Abstract
We investigated an outbreak of Acinetobacter baumannii in an intensive care unit and in the surgery, medicine, neurology, and urology wards of the Kosin University Gospel Hospital in Busan, Korea. The outbreak involved 36 cases of infection by A. baumannii producing the OXA-23 β-lactamase over an 8-month period and was caused by a single pulsed-field gel electrophoresis clone. The epidemic isolates were characterized by a modified cloverleaf synergy test. Isoelectric focusing of crude bacterial extracts detected one nitrocefin-positive band with a pI value of 6.65. PCR amplification and characterization of the amplicons by direct sequencing indicated that the epidemic isolates carried a blaOXA-23 determinant. The epidemic isolates were characterized by a multidrug resistance phenotype that remained unchanged over the outbreak, including penicillins, cephamycins, extended-spectrum cephalosporins, carbapenems, monobactams, and aminoglycosides. This study shows that the blaOXA-23 resistance determinant may become an emerging therapeutic problem.
Acinetobacter baumannii has emerged as an important nosocomial pathogen in outbreaks of hospital infections and is ranked second after Pseudomonas aeruginosa among nosocomial pathogens of aerobic nonfermentative gram-negative bacilli (17, 19). A. baumannii causes respiratory and urinary tract infections, meningitis, endocarditis, burn infections, and wound sepsis, especially in intensive care units (ICUs) (4). A. baumannii infections are often difficult to eradicate due to high-level resistance to many antibiotics as a result of both intrinsic and acquired mechanisms. β-Lactamase production is the most important mechanism of acquired β-lactam resistance in gram-negative pathogens (21). Carbapenems (e.g., imipenem and meropenem) have become the drugs of choice against Acinetobacter infections in many centers but are being compromised by the emergence of carbapenem-hydrolyzing β-lactamase (carbapenemase) of molecular classes B and D (14). Class B carbapenemases found thus far in Acinetobacter spp. include various IMP- and VIM-type metallo-β-lactamases (http://www.lahey.org/studies/webt.asp), but most Acinetobacter spp. produce zinc-independent members of β-lactamase molecular class D (1). Sequenced carbapenemases of this latter class from that species include the following two distinct clusters: (i) the OXA-23-like cluster (OXA-23 and -27) and (ii) the OXA-24-like cluster (OXA-24, -25, -26, and -40). OXA-23 and OXA-27 have 99% amino acid identity, whereas they have only 60% identity with those of OXA-24-like cluster (1, 3, 6, 7).
Over an 8-month period from January to August 2003, 193 A. baumannii isolates were isolated from 193 patients hospitalized at the Kosin University Gospel Hospital. The purpose of the present study was to investigate an outbreak of A. baumannii in Korea and to characterize the imipenem resistance mechanism of the outbreak isolates.
MATERIALS AND METHODS
Bacterial strains and susceptibility tests.
A total of 193 nonrepetitive clinical isolates of A. baumannii were isolated from January to August 2003 in the Kosin University Gospel Hospital (Busan, Republic of Korea) with 1,300 beds. These isolates were collected from different patients hospitalized at ICUs and in surgery, medicine, neurology, and urology wards. The isolates were identified by using conventional techniques (17) and/or Vitek GNI card (bioMérieux Vitek, Inc., Hazelwood, Mo.). A. baumannii (YMC02/8/P535) (21) was used as the recipient strain for transfer by transconjugation. Escherichia coli DH5α and E. coli ATCC 25922 were used as the host strain for transformation and the MIC reference strain, respectively.
Antibiotic susceptibility was determined by disk diffusion tests that were performed according to the manufacturer's instructions with BBL (Cockeysville, Md.) disks impregnated with amikacin (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), tetracycline (30 μg), tobramycin (10 μg), trimethoprim-sulfamethoxazole (1.25 and 23.75 μg, respectively), ampicillin (10 μg), ampicillin-sulbactam (10 and 10 μg, respectively), piperacillin (100 μg), piperacillin-tazobactam (100 and 10 μg, respectively), cephalothin (30 μg), cefoxitin (30 μg), cefoperazone (75 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), aztreonam (30 μg), and imipenem (10 μg). Disks were dispensed with a BBL Sensi-Disk 12-place dispenser. MICs were determined by the agar dilution technique on Muller-Hinton agar plates (Becton-Dickinson, Sparks, Md.) containing serially twofold-diluted β-lactams as described previously (15).
Microbiological tests of carbapenemase activity.
In order to study the inactivation of imipenem by the A. baumannii OXA-type β-lactamase, a microbiological disk synergy test was performed. The cloverleaf test of Hornstein et al. (8) was modified by substituting E. coli ATCC 25922 for imipenem-susceptible Micrococcus luteus and using Acinetobacter genomospecies 3 YMC 99/11/160 as a VIM-2-producing positive control. The surface of a Muller-Hinton agar plate was inoculated evenly by using a cotton swap with an overnight culture suspension of E. coli, which was adjusted to the turbidity of the McFarland no. 0.5 tube. After brief drying, an imipenem disk was placed at the center of the plate, and imipenem-resistant test strains from the overnight culture plates were streaked heavily from the edge of the disk to the periphery of the plate. The presence of a distorted inhibition zone after overnight incubation was interpreted as a positive modified cloverleaf synergy test showing the inactivation of imipenem by carbapenemase (class B and/or D).
Microbiological testing of metallo-β-lactamase activity was performed with an EDTA-disk test synergy test (11) modified as follows. An overnight culture of the test strain was suspended to the turbidity of the McFarland no. 0.5 tube and used to swab inoculate a Muller-Hinton agar plate. After drying, a 30-μg imipenem disk (BBL) and a blank filter paper disk were placed 15 mm apart from edge to edge, and 10 μl of 0.5 M EDTA solution was then applied to the blank disk, which resulted in ca. 1.5 mg/disk. After overnight incubation, the presence of enlarged zone of inhibition was interpreted as EDTA-disk synergy test positive showing the inactivation of metallo-β-lactamase (class B) activity by EDTA.
Transconjugation experiments and IEF analysis.
Curing was attempted by growing cultures overnight in nutrient broth containing ethidium bromide (Sigma-Aldrich, Louis, Mo.) at 0.25 to 0.5 times the MIC, followed by replica plating on to Muller-Hinton agar plates with or without imipenem at 2 or 10 mg/liter. Isolation of plasmid DNA was performed as described by Sambrook and Russell (16) with plasmid-safe ATP-dependent DNase (Epicentre Technology, Madison, Wis.) for removing contaminated bacterial chromosomal DNA. Transconjugation experiments were performed as described previously (21) with rifampin-resistant A. baumannii (YMC02/8/P535) as the recipient. Transconjugants were selected on Muller-Hinton agar supplemented with rifampin (100 mg/liter) to inhibit the growth of the donor strain and with imipenem (1 mg/liter) to inhibit the growth of the recipient strain. Crude bacterial extracts were obtained from clinical isolates after centrifugation of sonicated culture as previously described (12). Sonic extracts were used for the determination of isoelectric points (pIs) and β-lactamase activity. Isoelectric focusing (IEF) was performed in Ready Gel precast IEF polyacrylamide gels (Bio-Rad, Hercules, Calif.) as previously described (12). Gels were developed with 0.5 mM nitrocefin (Merck, Whitehouse Station, N.J.).
Molecular studies.
Unless otherwise stated, molecular biological reagents and restriction enzymes were obtained from Sigma-Aldrich. Genomic DNA of clinical isolates were prepared with Wizard Genomic DNA Purification Kit (Promega, Madison, Wis.) and used as template DNA in PCR. The primers for PCR amplification were designed by selecting consensus sequences in multiple-nucleotide alignment of six OXA-type β-lactamase genes (blaOXA), five IMP-type β-lactamase genes (blaIMP), and six VIM-type β-lactamase genes (blaVIM) by using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The primers are described in Table 1. PCR amplifications were carried out as described previously (9, 10). For the cloning of blaOXA-23 gene, the PCR products were cloned into pGEM-T (Promega), and the recombinant plasmids were transformed into chemically competent cell of E. coli DH5α by heat shock as detailed in the supplier's instructions. Transformants were selected and subcultured on nutrient agar plates containing ampicillin (100 mg/liter).
TABLE 1.
Nucleotide sequences of oligonucleotides used for PCR amplifications and sequencing of blaOXA-23-like, blaOXA-24-like, blaIMP-type, and blaVIM-type genes
| Primer (orientation)a | Accession no. of blab | Sequence (5′→3′) | Annealing sitec | Amplicon size (bp)d |
|---|---|---|---|---|
| OXA-23F (F) | AJ132105 | GATGTGTCATAGTATTCGTCGT | −108 to −87 | 1,058 |
| OXA-23R (R) | AF201828 | TCACAACAACTAAAAGCACTGT | +929 to +950 | (OXA-23F/-23R) |
| OXA-24F (F) | AJ239129, AF201826 | ATGAAAAAATTTATACTTCCTATATTCAGC | +1 to +30 | 825 |
| OXA-24R (R) | AF201827, AF509241 | TTAAATGATTCCAAGATTTTCTAGC | +801 to +825 | (OXA-24F/-24R) |
| IMP-F (F) | S71931, AB010417, AF290912 | CATGGTTTGGTGGTTCTTGT | +154 to +173 | 488 |
| IMP-R (R) | AB040994, AB074433 | ATAATTTGGCGGACTTTGGC | +582 to +601 | (IMP-1F/-1R) |
| VIM-F (F) | AF191564, AF300454, AY165025 | ATTGGTCTATTTGACCGCGTC | +24 to +44 | 780 |
| VIM-R (R) | AY524987, AY524988, AY524989 | TGCTACTCAACGACTGAGCG | +784 to +803 | (VIM-2F/-2R) |
Orientation of each primer: F, forward; R, reverse.
β-lactamase genes (bla) used in the multiple sequence alignment for designing each primer pair.
Positions of annealing sites are with respect to the first nucleotide of the coding region (+1).
The primer pair for PCR amplification is indicated in parentheses.
DNA sequencing was performed by the direct sequencing method with an automatic sequencer (ABI Prism3100; Applied Biosystems, Weiterstadt, Germany) as previously described (13). DNA sequence analysis was performed with DNASIS for Windows (Hitachi Software Engineering America, Ltd., San Bruno, Calif.). Database similarity searches for both the nucleotide sequences and deduced protein sequences were performed with BLAST at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
For pulsed-field gel electrophoresis (PFGE), SmaI-digested genomic DNA was prepared according to the instruction of Bio-Rad (Hercules, Calif.), and fragments were separated for 20 h at 6 V/cm at 11°C by using a CHEF-DRII system (Bio-Rad), with initial and final pulse times of 0.5 and 60 s, respectively. DNA fingerprints were interpreted as recommended by Tenover et al. (20)
Transfer of SmaI-digested genomic DNA to nylon membrane (Hybond-N; Amersham International, Buckinghamshire, England) was performed essentially as described by Sambrook and Russell (16). Labeling of DNA (PCR product between OXA-23F primer and OXA-23R primer) probe was performed with digoxigenin as described by the manufacturer (Roche Diagnostics GmbH, Mannheim, Germany). Southern hybridization was performed at 68°C with the buffers recommended in the instructions included in the digoxigenin kit from Roche.
RESULTS
Phenotypic properties of imipenem-resistant isolates.
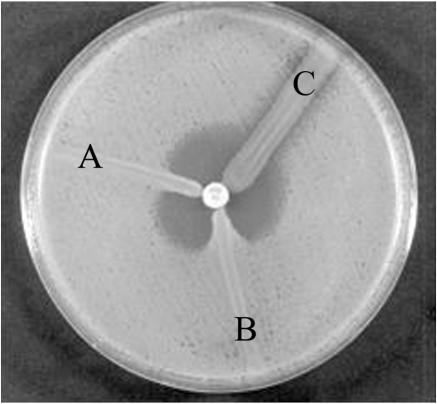
The results of antimicrobial susceptibility testing of 193 nonrepetitive A. baumannii isolates showed a significant detection (26.9%, 52 of 193) of imipenem-resistant isolates. A total of 52 isolates of imipenem-resistant A. baumannii were studied by using the modified cloverleaf synergy test and the EDTA-disk synergy test. Metallo-β-lactamase-producing isolates (EDTA-disk synergy test positive) were not detected. The prevalence of carbapenemase-producing isolates (modified cloverleaf synergy test positive) was 69.2% (36 of 52) (Fig. 1 and Table 2). These results were confirmed with the carbarpenemase activity data by spectrophotometric assays in the presence or absence of EDTA. The results suggest that 36 imipenem-resistant A. baumannii isolates do not produce class B metallo-β-lactamases but produce other type β-lactamases. Thirty-six β-lactamase-producing A. baumannii were isolated from 36 different patients hospitalized in nine wards (cardiac internal medicine, cardiac surgery, general surgery, kidney internal medicine, neurology, neurosurgery, orthopedic surgery, pulmonary internal medicine, and urology) and showed high levels of resistance to amikacin, gentamicin, tetracycline, tobramycin, ampicillin, piperacillin, piperacillin-tazobactam, cephalothin, cefoxitin, cefoperazone, ceftazidime, cefotaxime, cefepime, aztreonam, and imipenem (Table 2). Eight β-lactamase-producing A. baumannii isolates were only sensitive to ampicillin-sulbactam of the antibiotics tested (22.2%, 8 of 36). All 36 clinical isolates and their transformants produced a β-lactamase with an apparent pI of 6.65 (data not shown). Based on IEF results and on the previous reports of OXA-23-producing A. baumannii (6) and Proteus mirabilis (2), we suspected that the OXA-23 carbapenemase (class D) could be involved in imipenem resistance.
FIG. 1.
Modified cloverleaf test results for a representative OXA-23-producing isolate (A. baumannii K10708) (A), a VIM-2-producing positive control (A. genomospecies 3 YMC 99/11/160) (B), and a non-OXA-23-producing strain (A. baumannii ATCC 19606) (C). A Mueller-Hinton agar plate was incubated with E. coli ATCC 25922. An imipenem disk was put in the center, and test isolates were streaked from the edge of the disk to the periphery of the plate, followed by overnight incubation.
TABLE 2.
Characterization of imipenem-resistant A. baumannii isolates harboring the blaOXA-23 gene
| Isolate | Datea of isolation | Wardb | Isolate sourcec | Isolate resistance, non-β-lactamsd | Synergy test
|
|
|---|---|---|---|---|---|---|
| Clover leaf | EDTA disk | |||||
| K10708 | 16/1/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K11468 | 30/1/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K12817 | 28/2/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm | + | − |
| K13710 | 15/3/2003 | GS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K13994 | 20/3/2003 | PI | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14190 | 25/3/2003 | UR | Pus | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14249 | 27/3/2003 | NS | Urine | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14615 | 3/4/2003 | CS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14910 | 10/4/2003 | CS | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14911 | 11/4/2003 | NS | Sputum | Amik, Cpfx, Gm, Tet, Tm | + | − |
| K15034 | 13/4/2003 | KI | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15289 | 17/4/2003 | PI | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15348 | 18/4/2003 | NS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15657 | 25/4/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15929 | 1/5/2003 | NS | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K14687 | 4/4/2003 | GS | Wound | Amik, Gm, Tet, Tm, Tp/Sx | + | − |
| K16386 | 19/5/2003 | OS | Wound | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K18031 | 9/6/2003 | NS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K17885 | 10/6/2003 | PI | SCVP tip | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K17877 | 8/6/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K18909 | 26/6/2003 | NS | Urine | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K19654 | 11/7/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K19706 | 12/7/2003 | NE | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K20180 | 21/7/2003 | NS | IV catheter | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15173 | 14/4/2003 | GS | Wound | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15382 | 18/4/2003 | PI | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K15647 | 24/4/2003 | GS | Wound | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21090 | 4/8/2003 | CS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21161 | 5/8/2003 | OS | Wound | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21115 | 6/8/2003 | NE | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21312 | 7/8/2003 | NS | Urine | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21503 | 13/8/2003 | NS | Urine | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K21846 | 20/8/2003 | CI | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K22202 | 27/8/2003 | NS | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K22222 | 27/8/2003 | NS | E-tube | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| K16544 | 15/5/2003 | NS | Sputum | Amik, Cpfx, Gm, Tet, Tm, Tp/Sx | + | − |
| YMCe | + | + | ||||
| 19606f | − | − | ||||
Dates are presented as day/month/year.
CI, cardiac internal medicine; CS, cardiac surgery; GS, general surgery; KI, kidney internal medicine; NE, neurology; NS, neurosurgery; OS, orthopedic surgery; PI, pulmonary internal medicine; UR, urology.
E-tube, endotracheal tube; IV, intravenous; SCVP, subclavian vein catheter.
Amik, amikacin; Cpfx, ciprofloxacin; Gm, gentamicin; Tet, tetracycline; Tm, tobramycin; Tp/Sx, trimethoprim-sulfamethoxazole (1:19).
Imipenem-sensitive A. baumannii ATCC 19606.
VIM-2-producing A. genomospecies 3 YMC 99/11/160.
Molecular characterization of OXA-23-producing A. baumannii isolates.
Neither transfer nor curing of imipenem resistance was achieved for any imipenem-resistant isolate, despite multiple attempts. blaOXA-carrying plasmid DNA was apparently not detectable either in plasmid preparation without contaminated bacterial chromosomal DNA by plasmid-safe ATP-dependent DNase or in whole genomic DNA preparations from 36 isolates. In a Southern blot experiment carried out with the genomic DNA of 36 isolates, the blaOXA-23 probe hybridized to the band of chromosomal DNA. Thirty-six clinical isolates gave a PCR product with the OXA-23F/OXA-23R primer pair but not with other primer pairs (OXA-24F/OXA-24R, IMP-F/IMP-R, and VIM-F/VIM-R). A 1,058-bp PCR product (between OXA-23F and OXA-23R primer) for blaOXA-23-like cluster genes was obtained. Taking into account the resistance phenotypes of 36 clinical isolates, the resistance genotypes of these isolates were analyzed by direct sequencing of the PCR-amplified fragments specific for blaOXA-23-like cluster genes. On the basis of DNA sequencing, 36 clinical isolates and their transformants were determined to harbor the blaOXA-23 gene. The OXA-23 carbapenemase was involved in the imipenem resistance as previously described (5, 6).
PFGE analysis.
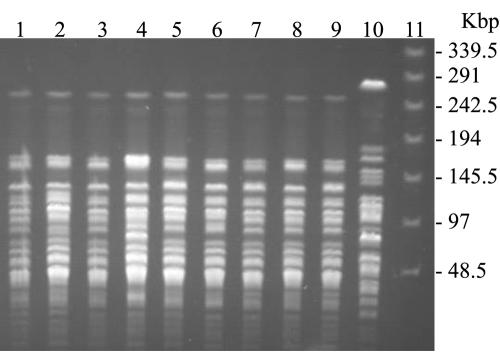
SmaI macrorestriction analysis was conducted on 36 OXA-23-producing isolates obtained from the 36 patients hospitalized in nine different wards. The macrorestriction pattern of the isolates consisted of 22 discernible bands ranging from 10 to 260 kbp (Fig. 2). Thirty-six isolates showed very closely related patterns with one- or two-fragment variations. According to the report of Zarrilli et al. (22), the PFGE analysis showed the presence of an epidemic clone and that the OXA-23-producing isolates recovered from January to August 2003 were of a clonal origin.
FIG. 2.
Genotyping of A. baumannii clinical isolates. PFGE fingerprints of A. baumannii isolates are shown. Lanes 1 to 9, isolates from nine different wards (K10708 from neurology, K13994 from pulmonary internal medicine, K14190 from urology, K14910 from cardiac surgery, K15034 from kidney internal medicine, K18909 from neurosurgery, K15173 from general surgery, K21161 from orthopedic surgery, and K21846 from cardiac internal medicine, respectively); lane 10, imipenem-sensitive A. baumannii ATCC 19606; lane 11, multimer of phage lambda DNA (48.5 kbp) molecular mass markers. The sizes in kilobase pairs of lambda DNA molecular mass markers are indicated on the right of the panel.
DISCUSSION
Since 2003, a high degree of imipenem resistance incidence has been observed among nosocomial A. baumannii isolates in Korea. Therefore, it was our purpose to ascertain whether the resistance was due to the dissemination of an imipenem resistance determinant and/or to the spread of one A. baumannii clone. Microbiological assays showed that imipenem was inactivated by imipenem-resistant A. baumannii isolates. An alteration of carbapenemase activity was not observed in the presence of EDTA, suggesting the production of a non-metallo-β-lactamase. OXA class D carbapenemases have been identified in A. baumannii collected in UK (OXA-23) (6), Spain (OXA-24, -25, and -40) (1, 3, 6, 7), Belgium (OXA-26) (18), and Singapore (OXA-27) (18). A. baumannii is an emerging nosocomial pathogen that is in part due to the capacity of acquiring resistance to multiple antimicrobial agents. Because OXA-23-producing A. baumannii confers resistance to most β-lactams, such as imipenem, aztreonam, ceftazidime, and cefepime, a limited number of antimicrobial agents maintain reliable activity against OXA-23-producing A. baumannii, including polymyxin, sulbactam, and minocycline (5). However, our studies revealed that 78% of OXA-23-producing A. baumannii isolates showed resistance to ampicillin-sulbactam. Thus, it is important to monitor and control the spread of OXA-23-producing A. baumannii conferring resistance to most β-lactams.
We detected 52 different isolates of imipenem-resistant A. baumannii from Korean patients. Of 52 isolates, 36 produced OXA-23. The imipenem resistance of the remaining isolates may be due to reduced permeability of the outer membrane, alteration in penicillin biding proteins (18), and/or recently reported OXA derivatives such as OXA-58 (GenBank accession number AY570763). The clonal relatedness of A. baumannii Korean isolates was evaluated by PFGE. All of the isolates presented a very similar antibiotic resistance profile and a very similar DNA fingerprinting pattern. Such a correspondence of phenotypic and genotypic characteristics can be explained by a common clonal origin. Our results indicate that the observed imipenem resistance among 36 Korean A. baumannii isolates is due to the spread of an OXA-23-producing clone. Although a clonal outbreak due to an OXA-23-producing strain has been reported in Brazil (5), this is the first report of an OXA-23-producing clone in Asia. The clinical significance of these isolates, which were widespread in Korea, is of great importance, since clinicians are advised against the use of extended-spectrum cephalosporins, aztreonam, cephamycins, imipenem, and aminoglycosides. This observation emphasizes the importance of having effective control measures in Asian hospitals, such as early detection of colonized patients, isolation procedures, and a judicious use of antibiotics.
Acknowledgments
This study was supported by a grant from BioGreen 21 Program, Rural Development Administration, Republic of Korea; by a research grant from Next-Generation Growth Engine R&D Program, Kyeonggi Provincial Government; and by Korea Research Foundation Grant KRF-2004-042-E00117.
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27 molecular class D β-lactamase associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou, G., A. Oliver, and J. Martínez-Beltrán. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastre, J., and J. L. Trouillet. 2000. Problem pathogens (Pseudomonas aeruginosa and Acinetobacter). Semin. Respir. Infect. 15:287-298. [DOI] [PubMed] [Google Scholar]
- 5.Dalla-Costa, L. M., J. M. Coelho, H. A. P. H. M. Souza, M. E. S. Castro, C. J. N. Stier, K. L. Bragagnolo, A. Rea-Neto, S. R. Penteado-Filho, D. M. Livermore, and N. Woodford. 2003. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J. Clin. Microbiol. 41:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donald, H. M., W. Scaife, S. G. B. Amyes, and H.-K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornstein, M., C. Sautjeau-Rostoker, J. Péduzzi, A. Vessières, L. T. H. Hong, M. Barthélémy, M. Scavizzi, and R. Labia. 1997. Oxacillin-hydrolyzing β-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol. Lett. 153:333-339. [DOI] [PubMed] [Google Scholar]
- 9.Jeong, S. H., I. K. Bae, J. H. Lee, S. H. Sohn, G. H. Kang, G. J. Jeon, Y. H. Kim, B. C. Jeong, and S. H. Lee. 2004. Molecular characterization of extended-spectrum β-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean nationwide survey. J. Clin. Microbiol. 42:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong, S. H., K. Lee, Y. Chong, J. H. Yum, S. H. Lee, H. J. Choi, J. M. Kim, K. H. Park, B. H. Han, S. W. Lee, and T. S. Jeong. 2003. Characterization of a new integron containing VIM-2, a metallo-β-lactamase gene cassette, in a clinical isolate of Enterobacter cloacae. J. Antimicrob. Chemother. 51:397-400. [DOI] [PubMed] [Google Scholar]
- 11.Lee, K., Y. Chong, H. B. Shin, Y. A. Kim, D. Young, and J. H. Yum. 2000. Modified Hodhe and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88-102. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S. H., J. Y. Kim, G. S. Lee, S. H. Cheon, Y. J. An, S. J. Jeong, and K. J. Lee. 2002. Characterization of blaCMY-11, an AmpC-type plasmid-mediated β-lactamase gene in a Korean clinical isolate of Escherichia coli. J. Antimicrob. Chemother. 49:269-273. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. H., J. Y. Kim, S. H. Shin, S. K. Lee, M. M. Choi, I. Y. Lee, Y. B. Kim, J. Y. Cho, W. Jin, and K. J. Lee. 2001. Restriction fragment length dimorphism-PCR method for the detection of extended-spectrum β-lactamases unrelated to TEM- and SHV-types. FEMS Microbiol. Lett. 200:157-161. [DOI] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 2002. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs 3:218-224. [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standards, 6th ed. Document M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 16.Sambrook, J., and D. W. Russell. 2001. Extraction and purification of plasmid and screening of bacterial colonies by hybridization, p. 1.40-1.92. In J. Argentine, N. Irwin, K. A. Jassen, S. C. M. Zierler, N. Mclnerny, D. Brown, and S. Schaefer (ed.), Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Schreckenberger, P. C., and A. Von Graevenitz. 1999. Acinetobacter, Achromobacter, Alcaligenes, Moraxella, Methylobacterium, and other non-fermentative gram-negative rods, p. 539-560. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 18.Silva, G. J., S. Quinteira, E. Bértolo, J. C. Sousa, L. Gallego, A. Duarte, and L. Peixe. 2004. Long-term dissemination of an OXA-40 carbapenemase-producing Acinetobacter baumannii clone in the Iberian Peninsula. J. Antimicrob. Chemother. 54:255-258. [DOI] [PubMed] [Google Scholar]
- 19.Simor, A. E., S. F. Bradley, L. J. Strausbaugh, K. Crossley, and L. E. Nicolle. 2002. An outbreak due to multiresistant Acinetobacter baumannii in a burn unit: risk factors for acquisition and management. Infect. Control Hosp. Epidemiol. 23:261-267. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yong, D., J. H. Shin, S. Kim, S. Kim, Y. Lim, H. H. Yum, K. Lee, Y. Chong, and A. Bauernfeind. 2003. High prevalence of PER-1 extended-spectrum β-lactamase-producing Acinetobacter spp. in Korea. Antimicrob. Agents Chemother. 47:1749-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarrilli, R., M. Crispino, M. Bagattini, E. Barretta, A. D. Popolo, M. Triassi, and P. Vollari. 2004. Molecular epidemiology of sequential outbreaks of Acinetobacter baumannii in an intensive care unit shows the emergence of carbapenem resistance. J. Clin. Microbiol. 42:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]