Abstract
A liquid culture followed by molecular confirmation was evaluated for potential to improve sensitivity and reduce time to diagnosis of Mycobacterium avium subsp. paratuberculosis infection. Fecal samples from 240 animals from Ohio farms were assessed for presence of M. avium subsp. paratuberculosis using four different protocols: (i) sedimentation processing followed by inoculation on Herrold's Egg Yolk media (HEYM) slants (monitored biweekly up to 16 weeks), (ii) double centrifugation processing followed by inoculation on HEYM slants (monitored biweekly up to 16 weeks), (iii) liquid-solid double culture method using modified 7H9 broth (8 weeks) followed by subculture on HEYM slants (monitored up to 8 weeks), and (iv) liquid culture using modified 7H9 broth (8 weeks) followed by molecular assays for the presence of two M. avium subsp. paratuberculosis-specific targets. The number of positive samples detected by each protocol was 37, 53, 65, and 76, respectively. Twenty-seven samples were positive by all four methods. Based on samples positive by at least one method (n = 81), the sensitivities for sedimentation processing, double centrifugation processing, liquid-solid double culture, and liquid culture followed by molecular confirmation were 46%, 65%, 80%, and 94%, respectively. Fingerprinting of the positive samples using two polymorphic (G and GGT) short sequence repeat regions identified varying levels of within-farm and between-farm diversity. Our data indicate that liquid culture followed by molecular confirmation can significantly improve sensitivity and reduce time-to-diagnosis (from 16 to 8 weeks) of M. avium subsp. paratuberculosis infection and can also be efficiently employed for the systematic differentiation of M. avium subsp. paratuberculosis strains to understand the epidemiology of Johne's disease.
Johne's disease is a chronic granulomatous enteritis caused by infection with Mycobacterium avium subspecies paratuberculosis. Clinical signs of the disease in cattle include diarrhea, weight loss, fatigue, decreased milk production, and mortality. Johne's disease is now recognized to be of serious economic and animal health consequences in domesticated ruminant species (primarily dairy and beef cattle, sheep, and goats) throughout the world (28, 47). Johne's disease has the greatest economic impact in dairy cattle, where premature culling, reduced carcass value, decreased weight gain, and milk production result in estimated losses up to $250 million annually (31). The growing recognition of M. avium subsp. paratuberculosis infection in wild ruminant and nonruminant species is also of great concern, as it may limit opportunities to control or eradicate Johne's disease from domesticated animals (13, 14). In addition, M. avium subsp. paratuberculosis has been implicated in the etiology of Crohn's disease in humans (5, 16, 22, 34). This has served as a further impetus to control Johne's disease.
Prevention and control of Johne's disease is severely hindered due to its long incubation period, presence of undetected subclinical cases, absence of rapid M. avium subsp. paratuberculosis-specific diagnostic tools and efficacious vaccines, and lack of knowledge of strain diversity (28, 40, 44). Isolation of M. avium subsp. paratuberculosis in conjunction with histopathological lesions is regarded as the gold standard for diagnosis of Johne's disease (28). However, fecal culture alone is frequently used as an acceptable standard procedure for determining an animal's infection status. Although the specificity of the fecal culture method is high, the organism can take 12 to 16 weeks to grow to detectable levels, and even the most sensitive culture methods have less than 50% sensitivity (38, 45). Despite considerable research efforts aimed at development of better fecal processing and detection methods, the sensitivity of fecal cultures remains low. Serologic tests such as agar gel immunodiffusion (18), complement fixation (23), and enzyme-linked immunosorbent assay (ELISA) (9, 15, 23) are limited in their use because of both low specificity and sensitivity (7, 15, 47).
There is much to learn about the dynamics of transmission of infection within animal populations and the involvement of specific subtypes in determining the characteristics of the infections and the velocity of their spread. A comprehensive analysis of within-farm and between-farm diversity in M. avium subsp. paratuberculosis strains will augment our understandingof the distribution and natural history of M. avium subsp. paratuberculosis infections and also aid in the development of a population genetic framework for this economically important bacterium. A coherent framework is necessary for identifying genomic regions of evolutionary or functional importance and in determining how M. avium subsp. paratuberculosis may have evolved and adapted to its hosts.
This study was aimed at addressing two of the issues that hinder implementation of prevention and control strategies. First, liquid culture followed by molecular confirmation was evaluated for potential to improve sensitivity and reduce time to diagnosis of M. avium subsp. paratuberculosis in field samples. Second, short sequence repeat (SSR) analysis of the M. avium subsp. paratuberculosis strains was undertaken to assess the distribution and molecular diversity among M. avium subsp. paratuberculosis strains from farms in Ohio.
MATERIALS AND METHODS
Fecal sample processing.
A total of 240 fecal samples were obtained from farms (n = 40) from several Ohio counties (n = 24) (Fig. 1). These samples represent a convenient subset of the field samples submitted to the Animal Disease Diagnostic Lab (ADDL), Ohio Department of Agriculture, over a 4-week period for routine M. avium subsp. paratuberculosis culture. Each fecal sample was processed using two different methods: (i) sedimentation (43) and (ii) double centrifugation (39).
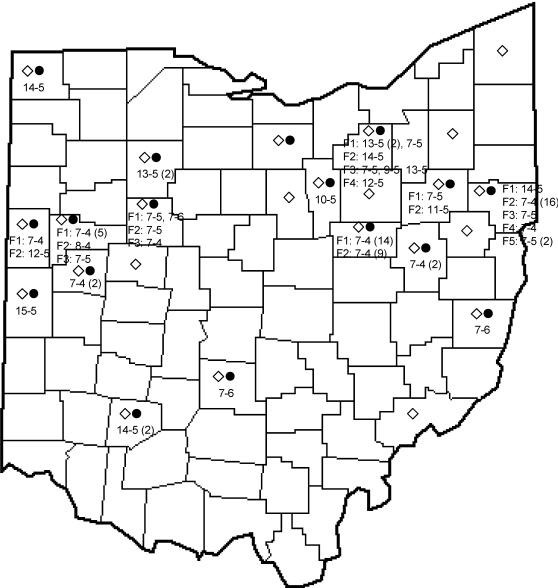
FIG. 1.
Distribution of short sequence repeat fingerprints of M. avium subsp. paratuberculosis isolates from Ohio. Open diamonds indicate the counties from which samples were obtained. Closed circles indicate the counties from which a sample was found to be M. avium subsp. paratuberculosis positive. The alleles are indicated by the number of G repeats followed by the number of GGT repeats. When samples were obtained from more than one farm in a county, they are indicated as F1, F2, and so on. The number of isolates from a particular farm is one unless otherwise indicated by a number in parentheses.
The sedimentation processing was performed as previously described (43). Briefly, 1 g of feces was mixed with 35 ml of sterile double-distilled water and placed on a shaker for 30 min. The sample was then allowed to settle for 30 min at room temperature. Five milliliters of the supernatant was then transferred to a tube containing 25 ml of 0.9% hexadecylpyridinium chloride and incubated overnight at room temperature. The sediment from the bottom of the tube was used to inoculate Herrold's egg yolk medium (HEYM) slants with (2 tubes) and without (1 tube) mycobactin J to assess the mycobactin J dependence commonly associated with M. avium subsp. paratuberculosis. The tubes were monitored biweekly for 16 weeks. A sample was considered positive if any one of the HEYM slants with mycobactin J had one or more colonies, while the HEYM slant without mycobactin J had no growth. A sample was considered negative if none of the HEYM slants had any growth at the end of 16 weeks or if HEYM slants, both with and without mycobactin J, had growth. A slant with fungal or nonmycobacterial contamination was also considered negative. The samples were characterized based on the average CFU/slant. Samples with 1 to 10, 11 to 50, and 51 or more CFU/slants were characterized as low, medium, and heavy shedders, respectively.
The double centrifugation processing was performed as previously described (39). Briefly, 1 g of feces was mixed with 35 ml of sterile double distilled water and placed on a shaker for 30 min. The sample was then allowed to stand for 30 min. The supernatant (25 ml) was poured into a sterile centrifuge tube and centrifuged at 4,000 × g for 20 min. The sediment was resuspended in 25 ml of 0.9% hexadecylpyridinium chloride in 0.5× brain heart infusion (BHI) broth and incubated overnight at 37°C. The suspension was centrifuged again, and the pellet resuspended in 0.5× BHI broth with amphotericin B (50 μg/ml), vancomycin (100 μg/ml), and naldixic acid (100 μg/ml) and incubated overnight at 37°C. The processed samples were used to inoculate HEYM slants with and without mycobactin J as described above. The tubes were monitored biweekly for 16 weeks and assessed using the same criteria described above.
Liquid-solid double culture method.
Fecal samples, processed using the double centrifugation method, were also used to inoculate MB/BacT broths. The MB/BacT system (BioMerieux, Durham, N.C.) utilizes a modified Middlebrook 7H9 broth medium and a proprietary nonradioactive growth detection system. Broth bottles were incubated in a specialized incubator that monitors changes in CO2 production every 10 min. Detection of organism growth was based upon a programmed algorithm. Broth cultures were incubated until the system indicated growth. Five-hundred microliters of the positive broth was subcultured onto HEYM slants with (2 tubes) and without (1 tube) mycobactin J as described above. The remaining broth was frozen at −20°C until use for molecular detection and short sequence repeat analysis. At the end of 8 weeks, all remaining bottles were removed and subcultured onto HEYM slants with and without mycobactin J. The subcultures were monitored for 8 weeks and assessed using the same criteria as described above.
Molecular detection.
An independent laboratory performed the molecular detection from the broth cultures in a blinded fashion. The culture results from sedimentation and double centrifugation processing, and HEYM subcultures were revealed after the molecular analyses were complete. The short sequence repeat analysis was then performed on all the samples that were positive by any of the four protocols.
Cell pellet from 1 ml of broth culture was used for DNA extraction with a QIAamp DNA Blood Mini kit (QIAGEN Inc., Valencia, CA) as described previously (27). The extracted DNA was used for detection of two well-defined molecular markers as previously described (27): (i) PCR amplification and hybridization for two integration loci (L1 and L9) of the insertion sequence IS900 regarded as diagnostically definitive for M. avium subsp. paratuberculosis and (ii) PCR amplification of a M. avium subsp. Paratuberculosis unique sequence (locus 251). Amplification of the hsp65 gene (27) was used to assess presence of PCR inhibitors in the DNA extracts. A sample was considered positive if it was either positive for both L1-L9 and 251 or positive for L1-L9 alone.
Since IS900 has been demonstrated in mycobacteria other than M. avium subsp. paratuberculosis (11, 17), its specificity is debatable. In a published analysis of 600 bacterial cultures, L1-L9 and 251 have detected M. avium subsp. paratuberculosis with 100% specificity (27). Hence, L1-L9 and 251 have been the markers of choice over IS900. However, there still exists the possibility that one or both of these loci may be absent in a rare M. avium subsp. paratuberculosis isolate.
Short sequence repeat analysis.
PCR amplification of two of the most discriminatory SSR loci, mononucleotide G and trinucleotide GGT repeats (3), was carried out as described previously (26) using primer sets (i) 5′-TCA GAC TGT GCG GTA TGG AA-3′ and 5′-GTG TTC GGC AAA GTC GTT GT-3′ and (ii) 5′-AGA TGT CGA CCA TCC TGA CC-3′ and 5′-AAG TAG GCG TAA CCC CGT TC-3′, respectively. Five microliters of the PCR product was electrophoresed at 125 V for 45 min in 1.5% agarose gels prestained with ethidium bromide and visualized on a UV transilluminator (Alpha Innotech Corporation, San Leandro, CA). For both the repeat loci under investigation, only those PCR products that were detectable on 1.5% agarose gels were sequenced for further analysis. The PCR products were purified with a QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA) and sequenced by using standard dye terminator chemistry, and the sequences were analyzed on an automated DNA sequencer (3700 DNA Analyzer, Applied Biosystems, Foster City, CA).
A subset of isolates (n = 16) representing a single farm in Columbiana county were further assessed using an additional SSR loci. This loci also contained mononucleotide G repeats and was the third most discriminatory loci described (3). PCR amplification and sequence analysis was performed as described above using primers 5′-GTG ACC AGT GTT TCC GTG TG-3′ and 5′-TGC ACT TGC ACG ACT CTA GG-3′
All chromatograms were visually inspected and sequences were edited with EditSeq (DNASTAR, Madison, WI) to correct ambiguities and then aligned using MegAlign (DNASTAR, Madison, WI) to identify the number of repeats in both loci for each isolate. The alleles were assigned a number congruent to the number of G and GGT residues.
RESULTS
M. avium subsp. paratuberculosis detection.
A total of 240 fecal samples were analyzed using four different protocols. Thirty-seven of the 240 samples were positive by sedimentation processing, 53 by double centrifugation processing, 65 by liquid-solid double culture, and 76 by liquid culture followed by molecular confirmation. Thirteen samples were considered negative due to contamination. Of these, two were contaminated in the double centrifugation method, seven were in the liquid-solid double culture method, and four were contaminated in both methods. Of the 37 samples that were positive by the sedimentation method, 7 were characterized as heavy shedders, 16 as medium shedders, and 14 as low shedders. Of the 53 samples that were positive by the double centrifugation method, 23 were characterized as heavy shedders, 12 as medium shedders, and 18 as low shedders. Of the 65 samples that were positive by the liquid-solid double culture method, 41 were characterized as heavy shedders, 13 as medium shedders, and 11 as low shedders. Of the 76 samples that were positive by molecular detection, 69 were positive for both L1-L9 and 251 while 7 were positive for L1-L9 alone.
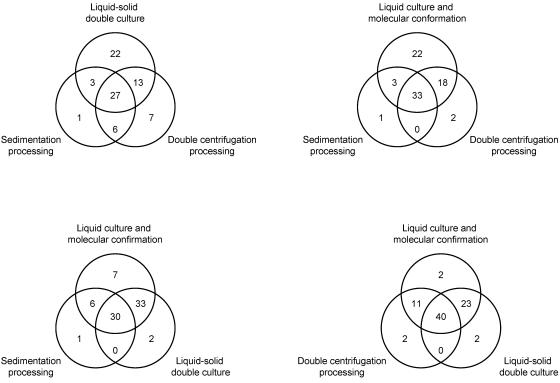
Twenty-seven samples were positive by all four protocols, while 81 samples were positive by at least one of the four methods (Fig. 2). Of the five samples that were negative by molecular detection, one was positive by sedimentation processing, two were positive by double centrifugation processing, and two were positive by liquid-solid double culture. All five of these samples were confirmed to be M. avium subsp. paratuberculosis positive using DNA extracts from slant cultures. Using the total number of samples positive by at least one method (n = 81), the sensitivities for sedimentation processing, double centrifugation processing, liquid-solid double culture, and liquid culture followed by molecular confirmation were 46%, 65%, 80%, and 94%, respectively. The kappa coefficients for sedimentation processing, double centrifugation processing, and liquid-solid double culture compared to liquid culture followed by molecular confirmation were 0.54, 0.72, and 0.84, respectively. Contingency tables for sedimentation processing, double centrifugation processing, and liquid-solid double culture compared to liquid culture followed by molecular confirmation are shown in Table 1.
FIG. 2.
Identification of M. avium subsp. paratuberculosis in fecal samples by sedimentation processing, double centrifugation processing, liquid-solid double culture, and liquid culture followed by molecular confirmation.
TABLE 1.
Contingency tables for sedimentation processing, double centrifugation processing, and liquid-solid double culture compared to liquid culture followed by molecular confirmationa
| Result of liquid culture and molecular confirmation | No. of samples
|
|||||
|---|---|---|---|---|---|---|
| Sedimentation processing
|
Double centrifugation processing
|
Liquid-solid double culture
|
||||
| Positive | Negative | Positive | Negative | Positive | Negative | |
| Positive | 36 | 40 | 51 | 25 | 63 | 13 |
| Negative | 1 | 163 | 2 | 162 | 2 | 162 |
The respective kappa coefficients were as follows: for sedimentation processing, 0.54; for double centrifugation processing, 0.72; and for liquid-solid double culture, 0.84.
Short sequence repeat analysis.
To demonstrate the feasibility of using broth cultures for molecular epidemiology studies, two regions within the M. avium subsp. paratuberculosis genome that carry varying numbers of G and GGT residues were amplified from all samples that were positive for M. avium subsp. paratuberculosis (3). Of the 81 samples analyzed, only those (n = 80) with detectable G and GGT repeat products (approximately 425 bp each) were sequenced for further analysis. These samples represented 31 farms from 16 Ohio counties. The fingerprints were designated by the number of G repeats followed by the number of GGT repeats. A total of 11 fingerprints with 7 to 15 G-residue repeats and 4 to 6 GGT-residue repeats were identified among the 80 M. avium subsp. paratuberculosis samples included in the analysis. The distribution of the fingerprints by the farm and county are shown in Fig. 1. Sixty-four percent of the M. avium subsp. paratuberculosis isolates (51 of 80) had the 7g-4ggt repeat fingerprint. Fingerprints of isolates from the same farm were identical for all but three farms (Fig. 1). One farm, in Hardin county, had two isolates with different fingerprints: 7g-5ggt and 7g-6ggt. The second farm, in Medina county, had three isolates with different fingerprints: 7g-5ggt, 9g-5ggt, and 13g-5ggt. The third farm, also in Medina county, had three positive samples, two of which had the 13g-5ggt fingerprint profile, while the third one had the 7g-5ggt profile. All isolates (n = 16) from a single farm in Columbiana county that were analyzed using the additional G-repeat loci contained nine G-residue repeats.
DISCUSSION
Johne's disease is a chronic and progressive intestinal disease in ruminants caused by M. avium subsp. paratuberculosis (6). This disease has emerged as one of the most prevalent and costly infectious diseases of dairy cattle in countries around the world (28-31). In addition, controversy regarding potential zoonotic association between M. avium subsp. paratuberculosis and Crohn's disease persists (22, 34). Due to concerns about animal health, economic considerations, and zoonotic potential of paratuberculosis, several countries and many states within the United States have instituted Johne's disease certification programs for prevention and control of the disease (2, 21, 24, 37, 42). Adequate diagnostic tests are an essential element of a control program. This necessitates the development of high-throughput, sensitive diagnostic methods for the detection of infected and subclinical animals. Knowledge of the extent of strain sharing across different host species is vital to understanding the dynamics of M. avium subsp. paratuberculosis transmission. Methods for differentiation or subtyping of bacterial strains provide important information for molecular epidemiological analyses and help provide an understanding of the population genetics of the organism.
Diagnostics.
Diagnosis of Johne's disease presents a major problem in its prevention and control. Currently, cultivation of M. avium subsp. paratuberculosis from fecal and tissue specimens remains the most definitive method for detecting animals with Johne's disease. However, culturing for M. avium subsp. paratuberculosis has several disadvantages, such as long growth time, long processing time, difficulties in control of contamination on culture media, and need for storage of fecal specimens for delayed processing. Serological assays such as ELISA, agar gel immunodiffusion assay, and complement fixation test are also commonly used to diagnose paratuberculosis in a herd because of the low cost of the test and rapid availability of test results. However, serological tests are of limited value due to low specificity and sensitivity, since antibodies may not be detectable either due to anergy or until their late appearance in the pathogenesis of Johne's disease (7, 47).
The results of this study indicate that liquid culture followed by molecular confirmation was the most effective approach to detecting and typing M. avium subsp. paratuberculosis isolates in feces from naturally infected cattle. In our analysis, only 27 samples tested positive by all four protocols. Seventy-six of the total 81 positive samples were detected with liquid culture followed by molecular confirmation. All but two of the broth enriched-molecular assay-positive samples were positive by at least one of the other three protocols. This indicates that the 76 samples detected by molecular analyses were true positives and that the other three protocols are less sensitive than molecular assay of liquid cultures. In addition, the liquid culture-molecular assay-positive samples were detected by 8 weeks postinoculation, which is considerably faster than the 16-week incubation period required for the other three procedures.
In the sedimentation procedure, 5 ml of the suspension, representing approximately 0.14 g, is processed, while in the double centrifugation protocol, 25 ml of the suspension, representing approximately 0.71 g, is processed. This difference in the amount of fecal sample processed is reflected in the total number of positive samples by each protocol. Our results support previous observation that the double centrifugation method was a major improvement in sample processing over the sedimentation method (39). Of the 76 samples that were positive by molecular detection, 69 were positive for both L1-L9 and 251 while 7 were positive for L1-L9 alone. Since L1-L9 is a PCR-hybridization assay, its sensitivity is at least two logs greater than that of a PCR assay. Hence, it is expected that L1-L9 assay will detect more positive samples than 251 assay.
The MB/BacT system used in this study was designed for detection of M. tuberculosis from human hosts (36). A recent study has indicated that the sensitivity of detection of M. avium subsp. paratuberculosis in bovine feces by this system using the double centrifugation processing method was approximately 103 CFU/g of feces (41). However, the efficacy of the MB/BacT system in detection of M. avium subsp. paratuberculosis in field fecal specimens has not been reported. Although the purpose of this study was not to assess if this system can be used for M. avium subsp. paratuberculosis identification in fecal samples, it is worth noting that a nonradiometric broth-based system (20, 41) provides an easier, safer, and more cost-effective method for detection of mycobacteria than conventional culturing on solid media.
An advantage of fecal and tissue culture is isolation of specific M. avium subsp. paratuberculosis strains causing the infections. Thus, further studies can be performed to facilitate understanding of the biology and molecular epidemiology of this pathogen. An added advantage of broth over solid media-based culture is the relatively short time required to detect M. avium subsp. paratuberculosis (8, 10, 25, 41, 46). Some studies have identified M. avium subsp. paratuberculosis strains that do not show mycobactin J dependency (1, 26), indicating that diagnostic tests that rely on mycobactin J dependency alone need to be interpreted with caution. Use of molecular assays to confirm the identity of the isolates instead of mycobactin J dependency resolves this discrepancy. Although significant increase in sensitivity was achieved by coupling early broth cultures with molecular assays, there is a need for alternative processing procedures. For example, several studies have reported approximately two log-fold loss of M. avium subsp. paratuberculosis during fecal sample processing (32, 35, 41). Further investigation for alternative methods to process feces prior to culture may result in improvement of sensitivity of the reported assays.
Molecular subtyping.
Critical to the long-term goal of prevention and control of Johne's disease is the control of transmission of M. avium subsp. paratuberculosis on dairy and beef operations, resulting in improved animal (and potentially public) health. Molecular epidemiologic applications provide unique and powerful tools to assist in this endeavor. This study evaluated the potential of short sequence repeat analysis to aid in the characterization of transmission of Johne's disease on dairy farms, a step critical to advancements in paratuberculosis research.
This study has identified the existence of at least one dominant M. avium subsp. paratuberculosis subtype (7g-4ggt) in cattle herds in Ohio as well as the existence of multiple subtypes of M. avium subsp. paratuberculosis on three of the dairy farms with infected cattle. In an attempt to further dissect the M. avium subsp. paratuberculosis subtypes, all isolates from a single farm in Columbiana county were assessed using an additional G-repeat loci. All 16 of these isolates which had the 7g-4ggt profile also carried identical numbers of G repeats in the third SSR loci. This suggests that these isolates may be clonal and that there may be occurrence of intraherd transmission on this particular farm. Although the addition of the third locus did not break down the alleles identified by the first two loci, there may be situations where addition of other SSR loci into the analysis may provide meaningful diversity information.
It is expected that herds with a higher percentage of purchased cattle will have a more diverse subtype population than herds with fewer cattle introductions. However, the information about animal purchases was not available. Also, the movement of the dairy industry toward calf-heifer raising operations has increased the possibility of introduction of infected heifer into an uninfected herd. Some of these calf-heifer raising operation may receive infected animals from a variety of sources that could result in introduction of multiple strains. Application of SSR analysis for strain differentiation of M. avium subsp. paratuberculosis and development of population genetic frameworks could lead to better understanding of the role of cow to calf and other routes of transmission.
Other DNA-based molecular subtyping techniques, such as multiplex PCR for IS900 integration loci (4, 27), restriction fragment length polymorphism analysis (12, 33), and amplified fragment length polymorphism analysis (27) have been unable to resolve M. avium subsp. paratuberculosis isolates into meaningful epidemiologic groups due to the apparent restricted genetic diversity within the subspecies. In contrast, SSR analyses enabled high-resolution subtyping of M. avium subsp. paratuberculosis isolates from domestic and wild animal species and has opened new possibilities for a better understanding of transmission of M. avium subsp. paratuberculosis clonal populations (19, 26). Since the analysis is based on DNA sequencing, the results are unambiguous, reproducible, and amenable to adaptation for high-throughput analysis. Furthermore, SSR analyses will enable accurate interlaboratory comparisons to be made and the information used in the development of SSR databases for further molecular epidemiologic studies.
Since the SSR analysis was performed on a convenient subset of the field samples submitted to the Ohio ADDL over a 4-week period for routine M. avium subsp. paratuberculosis culturing, the subtypes do not necessarily represent the natural distribution in Ohio. A more systematic prospective study involving multiple herds and operation types needs to be undertaken to evaluate the associations between subtype status and dam-daughter status or other environmental associations and between introduction of new cattle and change in subtype distribution through time.
In conclusion, early broth cultures coupled with molecular assay not only improve sensitivity and reduce time to diagnosis of M. avium subsp. paratuberculosis from bovine feces but can potentially be used to aid in characterization of the transmission of Johne's disease on dairy farms. Our preliminary analyses suggest that this approach will be of considerable utility in enabling detailed molecular epidemiologic and population genetic analyses of this important animal pathogen.
Acknowledgments
This study was supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center (OARDC), including an OARDC Competitive Research Enhancement Seed Grant awarded to S. Sreevatsan.
REFERENCES
- 1.Aduriz, J. J., R. A. Juste, and N. Cortabarria. 1995. Lack of mycobactin dependence of mycobacteria isolated on Middlebrook 7H11 from clinical cases of ovine paratuberculosis. Vet. Microbiol. 45:211-217. [DOI] [PubMed] [Google Scholar]
- 2.Allworth, M. B., and D. J. Kennedy. 2000. Progress in national control and assurance programs for ovine Johne's disease in Australia. Vet. Microbiol. 77:415-422. [DOI] [PubMed] [Google Scholar]
- 3.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:2185-2197. [DOI] [PubMed] [Google Scholar]
- 5.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 74:218-262. [PubMed] [Google Scholar]
- 7.Collins, M. T. 1996. Diagnosis of paratuberculosis, p. 357-371. In R. W. Sweeney (ed.), The veterinary clinics of North America food animal practice. Paratuberculosis (Johne's Disease). W. B. Saunders Co., Philadelphia, Pa. [DOI] [PubMed]
- 8.Collins, M. T., K. B. Kenefick, D. C. Sockett, R. S. Lambrecht, J. McDonald, and J. B. Jorgensen. 1990. Enhanced radiometric detection of Mycobacterium paratuberculosis by using filter-concentrated bovine fecal specimens. J. Clin. Microbiol. 28:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, M. T., and D. C. Sockett. 1993. Accuracy and economics of the USDA-licensed enzyme-linked immunosorbent assay for bovine paratuberculosis. J. Am. Vet. Med. Assoc. 203:1456-1463. [PubMed] [Google Scholar]
- 10.Cousins, D. V., R. J. Evans, and B. R. Francis. 1995. Use of BACTEC radiometric culture method and polymerase chain reaction for the rapid screening of faeces and tissues for Mycobacterium paratuberculosis. Aust. Vet. J. 72:458-462. [DOI] [PubMed] [Google Scholar]
- 11.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 12.Cousins, D. V., S. N. Williams, A. Hope, and G. J. Eamens. 2000. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp. paratuberculosis using IS900 RFLP. Aust. Vet. J. 78:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Daniels, M. J., M. R. Hutchings, P. M. Beard, D. Henderson, A. Greig, K. Stevenson, and J. M. Sharp. 2003. Do non-ruminant wildlife pose a risk of paratuberculosis to domestic livestock and vice versa in Scotland? J. Wildl. Dis. 39:10-15. [DOI] [PubMed] [Google Scholar]
- 14.Daniels, M. J., M. R. Hutchings, and A. Greig. 2003. The risk of disease transmission to livestock posed by contamination of farm stored feed by wildlife excreta. Epidemiol. Infect. 130:561-568. [PMC free article] [PubMed] [Google Scholar]
- 15.Dargatz, D. A., B. A. Byrum, L. K. Barber, R. W. Sweeney, R. H. Whitlock, W. P. Shulaw, R. H. Jacobson, and J. R. Stabel. 2001. Evaluation of a commercial ELISA for diagnosis of paratuberculosis in cattle. J. Am. Vet. Med. Assoc. 218:1163-1166. [DOI] [PubMed] [Google Scholar]
- 16.El Zaatari, F. A., M. S. Osato, and D. Y. Graham. 2001. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol. Med. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 17.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira, R., L. S. Fonseca, and W. Lilenbaum. 2002. Agar gel immunodiffusion test (AGID) evaluation for detection of bovine paratuberculosis in Rio de Janeiro, Brazil. Lett. Appl. Microbiol. 35:173-175. [DOI] [PubMed] [Google Scholar]
- 19.Ghadiali, A. H., M. Strother, S. A. Naser, E. J. Manning, and S. Sreevatsan. 2004. Mycobacterium avium subsp. paratuberculosis strains isolated from Crohn's disease patients and animal species exhibit similar polymorphic locus patterns. J. Clin. Microbiol. 42:5345-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant, I. R., R. B. Kirk, E. Hitchings, and M. T. Rowe. 2003. Comparative evaluation of the MGIT and BACTEC culture systems for the recovery of Mycobacterium avium subsp. paratuberculosis from milk. J. Appl. Microbiol. 95:196-201. [DOI] [PubMed] [Google Scholar]
- 21.Groenendaal, H., M. Nielen, and J. W. Hesselink. 2003. Development of the Dutch Johne's disease control program supported by a simulation model. Prev. Vet. Med. 60:69-90. [DOI] [PubMed] [Google Scholar]
- 22.Hermon-Taylor, J. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalis, C. H., H. W. Barkema, J. W. Hesselink, C. van Maanen, and M. T. Collins. 2002. Evaluation of two absorbed enzyme-linked immunosorbent assays and a complement fixation test as replacements for fecal culture in the detection of cows shedding Mycobacterium avium subspecies paratuberculosis. J. Vet. Diagn. Investig. 14:219-224. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, D. J., and M. B. Allworth. 2000. Progress in national control and assurance programs for bovine Johne's disease in Australia. Vet. Microbiol. 77:443-451. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 26.Motiwala, A. S., A. Amonsin, M. Strother, E. J. Manning, V. Kapur, and S. Sreevatsan. 2004. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis isolates recovered from wild animal species. J. Clin. Microbiol. 42:1703-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Academy of Sciences Report. 2003. Diagnosis and control of Johne's Disease http://books.nap.edu/openbook/0309086116/html/index.html.
- 29.National Animal Health Monitoring System. 1997. Johne's disease on U. S. dairy operations. Report N245.1087. USDA, APHIS, VS, CEAH, National Animal Health Monitoring System, Fort Collins, Colo.
- 30.Nielsen, S. S., C. Gronbaek, J. F. Agger, and H. Houe. 2002. Maximum-likelihood estimation of sensitivity and specificity of ELISAs and faecal culture for diagnosis of paratuberculosis. Prev. Vet. Med. 53:191-204. [DOI] [PubMed] [Google Scholar]
- 31.Ott, S. L., S. J. Wells, and B. A. Wagner. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179-192. [DOI] [PubMed] [Google Scholar]
- 32.Ozbek, A., F. C. Michel, M. Strother, A. S. Motiwala, B. R. Byrum, W. P. Shulaw, C. G. Thornton, and S. Sreevatsan. 2003. Evaluation of two recovery methods for detection of Mycobacterium avium subsp. paratuberculosis by PCR: direct-dilution-centrifugation and C(18)-carboxypropylbetaine processing. FEMS Microbiol. Lett. 229:145-151. [DOI] [PubMed] [Google Scholar]
- 33.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, M. R. Du, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 34.Quirke, P. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddacliff, L. A., A. Vadali, and R. J. Whittington. 2003. The effect of decontamination protocols on the numbers of sheep strain Mycobacterium avium subsp. paratuberculosis isolated from tissues and faeces. Vet. Microbiol. 95:271-282. [DOI] [PubMed] [Google Scholar]
- 36.Rohner, P., B. Ninet, C. Metral, S. Emler, and R. Auckenthaler. 1997. Evaluation of the MB/BacT system and comparison to the BACTEC 460 system and solid media for isolation of mycobacteria from clinical specimens. J. Clin. Microbiol. 35:3127-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sockett, D. C. 1996. Johne's disease eradication and control: regulatory implications. Vet. Clin. N. Am. Food Anim. Pract. 12:431-440. [DOI] [PubMed] [Google Scholar]
- 38.Sockett, D. C., T. A. Conrad, C. B. Thomas, and M. T. Collins. 1992. Evaluation of four serological tests for bovine paratuberculosis. J. Clin. Microbiol. 30:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabel, J. R. 1997. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J. Vet. Diagn. Investig. 9:375-380. [DOI] [PubMed] [Google Scholar]
- 40.Stabel, J. R., T. L. Bosworth, T. A. Kirkbride, R. L. Forde, and R. H. Whitlock. 2004. A simple, rapid, and effective method for the extraction of Mycobacterium paratuberculosis DNA from fecal samples for polymerase chain reaction. J. Vet. Diagn. Investig. 16:22-30. [DOI] [PubMed] [Google Scholar]
- 41.Stich, R. W., B. Byrum, B. Love, N. Theus, L. Barber, and W. P. Shulaw. 2004. Evaluation of an automated system for non-radiometric detection of Mycobacterium avium paratuberculosis in bovine feces. J. Microbiol. Methods 56:267-275. [DOI] [PubMed] [Google Scholar]
- 42.Tharaldsen, J., B. Djonne, B. Fredriksen, O. Nyberg, and O. Siguroardottir. 2003. The National Paratuberculosis Program in Norway. Acta Vet. Scand. 44:243-246. [PubMed] [Google Scholar]
- 43.Whipple, D. L., D. R. Callihan, and J. L. Jarnagin. 1991. Cultivation of Mycobacterium paratuberculosis from bovine fecal specimens and a suggested standardized procedure. J. Vet. Diagn. Investig. 3:368-373. [DOI] [PubMed] [Google Scholar]
- 44.Whitlock, R. H., and C. Buergelt. 1996. Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. N. Am. Food Anim. Pract. 12:345-356. [DOI] [PubMed] [Google Scholar]
- 45.Whitlock, R. H., S. J. Wells, R. W. Sweeney, and J. Van Tiem. 2000. ELISA and fecal culture for paratuberculosis (Johne's disease): sensitivity and specificity of each method. Vet. Microbiol. 77:387-398. [DOI] [PubMed] [Google Scholar]
- 46.Whittington, R. J., I. Marsh, M. J. Turner, S. McAllister, E. Choy, G. J. Eamens, D. J. Marshall, and S. Ottaway. 1998. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified BACTEC 12B radiometric culture and direct confirmation by IS900 PCR. J. Clin. Microbiol. 36:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whittington, R. J., and E. S. Sergeant. 2001. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J. 79:267-278. [DOI] [PubMed] [Google Scholar]