Abstract
An antigen detection assay for severe acute respiratory syndrome (SARS) coronavirus was established in this study by an indirect immunofluorescence test, which utilized cells derived from throat wash samples of patients with SARS and a rabbit serum that recognized the nucleocapsid protein of SARS-associated coronavirus (SARS-CoV) but not that of other human coronavirus tested. It detected SARS-CoV in 11 of 17 (65%) samples from SARS patients as early as day 2 of illness but in none of the 10 samples from healthy controls. Compared with other diagnostic modalities for detecting SARS-CoV, this assay is simpler, more convenient, and economical. It could be an alternative for early and rapid diagnosis, should SARS return in the future.
Severe acute respiratory syndrome (SARS) is an emerging infectious disease that spread in 2003 in >30 countries, including China, Singapore, Vietnam, Canada, and Taiwan (11, 23). The etiological agent is a novel coronavirus (CoV), the SARS-associated CoV (SARS-CoV) (3, 6, 11, 13). Because of the relatively high transmissibility of SARS, early identification and prompt isolation of cases is one of the most important measures for controlling the disease (11, 26). Nowadays, various laboratory diagnostic modalities such as virus isolation, reverse transcriptase-PCR (RT-PCR), antigen detection, and serological tests have been developed for the diagnosis of SARS-CoV infection (11, 27). Since antibody against SARS-CoV was found to be detectable at least 10 to 28 days after the onset of illness, detection of viral components appears to be the best option for early diagnosis (5, 12, 22, 27). Virus isolation is insensitive and time consuming, and it requires special expertise and a biosafety level 3 facility (3, 6, 13, 19). The RT-PCR assay is sensitive, but it requires a thermal cycler for conventional PCR or more sophisticated machines for real-time PCR (1, 7-9, 14, 16, 21, 28, 29). Recently, the nucleocapsid (N) protein of SARS-CoV was reported to be detectable in sera of SARS patients by a capture enzyme immunoassay, demonstrating the feasibility of an antigen detection assay for SARS-CoV (2). Previously, we reported that SARS-CoV RNA could be detected in cells derived from throat wash samples of patients with SARS at an early stage of infection (21). In this study, we used the cells derived from throat wash samples of SARS patients and the polyclonal serum from a rabbit immunized with the N protein to establish an antigen detection assay for SARS-CoV.
The study included 17 adult patients, who met the clinical case definition of probable SARS and were admitted to the emergency department of the National Taiwan University Hospital between 16 April 2003 and 1 May 2003, during a 2-week period of the SARS outbreak in Taipei, Taiwan (21, 24). The diagnoses of all patients were confirmed by laboratory tests as described previously (21). The first day of fever is defined as day 1 of illness. With the patient's consent, throat wash samples with gargling of 10 ml normal saline were collected in an airborne isolation room according to the guidelines for aerosol-generating procedures (18). All samples were transferred to a biosafety level 3 laboratory and stored at −80°C until use (19).
After thawing, 5 ml of the throat wash samples was centrifuged at 1,500 rpm for 15 min to separate the supernatant and the mucus cell pellet. After the supernatant was collected, the remaining 1-ml portion of the mucus cell pellet was treated with an equal volume of 1% N-acetyl-l-cysteine (NAC) (Sigma, Saint Louis, MO) at room temperature for 25 min and centrifuged at 1,500 rpm for 15 min to further separate the cell pellet from the supernatant. The supernatant was removed, and the remaining 0.88-ml portion was collected as the NAC-treated cells of the throat wash sample, which were then spotted onto 12-well slides. After being air dried in the biosafety cabinet, fixed with cold acetone at −20°C for 20 min, blocked with 1× phosphate-buffered saline (PBS) containing 10% bovine serum albumin (USB, Cleveland, OH) at 37°C for 40 min, and washed with 1× PBS, the slides were incubated with the first antibody at 37°C for 1 h. After being washed with 1× PBS, the slides were incubated with the second antibody at 37°C for 1 h. Details will follow regarding the antibodies used. After a final washing, drying, and mounting, the slides were observed under a fluorescence microscope.
NAC is a mucolytic agent and is commonly used in treating mucins, which are large glycoproteins in the mucus and are known to be associated with cells derived from the respiratory tract (20). To examine the effect of NAC treatment, aliquots of untreated mucus cell pellets from some throat wash samples were prepared on spot slides and incubated with a monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) directed against mucin 5AC, which is one of the major secreted mucin glycoproteins in the respiratory tract (20), followed by the second antibody. As shown in Fig. 1 A to C, large amounts of mucin were readily detected by the anti-mucin 5AC monoclonal antibody in the untreated cells of throat wash samples (Fig. 1B). After treatment with NAC, only small amounts of mucin were seen, demonstrating the mucolytic effect of NAC (Fig. 1C). The NAC-treated cells of throat wash samples were thus used in the subsequent analysis.
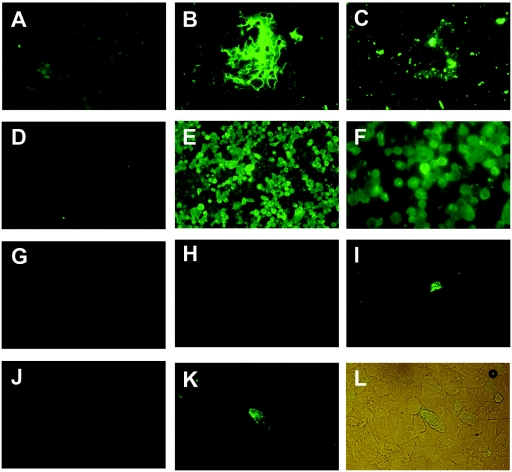
FIG. 1.
Detection of SARS-CoV in the cells derived from throat wash samples of SARS patients by an indirect immunofluorescence assay. (A to C) Untreated (A and B) and NAC-treated (C) cells derived from throat wash samples from a healthy control were incubated with 1× PBS (A) or anti-mucin 5AC monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) (B and C), followed by FITC-conjugated anti-mouse immunoglobulin G. (D and E) SARS-CoV-infected Vero E6 cells were incubated with preimmune (D) or postimmune (E and F) serum from a rabbit immunized with the recombinant nucleocapsid protein of SARS-CoV (5), followed by FITC-conjugated anti-rabbit immunoglobulin G. (F) Magnification (×400) of panel E. (G to L) NAC-treated cells of throat wash samples from a healthy control (G) and two patients (H to K) were incubated with the preimmune (H and J) or postimmune (G, I, and K) rabbit serum described above. (L) Light microscopic picture of panel K, taken with the fluorescent light on.
To develop an antigen detection assay for SARS-CoV, we used the previously described polyclonal serum from a rabbit immunized with the recombinant N protein of SARS-CoV as the first antibody in the indirect immunofluorescence assay (IFA) (5). As the reagent control, spot slides prepared from SARS-CoV-infected Vero E6 cells were incubated with the pre- or postimmune serum, followed by the second antibody, a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit gamma globulin (Pierce Biotechnology, IL). The results revealed that SARS-CoV-infected VeroE6 cells can be detected by the postimmune serum but not by the preimmune serum (Fig. 1D to F), demonstrating the specificity of the rabbit serum. NAC-treated cells of throat wash samples from two SARS patients, subjects no. 1 and no. 6, were next subjected to IFA. As shown in Fig. 1H to K, the postimmune serum but not the preimmune serum reacted with the cells with a speckled pattern. The identity of these cells as epithelial cells was supported by their size and morphology under light microscope (Fig. 1L). Only background signal was observed in the cells prepared from a healthy control, subject no. 18 (Fig. 1G).
The IFA was then employed in the NAC-treated cells of throat wash samples from other 15 SARS patients and 10 healthy controls. Compared with the cells incubated with the preimmune serum, cells incubated with the postimmune serum that showed typical apple-green intracellular fluorescence of a speckle pattern were scored as positive cells. For each sample, 10 fields were examined under a magnification of ×200, corresponding to approximately 300 cells, and 3 or more positive cells were found in each positive sample. The results are summarized in Table 1. As expected, SARS-CoV was not detected in cells derived from healthy controls. In contrast, SARS-CoV was detected in cells of throat wash samples in 11 of the 17 patients, corresponding to a positive rate of 65%. The sampling day of the 11 positive specimens ranged from day 2 to day 9 of illness. There was no correlation between the IFA positivity and sampling day (P = 0.202; Mann-Whitney test). Since the virus load in throat wash samples of these 17 cases has been reported previously (21), the relationship between the IFA positivity and virus load in throat wash samples was compared (Table 1). While the IFA positivity was not significantly correlated with virus load (P = 0.078; Mann-Whitney test), there was a trend of increasing IFA positivity as the virus load increased.
TABLE 1.
Detection of SARS-CoV in cells derived from throat wash samples by an indirect immunofluorescence assay
| Subject no. | Diagnosisa | Sampling dayb | Result of IFAc | Virus load in throat wash samples (copies/ml)d |
|---|---|---|---|---|
| 1 | Probable SARS | d2 | Positive | 1.58 × 105 |
| 2 | Probable SARS | d3 | Positive | 4.69 × 103 |
| 3 | Probable SARS | d3 | Positive | 1.56 × 103 |
| 4 | Probable SARS | d3 | Positive | 2.39 × 104 |
| 5 | Probable SARS | d3 | Positive | 3.56 × 103 |
| 6 | Probable SARS | d4 | Positive | 2.88 × 103 |
| 7 | Probable SARS | d4 | Negative | 1.32 × 103 |
| 8 | Probable SARS | d4 | Positive | 3.98 × 103 |
| 9 | Probable SARS | d4 | Negative | 3.56 × 103 |
| 10 | Probable SARS | d5 | Negative | 8.10 × 103 |
| 11 | Probable SARS | d5 | Positive | 4.10 × 105 |
| 12 | Probable SARS | d6 | Positive | 2.46 × 105 |
| 13 | Probable SARS | d6 | Negative | 2.22 × 103 |
| 14 | Probable SARS | d6 | Negative | 9.73 × 102 |
| 15 | Probable SARS | d7 | Negative | 1.74 × 103 |
| 16 | Probable SARS | d8 | Positive | 5.93 × 106 |
| 17 | Probable SARS | d9 | Positive | 9.58 × 102 |
| 18 | Healthy control | Negative | Undetectable | |
| 19 | Healthy control | Negative | Undetectable | |
| 20 | Healthy control | Negative | Undetectable | |
| 21 | Healthy control | Negative | Undetectable | |
| 22 | Healthy control | Negative | Undetectable | |
| 23 | Healthy control | Negative | Undetectable | |
| 24 | Healthy control | Negative | Undetectable | |
| 25 | Healthy control | Negative | Undetectable | |
| 26 | Healthy control | Negative | Undetectable | |
| 27 | Healthy control | Negative | Undetectable |
Patients of probable SARS were diagnosed according to WHO clinical definitions (24).
Sampling day 2 (d2) is the second day of fever.
An IFA was carried out on cells derived from throat wash samples using a rabbit serum against the N protein of SARS-CoV (5).
Virus load in throat wash was determined by a quantitative real-time RT-PCR assay as reported previously (21).
SARS is believed to be transmitted primarily by dispersal of and contact with respiratory droplets (24, 26). Several respiratory specimens, including nasopharyngeal aspirates or swabs, throat swabs, throat wash samples, bronchoaveolar lavage, and sputum, have been investigated by various groups for early detection of SARS-CoV (3, 6, 11-13, 16, 17, 21, 25, 28). The methodology utilized was primarily an RT-PCR assay, which requires expensive and sophisticated apparatus. An antigen detection assay for the respiratory specimens has not been reported previously. In this study, we developed a simple, convenient, and economical antigen detection assay for SARS-CoV by using cells derived from throat wash samples of SARS patients and a rabbit serum against N protein. The observation that the N protein can be detected in the epithelial cells from throat indicated that SARS-CoV can replicate in the upper respiratory tract. This finding resonates with previous observations of replication of SARS-CoV in the lower respiratory tract, including the bronchial tree and lung (6, 10, 13).
The sensitivity of various RT-PCR assays for SARS-CoV ranged from 32% to >79%, depending on the timing, type, and number of specimens collected (11, 12, 16, 21, 22, 28, 29). The sensitivity of detecting the N protein in sera was recently reported to be 50% and 71% for samples collected between day 3 and day 5 and between day 6 and day 10, respectively (2). Based on our study with a small sample size, this assay can detect SARS-CoV in 65% of samples collected between day 2 and day 9 of illness from SARS patients but in none of those from the healthy controls. Of note was that 7 of the 11 positive samples were collected between day 2 and day 4 with a detection rate of 78% (7 out of 9) during this period (Table 1), suggesting a promising feature of this assay in early diagnosis. Future studies with more cases as well as sequential samples are needed to further evaluate the sensitivity of this assay and determine the time period in which SARS-CoV can be detected by this assay. The specificity of the rabbit serum to the N protein of SARS-CoV was supported by the Western blot analysis, in which the recombinant N protein of SARS-CoV but not that of 229E, a common human coronavirus, was recognized by this serum (reference 5 and data not shown). This is also consistent with the low degree of amino acid similarity, 21% to 33%, between the N proteins of SARS-CoV and three other human coronaviruses (OC43, 229E, and NL [Netherlands] strains) (2, 4, 15, 22). Several rapid antigen detection assays using respiratory specimens have been successfully developed for other respiratory pathogens, such as respiratory syncytial virus, influenza virus, and Chlamydia pneumoniae. The turnaround time of the assay was estimated to be 5 h. Identification of monoclonal antibodies and direct conjugation with fluorescence dye in the future may improve this assay as a rapid diagnosis test by shortening the turnaround time. In brief, compared with the current methods for detecting SARS-CoV, such as virus isolation and RT-PCR, our IFA is simpler, more convenient, and less expensive. If SARS reemerges in the near future, our assay can be an alternative method for the early and rapid detection of SARS-CoV, especially in countries where RT-PCR or a virus isolation method is not available.
Acknowledgments
We are indebted to all the medical personnel at the National Taiwan University Hospital for taking care of SARS patients during the outbreak in Taipei. We thank all other members of the SARS research group of the National Taiwan University College of Medicine-National Taiwan University Hospital, including Ding-Shinn Chen, Yuan-Teh Lee, Hong-Nerg Ho, Chu-Min Teng, and Jin-Town Wang for discussion and coordination.
All authors declare no conflict of interest.
This work was supported by the National Science Council Taiwan (NSC93-2320-B-002-040).
REFERENCES
- 1.Bressler, A. M., and F. S. Nolte. 2004. Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Che, X. Y., L. W. Qiu, Y. X. Pan, K. Wen, W. Hao, L. Y. Zhang, Y. D. Wang, Z. Y. Liao, X. Hua, V. C. Cheng, and K. Y. Yuen. 2004. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 42:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 4.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, L. R., C. M. Chiu, S. H. Yeh, W. H. Huang, P. R. Hsueh, W. Z. Yang, J. Y. Yang, I. J. Su, S. C. Chang, and P. J. Chen. 2004. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J. Med. Virol. 73:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 7.Mahony, J. B., A. Petrich, L. Louie, X. Song, S. Chong, M. Smieja, M. Chernesky, M. Loeb, and S. Richardson. 2004. Performance and cost evaluation of one commercial and six in-house conventional and real-time reverse transcription-PCR assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 42:1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng, E. K., D. S. Hui, K. C. Chan, E. C. Hung, R. W. Chiu, N. Lee, A. Wu, S. S. Chim, Y. K. Tong, J. J. Sung, J. S. Tam, and Y. M. Lo. 2003. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin. Chem. 49:1976-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng, L. F., M. Wong, S. Koh, E. E. Ooi, K. F. Tang, H. N. Leong, A. E. Ling, L. V. Agathe, J. Tan, E. T. Liu, E. C. Ren, L. C. Ng, and M. L. Hibberd. 2004. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J. Clin. Microbiol. 42:347-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris, J. S. M., K. Y. Yuen, A. D. M. E. Osterhaus, and K. Stöhr. 2004. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 12.Peiris, J. S. M., C. M. Chu, V. C. C. Cheng, K. S. Chan, I. F. N. Hung, L. L. M. Poon, K. I. Law, B. S. F. Tang, T. Y. W. Hon, C. S. Chan, K. H. Chan, J. S. C. Ng, B. J. Zheng, W. L. Ng, R. W. M. Lai, Y. Guan, K. Y. Yuen, et al. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon, L. L., O. K. Wong, K. H. Chan, W. Luk, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2003. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin. Chem. 49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 16.Tsang, O. T. Y., T. N. Chau, K. W. Choi, E. Y. K. Tso, W. Lim, M. C. Chiu, W. L. Tong, P. O. Lee, B. H. S. Lam, T. K. Ng, J. Y. Lai, W. C. Yu, and S. T. Lai. 2003. Coronavirus-positive nasopharyngeal aspirate as predictor for severe acute respiratory syndrome mortality. Emerg. Infect. Dis. 9:1381-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.U.S. Centers for Disease Control and Prevention. 2003. Guidelines for collecting specimens from potential SARS patients. [Online.] http://www.cdc.gov/ncidod/sars/guidance/f/app4.htm.
- 18.U.S. Centers for Disease Control and Prevention. 2004. Interim domestic infection control precautions for aerosol-generating procedures on patients with severe acute respiratory syndrome (SARS). [Online.] http://www.cdc.gov/ncidod/sars/aerosolinfectioncontrol.htm.
- 19.U.S. Centers for Disease Control and Prevention. 2004. Laboratory biosafety guidelines for handling and processing specimens associated with SARS-CoV. [Online.] http://www.cdc.gov/ncidod/sars/guidance/f/app5.htm.
- 20.Voynow, J. A. 2002. What does mucin have to do with lung disease? Paediatr. Respir. Rev. 3:98-103. [DOI] [PubMed] [Google Scholar]
- 21.Wang, W. K., S. Y. Chen, I. J. Liu, Y. C. Chen, H. L. Chen, C. F. Yang, P. J. Chen, S. H. Yeh, C. L. Kao, L. M. Huang, P. R. Hsueh, J. T. Wang, W. H. Sheng, C. T. Fang, C. C. Hung, S. M. Hsieh, C. P. Su, W. C. Chiang, J. Y. Yang, J. H. Lin, S. Z. Hsieh, C. P. Hu, Y. P. Chiang, J. T. Wang, P. C. Yang, S. C. Chang,et al. 2004. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 10:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo, P. C., S. K. Lau, B. H. Wong, H. W. Tsoi, A. M. Fung, K. H. Chan, V. K. Tam, J. S. Peiris, and K. Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 42:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. 2003. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. [Online.] http://www.who.int/csr/sars/country/table2004_04_21.
- 24.World Health Organization. 2003. Case definitions for surveillance of severe acute respiratory syndrome (SARS). [Online.] http://www.who.int/csr/sars/casedefinition/en/.
- 25.World Health Organization. 2003. Sampling for severe acute respiratory syndrome (SARS) diagnostic tests. [Online.] http://www.who.int/csr/sars/sampling/en/.
- 26.World Health Organization. Severe acute respiratory syndrome (SARS).[Online.] http://www.who.int/csr/sars/en/.
- 27.World Health Organization. 2003. Severe acute respiratory syndrome (SARS): laboratory diagnostic test.[Online.] http://www.who.int/csr/sars/diagnostictests/en/.
- 28.Wu, H. S., S. C. Chiu, T. C. Tseng, S. F. Lin, J. H. Lin, Y. H. Hsu, M. C. Wang, T. L. Lin, W. Z. Yang, T. L. Ferng, K. H. Huang, L. C. Hsu, L. L. Lee, J. Y. Yang, H. Y. Chen, S. P. Su, S. Y. Yang, S. Y. Lin, T. H. Lin, and I. S. Su. 2004. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg. Infect. Dis. 10:304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yam, W. C., K. H. Chan, L. L. M. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. M. Peiris. 2003. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]