SLV and DLV, respectively, of ST177; Portugal19F-21) (40), ST1170 (SLV of ST236; Taiwan19F-14) (44), and ST1168 (DLV of ST315; Poland6B-20). Thus, a total of 45/60 (75%) erythromycin-resistant clinical isolates of S. pneumoniae in Norway were clonally related to pandemic resistant clones. The strains harboring allelic profiles identical to international pandemic clones displayed the serotypes and antimicrobial resistance profiles previously reported. Five of the strains that were SLVs or DLVs of pandemic clones showed a serotype or resistance pattern divergent from the original description of the corresponding pandemic clone (Table 5).
TABLE 4.
Distribution of STs among 60 strains of S. pneumoniae from Norway in relation to serotype, macrolide resistance determinants, sample site, and globally disseminated clones
| STa | Serotype | Genotype | No. of strains
|
Antibioticb resistance | Clonec | |
|---|---|---|---|---|---|---|
| Invasive | Noninvasive | |||||
| ST177 | 19F | erm | 1 | Portugal 19F-21 | ||
| ST1167 | 19F | erm | 1 | TET, CHL | Portugal 19F-21(SLV) | |
| ST1171 | 19F | erm | 1 | TET, SXT | Portugal 19F-21 (DLV) | |
| ST1162 | 6B | mef | 1 | |||
| ST1166 | 23F | mef | 1 | TET | Taiwan23F-15 (SLV) | |
| ST15 | 14 | mef | 1 | SXT, CHL | England14-9 (SLV) | |
| ST9 | 14 | mef | 19 | 4 | England14-9 | |
| ST1161 | 14 | mef | 1 | England14-9 (SLV) | ||
| ST1173d | 19F | 1 | ||||
| ST1159 | 19A | erm | 1 | TET, SXT | ||
| ST1160 | 14 | erm | 1 | TET, SXT | ||
| ST1157 | 23A | erm | 1 | TET | ||
| ST1169 | 23A | erm | 1 | TET | 1157 6/7 | |
| ST344 | Rough | erm | 1 | PEN, SXT | ||
| ST1158 | 6B | erm | 2 | PEN, TET, SXT | ||
| ST1165 | 3 | erm | 1 | PEN, TET, SXT | Spain6B-2 (SLV) | |
| ST90 | 6B | erm | 1 | PEN, TET, SXT, CHL | Spain6B-2 | |
| ST95 | 6B | erm | 1 | PEN, TET, SXT, CHL | Spain6B-2 (SLV) | |
| ST273 | 6B | erm | 2 | 3 | SXT or TET or TET, SXT, CHL | Greece6B-22 |
| ST1163 | 19F | mef | 1 | PEN, TET | ||
| ST1170 | 19F | mef | 1 | PEN, TET | Taiwan19F-14 (SLV) | |
| ST1164 | 15C | erm | 1 | PEN, TET | ||
| ST230 | 19F | mef | 2 | PEN, TET, SXT or TET, SXT | ||
| ST1168 | 6B | erm | 1 | PEN, TET | Poland6B-20 (DLV) | |
| ST315 | 6B | erm | 2 | PEN, TET, SXT or PEN, TET | Poland6B-20 | |
| ST1172 | 6A | mef | 1 | TET, SXT | ||
| ST469 | 6B | mef | 2 | 1 | Hungary19A-6 (SLV) | |
| ST490 | 6A | mef | 1 | CHL | ||
| ST63 | 15A | erm | 1 | PEN, TET | Sweden15A-25 | |
Assigned using the MLST website (http://www.mlst.net/).
Abbreviations: PEN, penicillin G; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol.
The Pneumococcal Molecular Epidemiological Network defined clones and related SLVs and DLVs.
A single strain with a weak cMLS phenotype and an A2058G mutation in the 23S rRNA gene.
FIG. 1.
MLST-based dendrogram of the genetic relationships among 60 macrolide-resistant isolates of S. pneumoniae from Norway. Genetic distances between STs are indicated.
TABLE 5.
New STs of S. pneumoniae isolates from Norway related to pandemic clonesa
| ST | Specimena | Genotype | Coresistanceb | Serotypec | Allele of indicated locusc,d
|
Clone | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpi | ddl | ||||||
| 1161 | Invasive | mef | 14 | 1 | 5 | 4 | 5 | 90 | 1 | 8 | England14-9 | |
| 1165 | Invasive | erm | PEN, TET, SXT | 3 | 5 | 6 | 1 | 8 | 6 | 3 | 4 | Spain6B-2 |
| 1166 | Noninv. | mef | TET | 23F | 11 | 29 | 4 | 21 | 30 | 1 | 14 | Taiwan23F-15 |
| 1167 | Noninv. | erm | TET, CHL | 19F | 7 | 14 | 4 | 12 | 1 | 1 | 8 | Portugal19F-21 |
| 1168 | Noninv. | erm | PEN, TET | 6B | 15 | 28 | 1 | 1 | 17 | 14 | 14 | Poland6B-20 |
| 1170 | Noninv. | mef | PEN, TET | 19F | 15 | 16 | 19 | 15 | 6 | 20 | 131 | Taiwan19F-14 |
| 1171 | Invasive | erm | TET, SXT | 19F | 1 | 14 | 40 | 12 | 1 | 1 | 14 | Portugal19F-21 |
Noninv., noninvasive.
Abbreviations: PEN, penicillin G; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol.
Serotypes and alleles that are different from those found in the major clones are underlined.
Assigned using the MLST website (http://www.mlst.net/).
The 11 unique strains differed at three or more loci from those described previously in the MLST database. Their allelic profiles and relevant characteristics are presented in Table 6. These “Norwegian” strains were recovered primarily from noninvasive isolates (n = 7). Seven strains carried erm(B), three strains harbored mef(A), and one strain carried the A2058G mutation in the 23S rRNA gene. Nine strains expressed resistance to at least one of the other antibiotics tested: penicillin (n = 4), tetracycline (n = 9), and trimethoprim-sulfamethoxazole (n = 5). These strains belonged to serotypes 6 (n = 4), 14 (n = 1), 19 (n = 3), 23 (n = 2), and 15C (n = 1).
TABLE 6.
Characteristics of 11 S. pneumoniae isolates with new STs, not related to known pandemic clones
| ST | Specimen | Genotype | Coresistancea | Serotype | Allele of indicated locusb
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpi | ddl | |||||
| 1157 | Noninvasive | erm | TET | 23A | 1 | 8 | 9 | 9 | 6 | 58 | 134 |
| 1158c | Invasive | erm | PEN, TET, SXT | 6B | 5 | 6 | 89 | 64 | 6 | 3 | 14 |
| 1159 | Noninvasive | erm | TET, SXT | 19A | 7 | 60 | 4 | 8 | 7 | 12 | 29 |
| 1160 | Invasive | erm | TET, SXT | 14 | 8 | 81 | 4 | 15 | 17 | 12 | 31 |
| 1162 | Noninvasive | mef | 6B | 7 | 25 | 4 | 2 | 48 | 1 | 28 | |
| 1163 | Noninvasive | mef | PEN, TET | 19F | 1 | 5 | 19 | 15 | 6 | 20 | 71 |
| 1164 | Noninvasive | erm | PEN, TET | 15C | 8 | 10 | 2 | 16 | 10 | 26 | 14 |
| 1169d | Noninvasive | erm | TET | 23A | 8 | 8 | 9 | 9 | 6 | 58 | 134 |
| 1172 | Invasive | mef | TET, SXT | 6A | 7 | 13 | 8 | 1 | 77 | 6 | 14 |
| 1173e | Noninvasive | 19F | 15 | 5 | 9 | 63 | 27 | 1 | 140 | ||
Abbreviations: PEN, penicillin G; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole.
Assigned using the MLST website (http://www.mlst.net/).
Two isolates.
SLV of ST1157.
A single strain with a weak cMLS phenotype and an A2058G mutation in the 23S rRNA gene.
Four clone complexes were represented by ≥3 strains. Strains in the first clone complex, comprising ST9 and its two SLVs, ST15 and ST1161, belonged to serotype 14, were mef(A) positive, and displayed no coresistance except for the single ST15 isolate, which was resistant to trimethoprim-sulfamethoxazole and chloramphenicol. This clone complex comprised 25/60 (42%) of the macrolide-resistant S. pneumoniae isolates in Norway and was dominant among the invasive isolates (21/33 [64%]). The clone was isolated in 15 laboratories covering all parts of the country. It first appeared in a noninvasive specimen in 1997. The second most prevalent clone in this study was ST273 (Greece6B-22) (49). This so-called “Mediterranean clone” was detected among invasive (n = 2) and noninvasive (n = 3) isolates, and all five isolates harbored the erm(B) gene. Two of the isolates expressed resistance to tetracycline, and another two were resistant to trimethoprim-sulfamethoxazole. The fifth strain was resistant to tetracycline, trimethoprim-sulfamethoxazole, and chloramphenicol. All five isolates were susceptible to penicillin and belonged to serotype 6B. This clone was first detected in 1993 in a noninvasive (throat) specimen from a patient in North Norway. The five strains were collected from five laboratories, covering different geographical regions of Norway, in 1993, 1997, 2001, and 2002. Finally, two clone complexes were represented by three strains: ST90, ST95, and ST1165 belong to the Spain6B-2 clone, and ST469 (n = 3) belongs to Hungary19A-6.
DISCUSSION
Norway has a low prevalence of antimicrobial-resistant bacteria, including MRSP. Here we present data from a nationwide molecular epidemiological study of clinical MRSP isolates. To our knowledge, this has not been performed previously in a country with a low prevalence of MRSP. The results may therefore contribute to our understanding of how resistant bacteria in general and MRSP in particular evolve and disseminate.
The overall prevalence of macrolide resistance was 2.7%. This is comparable to the figures reported for The Netherlands (5.2%), Sweden (4.7%), Denmark (4.1%), and the Czech Republic (2.0%) to the European Antimicrobial Resistance Surveillance System (12). In contrast, Belgium, France, Italy, and Spain reported resistance rates of >35% (15). Some studies have shown a direct association between the outpatient consumption of antibacterial agents and the prevalence of antimicrobial resistance in streptococci. MRSP is considered a marker of resistance to commonly used antibiotics in the treatment of respiratory tract infections (20, 32, 41). The prevalence of MRSP was recently shown to be directly associated with antibiotic selection pressure on a national level in 16 industrialized countries from 1994 to 2000 (1). Macrolide usage varied from 1.0 defined daily dose (DDD)/1,000 inhabitants per day in Sweden to 6.0 DDDs/1,000 inhabitants/day in France. The macrolide usage in Norway was 1.82 DDDs/1,000 inhabitants/day in 2002 (28), an increase of 32% from 1.4 DDDs/1,000 inhabitants/day in 1996. Thus, our relatively low prevalence of MRSP is in accordance with our national human consumption of macrolides.
The M type was the most common phenotype of macrolide resistance in Norwegian S. pneumoniae. This was due mainly to the mef-carrying ST9 (England14-9) clone, representing a majority of invasive isolates. The MLSB phenotype was most common among noninvasive isolates. These observations are consistent with the results of studies in other European countries, where MLSB- and M-type isolates predominate among noninvasive and invasive strains, respectively (3, 8, 24, 35). This distribution of different resistance phenotypes may be a clonal phenomenon, as observed in this study. In contrast, M-type strains have predominated in North America and Canada in invasive as well as noninvasive isolates (9, 18, 21).
Coresistance to other commonly used antibiotics was more frequent among noninvasive than invasive isolates and dominated in MLSB strains compared to the more susceptible M-type isolates. These observations can be explained by the predominant invasive ST9 clone, which was susceptible to all other antimicrobial agents tested. Tetracycline resistance was the most prevalent coresistance phenotype, and it was seen mainly in MLSB strains. This observation illustrates the genetic linkage between tet(M) and erm(B) resistance genes on the conjugative transposon Tn1545, originally described by Courvalin and Carlier (7) and by Clewell et al. (6). The chloramphenicol resistance determinant cat pC194 may also be genetically linked to Tn1545 according to Seral et al. (43). Penicillin resistance was observed primarily in noninvasive tetracycline-resistant strains with MLSB-type macrolide resistance.
MLST has proved useful for epidemiological and evolutionary studies of S. pneumoniae and other bacterial pathogens (11, 14, 54). We found that 75% of Norwegian MRSP isolates were indistinguishable from or related to pandemic resistant clones. The MLST data were supported by the results from serotyping and analysis of antimicrobial susceptibility patterns. The findings suggest that import of resistant strains may be the most important factor in the emergence of MRSP in Norway. In addition, four strains were assigned to three previously described STs that are not linked to pandemic clones: ST230 has been identified in Denmark and Italy, ST344 in Australia, and ST490 in Bulgaria, Finland, Greece, Greenland, Poland, and Sweden. The remaining 11 strains (18%) displayed 10 unique STs and were considered sporadic “Norwegian” strains. It should be noted that this interpretation is based on the S. pneumoniae MLST database, which still has a limited number of strains and is dominated by resistant lineages.
The most prevalent MRSP strain in Norway was the invasive serotype-14 clone defined as ST9 by MLST. This clone has been reported as one of the major global pneumococcal clones by the Pneumococcal Molecular Epidemiological Network and is often referred to as England14-9 (26). The ST9 clone has been identified as an important cause of meningitis throughout the United Kingdom (19) and is recognized as a cause of invasive disease in several other countries (http://www.mlst.net). Dissemination of invasive ST9 isolates has also been reported for Canada, Scotland, and, most recently, Greece (2, 17, 18). The mechanisms behind the impressive global spread of England14-9 remain unknown. The lack of information on the epidemiology of macrolide-susceptible ST9 precludes any speculations concerning the impact of the mef resistance determinant and/or linked genetic traits in the evolution of this invasive clone. The observed susceptibility to other commonly used antibiotics combined with the low macrolide consumption in Norway suggest that mechanisms other than antibiotic selection may play a part in the spread of the England14-9 clone in our country.
The MLST data also demonstrated the presence of several multidrug-resistant international S. pneumoniae clones such as Spain6B-2, also known as “the Icelandic” clone, and the recently recognized penicillin-susceptible multiresistant Greece6B-22 clone (49). This clone has already been described in three Mediterranean countries: Greece, Italy, and Israel. The other pandemic clones were represented with three or fewer strains in our study. ST469 (SLV of ST226; Hungary19A-6) was represented by two invasive isolates and one noninvasive mef(A)-positive isolate. The three isolates all belonged to serotype 6B and were susceptible to all other antimicrobials tested. ST226 has been reported earlier as a serotype-19A, multidrug-resistant clone containing erm(B) in Hungary. However, in the MLST database we found a recently submitted ST226 serotype-6B strain. No data on antibiotic resistance were accessible for this strain. ST230 was recently described as a novel, multiple-drug-resistant, serotype-24F lineage causing meningitis. It is genetically related to a penicillin-resistant serotype-14 lineage and is endowed with genetic traits shared with isolates of different serotypes (30). We found two serotype-19F mef-positive noninvasive ST230 isolates. One of these showed reduced susceptibility to penicillin. Moreover, the ST1165 (SLV of ST90; Spain6B-2) isolate belonged to serotype 3. These observations support the notions that capsular switching occurs regularly and that MLST is useful to identify clones emerging with different serotypes. The Norwegian MRSP isolates related to international clones were also shown to express coresistance to other commonly used antibiotics, thus supporting the role of antibiotic selection in the spread of these clones. The relatively low consumption of antibiotics by humans in Norway may contribute to the limited success of these clones in our country.
We detected 11 strains with 10 novel STs and consider them sporadic “Norwegian” MRSP strains, although the number of strains in the MLST database is still rather small. Interestingly, 9 out of these 11 strains were resistant to tetracycline, and several of them expressed reduced susceptibility to penicillin and trimethoprim-sulfamethoxazole. Seven of the tetracycline-resistant strains were shown to harbor the erm(B) gene, thus indicating the presence of Tn1545. If these isolates are considered nonimported, our observations suggest the presence of a Tn1545 reservoir that might be recruited within the S. pneumoniae population during antibiotic therapy. The molecular basis for tetracycline-induced conjugative transfer of Tn1545-Tn916-like elements has been elucidated (5). Norway has relatively high usage of tetracyclines (28), comprising 18% of total human systemic antibiotic consumption in 2002. Thus, cotransfer and coselection during tetracycline exposure may be involved in the emergence of sporadic Norwegian MLSB-type MRSP. It would be of interest to examine the reservoir and prevalence of Tn1545 elements within the human pharyngeal flora in Norway to investigate this hypothesis further.
In conclusion, we observed a low prevalence of macrolide resistance among invasive and noninvasive S. pneumoniae isolates in Norway. The majority (75%) of MRSP strains were genetically indistinguishable from or related to well-known pandemic antimicrobial-resistant clones, suggesting that the emergence of MRSP in Norway is largely due to imported strains from high-prevalence countries. The mef-positive England14-9 clone contributed significantly to the prevalence of MRSP. The mechanisms responsible for the spread of this invasive MRSP clone susceptible to other commonly used antimicrobial agents remains to be elucidated. The unique STs of MRSP strains considered sporadic “Norwegian” isolates were dominated by tetracycline-resistant MLSB strains. This may indicate a reservoir of Tn1545-related elements within the human pharyngeal flora in Norway, which is currently being examined.
Acknowledgments
We thank Jari Jalava and Marjo Haanperä, National Public Health Institute, Turku, Finland, for performing pyrosequence analysis of strain 01-41. We thank Torill Alvestad, Bjørg Haldorsen, Aase-Mari Kaspersen, and Jan Oknes for technical assistance. This publication made use of the Multi Locus Sequence Typing Web site (http://www.mlst.net), which was developed by Man-Suen Chan and is maintained by the Wellcome Trust Centre for the Epidemiology of Infectious Disease, University of Oxford, Oxford, United Kingdom.
Contributing members of the Norwegian Macrolide Study Group are Signe H. Ringertz and Bitten Rasmussen (Aker University Hospital), Martin Steinbakk and Siri Haug (Akershus University Hospital), Fredrik Müller and Miriam Sundberg (Bærum Hospital), Hjørdis Iveland and Ann Elise Johansen (Central Hospital of Buskerud), Liisa Mortensen and Karstein Korsvik (Central Hospital of Nordland), Arne Mehl and Eldbjørg Berg (Central Hospital of Nord-Trøndelag, Levanger), Gerd Skjervold and Lise Haaland (Central Hospital of Nord-Trøndelag, Namsos), Ingunn Haavemoen and Kari Ødegaard (Central Hospital of Oppland, Lillehammer), Linda Schildman and Lene Bjøntegård (Central Hospital of Hedmark, Elverum), Eivind Ragnhildstveit and Eva Madsen (Central Hospital of Østfold), Elisebet Haarr and Tone Roa (Central Hospital of Rogaland), Reidar Hjetland and Berit Ose (Central Hospital of Sogn og Fjordane), Sølvi Noraas and Torill S. Larsen (Central Hospital of Vest-Agder), Rolf Schøyen and Astrid Lia (Central Hospital of Vestfold), Liv J. Sønsteby and Pirrko-L. Kellokumpu (Central Hospital of Hordaland, Haugesund), Einar Vik and Margreet B. Sandhaug (County Hospital of Møre og Romsdal, Molde), Reidar Hide and Fillip Angeles (County Hospital of Møre og Romsdal, Ålesund), Asbjørn Digranes and Hilde Bekkeheien (Haukeland Hospital), Mette Walberg and Magli Bøvre (National Hospital, University of Oslo), Yngvar Tveten and Inger Johanne Lunde (Telelab A/S, Skien), Gaute Syversen and Thea Bergheim (Ullevål University Hospital), Gunnar S. Simonsen and Siv-H. Barkhald (University Hospital of North Norway), Trond Jacobsen and Mariann Hulsund (University Hospital of Trondheim), and Wibeke Aasnæs and Anne K. Andersen (Laboratory of Clinical Medicine, Oslo).
REFERENCES
- 1.Albrich, W. C., D. L. Monnet, and S. Harbarth. 2004. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg. Infect. Dis. 10:514-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angot, P., M. Vergnaud, M. Auzou, and R. Leclercq. 2000. Macrolide resistance phenotypes and genotypes in French clinical isolates of Streptococcus pneumoniae. Observatoire de Normandie du Pneumocoque. Eur. J. Clin. Microbiol. Infect. Dis. 19:755-758. [DOI] [PubMed] [Google Scholar]
- 4.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 6.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 7.Courvalin, P., and C. Carlier. 1986. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol. Gen. Genet. 205:291-297. [DOI] [PubMed] [Google Scholar]
- 8.Descheemaeker, P., S. Chapelle, C. Lammens, M. Hauchecorne, M. Wijdooghe, P. Vandamme, M. Ieven, and H. Goossens. 2000. Macrolide resistance and erythromycin resistance determinants among Belgian Streptococcus pyogenes and Streptococcus pneumoniae isolates. J. Antimicrob. Chemother. 45:167-173. [DOI] [PubMed] [Google Scholar]
- 9.Doern, G. V., and S. D. Brown. 2004. Antimicrobial susceptibility among community-acquired respiratory tract pathogens in the USA: data from PROTEKT US 2000-01. J. Infect. 48:56-65. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Antimicrobial Resistance Surveillance System. 2003. EARSS Annual Report 2003. European Antimicrobial Resistance Surveillance System, Bilthoven, The Netherlands (http://www.earss.rivm.nl/).
- 13.Farrell, D. J., I. Morrissey, S. Bakker, L. Morris, S. Buckridge, and D. Felmingham. 2004. Molecular epidemiology of multiresistant Streptococcus pneumoniae with both erm(B)- and mef(A)-mediated macrolide resistance. J. Clin. Microbiol. 42:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felmingham, D., D. J. Farrell, R. R. Reinert, and I. Morrissey. 2004. Antibacterial resistance among children with community-acquired respiratory tract infections (PROTEKT 1999-2000). J. Infect. 48:39-55. [DOI] [PubMed] [Google Scholar]
- 16.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50:25-37. [DOI] [PubMed] [Google Scholar]
- 17.Fotopoulou, N., P. T. Tassios, D. V. Beste, S. Ioannidou, A. Efstratiou, E. R. Lawrence, J. Papaparaskevas, R. C. George, and N. J. Legakis. 2003. A common clone of erythromycin-resistant Streptococcus pneumoniae in Greece and the UK. Clin. Microbiol. Infect. 9:924-929. [DOI] [PubMed] [Google Scholar]
- 18.Gay, K., W. Baughman, Y. Miller, D. Jackson, C. G. Whitney, A. Schuchat, M. M. Farley, F. Tenover, and D. S. Stephens. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417-1424. [DOI] [PubMed] [Google Scholar]
- 19.Hall, L. M., R. A. Whiley, B. Duke, R. C. George, and A. Efstratiou. 1996. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J. Clin. Microbiol. 34:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbarth, S., W. Albrich, D. A. Goldmann, and J. Huebner. 2001. Control of multiply resistant cocci: do international comparisons help? Lancet Infect. Dis. 1:251-261. [DOI] [PubMed] [Google Scholar]
- 21.Johnston, N. J., J. C. De Azavedo, J. D. Kellner, and D. E. Low. 1998. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2425-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristiansen, B. E., R. A. Sandnes, L. Mortensen, Y. Tveten, and L. Vorland. 2001. The prevalence of antibiotic resistance in bacterial respiratory pathogens from Norway is low. Clin. Microbiol. Infect. 7:682-687. [DOI] [PubMed] [Google Scholar]
- 23.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchese, A., S. Mannelli, E. Tonoli, F. Gorlero, M. Toni, and G. C. Schito. 2001. Prevalence of antimicrobial resistance in Streptococcus pneumoniae circulating in Italy: results of the Italian Epidemiological Observatory Survey (1997-1999). Microb. Drug Resist. 7:277-287. [DOI] [PubMed] [Google Scholar]
- 25.Marton, A., M. Gulyas, R. Munoz, and A. Tomasz. 1991. Extremely high incidence of antibiotic resistance in clinical isolates of Streptococcus pneumoniae in Hungary. J. Infect. Dis. 163:542-548. [DOI] [PubMed] [Google Scholar]
- 26.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing, 11th ed., vol. 21. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.NORM/NORM-VET. 2003. NORM/NORM-VET report 2002. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Norwegian Zoonosis Centre, Tromsø/Oslo, Norway. ISSN 1502-2307.
- 29.Overweg, K., P. W. Hermans, K. Trzcinski, M. Sluijter, R. de Groot, and W. Hryniewicz. 1999. Multidrug-resistant Streptococcus pneumoniae in Poland: identification of emerging clones. J. Clin. Microbiol. 37:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 31.Pihlajamaki, M., J. Jalava, P. Huovinen, and P. Kotilainen. 2003. Antimicrobial resistance of invasive pneumococci in Finland in 1999-2000. Antimicrob. Agents Chemother. 47:1832-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pihlajamaki, M., P. Kotilainen, T. Kaurila, T. Klaukka, E. Palva, and P. Huovinen. 2001. Macrolide-resistant Streptococcus pneumoniae and use of antimicrobial agents. Clin. Infect. Dis. 33:483-488. [DOI] [PubMed] [Google Scholar]
- 33.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinert, R. R., A. Al Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lutticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 49:61-68. [DOI] [PubMed] [Google Scholar]
- 35.Reinert, R. R., R. Lutticken, A. Bryskier, and A. Al Lahham. 2003. Macrolide-resistant Streptococcus pneumoniae and Streptococcus pyogenes in the pediatric population in Germany during 2000-2001. Antimicrob. Agents Chemother. 47:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronaghi, M., S. Karamohamed, B. Pettersson, M. Uhlen, and P. Nyren. 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242:84-89. [DOI] [PubMed] [Google Scholar]
- 38.Ruoff, K. L. 2003. Streptococcus, p. 405-421. In P. Murray, E. J. Baron, J. Jorgensen, M. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 39.Sa-Leao, R., A. Tomasz, and H. de Lencastre. 2001. Multilocus sequence typing of Streptococcus pneumoniae clones with unusual drug resistance patterns: genetic backgrounds and relatedness to other epidemic clones. J. Infect. Dis. 184:1206-1210. [DOI] [PubMed] [Google Scholar]
- 40.Sa-Leao, R., A. Tomasz, I. S. Sanches, A. Brito-Avo, S. E. Vilhelmsson, K. G. Kristinsson, and H. de Lencastre. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 182:1153-1160. [DOI] [PubMed] [Google Scholar]
- 41.Seppala, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenius, K. Lager, P. Huovinen, et al. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 42.Seppala, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 43.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Z. Y., M. C. Enright, P. Wilkinson, D. Griffiths, and B. G. Spratt. 1998. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J. Clin. Microbiol. 36:3514-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swedish Reference Group for Antibiotics. 2004. Report. [Online.] http://www.srga.org/MICTAB/MICTAB2.htm.
- 48.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syrogiannopoulos, G. A., F. Ronchetti, R. Dagan, I. Grivea, M. P. Ronchetti, N. Porat, T. A. Davies, R. Ronchetti, P. C. Appelbaum, and M. R. Jacobs. 2000. Mediterranean clone of penicillin-susceptible, multidrug-resistant serotype 6B Streptococcus pneumoniae in Greece, Italy and Israel. Int. J. Antimicrob. Agents 16:219-224. [DOI] [PubMed] [Google Scholar]
- 50.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mef(E) is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petitpas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, J., M. C. Enright, and B. G. Spratt. 2000. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J. Clin. Microbiol. 38:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]