Abstract
Forty-eight Streptococcus pneumoniae isolates recovered from sputum samples from 26 cystic fibrosis (CF) patients attending our CF unit (1995 to 2003) were studied. Mean yearly incidence of isolation was 5.5%, and all were strains recovered from young patients (≤12 years). The isolation was linked to clinical exacerbation in 35% of the cases, but only 27% of these were not accompanied by other CF pathogens. Fifty percent of the patients presented with two to four isolates over the studied period. Pulsed-field gel electrophoresis-SmaI digestion revealed a high heterogeneity (32 pulsotypes among 48 isolates) and the persistence over a 6-month period of a single clone (clone A) in two patients. This clone, presenting a varied multiresistance phenotype, was identified as the Spain23F-1 clone and was also recognized in six other patients, including two out of nine patients from the CF unit of Sant Joan de Dèu Hospital, Barcelona, Spain. In our isolates, 16 different serotypes were recognized, the most frequent being 23F (33.3%), 19F (18.8%), 6A (6.2%), and 6B (6.2%). High overall resistance rates were observed: to penicillin, 73%; to cefotaxime, 33%; to erythromycin, 42%; to tetracycline, 58%; to chloramphenicol, 48%; and to trimethoprim-sulfamethoxazole, 67%. Resistance to fluoroquinolones was not detected. Multiresistance was a common feature (60%). The percentage of S. pneumoniae strains with increased frequencies of mutation to rifampin resistance (≥7.5 × 10−8) was significantly higher (P = 0.02) in CF (60%) than among non-CF (37%) isolates in the same institution (M. I. Morosini et al., Antimicrob. Agents Chemother. 47:1464-1467, 2003). Even though a clear association with acute exacerbations could not be observed, long-term clonal persistence and variability, high frequency of antibiotic resistance, and hypermutability indicate the plasticity for adaptation of S. pneumoniae to the CF lung environment.
Streptococcus pneumoniae is the causative agent of severe infections, many of which may have strong epidemiological impacts, such as meningitis, otitis media, pneumonia, and sinusitis. The primary site of colonization of this pathogen is the nasopharyngeal epithelium, where it can persist as part of the commensal flora. The main cause of pneumonia is progression of the organism towards the lungs through aspiration, although its consolidation requires additional events (10, 38). The pathogenic role of S. pneumoniae in cystic fibrosis (CF) is controversial, and very few reports document its potential involvement in pulmonary damage (5, 15, 16). Gilligan (14) stated in 1991 that S. pneumoniae seems to play, if at all, a role secondary to that of other organisms that are clearly implicated in lung infections associated with CF. Studies in the early 1960s and 1970s established that S. pneumoniae isolates are occasionally recovered from the respiratory secretions of CF patients (15, 17, 18, 22). Isolation occurred predominantly in infants and young children but not at a frequency higher than in non-CF patients. The isolation of S. pneumoniae was associated with 5% of the acute-exacerbation episodes (16).
Although S. pneumoniae, in addition to nonencapsulated Haemophilus influenzae and respiratory viruses, has been considered to predispose patients to acute and chronic airway infections for other commonly encountered CF organisms, including mucoid Pseudomonas aeruginosa and Staphylococcus aureus, (19), recent studies and reviews have paid little attention to this pathogen (13, 21). Even the Cystic Fibrosis Foundation does not record the frequency of isolation of this organism in its annual reports (9). The population structure and the corresponding antibiotic susceptibility patterns of commonly encountered CF isolates have been repeatedly studied (30, 34) but have never included S. pneumoniae isolates from CF patients.
CF patients may be chronically colonized with mucoid Pseudomonas aeruginosa, Staphylococcus aureus, and nonencapsulated H. influenzae, but this does not seem to be the case with S. pneumoniae (14, 30). Chronic colonization, high bacterial loads, and exposure to frequent antibiotic courses have traditionally been considered as important factors for antimicrobial resistance development in CF pathogens. More recently, the role of mutator populations has been highlighted to accelerate this process in CF patients (27, 30, 31). We and other authors have found that P. aeruginosa, S. aureus, and H. influenzae from CF patients are more frequently hypermutable than those from non-CF patients, and an association with mutational antibiotic resistance has been demonstrated. Although the presence of mutators has also been studied in S. pneumoniae isolates in non-CF patients (23), their frequency in isolates from CF patients has not been documented.
The population structure of S. pneumoniae isolates consecutively recovered from CF patients is studied in this work to evaluate the possible persistence of the same strain and its potential role in pulmonary exacerbations. The serotype distribution, antimicrobial susceptibility pattern, and mutation frequencies of these isolates were also determined. This study was made possible by a long-term follow-up of patients (during 1995 through 2003).
MATERIALS AND METHODS
Bacterial isolates, patients, and medical chart review.
From 1995 to 2003, a total of 26 CF patients (10 females and 16 males), clinically and microbiologically followed in the CF unit of the Ramón y Cajal University Hospital in Madrid, Spain, had at least one positive sputum or respiratory secretion culture for S. pneumoniae. Medical charts were reviewed, demographics and antimicrobial use in the 6 months prior to pneumococcal isolation were determined, and clinical and microbiological data were collected. The potential correlation of S. pneumoniae colonization/infection with pulmonary exacerbation was studied. The presence of P. aeruginosa, as well as other commonly encountered CF pathogens, was also recorded as outcome criteria for the evaluation of the progression of pulmonary disease. Microbiological data were recovered from 26 patients, and clinical data were recovered from only 20 patients. The nine S. pneumoniae isolates obtained from eight CF patients followed at Sant Joan de Dèu Hospital in Barcelona, Spain, from 1997 to 2003 were included only in the population structure study.
Bacterial isolation and identification.
Respiratory samples collected during both scheduled clinical assessments and pulmonary exacerbations were homogenized with N-acetyl-cysteine and seeded in Columbia 5% blood, MacConkey, mannitol-salt, and a selective Burkholderia cepacia agar medium with a quantitative technique (37) and incubated in ambient air for 24 h at 35°C. In addition, bacitracin-chocolate agar was seeded and incubated in a 5% CO2 atmosphere for 48 h at 35°C. S. pneumoniae isolates were identified on the basis of standard laboratory procedures, including colony morphology on blood agar, the optochin test, and the sodium deoxycholate solubility test (33). Isolates were stored in skim milk at −70°C and reidentified when they were recovered from storage for further studies. The National Center of Microbiology of Spain (Instituto Carlos III, Majadahonda, Madrid) performed the serotyping with the Quellung reaction, using antisera from the Statens Seruminstitut, Copenhagen, Denmark.
Bacterial-population analysis.
Population structure was studied by pulsed-field gel electrophoresis (PFGE). Chromosomal DNA was prepared according to the classical gram-positive protocol with some modifications (R. Del Campo et al. unpublished data). DNA restriction was made with the endonuclease SmaI (Amersham Biosciences Europe GmbH, Freiburg, Germany). The electrophoresis was carried out in a CHEF DR-III apparatus (Bio-Rad, Birmingham, United Kingdom) for 23 h at 14°C, and the following settings were applied at 6 V/cm: 1 to 30 seconds. Wild-type laboratory R6 S. pneumoniae strain and a lambda ladder PFGE marker (New England Biolabs Inc., Beverly, MA) were used as an internal pneumococcal control and a molecular size standard, respectively. Genetic relatedness among the strains was analyzed visually and interpreted by the Phoretix 5.0 software (Nonlinear Dynamics Ltd., United Kingdom) using Dice's coefficient to construct the corresponding dendrogram.
Antimicrobial susceptibility testing.
MICs of penicillin, ampicillin, amoxicillin-clavulanic acid, cefuroxime, cefotaxime, cefepime, meropenem, vancomycin, teicoplanin, erythromycin, clindamycin, tetracycline, ciprofloxacin, levofloxacin, trimethoprim, chloramphenicol, and rifampin were determined by the standard NCCLS technique (25) using cation-adjusted Mueller-Hinton broth supplemented with 5% lysed horse blood (Oxoid, Basingstoke, United Kingdom). Susceptibility panels were incubated overnight at 35°C in ambient air after inoculation. S. pneumoniae ATCC 49619 and S. aureus 29213 were used as controls in each run. MIC breakpoints recommended by the NCCLS were considered for all antibiotics (26). For cefotaxime, the NCCLS meningitis criterion was applied.
Determination of mutation frequencies.
Three to five colonies from an overnight culture in 5% sheep blood agar plates (Oxoid) were resuspended in 10 ml of brain heart infusion (BHI) broth (Oxoid) and incubated for 6 to 8 h at 35°C without shaking. For total viable count determination, an aliquot of 100 μl was diluted in BHI broth and plated onto 5% sheep blood agar plates. The remaining culture was centrifuged for 5 min at 2,500 × g. The pellet was resuspended in 500 μl of BHI broth and seeded on 5% sheep blood agar plates containing 2 μg/ml of rifampin. Mutation frequency values are reported as the proportion of rifampin-resistant colonies (detected after 48 to 72 h of incubation in a 5% CO2 atmosphere) versus total viable-cell counts. Results correspond to the mean value obtained in duplicate experiments, each with duplicate colony counts.
We estimated only mutation frequencies exceeding 5 × 10−9, considering lower frequencies of mutation as possibly artifactual, due to the impaired growth of cells under the applied selective conditions. A strain was considered a mutator strain when its frequency was equal to or higher than 7.5 × 10−8. This breakpoint was derived from the value which separated the two main populations (normomutable and hypermutable) in a previous study of 200 non-CF isolates (23). The DNA repair-proficient normomutable (2.6 × 10−8) strain R800 (ami+ aliA+ aliB+ hex+) and its DNA repair-deficient hypermutable (1.06 × 10−7) derivative strain R310 (hexA::synSpc) (1) were used as controls for normo- and hypermutator populations, respectively.
Statistic analysis.
Statistical significance for comparison proportions was calculated by the chi-square test, and quantitative values were compared by Student's t test. A P of <0.05 was considered statistically significant.
RESULTS
Epidemiological background.
A total of 48 S. pneumoniae isolates were recovered from 26 CF patients (one to four isolates per patient) who attended the CF unit of the Ramón y Cajal University Hospital (Madrid, Spain) from 1995 to 2003. The dates of isolation, genders, and ages of patients are presented in Table 1. The yearly incidence of patients infected or colonized with S. pneumoniae ranged from 2.4% to 13.5% (mean value from 1995 to 2003, 5.5%). For six patients, medical charts were not available and only data from microbiology laboratory files were used. S. pneumoniae isolates from each patient were always recovered during infancy or early childhood (range, <1 to 12 years; mean ± standard deviation, 5.3 ± 3.6 years). Thirteen patients (50%) presented with a single episode of pneumococcal colonization or infection over the studied period. For six patients, S. pneumoniae was isolated two different times (23%), for five other patients, it was isolated three different times (19%), and finally, for two patients it was isolated four different times (8%). No differences in the median ages of acquisition for patients with single or repetitive episodes were observed. Repetitive episodes were separated for a period of time ranging from 1 month to 53 months (mean ± standard deviaition, 16 ± 16 months). Microbiological data revealed that S. pneumoniae isolates in our CF patients were recovered without the concomitant presence of other CF pathogens in 18 episodes (18/48, 37.5%). The corresponding figures of isolation with other pathogens were 16/48 (33.3%) for S. aureus, 15/48 (31.2%) for H. influenzae, 11/48 (22.9%) for P. aeruginosa, 4/48 (8.3%) for Aspergillus spp., and 2/48 (4.2%) for Achromobacter xylosoxidans.
TABLE 1.
Demographic data, cocolonizing organisms, and characteristics of S. pneumoniae isolates from CF patients followed at Ramón y Cajal University Hospital in Madrid, Spain
| Patient (gender) | Age (yrs) | Exacerbation (bacterial count)a | Cocolonization organism(s)b | Isolate | Mo/yr of isolation | PFGE pulsotype | Serotypec | Antimicrobial resistance phenotyped |
|---|---|---|---|---|---|---|---|---|
| 1 (M) | 4 | Yes (2 × 107) | 022507 | 04/1995 | AA-1 | 37 | ||
| 2 (F) | NAe | NA (9 × 106) | Sa | 043433 | 06/1995 | R | 9V | Pen, SxT |
| 3 (M) | NA | NA (2 × 105) | 053116 | 07/1995 | A-4 | 23F | Pen, Ery, Tet, SxT, Clo | |
| 4 (F) | 11 | No (5 × 105) | Sa, Ax | 098489 | 10/1995 | A-4 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo |
| 5 (M) | NA | NA (2 × 104) | 070356 | 06/1996 | A-5 | 23F | Pen, SxT, Clo | |
| 6 (F) | 5 | No (9 × 105) | 080034 | 07/1998 | G | 19F | Tet, SxT | |
| 7 (M) | 2 | No (1 × 105) | Hi | 053437 | 03/1999 | B | 23F | Pen, Ctx, Tet, SxT, Clo |
| 8 (M) | NA | NA (3 × 106) | 111007 | 11/1999 | AC | NT | Pen, Ery, Tet, SxT | |
| 9 (M) | NA | NA (2 × 105) | 056078 | 02/2000 | O | 6B | Clo | |
| 10 (M) | 6 | No (5 × 103) | Hi | 065922 | 04/2000 | C | 23F | |
| 11 (F) | 2 | No (7 × 105) | 076449 | 06/2000 | H | 19F | Ery, Tet, Clo | |
| 12 (F) | 3 | No (1 × 107) | Pa, Hi | 063986 | 03/2001 | J | 19F | Pen, Ery, Tet, SxT |
| 13 (F) | 11 | No (1 × 106) | Asp, Hi | 113881 | 11/2001 | L | 3 | |
| 14 (M) | 9 | Yes (5 × 107) | Sa, Asp | 082160 | 08/1996 | N | 6B | Tet, SxT, Clo |
| No (4 × 106) | Pa, Asp | 087207 | 09/2000 | AA | 37 | |||
| 15 (M) | 5 | No (4 × 103) | 052740 | 02/1997 | K | 3 | ||
| No (1 × 105) | Sa | 107347 | 12/1999 | U | 14 | |||
| 16 (F) | 8 | No (1 × 106) | Sa | 053199 | 02/2000 | M | 6A | |
| No (4 × 105) | Hi | 075528 | 06/2000 | M-1 | 6A | |||
| 17 (M) | 1 | Yes (3 × 105) | 056761 | 02/2001 | I | 19F | Pen, Ery, Tet, SxT, Clo | |
| No (3 × 104) | 059891 | 03/2001 | I-1 | 19F | Pen, Ery, Tet, SxT | |||
| 18 (F) | 12 | No (2 × 103) | Pa, Hi | 026391 | 01/2002 | Q | 7 | |
| Yes (1 × 108) | Pa, Sa | 022494 | 01/2003 | AB | NT | Pen, Ctx, Ery, Tet, SxT, Clo | ||
| 19 (M) | 10 | No (ND) | Sa, Asp | 115097 | 10/2002 | P | 6B | Pen, Ctx, Ery, Tet |
| No (2 × 106) | Sa | 024689 | 01/2003 | A-1 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo | ||
| 20 (F) | 3 | No (5 × 108) | Pa, Hi | 017958 | 03/1995 | A-4 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo |
| No (8 × 107) | Sa, Hi | 032605 | 05/1995 | A-4 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo | ||
| Yes (6 × 107) | Sa, Hi | 098347 | 10/1995 | A-4 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo | ||
| 21 (M) | 4 | No (1 × 107) | Sa, Hi | 020520 | 03/1995 | D | 19F | Pen, Ery, Tet, SxT, Clo |
| No (3 × 104) | 093842 | 11/1996 | X | 23B | ||||
| No (7 × 105) | Hi | 049800 | 01/1997 | Z | 31 | |||
| 22 (F) | 4 | NA (4 × 105) | 046932 | 06/1995 | Y | 24 | ||
| NA (6 × 105) | Sa, Hi | 066486 | 05/1996 | S | 11 | SxT | ||
| NA (2 × 107) | 050045 | 01/1998 | AD | 6A | Pen, Ctx, Ery, Tet, SxT, Clo | |||
| 23 (M) | 3 | No (ND) | Pa | 032740 | 01/1998 | A-2 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo |
| No (1 × 105) | Pa, Sa | 057075 | 03/1998 | T | 14 | Pen, Ctx, Ery, SxT | ||
| No (9 × 105) | Pa | 059480 | 02/2001 | A-3 | 23F | Pen, Ctx, Tet, SxT, Clo | ||
| 24 (M) | 2 | No (3 × 107) | Pa, Ax | 083346 | 07/1997 | W | 19A | SxT |
| No (7 × 105) | Hi | 052376 | 02/1998 | F | 19F | Pen, Ery, Tet, SxT, Clo | ||
| No (2 × 106) | 081643 | 07/1998 | A | 23F | Pen, Ctx, Tet, SxT, Clo | |||
| 25 (M) | 7 | Yes (1 × 108) | Sa, Hi | 036152 | 05/1995 | E | 19F | Tet, SxT, Clo |
| Yes (3 × 108) | 111939 | 11/1995 | AE | 22 | Pen, SxT, Clo | |||
| Yes (1 × 107) | 095838 | 10/1997 | V | 15B | ||||
| Yes (4 × 107) | 054335 | 01/2001 | AF | 19F | ||||
| 26 (M) | <1 | Yes (2 × 107) | 075357 | 07/1996 | A-5 | 23F | Pen, Ctx, Tet, SxT, Clo | |
| Yes (5 × 105) | Pa, Sa | 092170 | 10/1996 | A | 23F | Pen, Tet, SxT, Clo | ||
| Yes (7 × 105) | Pa, Sa | 093884 | 11/1996 | A | 23F | Pen, Tet, SxT, Clo | ||
| No (4 × 105) | Hi | 105118 | 12/2000 | A-3 | 23F | Pen, Ctx, Ery, Tet, SxT, Clo |
Number of CFU per milliliter of sputum. ND, not done.
Sa, S. aureus; Ax, A. xylosoxydans; Hi, H. infuenzae; Pa, P. aeruginosa; Asp, Aspergillus spp.
NT, nontypeable.
Pen, penicillin; SxT, trimethoprim-sulfametoxazole; Ery, erythromycin; Tet, tetracycline, Clo, chloramphenicol; Ctx, cefotaxime.
NA, not available.
Clinical findings.
The retrospective analysis of medical charts from 20 patients revealed that only 7 presented evidence of acute exacerbation at the time of isolation of S. pneumoniae (35%). One of these patients had a single S. pneumoniae isolate along the entire period studied, and six patients had more than one isolate. In one of these patients acute exacerbations were consistently associated with the repetitive isolation of S. pneumoniae. When exacerbation was present (12 episodes for seven patients), S. pneumoniae was not recovered with other pathogens in 6 episodes (6/12, 50%), whereas it coexisted with S. aureus and P. aeruginosa in 3 episodes (3/12, 25%), with S. aureus and H. influenzae in two episodes (2/12, 17%), and with S. aureus and Aspergillus spp. in one episode (1/12, 8%) (Table 1). Bacterial counts (Table 1) showed that the bacterial loads of S. pneumoniae during exacerbation episodes ranged from 1 × 107 to 3 × 108 CFU per ml of sputum sample (geometric mean, 1 × 107 CFU/ml) and from 2 × 103 to 5 × 108 (geometric mean, 5 × 105 CFU/ml) during the nonexacerbation periods. Despite clear differences in geometric mean values, no statistical difference was observed.
On the other hand, chart review revealed that during the 6 months prior to pneumococcal isolation, patients received in monotherapy or in combination a β-lactam (60%), fluoroquinolone (ciprofloxacin) (40%), or macrolide (20%) antibiotic.
Population structure.
The serotype distribution determined for 48 S. pneumoniae isolates from Ramón y Cajal University Hospital and PFGE results are presented in Table 1. Serotype 23F was the most frequently observed (16 isolates), followed by serotypes 19F (9 isolates), 6A (3 isolates), and 6B (3 isolates). Other serotypes were represented by one or two isolates. PFGE-SmaI digestion and the subsequent computer analysis clustered the 48 CF isolates in a total of 32 different pulsotypes. In 28 out of the 32 pulsotypes, a single isolate was present, whereas pulsotypes I, M, and AA (serotypes 19F, 6A, and 37, respectively) comprised two CF S. pneumoniae isolates each, and pulsotype A (serotype 23F) represented a total of 14 isolates (obtained from eight CF patients from 1995 to 2003). Within pulsotype A, five different genetically close related variants (A-1, A-2, A-3, A-4, and A-5) were observed. The PFGE-SmaI digestion-based genetic relationships were also investigated in the Sant Joan de Dèu Hospital CF S. pneumoniae collection (nine isolates from eight patients). Interestingly, two isolates from two patients corresponded also to two different variants of pulsotype A (A-6 and A-7), denoting a close relatedness with those isolates previously characterized in the Ramón y Cajal University Hospital collection (Fig. 1). It is of note that this pulsotype A, detected in both hospitals (which are geographically separated), corresponded to the major clone Spain23F-1 and presented a uniform resistance phenotype. This included resistance to penicillin, chloramphenicol, and trimethoprim-sulfamethoxazole. Other subtypes within this clone presented various susceptibility patterns to other antimicrobials (see below).
FIG. 1.
Dendrogram of genetic relatedness among CF S. pneumoniae isolates. aHSJD, isolates from the Hospital Sant Joan de Dèu collection; bNT, nontypeable isolate.
Table 1 also shows the evolution of the different pulsotypes in each patient. The evolution of pulsotype A was particularly interesting in one patient (patient 26), in whom the original clone and subtype A-5 coexisted in 1996, whereas after nearly 5 years, in 2000, the A-3 variant was detected.
Antimicrobial susceptibility and multiresistance.
Susceptibility results and the different antibiotic resistance combinations are shown in Tables 1. High rates of antibiotic resistance (intermediate plus resistant) were observed for antibiotics commonly used for the treatment of infections due to S. pneumoniae, such as penicillin (73.0%), cefotaxime (33.4%), erythromycin (41.7%), tetracycline (58.4%), chloramphenicol (48.0%), and trimethoprim-sulfamethoxazole (66.6%) (Table 2). Resistance to glycopeptides and quinolones was not observed, and only one isolate was resistant to rifampin. To analyze the multiresistance status, combinations of phenotypic resistance markers to penicillin, cefotaxime, erythromycin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole were considered. More than 60% of isolates were resistant to three or more antibiotics, whereas 30% were susceptible to all antibiotics tested. It should be noted that nearly 50% of isolates were resistant to four or more antibiotics. Among these isolates the most frequent multiresistance combination was penicillin, tetracycline, trimethoprim-sulfamethoxazole, and chloramphenicol.
TABLE 2.
Antimicrobial susceptibilities of S. pneumoniae isolates from CF patients followed at Ramón y Cajal University Hospital in Madrid, Spain
| Antimicrobial agent | MIC (μg/ml)
|
% of isolates
|
||||
|---|---|---|---|---|---|---|
| Range | 50 | 90 | Susceptible | Intermediate | Resistant | |
| Penicillin | ≤0.03-4 | 1 | 2 | 27.0 | 27.0 | 46.0 |
| Ampicillin | ≤0.06-4 | 1 | 4 | —a | — | — |
| Amoxicillin-clavulanate | ≤0.5->4/2 | ≤0.5 | 1/0.5 | 94.0 | 2.0 | 4.0 |
| Cefuroxime | ≤0.12->8 | 2 | 4 | 37.5 | 12.5 | 50.0 |
| Cefotaxime | ≤0.03->4 | 0.5 | 1 | 66.6 | 29.0 | 4.4 |
| Cefepime | ≤0.06->2 | 1 | 2 | 43.8 | 31.2 | 25.0 |
| Meropenem | ≤0.12-1 | 0.5 | 1 | 48.0 | 39.5 | 12.5 |
| Vancomycin | ≤0.25-0.5 | ≤0.25 | 0.5 | 100.0 | 0 | 0 |
| Teicoplanin | ≤0.25 | ≤0.25 | ≤0.25 | 100.0 | 0 | 0 |
| Erythromycin | ≤0.25->1 | >1 | >1 | 58.4 | 0 | 41.6 |
| Clarithromycin | ≤0.25->16 | ≤0.25 | >16 | 62.5 | 2.0 | 35.5 |
| Clindamycin | ≤0.12->1 | ≤0.12 | >1 | 70.9 | 4.1 | 25.0 |
| Tetracycline | ≤0.25->4 | >4 | >4 | 41.7 | 0 | 58.3 |
| Ciprofloxacin | ≤0.06-2 | 1 | 1 | — | — | — |
| Levofloxacin | ≤0.25-2 | 1 | 1 | 100.0 | 0 | 0 |
| SxTb | ≤0.5/9->2/38 | >2/38 | >2/38 | 33.4 | 4.1 | 62.5 |
| Chloramphenicol | ≤2->4 | >4 | >4 | 52.0 | 0 | 48.0 |
| Rifampin | ≤0.5->2 | <0.5 | <0.5 | 98.0 | 0 | 2.0 |
—, no NCCLS criteria available.
SxT, trimethoprim-sulfametoxazole.
Several antibiotic resistance phenotypes were observed within isolates belonging to clone A (Table 1). The international clone Spain23F-1 uniformly shows resistance to penicillin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole (11). Among the clone A isolates, this resistance pattern was detected in two out of three isolates; other isolates also showed resistance to cefotaxime. Other variants also included resistance to erythromycin and tetracycline.
Mutation frequency studies.
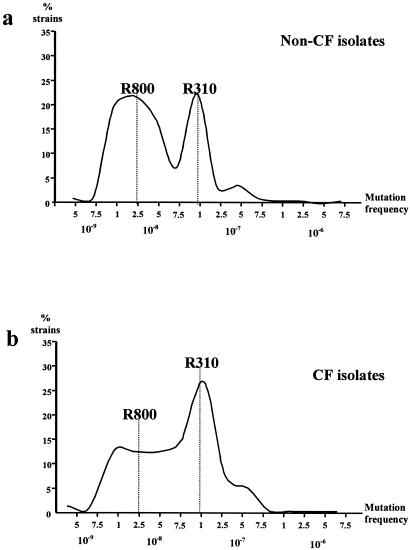
Frequencies of mutation to rifampin in the CF S. pneumoniae isolates are shown in Fig. 2. Considering all isolates, we obtained frequencies of mutation ranging from 4.5 × 10−10 to 6.8 × 10−7, but only mutation frequencies exceeding 5.10−9 were considered reliable for further calculations. Very low frequencies of mutation are possibly artifactual in S. pneumoniae, due to the impaired growth of cells under the applied selective conditions or autolysis during extended incubation. Within the accepted range, we obtained two main peaks, which corresponded to frequencies of 1 × 10−8 and 1 × 10−7 (10-fold difference), identical to those previously found in pneumococci from non-CF patients (23). Normomutable and hypermutable control strains had mutation frequencies around the corresponding peak distributions. Forty percent of the strains within the accepted range were included around the normal mutation frequency peak of the population distribution (5 × 10−9 to 7.4 × 10−8), whereas 60% were distributed within the high mutation frequency peak (7.5 × 10−8 to 1 × 10−7). In our previously published series of non-CF patients, the proportions of strains corresponding to the peaks of normal and high mutation frequency were 62% and 37%, respectively (23). These data suggest that the proportion of strains with increased mutation rates is higher among CF patients (P = 0.02).
FIG. 2.
Percentages of isolates exhibiting a given mutation frequency to rifampin resistance among (a) 195 S. pneumoniae isolates from non-CF patients (data are from reference 23) and (b) from 37 S. pneumoniae isolates from CF patients. The dotted lines show mutation frequencies of control strains as follows: for strain R800, normomutable (2.6 × 10−8), and for R310, hypermutable (1.06 × 10−7).
A possible relationship between antibiotic resistance and hypermutable phenotypes was analyzed by the chi-square statistical method, but no significant correlation was observed. Similar antibiotic resistance percentages were observed among all categories of mutation frequency. We also investigated the distribution of the mutation figures among the pulsotypes and the serotypes of the isolates, corroborating a homogeneous distribution. When isolates with pulsotype A (corresponding to the serotype 23F) were taken into account, the mean mutation frequency was 7.53 × 10−8, ranging from 1.07 × 10−7 to 9.4 × 10−8. Four isolates were located in the 10−7 category (mutator category), while the other seven isolates were in the 10−8 category.
DISCUSSION
Unlike that of P. aeruginosa, the role of S. pneumoniae in pulmonary damage occurring in CF patients is uncertain (14). Although this organism can be recovered from respiratory samples from CF patients in early stages of the disease (5, 16), there is no clear relationship with severe bacterial complications or with pulmonary deterioration (14). Older studies demonstrated that the level of isolation of S. pneumoniae from CF patients was not higher than from non-CF patients (17, 18, 22). More recently it has been shown that the majority of CF patients, like healthy individuals, have protective levels of antipneumococcal antibodies against the most-common circulating serotypes (20). From these data it could be considered that the occurrence and epidemiology of S. pneumoniae in the nasopharynxes of CF patients might be similar to those of healthy carrier children. However, the results obtained in our study suggest that CF isolates present singular characteristics, including high antibiotic resistance rates, specific clonal relationships (at least in our isolates), and particularly increased rates of mutation frequencies compared with non-CF S. pneumoniae isolates previously described from the same institution (23).
The yearly occurrence of S. pneumoniae in CF patients ranged in our series from 2.4% to 13.5%, with a mean value of 5.5%. This value is much lower than that for P. aeruginosa (57.8%), S. aureus (49.7%), and H. influenzae (16.3%) in the 2002 CF Foundation Annual Data Report (9). Other authors in Germany have found similar occurrence rates (6.9%), but in other studies from Spain this value was higher (28.3%) (12), particularly when only a CF pediatric population was included in the study (50%) (24). In the United States, during a phase III national collaborative study of aerosolized tobramycin, the percentage of CF patients colonized with S. pneumoniae was 3.4% (6).
As in other studies, we cannot rule out the possibility that the isolation of S. pneumoniae was due to contamination of sputum samples with isolates from the oropharyngeal cavity. Doubtlessly the conditions of the bronchial epithelia of CF patients might facilitate S. pneumoniae deep colonization of the lung in the early stages of the process (8). Pneumococcal adherence might be facilitated by the increased exposure of cellular receptors, such as asialo-GM2, in damaged bronchial epithelia of CF patients, as a consequence of initial inflammatory events, following infections by respiratory viruses or mycoplasmas, or the actions of inflammatory cytokines (interleukin 1 [IL-1], IL-8, and tumor necrosis factor alpha), with a decreased production of the anti-inflammatory IL-10 (19, 38).
All S. pneumoniae isolates in our study were recovered from young patients, but only 35% of the patients had an exacerbation of their pulmonary symptoms at the time of isolation of this pathogen. The coexistence of this organism with others in 50% of exacerbation episodes, mainly S. aureus (50%), P. aeruginosa (25%), H. influenzae (16.7%), and Aspergillus spp. (8.3%), makes it difficult to evaluate its potential role in the exacerbation episodes of the studied CF patients. However, it may be possible that S. pneumoniae plays a role due to the nearly 100-fold difference that we found between geometric mean values of S. pneumoniae bacterial loads in sputum samples obtained from exacerbation and nonexacerbation episodes, although statistical signification was not reached.
Unlike other typical CF pathogens, S. pneumoniae is not known as a persistent colonizer and is considered as a transient organism in these patients (30). The evolution of individual clones with a theoretical new immunological aggression or acquisition of new strains may also suggest that in certain CF patients, S. pneumoniae may play a role in CF exacerbations favoring antimicrobial treatment. Eventually, the involvement of an S. pneumoniae strain in acute exacerbations of a particular patient may facilitate its replacement by newly acquired strains (36), as occurred in several patients in our study (Table 1). Nevertheless, the possibility of persistence of certain clones for years in the same patient was also suggested by our results. This is the case for the serotype 23 multiresistant clone A (patient 26) (Table 1). Interestingly, this clone was widely disseminated among the studied patients (8 out of 26 patients). Two out of nine isolates collected in the Sant Joan de Dèu Hospital in Barcelona also corresponded to this clone. It is known that S. pneumoniae strains of the 23F serotype are widespread among children attending different day care centers in Portugal (35). Dissemination of P. aeruginosa clones among CF patients has previously been documented, even in very distant geographic areas (34), but to the best of our knowledge this has not been previously shown for S. pneumoniae.
Considering the possibility that certain clones of S. pneumoniae may be involved in early stages of lung inflammation in CF patients, a preventive therapeutic attitude could eventually be advisable to ensure eradication, as is recommended for P. aeruginosa (32). Data about the antibiotic resistance of S. pneumoniae strains isolated from CF patients remain scarce (7). Our CF pneumococcal pediatric isolates were unusually nonsusceptible to antibiotics, if they are compared with those isolated from non-CF patients (11, 28), particularly to penicillin (75% versus 48.5%) and cefotaxime (33% versus 6.2%). We have previously reported similar differences for oropharyngeal streptococci belonging to the viridans group, which are significantly more resistant to penicillin and cefotaxime in CF patients than those of healthy children (2). Such observations suggest that the extended use of antimicrobial agents (including cefuroxime, ceftazidime, cefepime, and carbapenems) in CF patients might select for β-lactam-resistant pneumococci, including those belonging to particularly resistant clones. Unlike other pathogens from CF patients (7, 31), no fluoroquinolone-resistant S. pneumoniae isolates were observed. This fact may be related to the lower persistence of this pathogen in the pulmonary compartment in these patients and the lack of frequent administration of fluoroquinolones in monotherapy in our CF patients.
Our group and other authors have shown that increased rates of resistance in CF patients may be related to the high frequency of hypermutable P. aeruginosa, S. aureus, and H. influenzae CF isolates (27, 30, 31). The present study indicated that the frequency of pneumococcal strains with increased frequencies of mutation was also significantly (P = 0.02) higher in isolates from CF patients (62%) than in those from non-CF patients in the same institution (37%) (23). As in our previously published work on hypermutation in S. pneumoniae (23), we did not demonstrate any significant association of CF mutator strains with antibiotic resistance.
The knowledge of serotype distribution in S. pneumoniae isolates may have epidemiological and clinical implications. It has been recommended that all CF patients older than 2 years should receive the 23-valent polysaccharide vaccine against S. pneumoniae (3). Recent data from the Spanish Pneumococcal Reference Laboratory show that the most-prevalent serotypes accounting for nearly 60% of all pathogenic isolates are serotypes 14, 19F, 1, and 6B (28). In our CF series, 64.5% of the isolates correspond to serotypes 23F (33.3%), 19F (18.8%), 6A (6.2%), and 6B (6.2%), but 16 different serotypes were detected. PFGE-SmaI digestion demonstrated a genetic diversity among isolates of all serotypes, with the exception of the predominant clonal structure of serotype 23F (clone Spain23F-1), the serotype most frequently involved in high-level penicillin resistance (MIC ≥ 2 μg/ml) (28). The use of the seven-valent conjugate vaccine in CF patients may also reduce the carriage of the 23F serotype, eventually reducing the high rates of β-lactam resistance.
In summary, S. pneumoniae might play a role in the early stages of lung colonization or infection in CF patients, and eventually some clones could persist and evolve within the host during prolonged periods of time and consequently facilitate the acquisition of high levels of resistance to β-lactam agents. The early use of vaccines and the application of antibiotics with low resistance rates in CF isolates may be suggested as measures to limit the possible impact of S. pneumoniae in CF patients.
Acknowledgments
Rosa del Campo is a recipient of a contract from the Spanish Pneumococcal Infection Study Network (G03/103). This work was partially supported by research grants from the Spanish Pneumococcal Infection Study Network (G03/103) and the Microbial Sciences Foundation.
General coordination for the Spanish Pneumococcal Infection Study Network (G03/103) was provided by Román Pallarés. The following Spanish Pneumococcal Infection Study Network (G03/103) members and centers participated in this study: Ernesto García, Centro de Investigaciones Biológicas, Madrid; Julio Casal, Asunción Fenoll and Adela G. de la Campa, Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid; Emilio Bouza, Hospital Gregorio Marañón, Madrid; Fernando Baquero, Hospital Ramón y Cajal, Madrid; Francisco Soriano and José Prieto, Fundación Jiménez Díaz y Facultad de Medicina de la Universidad Complutense, Madrid; Román Pallarés and Josefina Liñares, Hospital Universitario de Bellvitge, Barcelona; Javier Garau and Javier Martínez Lacasa, Hospital Mutua de Terrassa, Barcelona; Cristina Latorre, Hospital Sant Joan de Déu, Barcelona; Emilio Pérez-Trallero, Hospital Donostia, San Sebastián; Juan García de Lomas, Hospital Clínico, Valencia; and Ana Fleites, Hospital Central de Asturias.
REFERENCES
- 1.Alloing, G., B. Martin, G. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Álvarez, M., M. E. Álvarez, L. Máiz, A. Asensio, F. Baquero, and R. Cantón. 1998. Antimicrobial susceptibility profiles of oropharyngeal viridans group streptococci isolates from cystic fibrosis and non-cystic fibrosis patients. Microb. Drug Resist. 4:123-128. [DOI] [PubMed] [Google Scholar]
- 3.American Academy of Pediatrics. 2000. Pneumococcal infection, p. 458-459. In L. K. Pickering (ed.), Red book: report of the Committee in Infectious Diseases. American Academy of Pediatrics, Elk Grove Village, Ill.
- 4.Balough, K., M. McCubbin, M. Weinberger, W. Smits, R. Ahrens, and R. Fick. 1995. The relationship between infection and inflammation in the early stages of lung disease from cystic fibrosis. Pediatr. Pulmonol. 20:63-70. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., R. M. Bertele, K. Harms, G. Horl, R. Jungwirth, C. Petermuller, B. Przyklenk, and C. Weisslein-Pfister. 1987. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15:270-277. [DOI] [PubMed] [Google Scholar]
- 6.Burns, J. L., J. Emerson, J. R. Stapp, D. L. Yim, J. Krzewinski, L. Louden, B. W. Ramsey, and C. R. Clausen. 1998. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin. Infect. Dis. 27:158-163. [DOI] [PubMed] [Google Scholar]
- 7.Cantón, R., R. Girón, A. Martínez-Martínez, A. Oliver, A. Solé, S. Valdezate, and L. Máiz. 2002. Patógenos multiresistentes en la fibrosis quística. Arch. Bronconeumol. 38:376-385. [DOI] [PubMed] [Google Scholar]
- 8.Casal, J., and D. Tarragó. 2003. Immunity to Streptococcus pneumoniae: factors affecting production and efficacy. Curr. Opin. Infect. Dis. 16:219-224. [DOI] [PubMed] [Google Scholar]
- 9.Cystic Fibrosis Foundation. 2003. Cystic Fibrosis Foundation patient registry: annual data report 2002. Cystic Fibrosis Foundation, Bethesda, Md.
- 10.Feldman, C., and K. P. Klugman. 1997. Pneumococcal infection. Curr. Opin. Infect. Dis. 10:109-115. [Google Scholar]
- 11.Fenoll, A., G. Asensio, I. Jado, S. Berrón, M. T. Camacho, M. Ortega, and J. Casal. 2002. Antimicrobial susceptibility and pneumococcal serotypes. J. Antimicrob. Chemother. 50(Suppl. 2):S13-S19. [DOI] [PubMed] [Google Scholar]
- 12.Ferrer Marcelles, A., P. Bellver Moreira, N. Cobos Barroso, S. Linan Cortés, G. Codina Grau, and F. Fernández Pérez. 1995. Cystic fibrosis: a microbiological study over an 8-year period. Arch. Bronconeumol. 31:494-500. [PubMed] [Google Scholar]
- 13.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 14.Gilligan, P. H. 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 4:35-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Høiby, N. 1974. Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol. Microbiol. Scand. B 82:541-550. [PubMed] [Google Scholar]
- 16.Hoiby, N. 1982. Microbiology of lung infection in cystic fibrosis. Acta Pediatr. Scand. 301(Suppl.):33-54. [Google Scholar]
- 17.Huang, N. N., E. L. Van Loon, and K. T. Sheng. 1961. The flora of the respiratory tract of patients with cystic fibrosis of the pancreas. J. Pediatr. 59:512-521. [DOI] [PubMed] [Google Scholar]
- 18.Iacocca, V. F., M. S. Sibinga, and G. J. Barbero. 1963. Respiratory tract bacteriology in cystic fibrosis. Am. J. Dis. Child. 106:315-324. [DOI] [PubMed] [Google Scholar]
- 19.Konstan, M. W., and M. Berger. 1993. Infection and inflammation of the lung in cystic fibrosis. Lung Biol. Health Dis. 64:219-276. [Google Scholar]
- 20.Lahiri, T., and D. A. Waltz. 2001. Preimmunization anti-pneumococcal antibody levels are protective in a majority of patients with cystic fibrosis. Pediatrics 108:E62. [Online.] http://www.pediatrics.org/cgi/content/full/108/4/e62. [DOI] [PubMed] [Google Scholar]
- 21.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May, J. R., N. C. Herrick, and D. Thompson. 1972. Bacterial infections in cystic fibrosis. Arch. Dis. Child. 47:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morosini, M. I., M. R. Baquero, J. M. Sánchez-Romero, M. C. Negri, J. C. Galán, R. del Campo, J. C. Pérez-Díaz, and F. Baquero. 2003. Frequency of mutation to rifampin resistance in Streptococcus pneumoniae clinical strains: hexA and hexB polymorphisms do not account for hypermutation. Antimicrob. Agents Chemother. 47:1464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muñoz, C., T. Juncosa, A. Gené, J. Fortea, J. L. Séculi, and C. Latorre. 1996. Estudio microbiológico del tracto respiratorio en niños afectados de fibrosis quística. Enferm. Infecc. Microbiol. Clin. 14:142-144. [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 6th ed. Approved standard. Document M7-A6, vol. 320, no. 2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. 14th informational supplement. Document M100-S14, vol. 24, no. 1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Oliver, A., R. Cantón, P. Campos, F. Baquero, and J. Blázquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 28.Pallarés, R., A. Fenoll, J. Liñares, and the Spanish Pneumococcal Infection Study Network. 2003. The epidemiology of antibiotic resistance in Streptococcus pneumoniae and the clinical relevance of resistance to cephalosporins, macrolides and quinolones. Int. J. Antimicrob. Agents 22(Suppl. 1):S15-S24. [DOI] [PubMed] [Google Scholar]
- 29.Prunier, A. L., B. Malbruny, M. Laurans, J. Brouard, J. F. Duhamel, and R. Leclercq. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187:1709-1716. [DOI] [PubMed] [Google Scholar]
- 30.Renders, N., H. Verbrugh, and A. Van Belkum. 2001. Dynamics of bacterial colonisation in the respiratory tract of patients with cystic fibrosis. Infect. Genet. Evol. 1:29-39. [DOI] [PubMed] [Google Scholar]
- 31.Román, F., R. Cantón, M. Pérez-Vázquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosendfeld, M., B. W. Ramsey, and R. L. Gibson. 2003. Pseudomonas acquisition in young patients with cystic fibrosis: pathophysiology, diagnosis, and management. Curr. Opin. Pulm. Med. 9:492-497. [DOI] [PubMed] [Google Scholar]
- 33.Ruoff, K. L., R. A. Whiley, and D. Beighton. 2003. Streptococcus, p. 405-421. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 34.Saiman, L., and, J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sá-Leáo, R., A. Tomasz, I. S. Sanches, A. Brito-Avo, S. E. Vilhelmsson, K. G. Kristinsson, and H. de Lencastre. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 182:1153-1160. [DOI] [PubMed] [Google Scholar]
- 36.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 37.Wong, K., M. C. Roberts, L. Owens, M. Fife, and A. L. Smith. 1984. Selective media for the quantitation of bacteria in cystic fibrosis sputum. J. Med. Microbiol. 17:113-119. [DOI] [PubMed] [Google Scholar]
- 38.Zeiher, B. G., and D. B. Hornick. 1996. Pathogenesis of respiratory infections and host defenses. Curr. Opin. Pulm. Med. 2:166-173. [DOI] [PubMed] [Google Scholar]