Abstract
Recently, evidence has indicated that CTNNB1 is important in a variety of malignancies. However, how CTNNB1 interacts with immune cell infiltration remains to be further investigated. In this study, we focused on the correlations between CTNNB1 and tumorigenesis, tumor progression, mutation, phosphorylation, and prognosis via gene expression profiling interaction analysis; TIMER 2.0, cBioPortal, GTEx, CPTAC, and GEPIA2 database analyses; and R software. CTNNB1 mutations are most found in uterine endometrioid carcinoma and hepatocellular carcinoma. However, no CTNNB1 mutations were found to be associated with a poor prognosis. In addition, CTNNB1 DNA methylation levels were higher in normal tissues than in tumor tissues in cancer except for breast invasive carcinoma, which had higher methylation levels in tumor tissues. The phosphorylation level of the S675 and S191 sites of CTNNB1 was greater in the primary tumor tissues in the clear cell renal cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, pancreatic adenocarcinoma, and breast cancer datasets but not in the glioblastoma multiform dataset. As for, with respect to immune infiltration, CD8 + T-cell infiltration was negatively correlated with the expression of CTNNB1 in thymoma and uterine corpus endometrial carcinoma. The CTNNB1 level was found to be positively associated with the infiltration index of the corresponding fibroblasts in the TCGA tumors of colon adenocarcinoma, human papillomavirus-negative head and neck squamous cell carcinoma, mesothelioma, testicular germ cell tumor, and thymoma. We also identified the top CTNNB1-correlated genes in the TCGA projects and analyzed the expression correlation between CTNNB1 and selected target genes, including PPP4R2, RHOA, and SPRED1. Additionally, pathway enrichment suggested that NUMB is involved in the Wnt pathway. This study highlights the predictive role of CTNNB1 across cancers, suggesting that CTNNB1 might serve as a potential biomarker for the diagnosis and prognosis evaluation of various malignant tumors.
Keywords: cancer, CTNNB1, phosphorylation, prognosis
1. Introduction
Tumorigenesis is a very sophisticated process accompanied by enhanced proliferation, apoptosis inhibition, enhanced angiogenesis, and evasion of immunity. Pancancer expression research on target genes and their relationships with prognosis as well as the relevant mechanisms is greatly needed. Moreover, the public TCGA and GEO databases provide genomic datasets for various tumors and support for performing such studies. The tumor microenvironment (TME) includes many kinds of cells. The TME is very important in the development of human cancer.
With the emergence of immune checkpoint inhibitors (ICIs) and other therapeutic options, the relationship between tumors and immune cells has become a hot topic. Recently, immunotherapy has attracted widespread attention in a variety of cancer treatment fields. Several immune checkpoint blockers, such as anti-CTLA-4 and anti-PD-L1, have emerged.[1,2] Therefore, exploration of the tumor-immune interaction mechanism and investigation of new immunotherapeutic targets in cancer are needed.
The protein β-catenin is encoded by the CTNNB1 gene, which forms adherens junctions together with calmodulin and α-catenin. Adherens junctions are essential in the construction of the epithelial cell layer because they regulate intercellular adhesion.[3] Research has revealed a specific role for CTNNB1 in different cancers, and this role varies across cancers. Cell cycle blockade in granulosa cells is associated with the downregulation of WNT/CTNNB1 signaling, which promotes ovarian follicle maturation.[4] The CTNNB1 protein promotes ovarian cancer metastasis by targeting the downregulation of Dicer to attenuate microRNA biosynthesis.[5] WNT/CTNNB1 signaling is a mediator of lncRNA SNHG20 and lncRNA SNHG8 to promote ovarian cancer progression.[6,7] In mice, relevant experimental studies revealed that when exon 3 of CTNNB1 is deleted, the new glandular pathway is activated, which significant suppresses the proliferation of PI3K GOF-mutant cells, increasing the severity of the disease.[8] Moreover, mutation of exon 3 of CTNNB1 plays an important role in regulating the myometrial invasion of endometrial carcinoma in postmenopausal women. Some scholars’ experimental studies have shown that MyD88 deletion can affect the proliferation of intestinal epithelial tumor cells via Wnt/CTNNB1 signaling.[9] The mechanism of chemoresistance in rectal cancer is very complex; the main mechanism involves the activation of the SOX 2-β-catenin/Beclin-1/autophagy signaling axis, and the EMT process in tumors is also accelerated when this pathway is activated.[10] Additionally, in hepatocellular carcinoma, CTNNB1 mutations lead to reduced chemokine levels via modulation of a feature miRNA–IRG–CK axis.[11] In addition, sFRP-4 prevents the development of carcinoma cells by inhibiting the WNT/CTNNB1 signaling pathway.[12] In addition, relevant laboratory studies have perfectly demonstrated the function of CTNNB1 in medulloblastoma, human melanoma, and neurodevelopmental disorders.[13–17] Therefore, the relationship between aberrant CTNNB1 expression and tumor prognosis is not clear, and the immunological function of CTNNB1 in a pancancer context still needs to be clarified.
In the present study, we performed bioinformatics analysis of the expression of CTNNB1 and its relationship with survival conditions, DNA methylation, tumor-infiltrating immune cells (TIICs), and related cellular pathways via different databases and revealed its prognostic value.
The workflow of the study is shown in Figure S1, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows the flowchart illustrating the pancancer analysis.
2. Materials and methods
2.1. Analysis of gene expression patterns
Tissue samples from the TCGA project were examined for CTNNB1 levels in tumors versus neighboring normal tissues via the “Gene DE” module of TIMER. The GEPIA2 web server provided box plots of the expression differences in the GTEx database for certain tumors that did not have any or only a small number of normal tissues (e.g., TCGA-glioblastoma multiform (GBM), TCGA-acute myeloid leukemia). The “Pathological Stage Plot” module in GEPIA2 also provided violin plots of CTNNB1 levels in various stages of all TCGA tumors. Data for the box were converted as follows: log2 [transcripts per million (TPM) + 1].
Protein expression analysis of the CPTAC dataset was performed via the UALCAN portal. CTNNB1 (NP 001091679.1) has been shown to be differentially expressed between primary tumors and normal tissues in terms of total protein and phosphoprotein expression. Available datasets of 6 tumors (including clear cell renal cell carcinoma [RCC] and lung adenocarcinoma [LUAD]) were used. Additionally, the UALCAN tool was used to assess CTNNB1 methylation levels in 23 kinds of cancers.
2.2. Survival prognosis analysis
In this research process, the “survival map” module of GEPIA2 was applied to obtain the overall survival (OS) significance map data of CTNNB1 across all TCGA tumors. The expression thresholds of the high- and low-expression groups were set as the cutoff high (50%) and cutoff low (50%), respectively. The logarithmic rank test was applied in the inspection process, and the survival chart was generated via the “survival analysis” module of GEPIA2.
2.3. Genetic alteration analysis
When looking for information on CTNNB1 genetic alterations, we used the “TCGA Pan-Cancer Atlas Studies.” In the “cancer types summary” module, copy number alterations (CNAs) were displayed. The “mutations” module in the protein 3D structure showed mutation site information for CTNNB1 in the schematic diagram. Additionally, we utilized the “comparison” module to determine how TCGA cancer patients with and without CTNNB1 mutations differed in terms of overall, disease-free, and survival rates.
2.4. Analysis of immune infiltration
The CTNNB1 expression level was examined via the “immune-gene” module of the TIMER2 web server. CD8+ T cells were chosen for the study. A variety of methods were used to determine immune cell infiltration levels. These included the TIMER, CIBERSORT, and XCELL databases. These values were derived by adjusting the results of Spearman rank test for purity. A heatmap was used to visualize the data intuitively.
To predict the efficacy of immunotherapy, the tumor mutation burden (TMB) was used as a powerful biomarker. The variants that have been identified thus far can be classified as insertions or deletions across multiple bases. Molecular tumor phenotype microsatellite instability (MSI) refers to the loss or gain of nucleotides from relevant repetitive DNA regions. Research on the association between the TMB and MSI was carried out via Sangerbox.
The immunological microenvironment of tumors has emerged as a significant factor in the progression of malignancies. To establish the link between immune and stromal scores and CTNNB1 levels, this study employed the Sangerbox tool to compute 3 scores (the immune score, the stromal score, and the ESTIMATE score) for the TME. These scores indicate immunological and stromal functions. A higher immune score or stromal score indicates that a larger proportion of a certain cell type (immune or stromal) is present in the TME. The ESTIMATE score refers to the ratio of these components in the TME.
2.5. Enrichment analysis
The STRING database was searched using “CTNNB1” and “Homo sapiens.” Our next step was to set the following parameters: the meaning of network edges (“evidence”), the maximum number of participants to display (“≤50 participants” in the 1st shell), and the active interactions (“experiments”) that are to be used in the visualization. Finally, proteins experimentally validated to bind to CTNNB1 were obtained.
To identify the top 100 CTNNB1-related genes, we utilized GEPIA2 to analyze the TCGA tumor dataset and corresponding healthy tissue dataset. A Pearson correlation study of CTNNB1 and selected genes was also conducted in GEPIA2. Using the log2 TPM, we created a dot plot P value, and R2 information was provided in the table. We subsequently calculated the coexpression relationships of the 3 genes that were the genes most strongly linked to each other in the TCGA, GTEx, and CCLE databases with CTNNB1. In addition, we used TIMER2’s “Gene Corr” module, which provides a heatmap of the selected genes and includes the P value for adjusting Spearman correlation for purity. It was ultimately possible to identify enriched pathways via the R packages “tidyr” and “ggplot2.” To perform GO (Gene Ontology) enrichment analysis, we used the “clusterProfiler” R package. We created cnetplots (circular = F, colorEdge = T, node label = T) from the data. The relevant indexes of the net enrichment score, gene ratio, and P value were used to identify KEGG pathways with substantial enrichment. The R programming language [R-3.6.3, 64-bit] was used for this study, and P < .05 was considered to indicate statistical significance in two-tailed analyses.
3. Results
3.1. Analysis of gene expression
CTNNB1 was first studied in a variety of cell types and nontumor tissues. The mRNA and protein sequences of CTNNB1 are shown in Figure S2, Supplemental Digital Content, http://links.lww.com/MD/N828. Some scholars have reported that the multifunctional CTNNB1 protein is highly conserved according to the “HomoloGene” database, which indicates that the normal physiological role of CTNNB1 may be similar across a wide variety of species (see Figure S3, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows HomoloGene and phylogenetic tree analysis). The placenta expressed the highest levels of CTNNB1, followed by the ovary and endometrium (see Figure S4, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows CTNNB1 expression in different datasets), and was derived from the HPA and GTEx datasets. All observed tissues (consensus normalized expression levels >10) displayed CTNNB1 expression, suggesting poor RNA tissue specificity. Similarly, the HPA/Monaco/Schmiedel datasets lacked RNA blood cell type features when the CTNNB1 levels of various blood cells were evaluated. The CTNNB1 protein content in plasma was 2 µg/L, which is deemed physiological leakage of the intracellular CTNNB1 protein according to the PeptideAtlas. Immunohistochemical staining of CTNNB1 in different tumor tissues and normal tissues (see Figure S5, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows immunohistochemical staining of CTNNB1 in different tumor tissues and normal tissues).
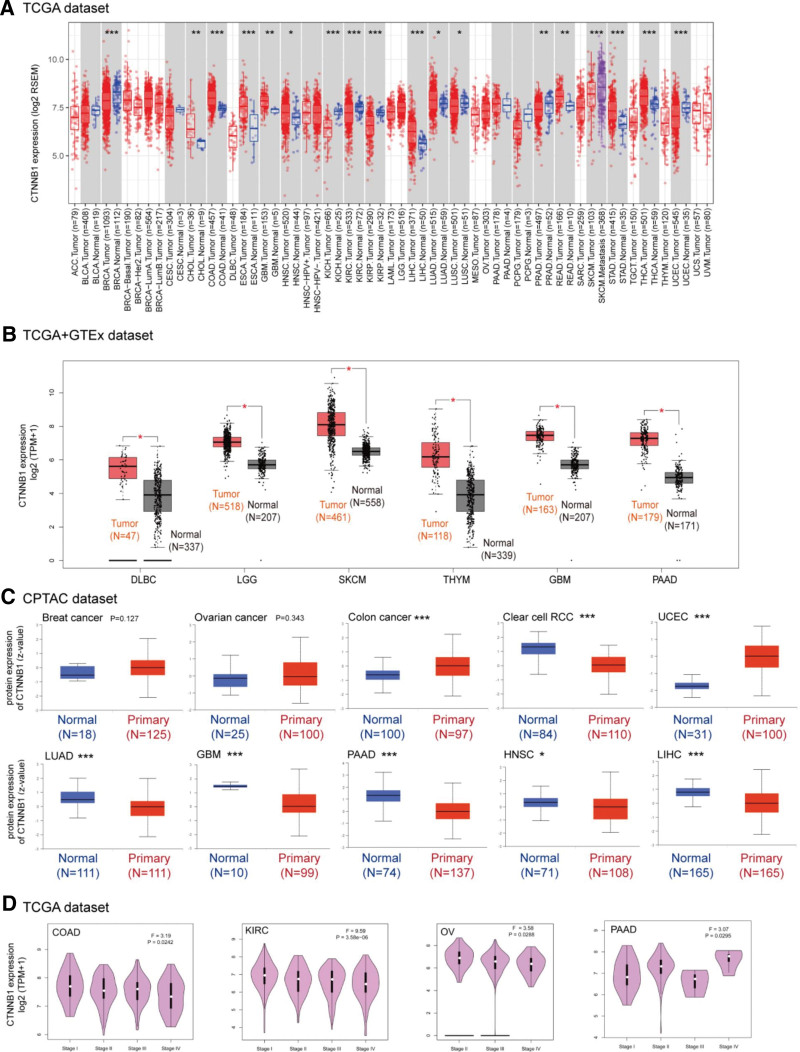
We used the TIMER2 method to examine CTNNB1 expression in TCGA samples from various cancers. As shown in Figure 1A, CTNNB1 expression levels in breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), kidney chromophobe, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma, liver hepatocellular carcinoma (LIHC), uterine corpus endometrial carcinoma (UCEC) (P < .001), cholangiocarcinoma, prostate adenocarcinoma, rectum adenocarcinoma (READ) (P < .01), head and neck squamous cell carcinoma (HNSC), LUAD, and lung squamous cell carcinoma (P < .05) tissues were greater than those in control tissues (Fig. 1B). The nonsignificant difference in CTNNB1 expression between tumor samples and normal tissues (see Figure S6, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows difference in CTNNB1 expression between tumor samples and normal tissues). We then examined the differences in protein levels between healthy and tumor tissues via the use of healthy tissues from the GTEx database as controls. However, for other malignancies, such as adrenocortical carcinoma (ACC), BRCA, acute myeloid leukemia, and ovarian serous cystadenocarcinoma (OV), we were unable to detect a significant difference (see Figure S4, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows CTNNB1 expression in different datasets).
Figure 1.
Expression level of the CTNNB1 gene in different tumors and pathological stages. (A) The expression status of the CTNNB1 gene in different cancers or specific cancer subtypes was analyzed via TIMER2. *P < .05; **P < .01; ***P < .001. (B) For the tumor types of lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), LGG, skin cutaneous melanoma (SKCM), THYM, GBM, and PAAD in the TCGA project, the corresponding normal tissues of the GTEx database were included as controls. The box plot data were supplied. *P < .05. (C) Based on the CPTAC dataset, we also compared the total protein expression level of SND1 between normal tissue and primary tissues of patients with breast cancer, ovarian cancer, colon cancer, clear cell RCC, UCEC, LUAD, GBM, PAAD, HNSC and LIHC. *P < .05; ***P < .001. (D) Based on the TCGA data, the expression levels of the SND1 gene were analyzed according to the main pathological stages of COAD, KIRC, OV, and PAAD. Log2 (TPM + 1) was applied for log-scale transformation.
The CPTAC dataset revealed significantly greater total CTNNB1 protein expression in tumors tissues versus normal tissues for colon cancer and other malignancies, such as colon cancer, UCEC, GBM, pancreatic adenocarcinoma (PAAD), HNSC, and LIHC (P < .001). However, there were no substantial differences between normal and malignant breast cancer tissues (Fig. 1C). Data on the correlation of pathological stage with CTNNB1 in different tumors is shown in Figure 1D.
The “pathological stage plot” module was used to examine the link between the CTNNB1 level and the stages of malignancies, such as COAD, OV, PAAD, and KIRC (Fig. 1D, P < .05).
3.2. Analysis of prognostic factors
The CTNNB1 level in cancer cases was compared with the prognosis of different malignancies, and we categorized the cases into two categories: high- and low-expression.
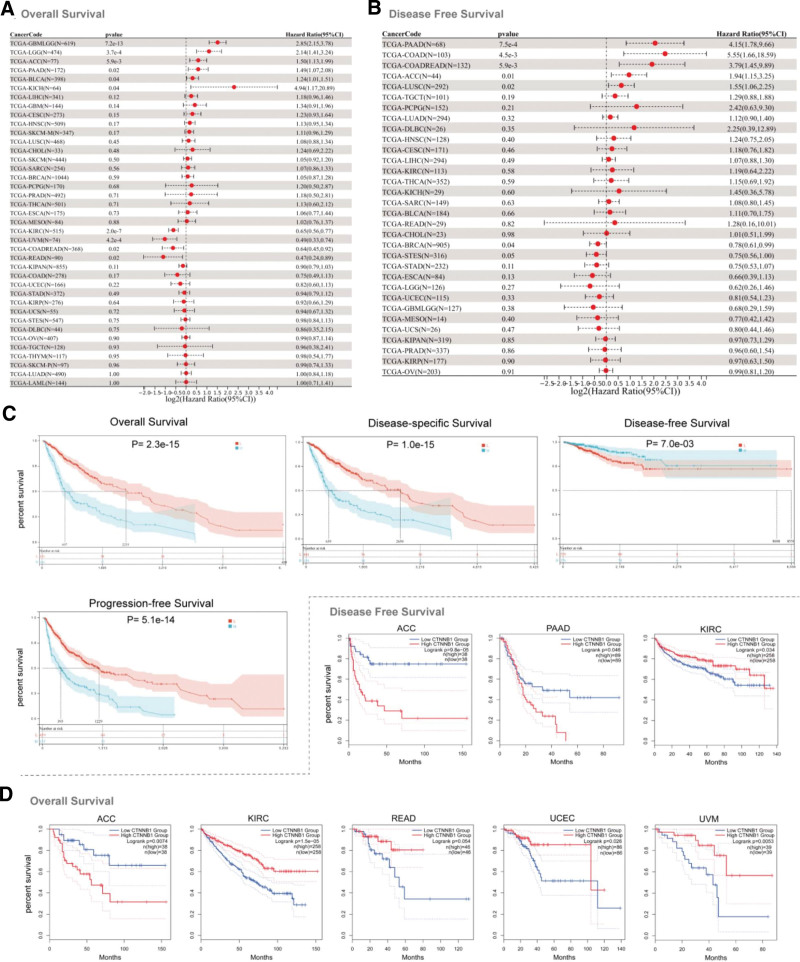
CTNNB1 overexpression was shown to be related to poor OS in the TCGA datasets for GBMLGG (P = 7.2e-13), brain lower-grade glioma (LGG) (P = 3.7e-04), ACC (P = 5.9e-03), PAAD (P = .02), bladder urothelial carcinoma (P = .04), and kidney chromophobe (P = .04), as shown in Figure 2A. In the TCGA datasets for PAAD (P = 7.5e-4), COAD (P = 4.5e-3), COADREAD (P = 5.9e-3), ACC (P = .01), and lung squamous cell carcinoma (P = .02), high CTNNB1 levels were correlated with a poor prognosis (Fig. 2B). CTNNB1 overexpression was associated with poor OS (P = 2.3e-15), disease-specific survival (P = 1.0e-15), progression-free survival (PFS) (P = 5.1e-14), and good DFS (P = 7.0e-3) (Fig. 2C).
Figure 2.
Correlation between CTNNB1 gene expression and prognosis of cancers in the TCGA database (A and B). We used the GEPIA2 tool to perform overall survival and disease-free survival analyses of different tumors in the TCGA database according to CTNNB1 gene expression (C). The survival map and Kaplan–Meier curves with positive results are shown (D).
A relationship between high CTNNB1 levels and poor OS (Fig. 2D, P = .0074) and DFS (P = 9.8e-05) in ACC patients was observed using the GEPIA tool. Low CTNNB1 gene expression was correlated with poor OS for KIRC (P = 1.5e-05), READ (P = .054), and uveal melanoma (P = .0053) patients and poor DFS for KIRC patients (P = .034) (Fig. 2D).
Moreover, the survival analysis via Kaplan–Meier (K–M) plotter revealed that low CTNNB1 expression was related to worse OS for kidney renal papillary cell carcinoma, testicular germ cell tumor (TGCT), stomach adenocarcinoma, READ, and KIRC patients and poor DFS for patients with stomach adenocarcinoma and breast cancer (see Figure S7, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows overall survival and disease-free survival were evaluated via the Kaplan–Meier plotter tool). In contrast, high CTNNB1 expression was associated with poor OS in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), HNSC, LIHC, OV, and PAAD patients and poor DFS in PAAD patients.
3.3. Genetic mutation analysis
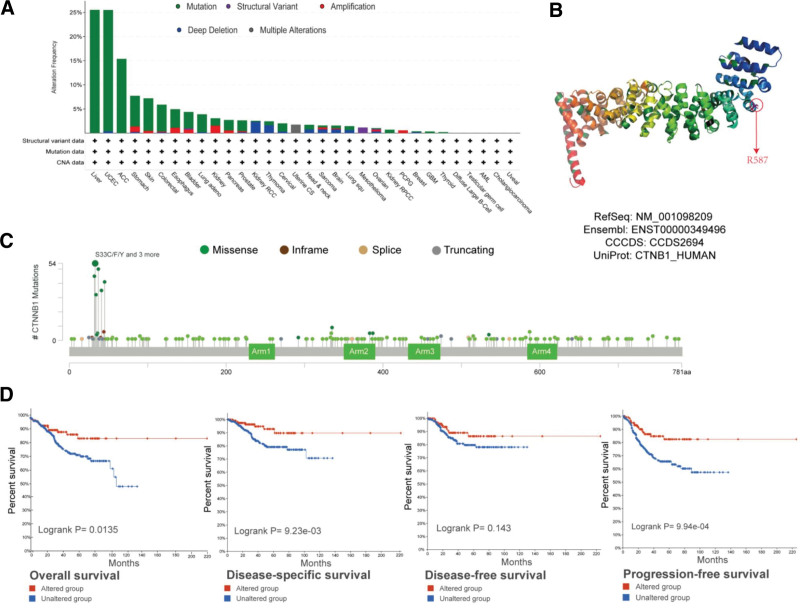
Figure 3A shows that 282 patients with “missense” mutations as the major category had the highest CTNNB1 mutation frequency.
Figure 3.
Mutation features of CTNNB1 in different tumors in the TCGA database. We analyzed the mutation features of CTNNB1 in TCGA tumors via the cBioPortal tool. The alteration frequencies with mutation type (A) and mutation site (C) are displayed. The mutation site with the highest alteration frequency in VUS in the 3D structure of CTNNB1 (B) is shown. We also analyzed the potential correlation between mutation status and overall, disease specific, disease-free, and progression-free survival in UCEC patients (D) via the cBioPortal tool.
Figure 3C shows the types, locations, and cases of the CTNNB1 genetic mutation. One of the most common types of genetic changes we discovered was a missense mutation in CTNNB1, which we found in 27 uterine endometrioid carcinoma cases, 17 hepatocellular carcinoma cases, 3 lung adenocarcinoma cases, 2 bladder urothelial carcinoma cases, 2 prostate adenocarcinoma cases, 1 papillary stomach case, and 1 COAD case; the change involved mutation of serine to cysteine/phenylalanine/tyrosine/proline/alanine/threonine. For the VUS gene, missense mutation was also the main alteration type, and R587Q mutation was detected in 3 cases of uterine endometrioid carcinoma, 1 case of tubular stomach adenocarcinoma, 1 case of rectal adenocarcinoma and 1 case of COAD, with translation from arginine to glutamine. The 3D structure of the CTNNB1 protein revealed R587 as the mutation site (Fig. 3B). We also investigated the possibility that a genetic change in CTNNB1 is linked to a poor prognosis for various types of cancer. In UCEC patients, altered CTNNB1 expression improved overall survival (P = .0135) but not disease-free survival (P = .143) (Fig. 3D).
3.4. Analysis of DNA methylation
Analysis of DNA methylation. With respect to the TCGA data, this study applied the MEXPRESS approach to identify a link between CTNNB1 DNA methylation and the etiology of various cancers (see Figure S8, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows association between CTNNB1 DNA methylation and etiology). The methylation levels of normal tissues in the COAD, bladder urothelial carcinoma, LIHC, PAAD, READ, TGCT, and UCEC datasets were greater than those of tumor tissues; however, for the BRCA dataset, the methylation levels were greater in tumor tissues (see Figure S9, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows CTNNB1 methylation levels in different tumors in the CPTAC database).
The GSE77871 dataset included CTNNB1 methylation levels in ACC and neighboring normal adrenocortical tissues. It shows the normalized methylation chip data that we used to identify differentially methylated probes and differentially methylated regions (see Figure S10, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows DNA methylation analysis: CTNNB1 methylation levels in ACC and neighboring normal adrenocortical tissues). The methylation level of CTNNB1 in normal tissues was not significantly different from that in malignant tissues.
3.5. Analysis of protein phosphorylation
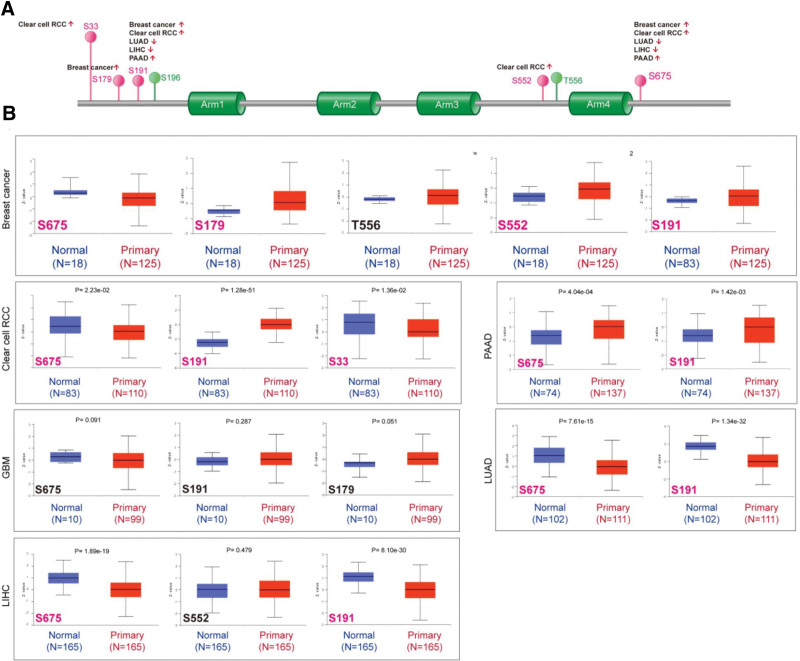
Normal tissues were compared to primary tumor tissues for changes in CTNNB1 phosphorylation levels. The CPTAC dataset was used to investigate the presence or absence of 6 tumor types (LUAD, LIHC, breast cancer, clear cell RCC, GBM, and PAAD). The CTNNB1 phosphorylation sites are summarized in Figure 4A, with important changes highlighted. S675 and S191 locus phosphorylation was greater in all primary tumor tissues except GBM (Fig. 4B, all P < .05), followed by the S179 and S552 loci for breast cancer and the S33 locus for clear cell RCC. Additional studies are needed to determine whether S426 phosphorylation and cell cycle regulation play a role in tumorigenesis, and we cannot exclude the conclusion that the high CTNNB1 phosphorylation level of S675 or S191 is merely a byproduct of dysregulated signaling.
Figure 4.
Phosphorylation analysis of the CTNNB1 protein in different tumors. On the basis of the CPTAC dataset, we analyzed the difference in the expression level of the CTNNB1 phosphoprotein between normal tissue and primary tissue of selected tumors via UALCAN. (A) The phosphoprotein sites with positive results are displayed in the schematic diagram of the CTNNB1 protein. (B) Box plots for different cancers, including breast cancer, clear cell RCC, LUAD, GBM, LIHC, and PAAD, are shown.
Additionally, we applied the relevant PhosphoNET tool to look at CTNNB1 phosphorylation at S191, which was empirically supported by a single paper, as well as S675 in the dataset.[18,19] S191 and S675 phosphorylation is very important in carcinogenesis; hence, further molecular studies are warranted.
3.6. Results of the study on immune infiltration
T cells in the TME have been found to have critical functions in cancer growth and metastasis. In the TME, cancer-associated fibroblasts have been found to influence the function of immune cells that have infiltrated the tumor.
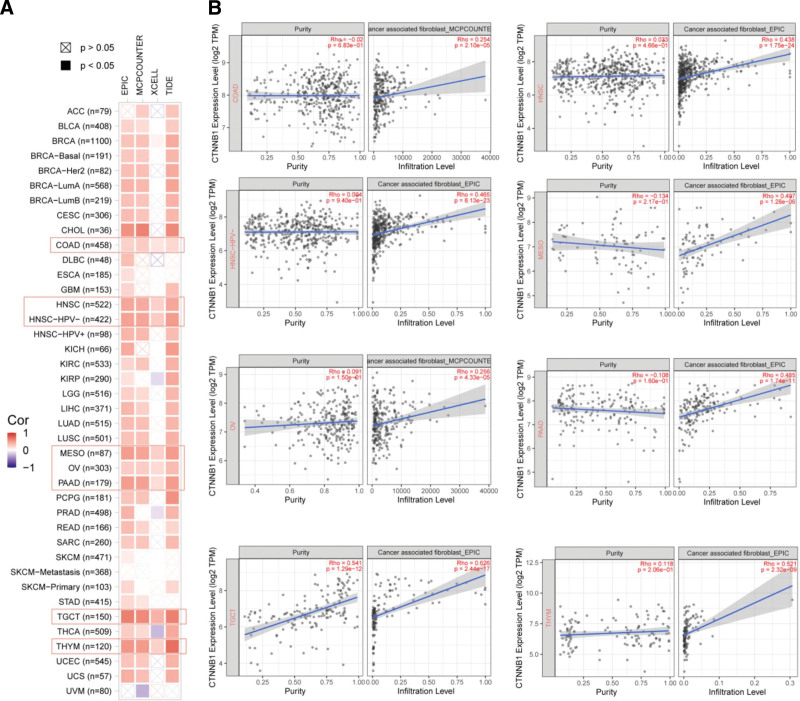
CD8 + T-cell immunological infiltration and CTNNB1 expression were found to be negatively correlated by all or most algorithms after a series of investigations in thymoma (THYM) and UCEC (see Figure S11, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows immunological role of CTNNB1). The CTNNB1 level and infiltration index of the corresponding fibroblasts were statistically linked in the TCGA tumors of COAD, HNSC-HPV−, mesothelioma, TGCT and THYM (Fig. 5A). Figure 5A and Figure S11, Supplemental Digital Content, http://links.lww.com/MD/N828 show the scatterplot data for the aforementioned tumors generated by 1 algorithm. On the other hand, the MCPCOUNTER algorithm revealed that the CTNNB1 expression level in COAD was positively associated with the infiltration level of relevant fibroblasts (Fig. 5B, cor = 0.254, P = 2.10e-05). Notably, CTNNB1 expression was highly associated with immune scores in the following tumors: GBMLGG, KIPAN and KIRC (stromal score); CESC, sarcoma and TGCT (immune score); and GBM, LGG, KIPAN and skin cutaneous melanoma-M (ESTIMATE score) (see Figure S11, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows immunological role of CTNNB1). The results revealed that CTNNB1 expression and immune infiltration in tumors were closely linked.
Figure 5.
Correlation analysis between CTNNB1 expression and immune infiltration of cancer-associated fibroblasts. (A) Different algorithms were used to explore the potential correlation between the expression level of the CTNNB1 gene and the infiltration level of cancer-associated fibroblasts across all types of cancer in the TCGA. (B) Significant relationship was found in particular tumors.
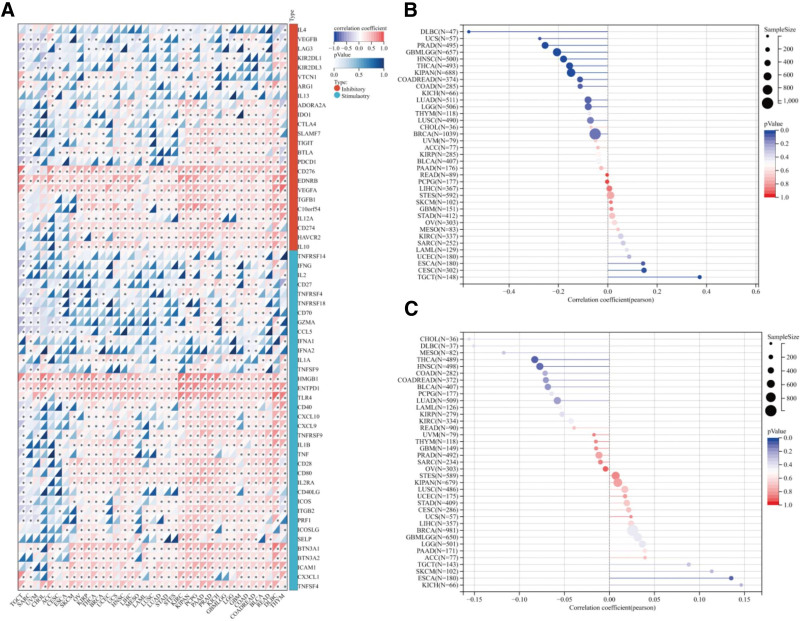
Immunosurveillance is largely acknowledged component in evaluating the prognosis of many malignancies. ICP genes, such as PD-1, can be exploited by tumors to elude detection. In terms of the connection between the CTNNB1 level and that of immunological genes. CTNNB1 expression was positively associated with the ICP gene level, except for IL1A, IFNA1, IFNA2, TNFRSF18, and VTCN1 in pheochromocytoma and paraganglioma (Fig. 6A). These results indicate that CTNNB1 could play a critical role in preventing the immune system from recognizing and responding to pathogens. The relationships between CTNNB1 levels and TMB/MSI in all cancers in the TCGA cohort were also investigated. We found no significant association between CTNNB1 expression and TMB in any of the 37 tumors examined, as shown in Figure 6B and C. Figure S10, Supplemental Digital Content, http://links.lww.com/MD/N828 shows that the CTNNB1 level was negatively related to MSI in GBM, HNSC, KIPAN, COAD, READ, and uterine carcinosarcoma (all P < .05) but positively related to MSI in TGCT and CESC (all P < .05) (see Figure S10, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows DNA methylation analysis: CTNNB1 methylation levels in ACC and neighboring normal adrenocortical tissues). These findings merit further investigation.
Figure 6.
Correlations between CTNNB1 expression and immune features, including immune marker sets, TMB and MSI, in cancers. (A) Correlations between CTNNB1 expression and immune marker sets. (B) Lollipop chart of the correlation between CTNNB1 expression and TMB. (C) Lollipop chart of the correlation between CTNNB1 expression and MSI.
3.7. Enrichment analysis of CTNNB1-related partners
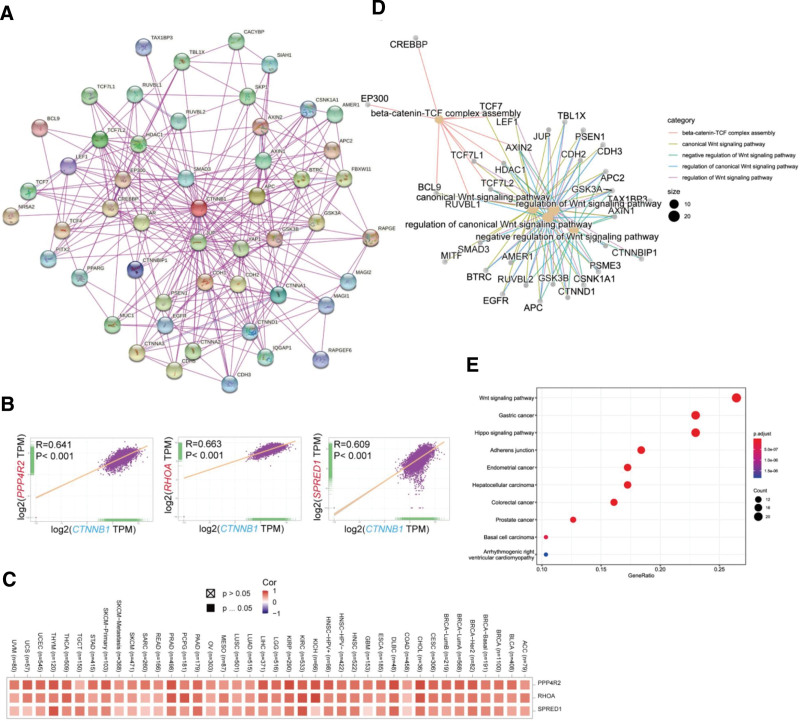
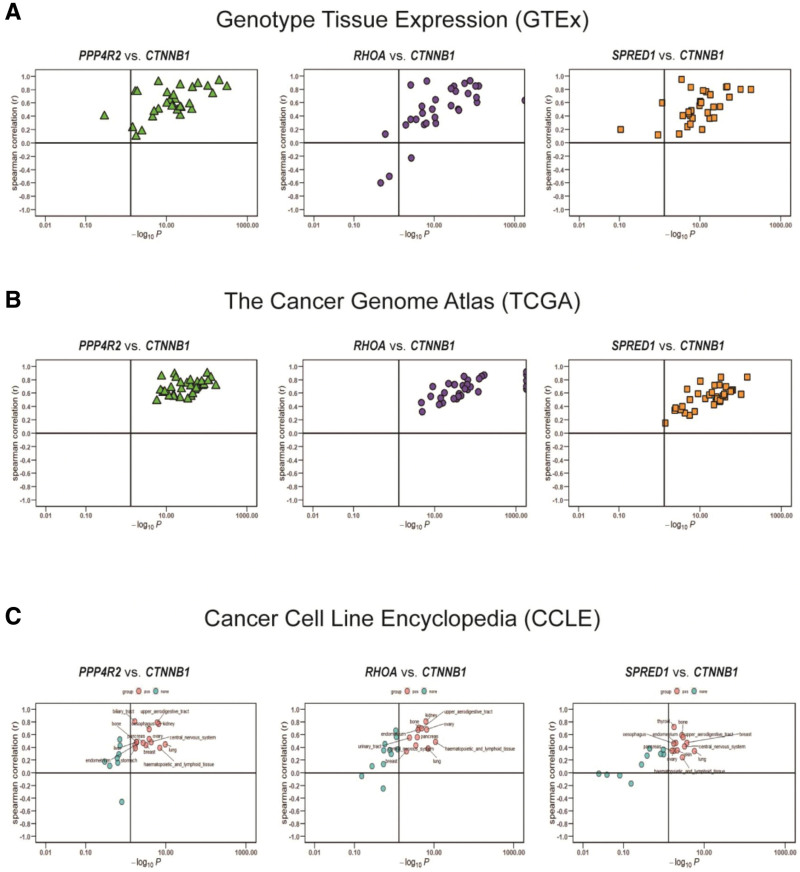
To clarify the molecular mechanism of the CTNNB1 gene in tumor pathology, we searched a database to identify the binding proteins targeting CTNNB1 and specific genes and then carried out enrichment analysis. On the basis of the STRING tool, 50 CTNNB1-binding proteins were identified, which was consistent with the experimental results. After the interactions of these proteins are analyzed, corresponding networks are established, as shown in Figure 7A below. During the study, the GEPIA2 tool was used to compare the expression data of various types of tumors in the TCGA, and genes related to the expression of CTNNB1 were obtained. After sequencing, the top CTNNB1-correlated genes in the TCGA projects were identified, and we analyzed the expression correlation between CTNNB1 and selected target genes. The correlation analysis results revealed a positive correlation between the expression levels of CTNNB1 and those of protein phosphatase 4 regulatory subunit 2 (PPP4R2) (R = 0.641), homolog family member A (RHOA) (R = 0.663) and sprouty-related EVH1 domain containing 1 (SPRED1) (R = 0.609) genes (all P < .001) (Fig. 7B and C). We additionally assessed the relationships between the above 3 genes and CTNNB1 in the GTEx, TCGA, and CCLE databases (Fig. 8A–C). The results of KEGG enrichment analysis are shown in Figure 7E and GO analysis in Figure 7D. According to the results shown in this figure, the “Wnt pathway” and “metabolic pathway” participate in the regulatory effect of CTNNB1 on pathological changes in tumors. The results of the GO enrichment analysis revealed that these genes were closely related to the negative regulation of the typical Wnt signaling pathway and participated in the pathological regulation of intestinal epithelioma via the Wnt signaling pathway. Pathways related to molecular function and cellular components (see Figures S12–S14, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows 3 signaling pathways associated with the enriched genes, diagram of the Wnt signaling pathway pattern, and the cnetplot for the biological process and cellular component results from the GO analysis).
Figure 7.
Enrichment analysis of CTNNB1-related genes. (A) We first obtained the available experimentally determined CTNNB1-binding proteins via the STRING tool. (B) Using the GEPIA2 approach, we also obtained the top 100 SND1-correlated genes in TCGA projects and analyzed the expression correlation between SND1 and selected target genes, including PPP4R2, RHOA, and SPRED1. (C) The corresponding heatmap of the detailed cancer types is shown. (D) The cnetplot for the molecular function data in the GO analysis is also shown. (E) KEGG pathway analysis of the CTNNB1-binding and interacting genes was performed.
Figure 8.
Correlation analysis between CTNNB1 and PPP4R2, RHOA and SPRED1 in the GTEx (A) and TCGA cohorts (B) and CCLE cell lines (C).
4. Discussion
Researchers have reported that CTNNB1 is linked to a wide range of clinical illnesses and cancers. CTNNB1 may share biological pathways with other cancers, but further research is needed to confirm this. No articles on the anticancer effects of CTNNB1 could be found, despite our best efforts.
In the absence of Wnt ligands, CTNNB1 interacts with other proteins, such as APC, Axin, CK1, and GSK3. Phosphorylation of CTNNB1 by CK1 (CK1) and GSK3 causes ubiquitination and protease degradation. The proteolytic activity of Wnt and GSK3 leads to the phosphorylation of LRP5/6, which in turn causes Axin to be transported to the cell membrane. However, the level of intracellular Axin is insufficient for CTNNB1. CTNNB1 is phosphorylated in the nucleus by T-cell factor (TCF). CTNNB1 and LEF1 in the nucleus inhibit the activity of E-transcriptional cadherin, causing epithelial–mesenchymal transition.
The relevant “HomoloGene” data in this study also revealed the stability of the CTNNB1 protein structure in various species (see Figure S3, Supplemental Digital Content, http://links.lww.com/MD/N828, which shows HomoloGene and phylogenetic tree analysis), revealing that similar mechanisms for the function of CTNNB1 exist. Clinical diseases, such as tumors, have been linked to CTNNB1 in recent studies. However, it remains to be determined whether CTNNB1 has similar biological mechanisms in various tumors. We searched for an article on the pancancer analysis of CTNNB1 but did not find 1.
CTNNB1 binds to APC and GSK3β among Wnt ligands.[20] GSK3β sequentially phosphorylates CTNNB1, leading to the ubiquitination of CTNNB1.[21] Among the Wnt ligands, CK1γ and GSK3β are responsible for mediating LRP5/6 and Dsh phosphorylation. This results in LRP5/6 and Dsh recruiting Axin as a complex to the cell membrane.
Because of the restricted level of intracellular Axin,[22] CTNNB1 expression is also very limited. When it forms complexes with other proteins belonging to the LEF family (LEF/TCF), the cytoplasmic unphosphorylated form of CTNNB1 translocates to the nucleus of the cell and works as a transcriptional activator there.[23] CTNNB1 is responsible for decreasing the transcriptional activity of E-cadherin. This occurs via the formation of a complex with LEF1 in the nucleus.[24]
The Wnt pathway is a dominant pathway related to CTNNB1 and is a highly conserved signaling pathway among species. If mutations in key proteins of this pathway alter signaling, cancer development may occur. For example, in normal colonic epithelial cells, secreted frizzled-related proteins (SFRPs) compete with Wnt for binding to the frizzled family receptor (FZD), thereby antagonizing Wnt signaling. When Wnt signaling is inactivated, APC phosphorylates β-catenin (CTNNB1), which degrades and prevents the deposition of β-catenin, resulting in a cell differentiation phenomenon, thus keeping the colonic epithelium in a state of dynamic equilibrium.[25] There are many studies on CTNNB1 mutations in colorectal cancer.[26–28] According to the relevant statistical results, the incidence of CTNNB1 mutation in adenoma (12.5%) is significantly greater than that in invasive cancer, but this mutation occurs in the early stage of colorectal cancer. Comparative analysis revealed that the effect of this mutation on cancer pathological processes is similar to that of APC mutation.[29,30]
First, we used the GEPIA2 tool to detect the relationship between the CTNNB1 level and the OS of ovarian cancer patients. Moreover, in this research, the OncoLnc tool was also used to conduct Cox regression analysis on the TCGA-OV cohort, and the results were statistically analyzed. No significant correlation was found (P = .55). In addition, we applied the K–M plotter method to perform a series of survival analyses in ovarian cancer patients in the GSE cohorts (GSE15622, GSE19829, etc). We found an obvious relationship between the CTNNB1 level and prognosis in terms of PFS (P = .034) and postprogression survival (PPS) (P = .011) but no significant correlation between the CTNNB1 level and OS (HR = 1.15, P = .064), which was different from the results of the pancancer survival analysis (OS for OV (HR = 1.37 P = .02)) via the K–M plotter tool. These results suggest that a low level of CTNNB1 is related to a worse prognosis in terms of OS for ovarian cancer patients in the “histology: serous,” “debulk: optimal” and “TP53 mutation status: wild type” subgroups.
In summary, current clinical databases do not provide enough evidence to verify the role of CTNNB1 in prognosis. In the binary staging of epithelial ovarian cancer, type II mainly exhibits BRCA1 or BRCA2 gene mutations, which are associated with DNA homologous recombination repair damage.[31] PARP inhibitors can bind to tumor cells during homologous recombination repair to cause their death.[32] BRAF (38%), KRAS (19%), p53 (8%), and PIK3CA mutations are present in clear cell carcinoma, and ARID1A (50%) mutations are present in low-grade plasmacytomas.[33,34] Endometrioid ovarian carcinoma has a high frequency of CTNNB1 mutations,[35] and the frequency of KRAS mutations in mucinous ovarian carcinoma is close to 100%.[36] As a result, the differences between OC subtypes may imply a relationship between etiologic and spectrum effects to improve the prognosis of OC patients.
Upregulation of CTNNB1 was related to a worse prognosis, especially in ACC. However, there were no data on ACC on the K–M plotter website. Additionally, our differential methylation analysis of ACC tumor tissue and adjacent normal tissue did not identify CTNNB1 as one of the differentially methylated genes. Thus, more clinical features should be considered. Second, more molecular experiments are needed to determine whether high CTNNB1 expression is a dominant factor in the development of ACC or is simply an effect of tumor changes.
Tumor immunotherapy can activate the body’s antitumor immune system. The related therapies mainly include monoclonal antibody ICIs, cancer vaccines, cell therapy and other methods, each of which has a certain scope of application. The results of relevant experimental studies have shown that TIIC has a significant effect on the prognosis of different cancers.[37] In this study, we analyzed 60 major immune checkpoint genes, investigated the correlation between gene expression and immune checkpoint expression, and then calculated and analyzed the correlation between gene expression and CTNNB1 expression. High expression of PD-1 and PD-L1 in TIICs generally indicates a poor prognosis.[38] CTNNB1 was positively correlated with tumor pathological stage and negatively correlated with TIIC infiltration levels. The above results suggest that the expression of CTNNB1 affects the degree of tumor invasion. The results of this study revealed a certain correlation between the expression of CTNNB1 and TMB in all TCGA tumors. MSI is associated with a greater risk of cancer, which is manifested mainly by increases in the TMB and lymphocyte count.[39] TMB can be considered a potential biomarker for predicting the response to immune checkpoint blockade[40,41] and a very valuable predictor of the outcome of ICI therapy.[42,43] Furthermore, Thomas reported that the TMB strongly influences the immune-related survival of lymphoid neoplasm diffuse large B-cell lymphoma patients.[44] Recent studies have identified a potential biomarker role for CTNNB1 mutation in predicting the immunotherapy response in HCC patients.[45,46] In this context, our study provides prospective suggestions on the function of CTNNB1 in tumor immunology.
5. Conclusion
GTEx and CCLE datasets were applied to calculate coefficients between the 3 genes with the highest correlations to the CTNNB1 gene. CTNNB1-binding genes and CTNNB1-related genes from all malignancies were analyzed to identify a Wnt signaling pathway that may contribute to carcinogenesis. For THYM and UCEC, there was a statistically significant correlation between CTNNB1 and CD8 + T-cell infiltration levels when deconvolution methods were applied. For the first time, CTNNB1 expression and cancer-associated fibroblasts were found to be connected in different cancers.
Although we analyzed and integrated data from several sources, the current study has several drawbacks. The bioinformatics analysis of CTNNB1 in malignancies provided us with some useful insights; however, biological experiments are necessary to verify the above results and improve its utility. A better understanding of how CTNNB1 operates at the cellular and molecular levels will require additional research in this area. The posttranslational changes in CTNNB1, which are crucial for altering the action of relevant adjusting factors, are not included in the above databases. Although the CTNNB1 level has been reported to be related to immunity and clinical survival in patients with malignancies, we are unsure whether CTNNB1 influences patient survival through immunological pathways. There is a clear relationship between CTNNB1 levels and prognosis, as researchers have concluded. CTNNB1 is related to poorer outcomes and has been shown to be elevated in a variety of cancers, making it a potential new target for cancer therapy. CTNNB1 plays a very dominant role in carcinogenesis and metastasis, suggesting that the expression of CTNNB1 may influence tumor immunology and metabolic activity. Future studies on the level of CTNNB1 and the corresponding immune milieu of tumors may reveal options for tumor immunotherapy.
Acknowledgments
The authors would like to thank the investigators and staff who contributed to this study.
Author contributions
Conceptualization: Xiaoyuan Xu, Na Cui.
Data curation: Xiaoyuan Xu, Aimin Yang, Siran Li.
Formal analysis: Aimin Yang, Yan Han, Siran Li, Guimin Hao.
Writing – original draft: Xiaoyuan Xu.
Writing – review & editing: Xiaoyuan Xu, Aimin Yang, Yan Han, Siran Li, Guimin Hao, Na Cui.
Supplementary Material
Abbreviations:
- ACC
- adrenocortical carcinoma
- BRCA
- breast invasive carcinoma
- CESC
- cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD
- colon adenocarcinoma
- GBM
- glioblastoma multiform
- HNSC
- head and neck squamous cell carcinoma
- ICI
- immune checkpoint inhibitors
- KIRC
- kidney renal clear cell carcinoma
- LGG
- brain lower-grade glioma
- LIHC
- liver hepatocellular carcinoma
- LUAD
- lung adenocarcinoma
- MSI
- microsatellite instability
- OS
- overall survival
- OV
- ovarian serous cystadenocarcinoma
- PAAD
- pancreatic adenocarcinoma
- RCC
- renal cell carcinoma
- READ
- rectum adenocarcinoma
- TGCT
- testicular germ cell tumor
- THYM
- thymoma
- TIICs
- tumor-infiltrating immune cells
- TMB
- tumor mutation burden
- TME
- tumor microenvironment
- UCEC
- uterine corpus endometrial carcinoma
This study was supported by the Hebei Province Medical Applicable Technology Tracking Project (GZ2022019) and the Hebei Provincial Science and Technology Program (20377714D).
This study did not involve human or animal experiments, so it does not require ethical approval.
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental Digital Content is available for this article.
How to cite this article: Xu X, Yang A, Han Y, Li S, Hao G, Cui N. Pancancer analysis of the interactions between CTNNB1 and infiltrating immune cell populations. Medicine 2024;103:44(e40186).
Contributor Information
Xiaoyuan Xu, Email: 17743760836@163.com.
Aimin Yang, Email: Aiminyang1984@outlook.com.
Yan Han, Email: hanyan1992@outlook.com.
Siran Li, Email: leesr6909@163.com.
Guimin Hao, Email: haoguimin@163.com.
References
- [1].Hong JY, Cho HJ, Sa JK, et al. Hepatocellular carcinoma patients with high circulating cytotoxic T cells and intra-tumoral immune signature benefit from pembrolizumab: results from a single-arm phase 2 trial. Genome Med. 2022;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xu H, Liang XL, Liu XG, Chen NP. The landscape of PD-L1 expression and somatic mutations in hepatocellular carcinoma. J Gastrointestinal Oncol. 2021;12:1132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ishiko A, Matsunaga Y, Masunaga T, Aiso S, Nishikawa T, Shimizu H. Immunomolecular mapping of adherens junction and desmosomal components in normal human epidermis. Exp Dermatol. 2003;12:747–54. [DOI] [PubMed] [Google Scholar]
- [4].Habara O, Logan CY, Kanai-Azuma M, Nusse R, Takase HM. WNT signaling in pre-granulosa cells is required for ovarian folliculogenesis and female fertility. Development (Cambridge, England). 2021;148:dev198846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].To SKY, Mak ASC, Eva Fung YM, et al. β-catenin downregulates Dicer to promote ovarian cancer metastasis. Oncogene. 2017;36:5927–38. [DOI] [PubMed] [Google Scholar]
- [6].Miao W, Lu T, Liu X, Yin W, Zhang H. LncRNA SNHG8 induces ovarian carcinoma cells cellular process and stemness through Wnt/β-catenin pathway. Cancer Biomarkers. 2020;28:459–71. [DOI] [PubMed] [Google Scholar]
- [7].He S, Zhao Y, Wang X, et al. Up-regulation of long non-coding RNA SNHG20 promotes ovarian cancer progression via Wnt/β-catenin signaling. Biosci Rep. 2018;38:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Terakawa J, Serna VA, Taketo MM, Daikoku T, Suarez AA, Kurita T. Ovarian insufficiency and CTNNB1 mutations drive malignant transformation of endometrial hyperplasia with altered PTEN/PI3K activities. Proc Natl Acad Sci USA. 2019;116:4528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kajino-Sakamoto R, Fujishita T, Taketo MM, Aoki M. Synthetic lethality between MyD88 loss and mutations in Wnt/β-catenin pathway in intestinal tumor epithelial cells. Oncogene. 2021;40:408–20. [DOI] [PubMed] [Google Scholar]
- [10].Zhu Y, Huang S, Chen S, et al. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021;12:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xiao X, Mo H, Tu K. CTNNB1 mutation suppresses infiltration of immune cells in hepatocellular carcinoma through miRNA-mediated regulation of chemokine expression. Int Immunopharmacol. 2020;89(Pt A):107043. [DOI] [PubMed] [Google Scholar]
- [12].Wu Q, Xu C, Zeng X, Zhang Z, Yang B, Rao Z. Tumor suppressor role of sFRP‑4 in hepatocellular carcinoma via the Wnt/β‑catenin signaling pathway. Mol Med Rep. 2021;23:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Surun A, Varlet P, Brugières L, et al. Medulloblastomas associated with an APC germline pathogenic variant share the good prognosis of CTNNB1-mutated medulloblastomas. Neuro-Oncology. 2020;22:128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zingg D, Debbache J, Peña-Hernández R, et al. EZH2-Mediated primary cilium deconstruction drives metastatic melanoma formation. Cancer cell. 2018;34:69–84.e14. [DOI] [PubMed] [Google Scholar]
- [15].Zhang W, Mao K, Liu S, Xu Y, Ren J. miR-942-5p promotes the proliferation and invasion of human melanoma cells by targeting DKK3. J Recept Signal Transduct Res. 2021;41:180–7. [DOI] [PubMed] [Google Scholar]
- [16].Zuluaga Gómez LM, Caballero Mojica SC, Vélez Rengifo GJ, Bravo Acosta JD, Montoya Villada JH. CTNNB1 gene mutation associated with neurodevelopmental disorder, microcephaly, and persistence of bilateral hyperplastic primary vitreous: A case report and literature review. Arch Soc Esp Oftalmol. 2022;97:44–7. [DOI] [PubMed] [Google Scholar]
- [17].Ho S, Tsang MH, Fung JL, et al. CTNNB1-related neurodevelopmental disorder in a Chinese population: a case series. Am J Med Genet A. 2022;188:130–7. [DOI] [PubMed] [Google Scholar]
- [18].Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25:9063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Muñoz JP, Huichalaf CH, Orellana D, Maccioni RB. cdk5 modulates beta- and delta-catenin/Pin1 interactions in neuronal cells. J Cell Biochem. 2007;100:738–49. [DOI] [PubMed] [Google Scholar]
- [20].Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Development. 2006;20:1394–404. [DOI] [PubMed] [Google Scholar]
- [21].Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci. 2007;120(Pt 19):3337–44. [DOI] [PubMed] [Google Scholar]
- [22].Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Monga SP. β-Catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology. 2015;148:1294–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kemler R, Hierholzer A, Kanzler B, et al. Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development (Cambridge, England). 2004;131:5817–24. [DOI] [PubMed] [Google Scholar]
- [25].Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–81. [DOI] [PubMed] [Google Scholar]
- [26].Yu J, Liu D, Sun X, et al. CDX2 inhibits the proliferation and tumor formation of colon cancer cells by suppressing Wnt/β-catenin signaling via transactivation of GSK-3β and Axin2 expression. Cell Death Dis. 2019;10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu H, Lu XX, Wang JR, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15:1506–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wen J, Min X, Shen M, et al. ACLY facilitates colon cancer cell metastasis by CTNNB1. J Exp Clin Cancer Res. 2019;38:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ji Y, Lv J, Sun D, Huang Y. Therapeutic strategies targeting Wnt/β‑catenin signaling for colorectal cancer (Review). Int J Mol Med. 2022;49:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].He L, Lu N, Dai Q, et al. Wogonin induced G1 cell cycle arrest by regulating Wnt/β-catenin signaling pathway and inactivating CDK8 in human colorectal cancer carcinoma cells. Toxicology. 2013;312:36–47. [DOI] [PubMed] [Google Scholar]
- [31].Braicu EI, Sehouli J, Richter R, Pietzner K, Denkert C, Fotopoulou C. Role of histological type on surgical outcome and survival following radical primary tumour debulking of epithelial ovarian, fallopian tube and peritoneal cancers. Br J Cancer. 2011;105:1818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Curtin NJ, Szabo C. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat Rev Drug Discovery. 2020;19:711–36. [DOI] [PubMed] [Google Scholar]
- [33].Takeda T, Banno K, Okawa R, et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol Rep. 2016;35:607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Heckl M, Schmoeckel E, Hertlein L, Rottmann M, Jeschke U, Mayr D. The ARID1A, p53 and ß-Catenin statuses are strong prognosticators in clear cell and endometrioid carcinoma of the ovary and the endometrium. PLoS One. 2018;13:e0192881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zyla RE, Olkhov-Mitsel E, Amemiya Y, et al. CTNNB1 mutations and aberrant β-Catenin expression in ovarian endometrioid carcinoma: correlation with patient outcome. Am J Surg Pathol. 2021;45:68–76. [DOI] [PubMed] [Google Scholar]
- [36].Chang KL, Lee MY, Chao WR, Han CP. The status of Her2 amplification and Kras mutations in mucinous ovarian carcinoma. Hum Genomics. 2016;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Becht E, Giraldo NA, Dieu-Nosjean MC, Sautès-Fridman C, Fridman WH. Cancer immune contexture and immunotherapy. Curr Opin Immunol. 2016;39:7–13. [DOI] [PubMed] [Google Scholar]
- [38].Mikami S, Mizuno R, Kondo T, et al. Clinical significance of programmed death-1 and programmed death-ligand 1 expression in the tumor microenvironment of clear cell renal cell carcinoma. Cancer Sci. 2019;110:1820–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sahin IH, Akce M, Alese O, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu L, Bai X, Wang J, et al. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin Cancer Res. 2019;25:7413–23. [DOI] [PubMed] [Google Scholar]
- [42].Klempner SJ, Fabrizio D, Bane S, et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncologist. 2020;25:e147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schrock AB, Ouyang C, Sandhu J, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096–103. [DOI] [PubMed] [Google Scholar]
- [44].Thomas A, Routh ED, Pullikuth A, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology. 2018;7:e1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen L, Zhou Q, Liu J, Zhang W. CTNNB1 alternation is a potential biomarker for immunotherapy prognosis in patients with hepatocellular carcinoma. Front Immunol. 2021;12:759565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang J, Zhu G. A precise prognostic signature in CTNNB1-mutant hepatocellular carcinoma: Prognosis prediction and precision treatment exploration. Heliyon. 2023;9:e22382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.