Abstract
Since the publication of the complete genomic sequence of Campylobacter jejuni NCTC 11168 in February 2000, evidence has been compiling that suggests C. jejuni strains exhibit high genomic diversity. In order to investigate this diversity, the unique genomic DNA sequences from a nonsequenced Campylobacter strain, C. jejuni 81-176, were identified by comparison with C. jejuni NCTC 11168 by using a shotgun DNA microarray approach. Up to 63 kb of new chromosomal DNA sequences unique to this pathogen were obtained. Eighty-six open reading frames were identified by the presence of uninterrupted coding regions encoding a minimum of 40 amino acids. In addition, this study shows that the whole-plasmid shotgun microarray approach is effective and provides a comprehensive coverage of DNA regions that differ between two closely related genomes. The two plasmids harbored by this Campylobacter strain, pTet and pVir, were also sequenced, with coverages of 2.5- and 2.9-fold, respectively, representing 72 and 92% of their complete nucleotide sequences. The unique chromosomal genes encode proteins involved in capsule and lipooligosaccharide biosynthesis, restriction and modification systems, and respiratory metabolism. Several of these unique genes are likely associated with C. jejuni 81-176 fitness and virulence. Interestingly, the comparison of C. jejuni 81-176 unique genes with those of C. jejuni ATCC 43431 revealed a single gene which encodes a probable TraG-like protein. The product of this gene might be associated with the mechanism of C. jejuni invasion into epithelial cells. In conclusion, this study extends the repertoire of C. jejuni genes and thus will permit the construction of a composite and more comprehensive microarray of C. jejuni.
Campylobacter jejuni causes an acute diarrheal disease with a variety of clinical symptoms, such as fever, diarrhea, headache, abdominal pain, myalgia, vomiting, and blood in feces (36). In addition, this bacterium is associated with the development of Guillain-Barré syndrome, an autoimmune-mediated disorder of the peripheral nervous system (36). Until now, Penner serotyping, flagellin typing, and pulsed-field gel electrophoresis (PFGE) have been the methods of choice for Campylobacter subtyping (37). The capsular polysaccharide is the major determinant for the heat-stable Penner serotyping scheme (18). Up to 60 Penner serotypes have been described, and certain serotypes appear to be preferentially associated with particular clinical outcomes (31). For instance, strains belonging to the O:19 serotype seem to predominate among C. jejuni strains isolated from Guillain-Barré patients (21). Epidemiological studies rely on these serotyping schemes. While these technologies are routinely used in clinical laboratories, there is a great interest in developing genome-based approaches for in-depth molecular typing.
The wealth of genome sequence information has driven the development and use of microarray approaches for the analysis of strain relatedness. In these genome-wide approaches, the “target” (not sequenced) genome is interrogated by the microarray for the presence or absence of genes belonging to the sequenced strain. Several reports, using this method, described an extensive variability in the gene content between the sequenced strain C. jejuni NCTC 11168 and strains of diverse origins (12, 23, 24, 30). While these studies proved to be extremely valuable for the identification of the minimum set of C. jejuni core genes, they failed to identify genotypic traits associated with phenotypic characteristics, such as serotypes, clinical outcomes, isolate origins, or virulence. Undoubtedly, the success of this methodology for strain typing or genotypic trait identification relies on the completeness and diversity of the arrayed “probe” for a particular species. Presently, only one strain of C. jejuni has been completely sequenced (C. jejuni NCTC 11168) (29), and another one (C. jejuni RM 1221) is being sequenced at the TIGR institute. The addition of new genome information from C. jejuni strains of different origins will facilitate the identification of genotypic markers and improve the power of this genotyping method.
Recently, we have described a novel approach to identify nonredundant sequences of C. jejuni ATCC 43431 by combining the construction of a shotgun library and the power of microarray technology (32). This genome-wide approach consists of five consecutive steps: (i) construction of a shotgun library representative of the size of the genome, (ii) arraying of the whole plasmids on glass slides, (iii) competitive genomic hybridization, (iv) identification of the plasmids carrying unique DNA segments, and (v) DNA sequencing. A total of 84 kb of new DNA sequences unique to ATCC 43431 were identified (32). These sequences carry up to 130 genes, representing an addition of approximately 7.5% of novel genes to the current set of known C. jejuni genes. These unique genes likely constitute ideal targets for molecular markers of ATCC 43431 and other closely related strains. However, in order to better define common and unique genes between C. jejuni strains, additional DNA sequence information from other C. jejuni isolates is needed.
In the present study, we propose to further increase the repertoire of genes belonging to the C. jejuni species by identifying the genes unique to C. jejuni 81-176. This isolate has been extensively studied and shown to carry two large plasmids, named pVir and pTet (1, 6). Recently, the DNA sequence of the pVir plasmid has been released (2). Interestingly, this plasmid harbors 54 predicted open reading frames (ORFs) encoding orthologs of components of the Helicobacter pylori type IV secretion apparatus (2). Mutational analysis of 21 of these genes identified 7 genes encoding proteins required for a high level of invasion into INT407 cells. Interestingly, the pVir plasmid alone is insufficient to increase the low C. jejuni NCTC 11168 invasion ability (2). Given that the plasmid pTet seems to be nonessential for C. jejuni 81-176 invasion potential, since pTet-deficient 81-176 is still invasive, the chromosome of this strain likely carries genes encoding proteins involved in the invasion process. The identification of this chromosomal set of genes will provide a valuable resource to study Campylobacter invasiveness.
MATERIALS AND METHODS
Campylobacter strains.
The two strains of C. jejuni used in this study, C. jejuni NCTC 11168 and 81-176, were cultured in Mueller-Hinton medium and incubated at 37°C under microaerophilic conditions (83% N2, 4% H2, 8% O2, and 5% CO2) maintained by using the MACS-VA500 variable atmosphere workstation (Don Whiteley, West Yorkshire, England). C. jejuni 81-176 was originally isolated from a human disease outbreak in 1985 and has been shown to cause inflammatory colitis in two human challenge studies (10, 20). Likewise, this isolate caused acute diarrheal illness in monkey (33), mouse (4), and ferret (1) animal models.
Construction of the C. jejuni 81-176 shotgun microarray.
The C. jejuni 81-176 shotgun microarray was constructed as previously described for C. jejuni ATCC 43431 (32). Briefly, the construction of this microarray involved four consecutive steps: (i) construction of a genomic library of C. jejuni 81-176, (ii) plasmid extraction and purification, (iii) assessment of the plasmid quality and concentration by agarose gel electrophoresis, and (iv) printing of the whole plasmids on microarray slides. The genomic library was constructed by cloning nebulized genomic DNA fragments of 400 to 800 bp into the pCR4Blunt-TOPO vector from Invitrogen, according to the manufacturer's protocol and as previously described (32). This library was produced in TOP10 Escherichia coli cells and replicated into 96-well plates containing Luria-Bertani broth and ampicillin (100 μg/ml). Plasmids from 9,600 clones were purified using the 96-well Millipore Multiscreen filter plates (Millipore MAFB NOB 50) and Costar polyvinylidene difluoride filter plates (0.2 μm; Costar) following the manufacturers' instructions. The yield of plasmid purification was estimated by agarose gel electrophoresis, and plasmid concentrations were adjusted to approximately 100 ng/μl by dilution in 3× SSC buffer (1× SSC buffer is 0.15 M NaCl plus 0.015 M sodium citrate). Whole plasmids were printed on aminosilane-coated glass slides (CMT GAPS-II from Corning Inc., Corning, N.Y.) using a Molecular Dynamic arrayer. Plasmids were immobilized on slides by baking at 80°C for 4 h. The diameter of each spot was about 200 μm, with a spacing of 250 μm between spot centers.
Preparation of fluorescently labeled probes, hybridization, scanning, and data analysis.
Fluorescently labeled probes were prepared from 2 μg of genomic DNA by random priming using the BioPrime DNA labeling system from Invitrogen in the presence of aminoallyl-dUTP, followed by coupling with either the Cy3 or Cy5 monoreactive dye (Amersham Pharmacia, N.J.) as previously described (32). Before hybridization, the slides were prehybridized in 1% bovine serum albumin, 25% formamide, 5× SSC buffer, and 0.1% sodium dodecyl sulfate (SDS) for 45 min at 42°C. Next, the slides were briefly rinsed with water and spun dry. Both labeled probes were combined in a 36-μl hybridization solution consisting of 25% formamide, 5× SSC buffer, 0.1% SDS, and 25 μg of salmon sperm. This hybridization solution was heated at 99°C for 2 min, cooled down to 42°C, and then placed on the microarray slide and covered by a coverslip (Fisher). The microarray was placed in a humidified chamber (ArrayIt) and incubated at 42°C. After 16 h, the slides were washed for 5 min with 2× SSC-0.1% SDS, which was preheated to 42°C. Next, the slides were washed at room temperature in 0.1× SSC-0.1% SDS for 10 min, followed by four washes of 1 min in 0.1× SSC. Finally, the slides were briefly rinsed in water and spun dry.
The microarray slides were scanned using a laser-activated confocal scanner (ScanArray 5000) at 532 and 635 nm. Fluorescent signal values were obtained by using the GenePix Pro4 software (Axon Instrument, Foster City, Calif.). Spots were removed from further analysis if they were localized in a region of hybridization abnormalities or if the signal intensity in the channel corresponding to C. jejuni 81-176 was 2 times below the standard deviation of the background. The fluorescence intensities were normalized by applying an iterative linear regression analysis as described previously (32). The microarray hybridization was repeated three times. The data from the three microarrays were merged, and the mean of the fluorescence ratio (Cy5/Cy3) for each spot was computed. Then, the spots were sorted by descending ratio (signal intensity corresponding to C. jejuni 81-176/signal intensity corresponding to C. jejuni NCTC 11168), and the DNA inserts were gradually sequenced in 96-well plate formats. The sequencing was stopped when the DNA sequence of at least 90% of the clones from a 96-well plate exhibited 100% identity with the genomic sequence of C. jejuni NCTC 11168.
DNA sequencing, ORF finding, and annotation.
DNA sequences were determined in both directions with the M13 universal primers by the DNA sequencing facility of the Laboratory of Genomics and Bioinformatics at University of Oklahoma Health Science Center (Oklahoma City). Vector trimming and sequence assembly were performed automatically by using the Lasergene software (DNAstar, Inc.). Each contig was manually edited and checked for residual error or misassembling. The sequences exhibiting 100% identity with C. jejuni NCTC 11168 were identified by BLASTN. These sequences corresponded to false positives and were therefore discarded from further analysis. Potential open reading frames were identified by using Artemis (available at http://www.sanger.ac.uk/Software/Artemis). Similarity searches were performed with the BLASTP and BLASTX programs against the NCBI nonredundant database, and the genes were grouped based on functional categories.
Plasmid extraction, labeling, and sequencing.
The plasmids pVir and pTet were extracted together using the QIAGEN plasmid Midi kit following the manufacturer's instructions. The plasmids were labeled with the Cy3 monoreactive dye and applied to the shotgun array as described above for genomic DNA. Probes with fluorescent signal intensities above 2 times the standard deviation of the background were considered to carry DNA fragments belonging to one of the two plasmids. The corresponding probes were retrieved from the clone library and the inserts sequenced. The DNA sequences, which belong to the pTet or pVir plasmids, were assembled into contigs using the Lasergene software (DNAstar, Inc.) as described above. The BLASTN program was used to identify the sequences exhibiting 100% identity with the pVir (NCBI accession number AF226280) or pTet (NCBI accession number AY394561) plasmids.
PFGE preparation and analysis.
Agarose plugs containing 81-176 and NCTC 11168 were prepared as described previously (33). Plugs with bacterial genomic DNA were digested with three restriction enzymes chosen based on an in silico digestion of NCTC 11168 to yield 10 or fewer fragments. Digests were performed with AsiSI, BsiWI, and SalI according to manufacturer's specifications, with AsiSI and SalI digests at 37°C and BsiWI at 55°C. Electrophoresis conditions were the same for the three digests and were as previously described for SalI, with an initial switch time of 50 s and a final switch time of 90 s (33). These conditions are optimized to separate the concatemerized lambda markers (molecular weight range, 48,500 to 1,000,000; Bio-Rad) used to estimate the bacterial genomic fragment sizes. Band sizes were determined as previously described (UV Doc It system; LabWorks Image Acquisition and Analysis Software) (33), and bands resulting from the two 81-176 plasmids were not visualized for any of the digests. The smallest fragment size seen for 81-176 was greater than 100 kb and thus larger than either plasmid. The amount of genomic DNA in the plugs was estimated to be no more than 0.3 μg, which contributed to the lack of visible plasmid bands. The average genome sizes for 81-176 and 11168 were estimated from the fragment sizes for the three enzymes and were 1.736 ± 0.0066 Mb for 81-176 and 1.735 ± 0.011 Mb for 11168.
Nucleotide sequence accession numbers.
The 58 chromosomal DNA fragments unique to C. jejuni 81-176 were submitted to the NCBI database and assigned accession numbers AY681239 to AY681296.
RESULTS
Construction of the C. jejuni 81-176 shotgun microarray.
Our objective was to identify the set of genes unique to C. jejuni 81-176 compared to the genome-sequenced strain C. jejuni NCTC 11168 (29). The approach adopted for this study was previously developed for the identification of C. jejuni ATCC 43431 unique genes (32). This approach combines the construction of a whole-plasmid DNA microarray obtained from a shotgun library of C. jejuni 81-176, as well as the competitive hybridization to the array of genomic DNA from strains C. jejuni 81-176 and NCTC 11168.
The sizes of the C. jejuni 81-176 and NCTC 11168 chromosomes were estimated to be similar in length (1,735 ± 10 kb) by PFGE analysis (data not shown). However, in contrast to C. jejuni 11168, the 81-176 strain carries two large plasmids named pVir and pTet (2, 6). While the complete DNA sequence of the 37.5-kb pVir plasmid has been previously reported (2), the genetic information of the pTet plasmid, with the exception of the presence of a gene conferring tetracycline resistance, was initially unknown. However, the complete pTet plasmid was sequenced by Batchelor et al. during the course of the present study (6). Its sequence and annotation were recently made accessible at the NCBI database under the accession number AY394561.
A shotgun library of the C. jejuni 81-176 genome consisting of 9,600 individual clones and comprising both the chromosomal and plasmidic DNAs was constructed in the E. coli strain TOP10 as previously described (32) and arrayed on glass slides. The mean DNA insert size was estimated (subsequently subjected to DNA sequencing and analysis) to be approximately 0.5 kb and ranged from 0.1 to 1.5 kb. Based on the size of the genome (1,724 kb, which corresponds to the sum of the sizes of the chromosome and the two plasmids), the insert (0.5 kb), and the shotgun library (9,600 clones), the probability of a DNA fragment to be present on our array is equal to 0.94 (P = 1 − e[9,600 × ln (1 − 0.5/1,724)]).
Identification of clones carrying unique DNA sequences.
The unique DNA sequences of C. jejuni 81-176 belonging to the chromosome and the two plasmids were identified by two different sets of microarray hybridization experiments.
The first set of experiments allowed the identification of every unique DNA sequence present on our shotgun array. These DNA sequences were identified by competitive hybridization on the array of genomic DNA from C. jejuni 81-176 with genomic DNA from C. jejuni NCTC 11168. The microarray hybridization was repeated three times. The microarray data were normalized using a linear regression analysis as previously described (32), and the mean of the fluorescent intensities ratio (fluorescent intensity corresponding to the genomic DNA of C. jejuni 81-176/fluorescent intensity corresponding to the genomic DNA of C. jejuni NCTC 11168) was computed. The ratios were sorted in descending number, and the inserts of the first 96 plasmids with the highest ratio values were sequenced. The DNA sequences obtained were compared to the C. jejuni NCTC 11168 genome using the BLASTN program, and the inserts of the next 96 plasmids with the highest ratio values were sequenced until more than 90% of the sequences within a block of 96 plasmids shared 100% identity over their entire length with C. jejuni NCTC 11168. A total of 1,056 plasmids were sequenced, generating approximately 130 kb of DNA sequences unique to C. jejuni 81-176.
The second set of microarray experiments allowed the identification of the unique DNA sequences belonging to the pVir and pTet plasmids. The plasmids were extracted from C. jejuni 81-176 using the QIAGEN plasmid Midi kit, labeled with the Cy3 fluorescent dye, and cohybridized on the shotgun array with Cy5-labeled genomic DNA. Spots with a signal intensity above 2 times the standard deviation of the background in the Cy3 channel were selected as containing DNA fragments belonging to one of the two plasmids. With this criterion, 388 clones were selected and their inserts sequenced. Of the 388 sequences obtained, 212 and 176 of them were identified as belonging to the pVir and pTet plasmids, respectively, by using the BLASTN program at the NCBI. Furthermore, given that 388 clones contained DNA inserts of plasmidic origin, 668 plasmids among the 1,056 selected by the first set of microarray experiments should contain DNA inserts of chromosomal origin. Note that this second set of microarray experiments permitted the efficient differentiation between chromosomic and plasmidic DNA sequences. Indeed, only one clone out of the 668 that were assigned as containing an insert of chromosomal origin was found to harbor a DNA sequence belonging to one of the plasmids.
Sequence assembling and assessment of the shotgun microarray approach to identify unique DNA fragments.
The DNA sequences were first split based on their plasmidic or chromosomic origins and then assembled into contigs using the SeqMan II program from Lasergene (DNASTAR, Inc.). A total of 86 contigs, yielding 130 kb of DNA sequences unique to C. jejuni 81-176, were generated by this assembly process. The size distribution of these contigs for the chromosome and the two plasmids is shown in Table 1. There are 12 and 13 gaps remaining in the sequences of the pTet and pVir plasmids, respectively. Based on the total number of base pairs sequenced (177.5 kb for the chromosome, 98.8 kb for pVir, and 82.3 kb for pTet), the DNA sequences identified in this study represent about 2.5, 2.9, and 2.8 times coverage for the sizes of pTet, pVir, and the chromosome of C. jejuni 81-176, respectively. The lower coverage obtained for the pTet plasmid is likely the result of an underrepresentation of its DNA sequences on the array. The genomic DNA used to construct the shotgun library was extracted from C. jejuni 81-176 grown in the absence of tetracycline. Given the observed high frequency (12%) of spontaneous loss of this plasmid in the absence of a selective pressure (tetracycline) (2), the DNA sequences from this plasmid are likely underrepresented in our library and therefore on the array. Concomitant to this study, the complete sequence of the pTet plasmid was released by Batchelor et al. to the scientific community at the NCBI database (6). This plasmid was found to be 45.2 kb in length. Therefore, a 2.5 times coverage of the size of the pTet plasmid yielded 72% of its 45.2 kb, while a 2.9 times coverage of the size of the pVir plasmid yielded 92% of its 37.5 kb. Consequently, a 2.8 times coverage of the unique DNA fragments from the chromosome of C. jejuni 81-176 likely yielded between 72 and 92% of the total unique nucleotides.
TABLE 1.
Size distribution of the assembled contigs for the unique regions from the chromosome and pTet and pVir plasmids
| Source and size range (kb) | No. of contigs | Total DNA (kb) |
|---|---|---|
| Chromosome | ||
| 4 to 5 | 1 | 4.4 |
| 3 to 4 | 2 | 6.3 |
| 2 to 3 | 1 | 2.3 |
| 1 to 2 | 22 | 29.7 |
| 0.5 to 1 | 21 | 15.5 |
| <0.5 | 10 | 4.1 |
| Total | 57 | 62.3 |
| pTet | ||
| >5 | 2 | 12 |
| 4 to 5 | 0 | 0 |
| 3 to 4 | 1 | 3.4 |
| 2 to 3 | 0 | 0 |
| 1 to 2 | 11 | 16 |
| <1 | 2 | 1.2 |
| Total | 16 | 32.6 |
| pVir | ||
| >5 | 2 | 12.5 |
| 4 to 5 | 2 | 8.5 |
| 3 to 4 | 1 | 3.7 |
| 2 to 3 | 2 | 5.2 |
| 1 to 2 | 2 | 2.8 |
| <1 | 4 | 1.8 |
| Total | 13 | 34.5 |
| Total | 86 | 130 |
Sequence and analysis of the DNA fragments unique to the chromosome of C. jejuni 81-176.
As shown in Table 2, 87 ORFs unique to the chromosome of C. jejuni 81-176 were identified. The only criterion for including an ORF in this group was the presence of an uninterrupted coding frame of at least 120 nucleotides in length. Of the 87 ORFs, 69 were assigned a potential function based on BLASTP findings. The remaining 18 were unknown. The mean G+C content of the unique ORFs was found to be equal to 28.3%, slightly lower than the genome of C. jejuni NCTC 11168 (30.6%). These unique genes can be grouped into seven functional categories: (i) cell envelope and surface structure (mainly capsule and lipooligosaccharide), (ii) restriction and modification systems, (iii) small-molecule metabolism, (iv) transport, (v) antibiotic resistance, (vi) others and hypothetical, and (vii) unknown.
TABLE 2.
Chromosomal DNA fragments present in C. jejuni 81-176 but not in C. jejuni NCTC 11168
| Function and ORF | Lengtha (bp) | % G+C content | Closest relationshipb | Identity (%)c | Accession no. |
|---|---|---|---|---|---|
| Capsule | |||||
| Cj81-045 | 1,0203′ | 26.6 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01891) | 334/340 (98) | AY681253 |
| Cj81-046 | 3193′ | 30.7 | Putative dTDP-4-dehydro rhamnose 3,5-epimerase C. jejuni ATCC 43456 (AAR01889) | 106/106 (100) | AY681254 |
| Cj81-047 | 5645′ | 19.1 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01890) | 139/177 (78) | AY681254 |
| Cj81-062 | 5943′ | 24.7 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01890) | 198/198 (100) | AY681260 |
| Cj81-063 | 1705′ | 30.4 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01891) | 44/56 (78) | AY681260 |
| Cj81-079 | 1,0583′ | 25.6 | Putative glycosyltransferase, C. jejuni CCUG 10954 (AAR01916) | 303/333 (90) | AY681267 |
| Cj81-080 | 1,0115′ | 21.9 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01894) | 547/603 (90) | AY681267 |
| Cj81-081 | 1,613 | 21.9 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01896) | 535/536 (99) | AY681267 |
| Cj81-102 | 5805′3′ | 21.6 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01892) | 176/192 (91) | AY681278 |
| Cj81-103 | 468 | 22.0 | Putative glycosyltransferase, C. jejuni ATCC 43456 (AAR01881) | 154/155 (99) | AY681279 |
| Cj81-128 | 5315′ | 34.0 | Putative GDP-mannoheptose-4,6 dehydratase, C. jejuni ATCC 43456 (AAR01886) | 142/143 (99) | AY681295 |
| Cj81-129 | 4585′ | 25.6 | Unknown, C. jejuni ATCC 43456 (AAR01888) | 151/151 (100) | AY681296 |
| Cj81-130 | 3843′ | 31.2 | Putative GDP-fucose synthase, C. jejuni ATCC 43456 (AAR01887) | 113/127 (88) | AY681296 |
| Lipooligosaccharide, cell envelope, and surface structures | |||||
| Cj81-003 | 1943′ | 26.3 | Putative acetyltransferase, C. jejuni ATCC 43449 (AAL06009) | 47/47 (100) | AY681239 |
| Cj81-008 | 6035′ | 22.2 | Beta-1,4-N-acetylgalactosaminyltransferase, C. jejuni OH4384 (AAF31769) | 200/200 (100) | AY681241 |
| Cj81-009 | 2805′ | 20.3 | Beta-1,3-galactosyltransferase, C. jejuni OH4384 (AAF31770) | 59/92 (64) | AY681241 |
| Cj81-010 | 3603′ | 27.1 | Beta-1,3-galactosyltransferase, C. jejuni OH4384 (AAF31770) | 120/120 (100) | AY681242 |
| Cj81-011 | 8533′ | 25.8 | Alpha-2,3-sialyltransferase, C. jejuni OH4384 (AAF34137) | 269/284 (94) | AY681242 |
| Cj81-030 | 1825′ | 25.7 | Putative amino acid-activating enzyme Cj1307, C. jejuni NCTC 11168 (NP_282453) | 48/60 (80) | AY681244 |
| Cj81-042 | 6465′3′ | 24.5 | Putative lipopolysaccharide A protein, Vibrio parahaemolyticus RIMD 2210633 (NP_796570) | 82/200 (41) | AY681251 |
| Cj81-043 | 8395′ | 24.3 | Putative glycosyltransferase, C. jejuni OH4384 (AAF31768) | 243/277 (87) | AY681252 |
| Cj81-044 | 1673′ | 19.8 | Beta-1,4-N-acetylgalactosaminyltransferase, C. jejuni OH4384 (AAF31769) | 46/50 (92) | AY681252 |
| Cj81-058 | 7783′ | 31.7 | Beta-1,4-N-acetylgalactosaminyltransferase, C. jejuni ATCC 43456 (AAL05993) | 259/259 (100) | AY681257 |
| Cj81-059 | 6535′ | 26.4 | NeuC1, C. jejuni 81-176 (AAL09370) | 215/217 (99) | AY681257 |
| Cj81-085 | 1,442 | 24.7 | Penicillin-binding protein 1A, Oceanobacillus iheyensis HTE831 (NP_692087) | 53/137 (38) | AY681270 |
| Cj81-104 | 4295′3′ | 34.0 | Probable periplasmic protein Cj0737, C. jejuni NCTC 11168 (NP_281909) | 84/131 (64) | AY681280 |
| Cj81-106 | 558 | 34.2 | Sialic acid synthase, C. jejuni ATCC 43449 (AAL06005) | 185/185 (100) | AY681282 |
| Cj81-119 | 853 | 33.0 | FlgE, C. jejuni 81-176 (AAP34261) | 284/284 (100) | AY681288 |
| Cj81-121 | 644 | 28.2 | Probable outer membrane protein Cj1721c, C. jejuni NCTC 11168 (NP_282847) | 140/214 (65) | AY681290 |
| Cj81-127 | 5403′ | 28.6 | a) Unknown, C. jejuni 81-176 (AAR82939) | 180/180 (100) | AY681294 |
| b) Putative glycosyltransferase WavM, Vibrio cholerae (AAL77351) | 67/187 (35) | ||||
| Restriction-modification, recombination, and repair (DNA modification) | |||||
| Cj81-032 | 2993′ | 31.4 | Type I site-specific restriction-modification system, Trichodesmium erythraeum IMS101 (ZP_00071585) | 31/86 (36) | AY681246 |
| Cj81-057 | 1,268 | 27.7 | Type I restriction-modification system S subunit, Synechocystis sp strain PCC 6803 (BAD02014) | 138/428 (32) | AY681256 |
| Cj81-060 | 6905′3′ | 34.0 | a) Type I site-specific restriction-modification system, Haemophilus influenzae (ZP_00321696) | 125/241 (51) | AY681258 |
| b) Type I restriction enzyme HsdR, putative Vibrio cholerae O1 biovareltor strain N16961 (NP_231400) | 122/244 (50) | ||||
| Cj81-061 | 3375′3′ | 30.6 | Putative type I restriction-modification system (HsdR), Acinetobacter sp. strain ADP1 (YP_047908) | 70/113 (61) | AY681259 |
| Cj81-087 | 4195′ | 26.4 | a) Probable type IIS restriction-modification enzyme, C-terminal half Cj0032 C. jejuni NCTC 11168 (NP_281254) | 89/141 (63) | AY681270 |
| b) Type I restriction enzyme HsdR, putative Vibrio cholerae O1 biovareltor strain N16961 (NP_231400) | 55/122 (45) | ||||
| Cj81-118 | 9413′ | 32.7 | Type I restriction-modification system methyltransferase subunit, Trichodesmium erythraeum IMS101 (ZP_00071587) | 183/318 (57) | AY681287 |
| Cj81-126 | 4485′3′ | 30.3 | a) Type I restriction-modification system specificity subunit, Methanosarcina mazei Go1 (NP_635005) | 69/128 (53) | AY681293 |
| b) Type I restriction-modification system methyltransferase subunit, Trichodesmium erythraeum IMS101 (ZP_00071587) | 183/318 (57) | ||||
| Small-molecule metabolism | |||||
| Cj81-005 | 8593′ | 34.8 | Cytochrome c family protein, Geobacter sulfurreducens PCA (NP_951335) | 128/281 (45) | AY681240 |
| Cj81-006 | 584 | 25.3 | a) Conserved hypothetical protein, Wolinella succinogenes DSMZ 1740 (NP_908252) | 66/184 (35) | AY681240 |
| b) Thiol-disulfide interchange protein HP0377, H. pylori 26695 (NP_207175) | 54/180 (30) | ||||
| Cj81-007 | 4925′ | 30.6 | Cytochrome c biogenesis protein (YCF5), Wolinella succinogenes DSMZ 1740 (NP_908253) | 70/152 (46) | AY681240 |
| Cj81-038 | 2065′ | 25.1 | Glutamate synthase (NADPH) large-chain Cj0007 precursor, C. jejuni NCTC 11168 (NP_281229) | 68/68 (100) | AY681247 |
| Cj81-041 | 7005′3′ | 26.0 | a) Hypothetical protein Cj1300, C. jejuni NCTC 11168 (NP_282446) | 126/229 (55) | AY681250 |
| b) 3-Demethylubiquinone-9 3-methyltransferase, Bacteroides thetaiotaomicron VPI-5482 (NP_811691) | 41/124 (33) | ||||
| Cj81-074 | 5133′ | 34.8 | Predicted acyl esterases, Rhodobacter sphaeroides (ZP_00007094) | 92/151 (60) | AY681266 |
| Cj81-083 | 6415′3′ | 30.9 | a) Hypothetical protein WS1432, Wolinella succinogenes DSMZ 1740 (NP_907593) | 87/211 (41) | AY681268 |
| b) Anaerobic DMSO reductase chain C, Haemophilus influenzae Rd KW20 (NP_439204) | 46/185 (24) | ||||
| Cj81-088 | 1,1675′3′ | 28.7 | Cytochrome c biogenesis protein YCF5, Wolinella succinogenes DSMZ 1740 (NP_908253) | 196/393 (49) | AY681271 |
| Cj81-095 | 1,0535′ | 30.4 | 8-Amino-7-oxononanoate synthase BioF, C. jejuni NCTC 11168 (CAB72773) | 243/350 (69) | AY681274 |
| Cj81-097 | 5793′ | 32.6 | Cytochrome c family protein, Geobacter sulfurreducens PCA (NP_951335) | 60/204 (29) | AY681275 |
| Cj81-098 | 443 | 30.2 | a) Cytochrome b subunit of formate dehydrogenase, Dechloromonas aromatica (ZP_00151826) | 59/189 (31) | AY681275 |
| b) Nitrate-TMAO reductases, membrane-bound tetraheme cytochrome c subunit, Desulfitobacterium hafniense taxon:49338 (ZP_00099300) | 48/133 (36) | ||||
| Cj81-099 | 1,1415′3′ | 35.7 | Cytochrome c, putative Shewanella oneidensis MR-1 (NP_716115) | 235/376 (62) | AY681276 |
| Cj81-105 | 1,6275′3′ | 35.1 | Anaerobic DMSO reductase chain A (DmsA), Wolinella succinogenes DSM 1740 (NP_907591) | 325/513 (63) | AY681281 |
| Cj81-109 | 8933′ | 25.1 | Acyl dehydratase, Azotobacter vinelandii (ZP_00089607) | 18/41 (43) | AY681284 |
| Cj81-110 | 227 | 23.2 | Probable acyl carrier protein Cj1299, C. jejuni NCTC 11168 (NP_282445) | 44/75 (58) | AY681284 |
| Cj81-111 | 7585′ | 28.2 | a) Hypothetical protein Cj1298, C. jejuni NCTC 11168 (NP_282444) | 110/242 (45) | AY681284 |
| b) Aminoglycoside N3′-acetyltransferase, Clostridium acetobutylicum (NP_348814) | 85/250 (34) | ||||
| Cj81-124 | 2215′ | 30.2 | Putative cytochrome c type biogenesis protein, Helicobacter pylori J99 (NP_223720) | 44/69 (63) | AY681291 |
| Transport and/or binding proteins, protein and peptide secretion | |||||
| Cj81-027 | 8223′ | 33.9 | Hypothetical protein Tgh073, C. jejuni TGH 9011 (AAS99026) | 26/26 (100) | AY681243 |
| Cj81-048 | 272 | 31.9 | a) Conserved hypothetical protein, Clostridium perfringens strain 13 (NP_561338) | 38/72 (52) | AY681255 |
| b) Putative transporter CgxT, E. coli taxon:562 (AAL61900) | 26/72 (36) | ||||
| Cj81-049 | 453 | 34.2 | Preprotein translocase chain Cj1688c, C. jejuni NCTC 11168 (NP_282814) | 116/117 (99) | AY681255 |
| Cj81-073 | 2975′ | 29.6 | a) Permeases of the major facilitator superfamily, Burkholderia cepacia R18194 (ZP_00217922) | 31/83 (37) | AY681266 |
| b) Putative transporter CgxT, E. coli taxon:562 (AAL61900) | 23/77 (29) | ||||
| Cj81-094 | 1,1033′ | 29.7 | Similar to di-tripeptide ABC transporter PLU1813, Photorhabdus luminescens subsp. laumondii (NP_929091) | 148/366 (40) | AY681273 |
| Cj81-107 | 4045′ | 29.3 | Probable efflux protein Cj1687, C. jejuni NCTC 11168 (NP_282813) | 88/129 (68) | AY681283 |
| Cj81-125 | 3515′3′ | 32.2 | Probable molybdenum transport ATP-binding protein Cj0300c, C. jejuni NCTC 11168 (NP_281491) | 77/117 (65) | AY681292 |
| Hypothetical and other | |||||
| Cj81-026 | 452 | 32.9 | a) Hypothetical protein Bucepa02004635, Burkholderia cepacia R1808 (ZP_00220764) | 60/137 (43) | AY681243 |
| b) Mating pair stabilization protein, Novosphingobium aromaticivorans (NP_049155) | 60/140 (42) | ||||
| Cj81-033 | 989 | 21.7 | Hypothetical protein, Leptospira interrogans serovar lai strain 56601 (NP_711975) | 117/309 (37) | AY681246 |
| Cj81-034 | 3265′ | 24.7 | Hypothetical protein STY3192, Salmonella enterica subsp. enterica serovar Typhi CT18 (NP_457434) | 42/58 (72) | AY681246 |
| Cj81-037 | 1,0743′ | 24.3 | Hypothetical protein jhp0174, H. pylori strain J99 (NP_222895) | 131/366 (35) | AY681247 |
| Cj81-068 | 847 | 23.5 | Hypothetical protein, Bacillus cereus ATCC 14579 (NP_831045) | 52/172 (30) | AY681264 |
| Cj81-069 | 1,319 | 24.5 | ATP/GTP-binding protein, Bacillus cereus ATCC 14579 (NP_831044) | 153/456 (33) | AY681264 |
| Cj81-070 | 4965′3′ | 24.2 | Conserved hypothetical protein, Porphyromonas gingivalis W83 (NP_905721) | 60/143 (41) | AY681265 |
| Cj81-086 | 894 | 23.8 | Hypothetical protein, Enterococcus faecalis V583 (NP_815563) | 43/140 (30) | AY681270 |
| Cj81-092 | 4203′ | 29.3 | Hypothetical protein Cj0741, C. jejuni NCTC 11168 (NP_281913) | 48/49 (97) | AY681272 |
| Cj81-096 | 611 | 27.3 | Hypothetical protein Cj0305c, C. jejuni NCTC 11168 (AL139074) | 128/203 (63) | AY681274 |
| Unknown | |||||
| Cj81-004 | 1233′ | 29.3 | No hit | AY681239 | |
| Cj81-028 | 1503′ | 30.7 | No hit | AY681244 | |
| Cj81-029 | 725 | 24.2 | No hit | AY681244 | |
| Cj81-040 | 1,1475′ | 29.4 | No hit | AY681249 | |
| Cj81-064 | 2985′ | 27.9 | No hit | AY681261 | |
| Cj81-065 | 4083′ | 31.0 | No hit | AY681261 | |
| Cj81-084 | 2755′ | 31.5 | No hit | AY681269 | |
| Cj81-093 | 1885′ | 32.8 | No hit | AY681272 | |
| Cj81-100 | 1723′ | 38.9 | No hit | AY681277 | |
| Cj81-101 | 2305′ | 37.7 | No hit | AY681277 | |
| Cj81-108 | 4465′ | 32.9 | No hit | AY681283 | |
| Cj81-115 | 3195′ | 38.0 | No hit | AY681285 | |
| Cj81-116 | 8843′ | 34.1 | No hit | AY681285 | |
| Cj81-117 | 7675′ | 26.8 | No hit | AY681286 |
The superscript numbers denote partial ORFs with the 3′ and/or 5′ region missing.
Database hits: a, first hit; b, other relevant hit.
Amino acid identity in the region of homology identified by BLASTP.
A large number of the unique genes encode putative proteins involved in the biosynthesis of the capsule and lipooligosaccharide (LOS). The entire capsular polysaccharide biosynthetic locus of C. jejuni 81-176 was previously identified and sequenced by Karlyshev et al. (GenBank accession number BX545858) (18). The identification of genes encoding the proteins involved in its biosynthesis is in agreement with the difference in Penner serotype of the two C. jejuni strains. C. jejuni 81-176 is Penner serotype O:23/36, while NCTC 11168 is Penner serotype O:2. It is noteworthy that the DNA sequence comparison of this locus with the unique genes identified here and the corresponding locus in C. jejuni NCTC 11168 revealed that the shotgun microarray approach employed in this study identified all of the expected unique genes, highlighting the power of the comparative shotgun microarray technology. The identification of C. jejuni 81-176 unique genes encoding putative proteins involved in LOS biosynthesis is in agreement with the reported high divergence of this locus between strains of C. jejuni (12, 23, 24, 30, 32). In fact, the C. jejuni NCTC 11168 genes associated with outer-core LOS biosynthesis were previously found to be absent or highly divergent in C. jejuni 81-176 by whole-genome comparison (12, 23).
Another functional group of unique C. jejuni 81-176 ORFs encodes several probable type I and type II restriction-modification (R-M) systems (Cj81-032, Cj81-057, Cj81-060, Cj81-061, Cj81-087, Cj81-118, and Cj81-126). As seen in Table 2, Cj81-118 and Cj81-126 exhibit identity with the same protein, a type I restriction-modification system methyltransferase subunit (HsdM) from Trichodesmium erythraeum, and thus they might be part of the same gene. Similarly, Cj81-060 and Cj81-061 might constitute a single gene encoding a protein with homology to a type I restriction-modification enzyme (HsdR) of Vibrio cholerae. A type I R-M system is constituted of three components: a restriction enzyme (HsdR), a methyltransferase (HsdM), and a site-specific subunit (HsdS) (27). Therefore, Cj81-60/Cj81-61 and Cj81-118/Cj81-126 might be part of the same R-M system, with the site-specific subunit encoded by Cj81-032 or Cj81-057.
A noteworthy set of unique C. jejuni 81-176 ORFs encodes proteins involved in bacterial respiratory metabolism, including several cytochrome c biogenesis proteins (Cj81-007, Cj81-097, Cj81-099, and Cj81-124), a cytochrome b subunit of formate dehydrogenase (Cj81-005), two components of a putative anaerobic dimethyl sulfoxide (DMSO) reductase (Cj81-083 and Cj81-105), and a nitrate-trimethylamine-N-oxide (TMAO) membrane-bound reductase (Cj81-098). These different proteins may allow C. jejuni to utilize electron acceptors other than oxygen under oxygen-restricted conditions, such as DMSO, TMAO, and nitrate. The DMSO reductase system is found in various bacteria, including E. coli (7), Actinobacillus pleuropneumoniae (3), Rhodobacter sphaeroides (17), Rhodobacter capsulatus (26), and Haemophilus influenzae (25). The E. coli anaerobic DMSO reductase is composed of three subunits, a catalytic subunit containing a molybdopterin cofactor (DsmA), a transmembrane electron carrier (DsmB), and a membrane anchor (DsmC) (8). Based on the BLASTP results, the C. jejuni 81-176 DsmA and DsmC proteins are encoded by Cj81-105 and Cj81-083, respectively. While a DmsB homolog was not recognized, several of the identified genes might encode a protein with a DmsB-like function, such as the cytochrome c or the nitrate-TMAO reductase. DmsA requires molybdenum for its catalytic activity; however, the C. jejuni NCTC 11168 genes (modABC) encoding the molybdenum transport apparatus were found to be missing in C. jejuni 81-176 (12). Interestingly, a gene encoding a probable molybdenum transport ATP-binding protein (a ModC homolog) was identified on the chromosome of C. jejuni 81-176 (Cj81-125), suggesting that this strain might be able to acquire molybdenum. However, homologs of ModA and ModC were not identified.
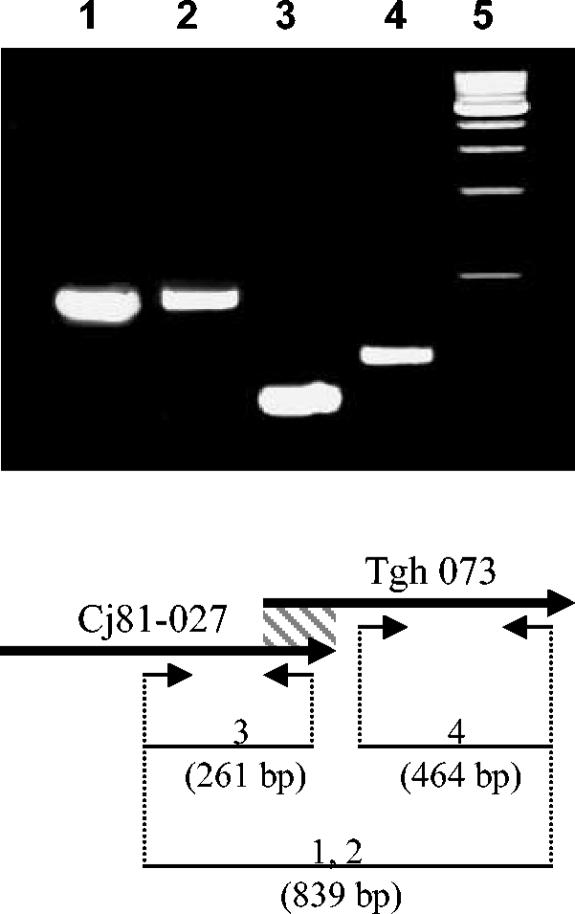
A notable ORF of C. jejuni 81-176 (Cj81-027) appears to be identical to a unique ORF from C. jejuni ATCC 43431 (Tgh073) (32). The sequences of these two ORFs overlap over a 78-bp nucleotide region with a 100% identity, suggesting they might be part of the same gene. In order to demonstrate that these two ORFs (Tgh073 and Cj81-027) are part of the same gene, a set of PCR primers was designed to amplify a DNA fragment encompassing both ORFs. As shown in Fig. 1, the PCR amplification using the chromosomal DNA of either C. jejuni 81-176 (Fig. 1, lane 1) or ATCC 43431 (Fig. 1, lane 2) yielded a product of the expected size, demonstrating that both ORFs belong to the same gene. Interestingly, Tgh073 was shown to encode a homologue of TraG from Neisseria gonorrhoeae (11). In other organisms, TraG participates in the formation of a conjugative DNA transfer system and a type IV secretion apparatus (14, 22). In fact, TraG-like proteins have been proposed to play an essential role in the secretion of substrates through these apparatus, such as the cytotoxin CagA protein in Helicobacter pylori (34).
FIG. 1.
PCR amplification and schematic outline of the chromosomal TraG-like region identified in C. jejuni ATCC 43431 and C. jejuni 81-176. The ORFs are denoted by boxed arrows. The slashed box indicates the region with 100% identity between both ORFs. Predicted PCR fragments are shown at the bottom. Gel lanes match the PCR product label. Lane 5 shows the 1-kb DNA ladder. PCR products 1 and 2 were obtained by using as template the chromosomal DNA from C. jejuni ATCC 43431 and C. jejuni 81-176, respectively. PCR products 3 and 4 were obtained by using the chromosomal DNA of C. jejuni ATCC 43431 as template.
Other unique C. jejuni 81-176 genes encode probable transporters, an efflux protein, and several hypothetical and unknown proteins.
DISCUSSION
Despite the absence of clear evidence for the existence of Campylobacter pathotypes, epidemiological and phenotypical studies suggest that strains of C. jejuni vary in their colonization and invasion abilities and thus likely in their virulence potential (13, 15, 16, 19, 28). Comparative genomic studies showed that there is substantial genetic diversity between isolates of C. jejuni (12, 23, 30). This diversity might enable Campylobacter to colonize various environmental niches and possibly contribute to its virulence. So far, these comparative genomic studies have been restricted to the investigation of the presence or absence of genes from the genome-sequenced strain C. jejuni NCTC 11168 in other Campylobacter isolates of various origins (12, 23, 30). Obviously, genes unique to nonsequenced strains cannot be identified by this classical approach. While the genomic subtractive hybridization procedure is the method of choice for the identification of strain-specific DNA sequences, the generation of comprehensive coverage is labor-intensive and tedious. As a result, this technology usually provides only a handful of new DNA fragments. Here, we describe and assess the use of a whole-plasmid shotgun microarray technology to identify DNA fragments of C. jejuni 81-176 absent in C. jejuni NCTC 11168. This technology presents several advantages for identifying unique genes. Whole-plasmid shotgun arrays are relatively simple and inexpensive to construct. These arrays are extremely versatile, and the technology could be adapted to virtually any organism. Given the sequence of a reference strain, this approach enables the identification of novel and unique DNA sequences on other strains without the need of their whole genome sequences.
The results presented here demonstrate the efficiency of the whole-plasmid shotgun microarray approach and add 87 novel chromosomal genes to the gene repertoire of C. jejuni. These additional genes will allow the construction of a more comprehensive microarray of C. jejuni, which will likely facilitate the identification of genotypic markers and then improve the power of genotyping methods. The genome coverage of this technology was assessed by three parameters: (i) the number of times that the same DNA fragment was sequenced, (ii) the effectiveness of identifying genes previously known to be unique to C. jejuni 81-176, and (iii) the nucleotide coverage of the two plasmids, pVir and pTet. Each chromosomal DNA fragment was sequenced on average 2.8 times, and among the 16 previously identified unique chromosomal C. jejuni 81-176 genes, only one was not detected by our study (NCBI accession number AAS46249). The failure to identify this gene is likely due to its relatively short length, 279 nucleotides, and the presence of flanking DNA sequences (in 5′ and 3′) with 100% identity to C. jejuni NCTC 11168. The two plasmids, pTet and pVir, were sequenced with coverages of 2.5- and 2.9-fold, respectively, representing 72 and 92% of their complete nucleotide sequences. Altogether, these results show that whole-plasmid shotgun microarray provides an effective and comprehensive approach to identify novel genes without whole-genome sequencing. Although this technology appears to be a powerful method for the identification of novel DNA fragments, this technique has its limitations. First, it is not possible to detect point mutations, gene location, or chromosomal rearrangements. Second, the boundaries of the novel genomic segments are not identified, probably due to a high level of cross-hybridization of these regions with the chromosomal DNA of C. jejuni NCTC 11168. Nevertheless, this technology allowed the comprehensive detection of the unique genes of the C. jejuni 81-176 strain and thus holds promise as an efficient tool for comparative genomics.
It should be noted that this study does not fully address the function of unique genes, and their possible role in virulence remains speculative. However, the in silico functional analysis of the unique C. jejuni 81-176 genes reveals several interesting traits of this isolate. The unique genes are characterized by three major functional groups: (i) capsule and LOS biosynthesis, (ii) restriction and modification systems, and (iii) respiratory metabolism. The C. jejuni NCTC 11168 capsule and LOS loci have been previously shown to be divergent in C. jejuni 81-176 (12, 23). This divergence has been attributed to the difference in Penner serotype between the two strains, O:23/36 and O:2 for C. jejuni 81-176 and NCTC 11168, respectively. Therefore, the identification of C. jejuni 81-176 unique genes encoding enzymes involved in the biosynthesis of these surface structures was expected and is in agreement with the diversity of these loci. It is noteworthy that the unique genes involved in the capsule biosynthesis encode proteins that exhibit high identity with proteins of similar function in C. jejuni ATCC 43456 (Table 2), which belongs to the same Penner serotype as C. jejuni 81-176 (Penner serotype O:36), corroborating the role of the capsule as the serodeterminant in the Penner serotyping scheme (18). A number of unique genes encode proteins with a high BLASTP match with components of type I R-M systems. Interestingly, the presence of genes encoding type I R-M systems was found to correlate with the extent of the host gastric response to H. pylori (9). Consequently, these enzymes have been proposed to affect H. pylori pathogenesis by modulating its gene expression. Based on this observation, it is tempting to propose an essential role for these R-M systems in the ability of Campylobacter to induce gastroenteritis. Namely, the presence or absence of a particular R-M system might modulate the virulence potential of Campylobacter strains. Another important group of genes identified in this study encodes proteins involved in respiratory metabolism. C. jejuni is a microaerophilic bacterium with a growth requirement for oxygen. Recently, oxygen has been proposed to be essential for DNA synthesis due to the use of an oxygen-dependent class I type ribonucleotide reductase (35). Nevertheless, C. jejuni NCTC 11168 has been shown to be able to carry out anaerobic-like respiration and use a wide range of electron acceptors other than oxygen, such as fumarate, nitrate, nitrite, TMAO, and DMSO (35). The genome of C. jejuni 81-176 has been shown to contain all of the NCTC 11168 genes that encode the predicted terminal reductases involved in these alternative respiratory pathways (12, 23). Therefore, the presence in C. jejuni 81-176 of additional genes encoding proteins with a potential function in electron transport suggests that this isolate might possess a capacity for adaptation to different ecological niches. In particular, the C. jejuni 81-176 genome contains an anaerobic DMSO reductase. In E. coli, the DMSO reductase allows cell respiration under reduced-oxygen or anaerobic conditions by utilizing a broad range of various N-oxide and sulfoxide compounds as terminal electron acceptors (38). This wide range of substrate specificity may allow C. jejuni 81-176 to grow under low oxygen level on a diverse array of N- and S-oxide compounds, such as TMAO and DMSO. Interestingly, the DMSO reductase of A. pleuropneumoniae has been shown to significantly contribute to the virulence of this pathogen (3). An A. pleuropneumoniae mutant lacking the DMSO reductase was attenuated in acute disease with a pig infection model, suggesting a role for alternative respiratory pathways in necrotic lung tissue (3). However, the nature of the substrate for this enzyme in the lung tissue is unclear, since DMSO and TMAO are essentially found in marine organisms (such as fish and aquatic invertebrates) and algae (5). Consequently, this enzyme might allow C. jejuni 81-176 to colonize seafood and/or to enhance gastrointestinal tract colonization if their hosts eat seafood.
Recently, we described the characterization of C. jejuni ATCC 43431 unique genes (32). The comparison of these unique genes with those of C. jejuni 81-176 uncovered some additional information. These two strains of C. jejuni appear to share only one unique gene. Interestingly, this gene encodes a protein with identity to TraG from Neisseria gonorrhoeae. This TraG-like protein has been shown to be involved in mating and to also function as an essential component of the type IV secretion apparatus in several pathogens, including H. pylori (14, 22, 34). The function of this gene in C. jejuni and its presence in both strains are intriguing. In contrast to C. jejuni NCTC 11168, both of these strains are highly invasive into intestinal epithelial cells (2, 32). The invasion efficiency of C. jejuni 81-176 has been shown to be dependent on a type IV secretion system encoded by the plasmid pVir (2). However, the pVir plasmid was found to be insufficient for the high level of invasion observed. Furthermore, the transformation of the pVir plasmid into C. jejuni NCTC 11168 did not result in an increase of its level of invasion into INT407 cells (2). Therefore, although the pVir plasmid was required for a high level of invasion, a chromosomally encoded protein(s) absent in NCTC 11168 was proposed to also be necessary (2). While C. jejuni ATCC 43431 does not harbor the pVir plasmid, its chromosome contains genes encoding components with homology to the type IV secretion system (32). Consequently, it is tempting to propose a role for the chromosomally encoded TraG protein in the formation of a type IV secretion system that might be involved in the invasion process into epithelial cells. We are currently investigating this possibility. Notably, a TraG homologue was also found on the pTet plasmid (6). This plasmid has been shown to be self-mobilizable and to carry 10 genes encoding proteins involved in conjugation (6). While these genes also encode a type IV secretion system, this system seems to serve a function different from the one encoded by pVir and to be required for DNA transfer. In addition, this plasmid was found to be fully dispensable for invasion into epithelial cells (2). Because of the sufficiency of the pTet plasmid-encoded DNA transfer functions, the chromosomally encoded TraG-like protein is unlikely to contribute to bacterial conjugation.
In conclusion, whole-plasmid shotgun microarray is a powerful and versatile method that allows the comprehensive identification of new genomic DNA sequences from a nonsequenced bacterial strain by comparison with a related species or strain of known sequence. In this study, we identified the repertoire of genes unique to C. jejuni 81-176 by comparison to C. jejuni NCTC 11168. Interestingly, several of these unique genes might be associated with C. jejuni 81-176 fitness and virulence. However, the true function of these genes needs to be addressed by targeted mutagenesis and phenotypic analysis. The results presented here demonstrate that there is a substantial genetic diversity among isolates of C. jejuni. Therefore, the study of additional strains of C. jejuni would be valuable to further increase the repertoire of genes. The identification of additional genes would allow the construction of a composite Campylobacter microarray. This microarray could then be used to identify unique and shared genes between strains of various origins. Ultimately, this type of comparative study may prove to be useful for the development of molecular typing methods. Finally, such microarrays could constitute a useful tool for epidemiological and evolutionary studies.
Acknowledgments
We thank I. Turcot for her contribution to the manuscript. We are grateful to all of the staff from OU Health Science Center Laboratory for Genomics and Bioinformatics and the University of Oklahoma's Microarray and Bioinformatics Core facilities.
This work was supported by the National Institutes of Health grants RO1-AI055612 and RR15564.
REFERENCES
- 1.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, L. Hu, T. E. Hickey, C. P. Ewing, R. A. Batchelor, T. J. Trust, and P. Guerry. 2002. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect. Immun. 70:6242-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltes, N., I. Hennig-Pauka, I. Jacobsen, A. D. Gruber, and G. F. Gerlach. 2003. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baqar, S., A. L. Bourgeois, L. A. Applebee, A. S. Mourad, M. T. Kleinosky, Z. Mohran, and J. R. Murphy. 1996. Murine intranasal challenge model for the study of Campylobacter pathogenesis and immunity. Infect. Immun. 64:4933-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett, E. L., and H. S. Kwan. 1985. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 39:131-149. [DOI] [PubMed] [Google Scholar]
- 6.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 7.Bilous, P. T., S. T. Cole, W. F. Anderson, and J. H. Weiner. 1988. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli Mol. Microbiol. 2:785-795. [DOI] [PubMed] [Google Scholar]
- 8.Bilous, P. T., and J. H. Weiner. 1988. Molecular cloning and expression of the Escherichia coli dimethyl sulfoxide reductase operon. J. Bacteriol. 170:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjorkholm, B. M., J. L. Guruge, J. D. Oh, A. J. Syder, N. Salama, K. Guillemin, S. Falkow, C. Nilsson, P. G. Falk, L. Engstrand, and J. I. Gordon. 2002. Colonization of germ-free transgenic mice with genotyped Helicobacter pylori strains from a case-control study of gastric cancer reveals a correlation between host responses and HsdS components of type I restriction-modification systems. J. Biol. Chem. 277:34191-34197. [DOI] [PubMed] [Google Scholar]
- 10.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 11.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 12.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everest, P. H., H. Goossens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 14.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanel, I., J. Muller, W. Muller, and F. Schulze. 2004. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni Vet. Microbiol. 101:75-82. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, J. L., N. R. Bastian, and K. V. Rajagopalan. 1990. Molybdopterin guanine dinucleotide: a modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. USA 87:3190-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 19.Konkel, M. E., and L. A. Joens. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 57:2984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecko, D. J., L. Hu, and K. J. Zaal. 2001. Campylobacter jejuni—microtubule-dependent invasion. Trends Microbiol. 9:389-396. [DOI] [PubMed] [Google Scholar]
- 21.Kuroki, S., T. Saida, M. Nukina, T. Haruta, M. Yoshioka, Y. Kobayashi, and H. Nakanishi. 1993. Campylobacter jejuni strains from patients with Guillain-Barre syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann. Neurol. 33:243-247. [DOI] [PubMed] [Google Scholar]
- 22.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 23.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 24.Leonard, E. E., II, L. S. Tompkins, S. Falkow, and I. Nachamkin. 2004. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barre syndrome and strains that cause enteritis by a DNA microarray. Infect. Immun. 72:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loosmore, S. M., J. M. Shortreed, D. C. Coleman, D. M. England, and M. H. Klein. 1996. Sequences of the genes encoding the A, B and C subunits of the Haemophilus influenzae dimethylsulfoxide reductase complex. Gene 169:137-138. [DOI] [PubMed] [Google Scholar]
- 26.McEwan, A. G., S. J. Ferguson, and J. B. Jackson. 1991. Purification and properties of dimethyl sulphoxide reductase from Rhodobacter capsulatus A periplasmic molybdoenzyme. Biochem. J. 274:305-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, N. E. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64:412-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newell, D. G., H. McBride, F. Saunders, Y. Dehele, and A. D. Pearson. 1985. The virulence of clinical and environmental isolates of Campylobacter jejuni J. Hyg. (London) 94:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 31.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poly, F., D. Threadgill, and A. Stintzi. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell, R. G., M. J. Blaser, J. I. Sarmiento, and J. Fox. 1989. Experimental Campylobacter jejuni infection in Macaca nemestrina. Infect. Immun. 57:1438-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd edition. American Society for Microbiology, Washington, D.C.
- 37.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner, J. H., D. P. MacIsaac, R. E. Bishop, and P. T. Bilous. 1988. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J. Bacteriol. 170:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]