Abstract
Nontyphoidal salmonellae are among the leading causes of food-borne disease in the United States. Because of the importance of Salmonella enterica in food-borne disease, numerous typing methodologies have been developed. Among the several molecular typing methods, pulsed-field gel electrophoresis (PFGE) is currently considered the “gold standard” technique in typing Salmonella. The aim of this study was to compare the discriminatory power of PFGE to multilocus sequence typing (MLST) in typing Salmonella enterica serovar Typhimurium clinical isolates. A total of 85 Salmonella Typhimurium clinical isolates from cattle were used in this study. PFGE using XbaI was performed on the 85 isolates by the Centers for Disease Control and Prevention method, and data were analyzed using the BioNumerics software package. Fifty PFGE profiles were observed among the isolates, and these grouped into three major clusters. For the MLST analysis, the manB, pduF, glnA, and spaM genes were amplified by PCR from the same 85 isolates. DNA sequencing of these four genes, manB, pduF, glnA, and spaM, showed no genetic diversity among the isolates tested, with a 100% identity in nucleotide sequence. Moreover, the DNA sequences of the aforementioned genes showed 100% identity to the sequence reported in GenBank for the S. enterica serovar Typhimurium LT2 strain. Therefore, MLST, using these genes, lacks the discriminatory power of PFGE for typing Salmonella enterica serovar Typhimurium.
Salmonella spp. are considered some of the major food-borne pathogens in the United States, causing an estimated 1.4 million cases of salmonellosis and over 500 deaths annually (18). A common serovar causing salmonellosis in humans is Salmonella enterica serovar Typhimurium, a globally distributed serotype that is frequently isolated from production animals, such as cattle (25). Several molecular biology techniques are used to discriminate between such strains on the DNA level, including macrorestriction analysis of chromosomal DNA by pulsed-field gel electrophoresis (PFGE), which is considered to be the method of choice (26). However, the discriminatory ability of PFGE is not absolute, and despite standardization, variation in the interpretations of PFGE results among various laboratories may exist (14).
Multilocus sequence typing (MLST), a recently developed methodology that requires minimal human input, has also been used to type Salmonella strains (14). This technique is based on determination of the DNA sequence of a series of selected housekeeping, ribosomal, and/or virulence-associated genes (for reviews, see references 6 and 28). MLST has been used to characterize several pathogenic bacteria, such as Neisseria meningitides (19), Streptococcus pneumoniae (8, 29), Staphylococcus aureus (7), Vibrio cholerae (15), Listeria monocytogenes (21), Escherichia coli O78 (1), and Campylobacter jejuni (22, 17, 4).
An MLST approach, based on the 16S RNA and the pduF, glnA, and manB genes, has recently been developed for Salmonella (14). The results of this study suggested that the discriminatory ability of MLST for the typing of Salmonella isolates is better than that of PFGE typing. Although the Salmonella isolates studied by Kotetishvili and colleagues (14) were primarily environmental in origin, it seems that MLST, using these genes, might have value in epidemiologic investigations of salmonellosis outbreaks. In the present study, the ability of MLST is compared to that of PFGE in typing a clinical set of Salmonella enterica serovar Typhimurium isolates using some of these same genes, pduF, glnA, and manB, as targets of the MLST procedure. Additionally, spaM of the inv-spa pathogenicity island of Salmonella enterica was used in the MLST procedure since this gene was previously reported to vary among the different subspecies (3).
MATERIALS AND METHODS
Bacterial strains.
A total of 85 Salmonella enterica serovar Typhimurium clinical isolates of cattle from different parts of the United States, which were collected at different times in the early 1990s, were characterized in this study. The isolates were obtained from the National Veterinary Service Laboratory at Ames, IA, where they were also serotyped. For simplicity, the isolates were given a serial designation from ST001 to ST085.
PFGE.
PFGE was performed according to the Centers for Disease Control and Prevention PulseNet protocol (24). Briefly, Salmonella isolates were grown overnight on MacConkey agar (Difco Becton Dickinson, MD) plates and then suspended in a cell suspension buffer (100 mM Tris-100 mM EDTA, pH 8.0) adjusted to an optical density at 610 nm of 1.35 using a SmartSpec 3000 spectrophotometer (Bio-Rad Laboratories, Hercules, CA). Ten microliters of proteinase K (20 mg/ml, stock) was added to 200 μl of the adjusted cell suspension and mixed gently with 200 μl of 1% SeaKem Gold-1% sodium dodecyl sulfate agarose, previously prepared in TE buffer (10 mM Tris-1 mM EDTA, pH 8.0) and kept at 55°C. Then, the mixture was immediately poured into disposable plug molds and left to cool. The bacteria were lysed within the plugs using a cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% Sarcosine, and 0.1 mg of proteinase K per ml). Plugs were incubated in this buffer for 2 h at 54°C in a shaking water bath. Plugs were then washed twice with water and four times with 1× TE buffer (10 mM Tris-1 mM EDTA, pH 8.0) at 50°C. Slices of plugs (2 millimeters wide) were incubated with XbaI (50 U/sample) in a 100 μl restriction mixture (10) for 4 hours at 37°C. The plugs were then loaded onto a 1% SeaKem Gold agarose gel. PFGE was performed with the CHEF-Mapper (Bio-Rad Laboratories, Hercules, CA) by using the following conditions: an initial switch time of 2.16 s, a final switch time of 63.8 s, and a run time of 18 h. The pulsed-field lambda ladder (Bio-Rad) was loaded onto all gels. The Centers for Disease Control and Prevention Salmonella serovar Branderup H9812 strain was used as the reference strain. After the electrophoresis was completed, gels were stained with ethidium bromide, and the images were captured with the ChemiImager 5500 gel documentation system (Alpha Innotech Corp., San Leandro, CA). Dendrograms and cluster analysis were performed using the BioNumerics software package (Applied Maths, Inc., Austin, TX). Similarity analysis was performed using the Dice coefficient, and clustering was created using the unweighted pair group method with arithmetic means.
MLST.
For MLST analysis, four Salmonella genes were selected: three housekeeping genes and one virulence gene. The three tested housekeeping genes were selected on the basis of a previous study (14) in which the authors proposed the first MLST approach for typing Salmonella strains. In the present study we selected the following three housekeeping genes: phosphomannomutase (manB), glutamate synthetase (glnA), and the 1,2-propanediol utilization factor (pduF) (14). The virulence gene spaM was also selected since it previously showed some diversity among Salmonella enterica subspecies (3) and some Salmonella enterica serovar Enteritidis strains (12). The PCR primers used to amplify internal fragments from the aforementioned genes are shown in Table 1. The same primers were used for both PCR amplification and sequencing.
TABLE 1.
Primers used for the MLST of the 85 Salmonella enterica serovar Typhimurium clinical isolates
| Gene | Direction | Primer sequence | Approx amplicon size (bp)a | Reference or source |
|---|---|---|---|---|
| spaM | Forward: | 5′-CGCTGTACGGTATTTCATT-3′ | 394 | 12 |
| Reverse: | 5′-CTGACTCGGCCTCTTCCTG-3′ | This studyb | ||
| manB | Forward: | 5′-CCGGCACCGAAGAGA-3′ | 893 | 14 |
| Reverse: | 5′-CGCCGCCATCCGGTC-3′ | 14 | ||
| pduF | Forward: | 5′-CTCAAAGTCGCYGGYGC-3′ | 518 | 14 |
| Reverse: | 5′-GGGTTCATTGCAAAACC-3′ | 14 | ||
| glnA | Forward: | 5′-CCGCGACCTTTATGCCAAAACCG-3′ | 474 | 14 |
| Reverse: | 5′-CCTGTGGGATCTCTTTCGCT-3′ | 14 |
The length of the sequenced DNA in both strands was slightly shorter.
This primer was designed using the DNA sequence of GenBank accession no. SEU43315.
Salmonella DNA was extracted from the plugs prepared for PFGE as described previously (14). Briefly, the bacterial DNA contained in the plugs was frozen and thawed twice, first at −70°C and then at 55°C, in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The samples were then centrifuged at 5,000 × g for 10 min, and the supernatants were collected. Aliquots (1 μl) of these supernatants were used as templates for PCR amplification. PCR amplification was performed using the Taq PCR master mix kit (QIAGEN Inc., Valencia, CA). Amplification conditions for the four tested genes were 94°C for 5 min, followed by 35 amplification cycles (94°C for 45 s, 55°C for 45 s, and 72°C for 1 min) and then a final extension at 72°C for 10 min. PCR products were purified using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA).
DNA sequencing of the amplified fragments was performed in both directions with the same primers used for amplification (Table 1) using the CEQ Dye Terminator cycle sequencing with Quick Start kit (Beckman Coulter, Inc., Fullerton, CA). The thermal cycling program for DNA sequencing was run according to manufacturer recommendations (96°C for 20 s, 50°C for 20 s, and 60°C for 4 min, for 30 cycles, followed by holding at 4°C). The sequencing products were run on a CEQ 2000XL DNA analysis system sequencer (Beckman Coulter, Inc., Fullerton, CA) located in the DNA sequencing facility at the Department of Plant Sciences, North Dakota State University. DNA sequence assembly was performed using the Staden Package software (http://staden.sourceforge.net/). All sequences were aligned and compared using the BioNumerics software package (Applied Maths, Inc., Austin, TX). A BLAST search was performed through the National Center for Biotechnology Information website.
RESULTS
PFGE analysis.
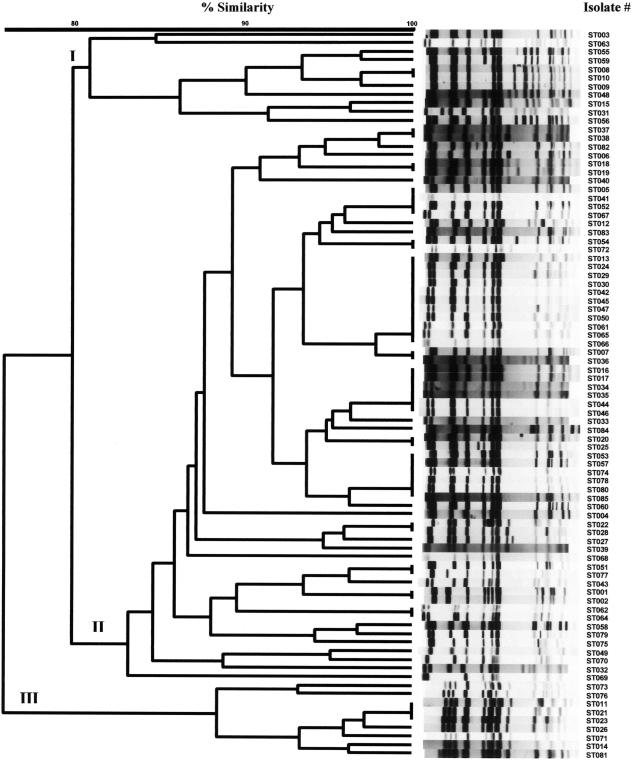
A total of 85 Salmonella enterica serovar Typhimurium isolates were used for PFGE analysis in this study. The XbaI PFGE patterns of these isolates are shown in Fig. 1. The genetic relatedness of these isolates ranged from 77 to 100%. Fifty distinct PFGE profiles were observed among the isolates (1 PFGE type per 1.7 strains), and these were categorized into three major clusters, I, II, and III (Fig. 1).
FIG. 1.
PFGE analysis of the 85 Salmonella enterica serovar Typhimurium clinical isolates digested with XbaI. Shown is the dendrogram and PFGE profiles of these isolates (ST001 to ST085). Fifty PFGE profiles were observed and categorized into three major clusters (I, II, and III). Similarity was determined by using the Dice coefficient, and clustering was created by the unweighted pair group method with arithmetic means.
MLST analysis.
Our aim here was to investigate the utility of the MLST approach in differentiating clinical isolates within the Typhimurium serovar. Three housekeeping genes, manB, pduF and glnA, were selected because of their previously reported utility as a target for the MLST procedure in typing Salmonella (14). The virulence gene spaM was selected since it previously showed some diversity among Salmonella enterica subspecies (3) and some Salmonella enterica serovar Enteritidis strains (12) and also since it encodes a protein predicted to be on the cell surface and, therefore, likely under a great selection pressure (3).
No nucleotide differences among the tested 85 Salmonella enterica serovar Typhimurium isolates for the four genes were detected. Moreover, when subjected to a BLAST search in GenBank, the DNA sequences of manB, pduF, glnA, and spaM showed 100% nucleotide identity to reported sequences for these genes from Salmonella enterica serovar Typhimurium LT2.
DISCUSSION
Our aim was to test the ability of MLST to discriminate between a set of clinical Salmonella isolates within the Typhimurium serovar and to compare this ability to that of PFGE. In a previous study, using the same primers and housekeeping genes as used in the present study, MLST was found to compare favorably to serotyping and/or PFGE in discriminating between various Salmonella serovars (14). However, the utility of the MLST procedure in discriminating clinical isolates of the Typhimurium serovar, using the manB, pduF, glnA, and spaM genes, is doubtful because of a complete lack of sequence diversity found for these genes among the 2,000 or so nucleotides that were sequenced per isolate in our study. Put another way, no sequence differences were found in a total of approximately 170,000 nucleotides sequenced in these 85 Salmonella isolates. The absolute nature of this result is a clear indication that the genes targeted here cannot be used in an MLST procedure to discriminate among such isolates. There is no contradiction here since the above-mentioned study (14) never indicated that MLST was discriminatory among the tested Typhimurium isolates, which were mostly environmental. So, while MLST might be helpful as a tool to differentiate among isolates of different Salmonella serovars and in some cases among isolates of the same serovar, this statement, according to our results, does not apply to serovar Typhimurium, at least when the sequenced fragments of the genes targeted here are used.
Similar results were obtained for Escherichia coli, in which a recent study demonstrated a striking lack of DNA sequence diversity (100% identity) among Escherichia coli O157:H7 isolates that were distinct by PFGE using seven different housekeeping genes (20). Also, in a global collection of Salmonella enterica serovar Typhi isolates, only three polymorphic sites were identified among seven housekeeping genes totaling 3,336 bp, resulting in a separation of the isolates into only four sequencing types (STs) (13). However, MLST may have greater utility for discriminating other bacterial species, since variation in nucleotide sequence among strains is commonly seen even within a single serovar (1, 23, 16).
Of special interest among our results is the homogeneity of the spaM sequences found among the isolates. This gene was chosen as a target because of its reputed variation among Salmonella enterica subspecies (3), which is thought to be associated with its location on the cell surface and, therefore, likely under selective pressure in the host to change (3). However, others have noticed similar sequence homogeneity among virulence genes. For example, DNA sequencing of several Salmonella virulence genes was unable to discern genetic differences among Salmonella enterica serovar Enteritidis phage types (12). In a more recent study, MLST using three virulence genes provided very poor discrimination among Escherichia coli O157:H7 isolates, with the authors concluding that PFGE remains the best method of discrimination among isolates of this pathogen (9).
The excellent discriminatory ability of PFGE among the 85 clinical isolates of Salmonella enterica serovar Typhimurium tested in our study (one PFGE type for 1.7 isolates) is not uncommon. PFGE was previously reported in several studies to be successful in the subtyping of Salmonella enterica serovar Typhimurium strains (2, 11, 25, 27). The conserved DNA sequence of the internal fragments of the four genes tested in this study, manB, glnA, pduF, and spaM, among the 85 tested isolates compared to the diversity shown using PFGE can be explained. PFGE does have the advantage over MLST since it involves random screening of the entire genome, whereas MLST analysis is limited to nucleotides within the targeted gene(s). Therefore, if there is little or no variation in the nucleotide sequence of the genes targeted by MLST, then MLST can provide little or no discrimination between strains tested. Analyzing multiple genes from various regions of the Salmonella chromosome might overcome this problem. Moreover, a recent comparison of the similarities and differences between five publicly available Salmonella genome sequences reveals extensive sequence conservation among the Salmonella serovars but variation in insertions and deletions (5). In our case, insertions, deletions, or the presence of plasmids can alter the PFGE pattern obtained without necessarily changing the DNA sequence of the targeted genes, resulting in a diversity in PFGE patterns in the face of homogeneity among MLST patterns obtained for the same isolates.
In conclusion, our study demonstrates that MLST, using the genes tested, lacks the ability to discriminate between Salmonella enterica serovar Typhimurium clinical isolates and that PFGE can still be considered the method of choice for the molecular typing of this serotype. For MLST to be useful as an epidemiological tool for the investigation of Salmonella enterica serovar Typhimurium outbreaks, genes with more-significant sequence variation than those targeted in the present study must be identified. Ongoing efforts to sequence entire genomes of multiple strains per species of pathogenic bacteria might help in choosing genes demonstrating the needed genetic diversity to be used in MLST.
Acknowledgments
We gratefully acknowledge the U.S. Department of Agriculture (TE no. 2002-06088) for the financial support of this project.
REFERENCES
- 1.Adiri, R. S., U. Gophna, and E. Z. Ron. 2003. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 222:199-203. [DOI] [PubMed] [Google Scholar]
- 2.Bender, J. B., C. W. Hedberg, D. J. Boxrud, J. M. Besser, J. H. Wicklund, K. E. Smith, and M. T. Osterholm. 2001. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N. Engl. J. Med. 344:189-195. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., J. Li, H. Ochman, and R. K. Selander. 1997. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J. Bacteriol. 179:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colles, F. M., K. Jones, R. M. Harding, and M. C. J. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. J. Day, C. E. Davie, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foley, S. L., S. Simjee, J. Meng, D. G. White, P. F. McDermott, and S. Zhao. 2004. Evaluation of molecular typing methods for Escherichia coli O157:H7 isolates from cattle, food, and humans. J. Food Prot. 67:651-657. [DOI] [PubMed] [Google Scholar]
- 10.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra, B., P. Schrors, and M. C. Mendoza. 2000. Application of PFGE performed with XbaI to an epidemiological and phylogenetic study of Salmonella serotype Typhimurium. Relations between genetic types and phage types. New Microbiol. 23:11-20. [PubMed] [Google Scholar]
- 12.Hudson, C. R., M. Garcia, R. K. Gast, and J. J. Maurer. 2001. Determination of close genetic relatedness of the major Salmonella enteritidis phage types by pulse-field gel electrophoresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis. 45:875-886. [PubMed] [Google Scholar]
- 13.Kidgell, C., U. Reichard, J. Wain, B. Linz, M. Torpdahl, G. Dougan, and M. Achtman. 2002. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect. Genet. Evol. 2:39-45. [DOI] [PubMed] [Google Scholar]
- 14.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotetishvili, M., O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. G. Morris, Jr. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden, M. C., J. A. Bygraves, E. Fiel, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and, B. G., Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCag, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolas, P., G. Raphenon, M. Guibourdenche, L. Decousset, R. Stor, and A. B. Gaye. 2000. The 1998 Senegal epidemic of meningitis was due to the clonal expansion of A:4:P1.9, clone III-1, sequence type 5 Neisseria meningitides strains. J. Clin. Microbiol. 38:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revazishvilli, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. G. Morris, and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 69:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamada, Y., Y. Nakaoka, K. Nishimori, A. Doi, T. Kumasi, N. Uemura, K. Tanaka, S.-I. Makino, T. Sameshima, M. Akiba, M. Nakazawa, and I. Uchida. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsen, H.-Y., J.-S. Lin, and H.-Y. Hsih. 2002. Pulse field electrophoresis for animal Salmonella enterica serovar Typhimurium isolates in Taiwan. Vet. Microbiol. 87:73-80. [DOI] [PubMed] [Google Scholar]
- 28.Urwin, R., and M. C. J. Maiden. 2003. Multilocus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 29.Zhou, J., M. C. Enright, and B. G. Spratt. 2000. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J. Clin. Microbiol. 38:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]