Abstract
Herpes simplex virus (HSV) is the most common cause of acquired, sporadic encephalitis in the United States. PCR identification of HSV in spinal fluid has become the diagnostic gold standard due to its sensitivity and potential for speed, replacing other methods such as culture. We developed a real-time PCR assay to detect HSV, using a new type of hybridization probe, the Eclipse probe. In this study, we ran 323 samples (171 positives and 152 negatives) with the Eclipse real-time PCR assay and compared these results with another PCR assay using gel detection. The real-time assay agreed with our reference method for 319 out of the 323 samples tested (99%). Using two different real-time PCR platforms, we discovered that SNPs within the amplicon's probe binding region that are used to distinguish HSV-1 from HSV-2 can decrease assay sensitivity. This problem is potentially a general one for assays using fluorescent probes to detect target amplification in a real-time format. While real-time PCR can be a powerful tool in the field of infectious disease, careful sequence evaluation and clinical validation are essential in creating an effective assay.
PCR is now accepted as the gold standard for diagnosis of herpes simplex virus (HSV) infections in the central nervous system (CNS), exhibiting a high degree of specificity and a sensitivity superior to that of culture or DFA (3, 7, 10, 11, 17, 18). While viral culture for HSV encephalitis is still routinely employed in the clinical lab, it is only capable of detecting virus in less than 5% of patients with CNS infections (10, 14). Detection of low viral titers often found in spinal fluid requires the high level of sensitivity that is provided by amplification-based assays (13).
In recent years, real-time PCR has made testing more convenient. Real-time PCR is more amenable to automation, allowing higher throughputs and decreased turnaround times. In addition, the implementation of a one-tube closed reaction greatly reduces the possibility for contamination. Besides the potential for sensitivity roughly equivalent to gel detection, the use of target-specific probes in real-time PCR offers an additional level of specificity. These probes also allow the detection of single nucleotide polymorphisms (SNPs) using melt curve analysis, permitting typing of different viral species.
While real-time PCR has been widely adopted for molecular infectious disease testing, real-time technology has often been compared to traditional methods such as culture, with a predictable enhancement in sensitivity (4, 6, 7, 12). The current literature is more limited on the comparative performance of real-time PCR with established PCR formats. There is little definitive published evidence that real-time detection using fluorescent probes is more sensitive than end detection by gel, raising the question of whether real-time PCR is necessarily more sensitive than conventional PCR with gel detection. Therefore, it is essential during the validation of any real-time assay to compare its performance to something in addition to culture, preferably another well-optimized amplification assay.
The presence of an SNP beneath a probe lowers the probe Tm relative to the consensus sequence (2, 15). However, when using hybridization probes to detect known sequence polymorphisms within the target, the potential exists for loss of sensitivity due to compromised annealing to mismatched targets. The presence of unexpected SNPs presents an additional challenge to maintaining desired sensitivity. We developed a multiplexed, real-time assay for the detection of HSV-1 and -2, using a novel type of hybridization probe termed the Eclipse probe (Epoch Biosciences) (1, 9). As a further assay improvement, we introduced an internal control into the sample extraction, allowing us to monitor nucleic acid extraction and detect inhibition of the PCR. In this report we describe the assay and its clinical validation. Furthermore, we present data from both our Eclipse probe assay and a Roche LightCycler assay that illustrate potential loss of sensitivity encountered when developing assays that attempt to distinguish sequence variants by melting analysis. This report is the first to demonstrate that sequence polymorphisms under the probe can negatively affect sensitivity in a real-time PCR assay. Our findings have general implications for this form of real-time testing.
MATERIALS AND METHODS
Specimens.
During the clinical validation of this assay, we tested 323 samples (amniotic fluids, spinal fluids, plasma samples, serum samples, bronchioalveolar lavage fluids, ocular fluids, swab samples, and tissues). Due to the infrequent number of positive samples obtained for ocular fluids, amniotic fluids, bronchioalveolar fluids, and tissues, most or all of the positive samples for these sample types were spiked. In these cases, positive control material was diluted into the appropriate HSV-negative sample type, at a ratio of 1:1. We obtained all unspiked positive samples from the clinical lab. These samples had previously been identified as HSV positive using the current PCR assay (gel detection). Positive control material consisted of cell culture material (mixture of HSV-1 and HSV-2 obtained from the ARUP Virology Lab; approximate concentration of 5,000 copies per ml) diluted to have a crossing threshold of approximately 35. A negative control of extracted water was included in each run.
Internal control.
The internal control for this assay is a plasmid containing a transcriptional fusion of green fluorescent protein (GFP) coding sequence downstream of a Caenorhabditis elegans promoter. The forward primer is within the C. elegans sequence with the reverse primer in the GFP gene, creating an amplicon that spans the breakpoint between the two regions.
Extractions.
With the exception of the tissue samples that were extracted with the Puregene tissue extraction kit, all samples were extracted using the QIAGEN 96-well blood kit, with a few minor modifications. Protease (25 μl), patient sample (200 μl), and prewarmed lysis buffer AVL (200 μl) were added to each well of the round well block in that order. For quality control of the extraction, the internal control was added to the lysis buffer at a concentration of 50,000 copies per ml (10,000 copies per extraction). The block was then shaken for 15 seconds and spun down briefly at 3,000 rpm. The block was incubated at 70°C for 10 min and spun down as before. A 200-μl volume of ethanol was added to each extraction, and the block was shaken for 15 seconds and spun down again briefly at 3,000 rpm. Samples were transferred to the filter plate, covered with airpore tape, and spun for 4 min at 6,000 rpm to apply the samples to the filter. The samples were washed with 500 μl of buffer AW1 (spun for 2 min at 6,000 rpm) and 500 μl of buffer AW2 (spun for 3 min at 6,000 rpm). The filter block was incubated for 10 min at 70°C. The filter plate was placed over the collection tubes, and 50 μl of prewarmed elution buffer (buffer AE; 70°C) was added. The plate was incubated at 70°C for 5 min and spun for 4 min at 6,000 rpm. An additional 50 μl of elution buffer was added to each sample well, and the plate spun another 4 min at 6,000 rpm.
PCR assay with gel detection.
The PCR assay amplifies a 179-bp region from the HSV DNA polymerase gene (5, 10, 12). Samples were extracted as described above. Ten microliters of extracted patient sample was added to 40 μl of master mix. The master mix contained primers (forward, ATCAACTTCGACTGGCCCTT; reverse, CCGTACATGTCGATGTTCAC) at 500 nM, deoxynucleoside triphosphates (dNTPs) at 250 μM with UTP at 500 μM, MgCl2 at 2.75 mM, 0.5 U of UNG, AmpliTaq LD, and TaqStart antibody in Tris-EDTA (TE) buffer. The PCRs were run using the following cycling parameters: 10 min at 50°C followed by 45 cycles of 94°C for 5 seconds, 64°C for 5 seconds, and 72°C for 20 seconds. These were followed by a 72°C hold until samples could be retrieved. Following amplification, 10 μl of each product was analyzed by gel electrophoresis.
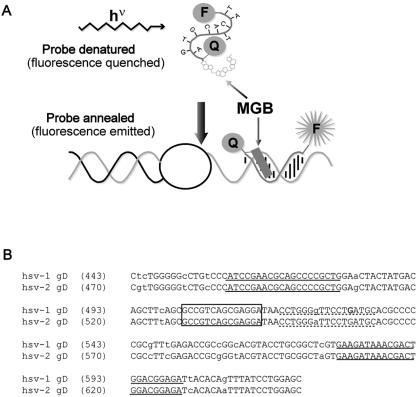
Eclipse probes. Eclipse probes are similar to TaqMan probes, both being dual labeled with a fluorescent dye and a quenching molecule, but differing in the orientation of the probe and quencher (Fig. 1a). Eclipse probes have the fluorescent dye attached to their 3′ end, with a nonfluorescent, dark quencher on the 5′ end, while TaqMan probes have the opposite orientation. Another key feature of the Eclipse probe is the incorporation of a minor groove-binding molecule (MGB) at the 5′ end, adjacent to the quencher. This minor groove binder, which is structurally similar to many naturally occurring antibiotics, serves a dual function. It stabilizes the binding between the probe and target molecule, allowing shorter probes to be used, and blocks probe hydrolysis that occurs when the advancing Taq polymerase encounters a TaqMan probe. Consequently, rather than being cleaved and consumed during the reaction, Eclipse probes remain available to be used in a dissociation analysis. In contrast to TaqMan hydrolysis probes, an Eclipse hybridization probe allows amplified products to be confirmed by their specific melting temperatures, as well as allowing different variants of target sequences (SNPs) to be identified. (Typing of variants can be accomplished with TaqMan probes, but this requires the use of two probes and two different fluorescent dyes.)
FIG. 1.
HSV Eclipse probe design. (a) Schematic diagram of an Eclipse hybridization probe. MGB indicates the minor groove-binding moiety. F indicates the fluorescent dye, and Q represents the quenching molecule. (b) Alignment of HSV-1 (gi:330064) and HSV-2 (gi:517467) glycoprotein D (gD) sequences surrounding the amplified fragment. Lowercase letters indicate polymorphisms between HSV-1 and -2. Primer sequences are underlined with solid lines. The dashed underline indicates the original probe sequence. Within this sequence, the bold G indicates the novel polymorphism that was identified. The final probe sequence is boxed.
Real-time HSV assay.
For the real-time HSV assay, 10 μl of extracted sample was added to 40 μl of reaction master mix. The master mix consisted of 5 μl Roche FastStart Master hybridization probes mix that contains dNTPs (with UTP), MgCl2, buffer, and Taq polymerase (Roche Diagnostics), HSV upstream primer at 1.2 μM, HSV downstream primer at 100 nM, internal control upstream and downstream primers at 300 nM each, HSV Eclipse probe at 200 nM, internal control Eclipse probe at 50 nM (Table 1), 5 μl PCR enhancer (Epicentre), ROX passive reference (Synthegen) at 60 nM, and 0.5 U of UNG (Perkin Elmer). This master mix was supplemented with 25 mM MgCl2 to obtain a final reaction Mg concentration of 4 mM.
TABLE 1.
Primer and probe sequences
| Primer or probe | Sequence (5′-3′)b |
|---|---|
| HSV MGBa Eclipse probe | TCC TCG CTG ACG GC-FAM |
| Internal control MGB Eclipse probe | TGC GGT ACG TGG TC-TET |
| HSV forward primer | ATC CGA ACG CAG CCC CGC TG |
| HSV reverse primer | TCT CCG TCC AGT CGT TTA TCT TC |
| Internal control forward primer | CTG CAC GGA CCA GTT ACT TTA CG |
| Internal control reverse primer | CTC ATT TTT TCT ACC GGA GAT CTT GT |
MGB, minor groove binding.
FAM, carboxyfluorescein; TET, tetrachlorofluorescein.
The Applied Biosystems HT7900 apparatus was programmed with the following amplification protocol: 2 min at 50°C for one cycle, 8 min at 95°C for one cycle, followed by 50 cycles of PCR (15 seconds at 95°C, 30 seconds at 58°C, and 30 seconds at 76°C). The amplification program was followed by a melting curve analysis consisting of 95°C for 15 seconds, 45°C for 15 seconds, and 95°C for 15 seconds. The temperature ramp rate was set to 100% for all steps except the final ramp between 45°C and 95°C, which was set to 5%. Fluorescence signal was acquired at the annealing stage during amplification (58°C) and during the final temperature ramp. The HT7900 amplifies and continuously detects the fluorescence of each well in the plate in real time. Amplification is detected by the hybridization and resulting fluorescence of a labeled Eclipse probe at the annealing step of each cycle. In the Molecular Infectious Disease lab at ARUP, all positive HSV samples are retested. The extracted sample is run again, and the original patient sample is reextracted and run in duplicate.
Roche LSR assay.
The Roche LSR experiments were performed on the LightCycler according to the package insert. Extractions were done using the QIAGEN 96-well blood kit protocol described above. Roche competitive internal control was added prior to extraction at a concentration of 5 μl per 50-μl extract volume (total of 10 μl added for each QIAGEN extract).
RESULTS
We initially evaluated a primer and Eclipse probe combination designed by Epoch Biosciences. This original version of the assay amplified a 142-bp segment of the glycoprotein D gene containing a single polymorphism beneath the probe to distinguish between HSV-1 and HSV-2 (Fig. 1b). The probe was a perfect match to the HSV-1 sequence and melted at approximately 63°C. The Tm for HSV-2 was about 58°C. However, during our preliminary validation of true HSV-positive patient samples from the clinical lab, an unacceptably large number of samples (9/22 on one run) failed to be detected (data not shown). Interestingly, samples that did not display amplification plots occasionally generated distinct dissociation curves. We evaluated the Tms of these discrepant samples and found that a majority (6/9) were HSV-2, melting at the annealing and data acquisition temperature for the assay (58°C). A few of these amplification reactions were removed from a 96-well plate and run on a gel to determine whether amplification had occurred. The gel results suggested that these samples had indeed amplified but failed to generate detectable fluorescence during probe annealing (data not shown). We conclude from these results that amplification occurred normally, but detection of the product by the probe was inefficient. Because fluorescence was being acquired at 58°C during the amplification (Tm for HSV-2), samples with low HSV titers were generating insufficient signal to be detected. However, during the dissociation analysis, the reaction temperature was lowered to 45°C (below the probe Tm), allowing efficient probe binding to the HSV-2 amplicon and consequently generating a dissociation curve.
One of the discrepants identified above revealed a third class of isolates that display melting temperatures at approximately 55°C, below those predicted for either HSV-1 or HSV-2. We tested a number of HSV-positive samples from the clinical lab and identified three additional examples of this class. Again, these isolates displayed the predicted amplicon size when run on a gel. We sequenced two of these amplicons and determined that both included an additional polymorphism within the region covered by the probe. Each contained a C instead of a G at position 13 of the sequence bound by the probe (Fig. 1b). Examination of the remainder of the amplicon sequence distinguished these isolates as variants of HSV-1 (data not shown). We conclude that, depending upon the design of the amplification reaction and the Tm of the probe for each variant, there exists the potential for a loss of sensitivity in assays that distinguish between variants based on sequence polymorphisms.
We initially tried lowering the annealing temperature of the assay to 56°C, hoping to achieve better probe binding to the HSV-2 target, but saw no improvement. Possibly, the attempt to improve probe binding may have deoptimized primer binding. Based on these observations, we chose to move the probe to a region that lacks known polymorphisms, resulting in a nontyping format.
In the modified version of this assay, we retained the same primers as the original assay but moved the probe just upstream (Fig. 1b). We also preserved the original PCR cycling conditions. When tested against commercially available DNA (Advanced Biotechnologies), amplification plots from serial dilutions of HSV-1 and HSV-2 were indistinguishable, as were the dissociation curves (data not shown).
We compared this Eclipse probe real-time assay with the PCR assay currently being performed in the Molecular Infections Disease Laboratory at ARUP. This assay uses gel electrophoresis as the detection method. Table 2 summarizes the results of this validation. We assayed a total of 323 samples, 171 negatives and 152 positives, comprising eight different sample types (amniotic fluid, spinal fluid, bronchioalveloar lavage fluid, ocular fluid, serum, plasma, tissue, and swab samples). The real-time assay agreed with the reference method for 319 out of the 323 samples tested (99%). Of the 43 spinal fluid samples tested (21 negatives and 22 positives), there were no discrepants. One patient that generated a weak band on a gel was detected in two out of three replicates on the Eclipse assay. We interpret this to indicate that the sample contained a very low titer of virus and detection was likely limited by sampling error. (This sample was not classified as a discrepant.) We assayed one serum sample that had been reported as negative in the clinical lab but was reproducibly positive by the real-time assay. There was insufficient sample to repeat analysis by the reference method. The three remaining discrepants were spiked, low-positive bronchioalveolar lavage samples. These were assayed on a run that displayed abnormal baseline drift. Although they generated detectable amplification on the multicomponent and/or amplification plots, the total fluorescence for these samples at the end of the reaction did not exceed the baseline fluorescence, and technically they had to be classified as negatives.
TABLE 2.
Validation data for the HSV Eclipse assay
| Sample typea | Positives (%) | Negatives (%) |
|---|---|---|
| Amniotic fluid | 20/20 (100) | 24/24 (100) |
| BAL fluid | 18/21 (86) | 20/20 (100) |
| CSF | 22/22 (100) | 21/21 (100) |
| Ocular fluid | 21/21 (100) | 23/23 (100) |
| Plasma | 22/22 (100) | 21/21 (100) |
| Serum | 22/22 (100) | 19/20 (95) |
| Swabs | 21/21 (100) | 21/21 (100) |
| Tissue | 3/3 (100) | 20/20 (100) |
| Total | 149/152 (98) | 170/171 (99) |
BAL, bronchoalveolar; CSF, cerebrospinal fluid.
Limits of detection for the test were determined by assaying serial twofold dilutions of positive control material in each sample type (except tissue). Our results indicated that HSV is detectable down to approximately 150 copies per ml in spinal fluid (equivalent to approximately 3 copies per reaction mixture), which is near the theoretical limit imposed by sampling error.
To determine the reproducibility of the assay, we performed both within-run and between-run precision studies. For between-run precision, we ran 10 samples (9 positives, 1 negative) once per day for three consecutive days. The standard deviation (SD) for the crossing thresholds was 1.7, with a coefficient of variation (CV) of 4.7%. The mean crossing thresholds for these samples ranged from 21 to 44. One outlier sample gave atypical replicates of 50, 21, and 36 (SD, 14.5; CV, 40.3%). If this sample is eliminated from the data set, the standard deviation is 0.3 with a CV of 0.7%. For within-run precision, we assayed 10 positive samples in triplicate on the same run. The standard deviation was 1.0 with a CV of 2.8%. The mean crossing thresholds for these samples ranged from 20 to 43.
Samples known to be positive for other infectious agents were run in this assay to determine whether there is any cross-reactivity with other targets. These samples were positive control materials (culture) from other assays currently being run in the clinical lab at ARUP. No cross-reactivity with Epstein-Barr virus, cytomegalovirus, varicella-zoster virus, Borrelia burgdorferi, enterovirus, parvovirus, Bordetella pertussis/parapertussis, Toxoplasma gondii, influenza virus A and B, Mycoplasma pneumoniae, human immunodeficiency virus (HIV), or adenovirus was seen in this assay.
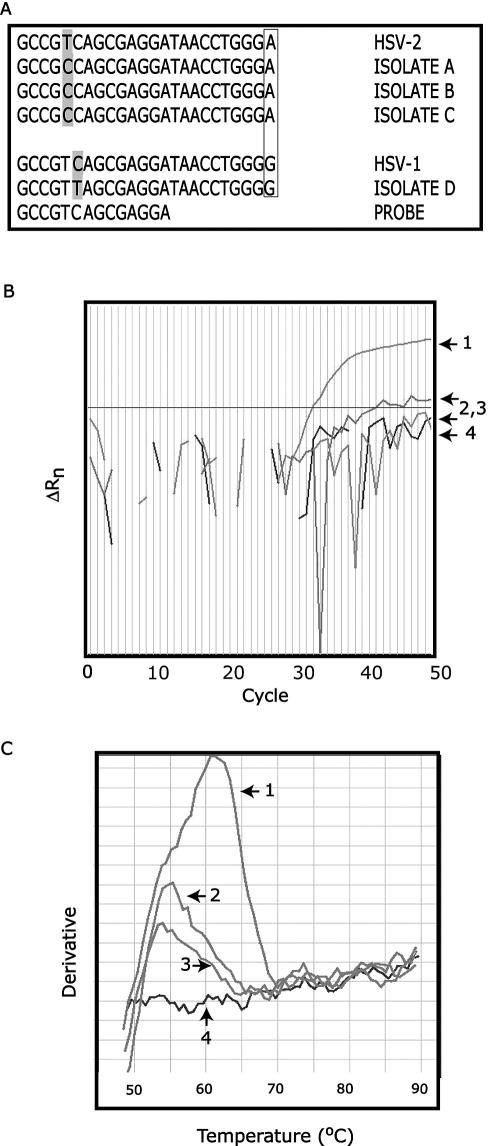
In the course of validating and running this modified assay, we again identified a small number of HSV-positive samples that either failed to be detected or generated weak amplification plots. These samples displayed melting curves that indicated a Tm much lower than what was predicted for this sequence. We sequenced the amplicons for four of these isolates and discovered they all contained a polymorphism within the probe-binding region, which was not predicted from the consensus sequence. Figure 2a shows the sequence in the probe-binding region for these four isolates, as well as the consensus sequences for HSV-1 and HSV-2 (the probe sequence is shown at the bottom). The final nucleotide in the sequence (A for HSV-2, G for HSV-1) is a polymorphism that allows HSV-1 and-2 to be differentiated. Clearly, unexpected polymorphisms appear in both HSV-1 and HSV-2 isolates, although at least in the cases we examined, they appeared in different positions. Figure 2b shows the amplification plot for isolate B. Curve 1 is a positive control, curves 2 and 3 are duplicate amplifications of isolate B, and curve 4 is a negative control. This atypical HSV isolate generated one weak amplification plot (2) and one that was indistinguishable from the negative control (3). However, when the dissociation curves were examined (Fig. 2c), it is clear that isolate B was amplified in both replicates (numbering is the same as in Fig. 2b). Specifically, the dissociation plots for isolate B (melting curves 2 and 3) should be compared with the plot for the negative control (curve 4). Furthermore, it is evident that the Tm for isolate B is significantly lower (approximately 55°C) than for the positive control sample (approximately 62°C). To ensure that we do not miss positive specimens of this type, at ARUP we routinely examine the dissociation curves of all samples that give a negative result on the amplification plot.
FIG. 2.
SNP detection. (a) Sequences of atypical HSV isolates. Locations of novel polymorphisms are shaded. Boxed nucleotides indicate known polymorphism that allows HSV-1 and HSV-2 to be distinguished. (b) Amplification plots for isolate B (2 and 3) with positive (1) and negative (4) controls. (c) Dissociation curves for isolate B (2 and 3) with positive (1) and negative (4) controls.
To evaluate the sensitivity of our Eclipse probe assay relative to another real-time test, we compared it with an LSR kit (life science research) distributed by Roche. The reagents in this kit are identical to those contained in Roche's HSV-1/2 ASR kit (analyte-specific reagent) that is currently available and being used in a number of labs in the United States. With this kit, we assayed serial dilutions of commercially available HSV-1 and HSV-2 DNA (Advanced Biotechnologies, Inc.) on the LightCycler. Dilutions ranged from 10−2 (calculated concentration of 100 copies per microliter, 500 copies per reaction mixture) down to 10−5 (0.1 copy per microliter, 0.5 copies per reaction mixture). Figure 3a shows the LightCycler amplification plots of HSV-1 (each dilution in duplicate) and HSV-2 (each dilution run singly). The amplification plots for the positive controls and the dilutions are marked on the right. The corresponding dissociation curves are shown in Fig. 3b. Our analysis of the LightCycler amplification plots reveals two interesting observations. First, the amplification curves for HSV-1 at 10−3 and HSV-2 at 10−4 are nearly identical, suggesting that the sensitivity for detecting HSV-1 in this assay is reduced approximately 10-fold relative to HSV-2. The lower melting temperature for HSV-1, as revealed by the dissociation curve, indicates that this variant carries at least one polymorphism beneath the probe. This loss of sensitivity is strikingly similar to what we observed with our original HSV Eclipse assay, where we tried identifying HSV-1 and 2 using a type-specific SNP. Our second finding reveals that this LightCycler assay is approximately 10-fold (for HSV-2) to 100-fold (for HSV-1) less sensitive than our reconfigured Eclipse assay, where we detected both variants down to the 10−5 dilution (Fig. 3c). We tested only the two most dilute extracted samples (10−4 and 10−5) with our Eclipse probe assay. (Since it is impossible to reproducibly detect a calculated 0.5 copies per reaction mixture, which corresponds to the 10−5 dilution, we presume that there is some level of inaccuracy in the quantitation of the commercial standard.)
FIG. 3.
Comparison of Eclipse and Roche assay sensitivities. (a) Amplification plots for Roche LSR kit. Dilutions are marked on the right. No-template control, 10−5 dilution for HSV-2 and the two lowest dilutions of HSV-1 all lie in the region marked with a star. (b) Dissociation curves for Roche LSR kit. Positive control contains both HSV-1 and HSV-2. (c) Sensitivity of Eclipse HSV assay, showing 10−4 and 10−5 dilutions of HSV-1 and HSV-2.
These differences in sensitivity prompted us to estimate the fraction of HSV-positive samples that would have gone undetected in the ARUP clinical lab, had we been using the Roche LightCycler assay. To make a conservative estimate, we defined the Roche assay's limit of detection as 10−3 for HSV-1 and 10−4 for HSV-2 (Fig. 3a), meaning that we would not detect HSV-1 samples that were less than the 10−3 dilution and HSV-2 samples that were less than the 10−4 dilution. We then examined the crossing thresholds for these dilutions on the Eclipse amplification plot (Fig. 3c). We estimated that any HSV-1 samples with Cts less than 40 would go undetected and any HSV-2 samples with Cts less than 42 would be missed. Examining 2 months of results from the ARUP clinical lab (HSV-positive samples only), the data suggest that between 7% (cutoff Ct of 42) and 15% (cutoff Ct of 40) of these positives samples would have gone undetected using the Roche assay.
DISCUSSION
There is currently a need for standardized commercially available kits to support HSV molecular testing, forcing labs to develop assays in-house. Designing and optimizing any new PCR assay can be a daunting task, requiring attention to a host of variables. In recent years, real-time PCR technology has been increasingly adopted in the field of clinical infectious disease, providing an attractive option for pathogen detection. Even so, replacing traditional gel detection methods with fluorescent probes in real-time PCR is no guarantee of improved performance. We observe that either method, when well optimized, can perform with good sensitivity. While real-time PCR provides a number of advantages, these must be balanced against any potential disadvantages. Benefits include a closed tube format that minimizes the possibility of amplicon contamination, the potential for higher throughput, lower cost, increased compatibility with robotics, and decreased hands-on time for the technologist. However, our experience in designing real-time tests reveals that polymorphisms within the target, both expected and unexpected, can seriously compromise the sensitivity of an assay.
The presence of base pair mismatches between the probe and target will lower the Tm of this interaction. For hybridization probes, if the annealing temperature of a reaction is at or above the Tm for the mismatch, probe binding is reduced and, as a consequence, the fluorescent signal is diminished. This reduction in fluorescence contributes to a lower assay sensitivity. A similar problem can potentially occur with TaqMan hydrolysis probes, which are displaced, rather than cleaved, when positioned over an SNP. Nucleic acid targets with unknown polymorphisms present an even more difficult challenge and may contribute to false-negative reporting.
Our experience with three different HSV assays indicates that polymorphisms beneath the probe reduce the ability to detect target that has been properly amplified. In the assay described here, we have addressed these difficulties in two ways. First, we chose to avoid known SNPs that distinguish HSV-1 from HSV-2, and we placed our probe in a region of the glycoprotein D gene where the two sequences are identical. Although this assay does not distinguish between these types, it has approximately equal sensitivity for both. Although there are circumstances where typing may be clinically useful, the overriding priority for this assay is extreme sensitivity, particularly with CSF samples. Second, we have chosen to use a hybridization probe that is not hydrolyzed during the reaction, allowing a postamplification dissociation analysis. During this melting phase of the reaction, lowering the temperature well below the predicted Tm for most potential variants will promote efficient probe binding, allowing the detection of PCR products that may be undetectable during the amplification phase. By examining the melting profiles of all samples that are negative on the amplification plot, we are able to detect most unknown sequence variants. In our hands, the comparative sensitivity of target detection using the dissociation curve versus the amplification plot varies among assays.
The HSV assay described in these studies employs the recently developed Eclipse probe system from Epoch Biosciences. Like molecular beacons, Eclipse probes require only a single-oligonucleotide probe reagent for hybridization-based interrogation of real-time amplification reactions, simplifying design, optimization, and subsequent quality control (QC) of test reagents compared to dual-oligonucleotide FRET probes. In addition, the presence of a 5′ minor groove-binding group at the 5′ end allows for the design of significantly shorter probes at a given Tm and affords good protection from exonuclease hydrolysis during PCR cycling. As demonstrated in these studies, the ability to perform high-quality melt analysis following amplification affords another level of interrogation compared to hydrolysis chemistries. Although Eclipse probes are theoretically compatible with a variety of amplification chemistries and platforms, care should be taken to ensure that the available fluorescent modifiers have spectral properties that are well suited for a given detection instrument.
A final consideration in the design of this HSV assay is the use of an internal control. The addition of an internal control to samples before extraction reveals whether the sample was extracted efficiently and monitors inhibition of the PCR (8). An internal control can be designed as either competitive or noncompetitive, depending on whether it is amplified with the same primer pair as the target. Competitive internal control systems exist that minimally effect target amplification (E. Konnick, S. Williams, E. Ashwood, and D. Hillyard, Abstr. Pan Am. Soc. Clin. Virol., 19th Ann. Clin. Virol. Symp., abstr. M40, 2003). In other assays however, significant competition between the target and internal control has been demonstrated (16). The potential for an internal control to compete with target amplification may be reduced when using a noncompetitive internal control. This potential competition is of great importance when analyzing samples with low target concentrations, such as HSV in spinal fluid. Another advantage of a heterologous target is that it can serve as a “universal” internal control for a wide variety of different assays. One disadvantage of a noncompetitive internal control is the potential for its amplification efficiency to differ from that of the target. Furthermore, amplification of the internal control may be affected differently by PCR inhibitors. These potential problems can generally be avoided by careful selection and validation of the internal control target. However, the advantages and disadvantages of either type of internal control should always be considered.
The concentration of internal control introduced into the reaction mixture is an important consideration when setting up a multiplex reaction. The copy number must be sufficient to consistently amplify, yet not so high that it significantly affects the sensitivity of the assay. When our internal control was multiplexed with the HSV reaction, no significant effects of on the HSV assay sensitivity were detected.
Here we describe a high-throughput real-time PCR assay for detecting HSV-1 and 2, utilizing the novel Eclipse hybridization probe system. Our results demonstrate that sequence polymorphisms within the probe-binding region of an amplicon can adversely effect the sensitivity of real-time PCR assays. We highlight some of the difficulties that SNPs can introduce into real-time assay design, with the goal of helping others become more aware of these issues and avoiding many of these pitfalls.
REFERENCES
- 1.Afonina, I. A., M. W. Reed, E. Lusby, I. G. Shishkina, and Y. S. Belousov. 2002. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. BioTechniques 32:940-944, 946-949. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, T. P., A. M. Werno, K. A. Beynon, and D. R. Murdoch. 2003. Failure to genotype herpes simplex virus by real-time PCR assay and melting curve analysis due to sequence variation within probe binding sites. J. Clin. Microbiol. 41:2135-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aurelius, E., B. Johansson, B. Skoldenberg, A. Staland, and M. Forsgren. 1991. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet 337:189-192. [DOI] [PubMed] [Google Scholar]
- 4.Burrows, J., A. Nitsche, B. Bayly, E. Walker, G. Higgins, and T. Kok. 2002. Detection and subtyping of herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, E. T., Jr., G. A. Short, A. R. Irvine, J. S. Duker, and T. P. Margolis. 1996. Acquired immunodeficiency syndrome-associated herpes simplex virus retinitis. Clinical description and use of a polymerase chain reaction-based assay as a diagnostic tool. Arch. Ophthalmol. 114:834-840. [DOI] [PubMed] [Google Scholar]
- 6.Espy, M. J., T. K. Ross, R. Teo, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Evaluation of LightCycler PCR for implementation of laboratory diagnosis of herpes simplex virus infections. J. Clin. Microbiol. 38:3116-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakeman, F. D., R. J. Whitley, et al. 1995. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. J. Infect. Dis. 171:857-863. [DOI] [PubMed] [Google Scholar]
- 11.Loeffelholz, M. J., C. J. Thompson, K. S. Long, and M. J. Gilchrist. 1999. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J. Clin. Microbiol. 37:2872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madhavan, H. N., K. Priya, A. R. Anand, and K. L. Therese. 1999. Detection of herpes simplex virus (HSV) genome using polymerase chain reaction (PCR) in clinical samples comparison of PCR with standard laboratory methods for the detection of HSV. J. Clin. Virol. 14:145-151. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell, P. S., M. J. Espy, T. F. Smith, D. R. Toal, P. N. Rys, E. F. Berbari, D. R. Osmon, and D. H. Persing. 1997. Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J. Clin. Microbiol. 35:2873-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nahmias, A. J., R. J. Whitley, A. N. Visintine, Y. Takei, and C. A. Alford, Jr. 1982. Herpes simplex virus encephalitis: laboratory evaluations and their diagnostic significance. J. Infect. Dis. 145:829-836. [DOI] [PubMed] [Google Scholar]
- 15.Schalasta, G., A. Arents, M. Schmid, R. W. Braun, and G. Enders. 2000. Fast and type-specific analysis of herpes simplex virus types 1 and 2 by rapid PCR and fluorescence melting-curve-analysis. Infection 28:85-91. [DOI] [PubMed] [Google Scholar]
- 16.Stocher, M., and J. Berg. 2002. Normalized quantification of human cytomegalovirus DNA by competitive real-time PCR on the LightCycler instrument. J. Clin. Microbiol. 40:4547-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang, Y. W., P. S. Mitchell, M. J. Espy, T. F. Smith, and D. H. Persing. 1999. Molecular diagnosis of herpes simplex virus infections in the central nervous system. J. Clin. Microbiol. 37:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilley, P. A., M. V. Kanchana, I. Knight, J. Blondeau, N. Antonishyn, and H. Deneer. 2000. Detection of Bordetella pertussis in a clinical laboratory by culture, polymerase chain reaction, and direct fluorescent antibody staining; accuracy, and cost. Diagn. Microbiol. Infect. Dis. 37:17-23. [DOI] [PubMed] [Google Scholar]