Abstract
One-hundred five influenza B-positive specimens obtained from southeast Asia in 2002 were categorized on the basis of DNA sequencing of HA1 gene as well as real-time PCR analysis of the NA gene. Phylogenetic analysis of the HA1 gene sequences showed that the majority of the viruses (96.2%) belonged to the B/Victoria/2/87 lineage, while a smaller percentage of the viruses (3.8%) belonged to the B/Yamagata/16/88 lineage. The B/Yamagata/16/88 viruses displayed significant antigenic drift in the deduced amino acid sequences of the HA1 protein, and the B/Victoria/2/87-like viruses consisted of B/Hong Kong/1351/02-like (72.3%) and B/Hong Kong/330/01-like (27.7%) viruses. The B/Hong Kong/1351/02-like viruses were reassortants with the HA gene belonging to the B/Victoria/2/87 lineage and the NA gene belonging to the B/Yamagata/16/88 lineage, whereas both the HA and NA genes of B/Hong Kong/330/01 virus belonged to the B/Victoria/2/87 lineage. In this study, however, all the B/Hong Kong/330/01-like isolates exhibited the B/Yamagata/16/88-like NA gene, which likely resulted from reassortment of B/Hong Kong/330/01 and B/Hong Kong/1351/02 viruses during coinfection. Additional molecular characterization of the six internal genes showed that the M, NS, PA, and PB2 genes of the new variants were B/Hong Kong/1351/02 in origin, whereas the NP and PA genes retained the B/Hong Kong/330/01 origin. Interestingly, these new variants all appeared late in the year 2002. These results support the notion that influenza B viruses continued to evolve through antigenic drift and shift.
Influenza is a major cause of respiratory illness in adults and children worldwide. Both influenza A and B viruses have a segmented genome consisting of eight negative-stranded RNA segments, six of which code for the internal proteins nucleoprotein (NP), matrix protein (M), nonstructural protein (NS), and RNA polymerase proteins (PA, PB1, PB2), and two of which code for the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). Viral infection is initiated by HA binding to the sialic acid-containing host cell receptor, while NA cleaves the sialic acid to promote the release of virus from infected cells. Functional coordination of these two proteins is required for efficient virus replication (14). However, the natural balance of virus infection and release can be disturbed by events such as reassortment.
The influenza HA surface antigen is synthesized as a single polypeptide that is subsequently cleaved into two chains, HA1 and HA2. Antigenic variation of the HA protein of influenza viruses occurs predominantly in the HA1 domain (15). The HA gene of influenza B viruses can also undergo antigenic drift, although this phenomenon occurs at a lower rate compared to that of influenza A viruses (18). In addition, influenza B viruses use other mechanisms, such as insertion/deletion and reassortment with antigenically and genetically distinct cocirculating viruses, to generate genetic diversity (8, 10, 12, 17, 18). Recent isolates of influenza B viruses consist of two major phylogenetic lineages: B/Victoria/2/87 and B/Yamagata/16/88. Circulation of the B/Yamagata/16/88 viruses was highly restricted in Asia during the 1990s but emerged during 2001 in various parts of the world (4). One of the strains, B/Hong Kong/1351/02, was determined to be a reassortant with the HA and NA genes arising from two distinct lineages (13). In this report, we studied the genetic diversity of influenza B viruses by molecular analysis of 105 influenza B-positive respiratory specimens collected from southeast Asia in 2002.
MATERIALS AND METHODS
Clinical specimens and RNA extraction.
Nasal specimens were collected from 6- to 36-month-old subjects with respiratory illness who enrolled in an experimental pediatric influenza vaccine trial conducted in the Philippines and Thailand during the year of 2002 (from March to November). Viral RNA was extracted from influenza B-positive specimens by an automated method as described previously (3). Briefly, viral RNA was extracted from 125 μl of clinical specimen or tissue culture supernatant with addition of 125 μl of 2× lysis buffer (Applied Biosystems, Foster City, CA) and 50 μg of carriers (Sigma, St. Louis, MO). A total of 250 μl of cell lysate was processed using the ABI Prism 6700 workstation (Applied Biosystems, Foster City, CA), and the RNA was eluted in a total volume of 150 μl of elution buffer (Applied Biosystems, Foster City, CA).
Amplification and sequencing of the HA1 gene.
HA1 cDNA fragments corresponding to nucleotides 98 to 836 were amplified by reverse transcription-PCR (RT-PCR) with oligos 5′-ATAACATCGTCAAACTCACC-3′ and 5′-GCACCATGTAATCAACAACA-3′ and sequenced as described previously (4). Multiple sequence alignment was performed to determine the phylogenetic relationship between the clinical virus isolates and the reference strains (B/Victoria/504/00, B/Hong Kong/330/01, and B/Hong Kong/1351/02) using MegAlign5.03 software (DNASTAR Inc., Madison, WI) and the ClustalV method.
Real-time PCR assay of NA gene.
A high-throughput TaqMan RT-PCR assay was designed to target the NA gene of influenza B virus. Each primer-probe set (Table 1) was chosen based on the different nucleotide composition of the NA gene in the B/Hong Kong/330/01 and B/Hong Kong/1351/02 viruses. Both sets of primer-probe were designed using Primer Express Software (Applied Biosystems, Foster City, CA). The primers were obtained from Invitrogen Life Technology (Carlsbad, CA), and the probes which contained the 5′ reporter dye 6-carboxyfluorescein (FAM) and the 3′ minor groove binder (MGB) were synthesized by Applied Biosystems (Foster City, CA). The TaqMan RT-PCR procedure was described previously (6), and the real-time RT-PCRs were carried out in an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA). The reverse transcription reactions were performed at 48°C for 30 min, followed by 10 min at 95°C to activate AmpliTaq DNA polymerase and then by 40 cycles of amplification, including 15 s at 95°C and 1 min at 60°C.
TABLE 1.
Oligonucleotide sequences for primers and probes of the NA genea
| Influenza B strain (accession no.b) | Primer or probe | Nucleotide sequence | Nucleotide positions |
|---|---|---|---|
| Hong Kong/330/01 (AY191502) | HK330P1c | 5′ CAGCCTTACTGTCATACTTACTGTATTCG | 81-109 |
| HK330P2c | 5′ GCGGAGTCCAACGACGTT | 154-171 | |
| HK330MGBd | 5′ CAACAAAAATAATTGCAC | 132-149 | |
| Hong Kong/1351/02 (AY139074) | HK1351P1c | 5′ GCTTCATTGTCATACTTACTATATTCGGATA | 83-113 |
| HK1351P2c | 5′ GATGCGTTTGCACAATCCAA | 161-180 | |
| HK1351MGBd | 5′ CAACAGAAATAACTGCAC | 132-149 |
Underlined nucleotides are different between B/Hong Kong/330/01 and B/Hong Kong/1351/02 NA sequences.
Accession numbers from GenBank.
Primer names.
Probe names.
Amplification and sequence of NA, NP, NS, M, PA, PB1, and PB2 gene fragments.
All gene fragments were amplified by RT-PCR and sequenced using an ABI 3100 Genetic Analyzer. Table 2 shows the oligo primers used for PCR and sequencing. A multiple sequence alignment was performed to determine the phylogenetic relationship (as described above).
TABLE 2.
Oligos for analyzing influenza B gene fragments
| Gene | Oligos for PCR or sequencing | Nucleotide positions | GenBank accession no. of reference sequence |
|---|---|---|---|
| NAa | 5′-GCTACCTTCAACTATACAAACG | 56-1549 | AY139074 |
| 5′-CAAGAGCATTTTTCAGAAAC | |||
| NP | 5′-GAAGTAGGTGGAGACGGAGGG | 1412-1802 | M20173 |
| 5′-GTAAACACCCACATTCCAAACG | |||
| NS | 5′-GAGCAGCTGAAACTGCGGTG | 810-1066 | M20224 |
| 5′-CACAAGCACTGCCTGCTGTAC | |||
| M | 5′-ATGTCGCTGTTTGGAGACAC | 25-320 | M20175 |
| 5′-GCTAGAATCAGGCCTTTCTT | |||
| PA | 5′-CCTGAATTACAACCAGCAATGC | 111-463 | M20171 |
| 5′-AGTTCCATGCTATTTCCCAG | |||
| PB1 | 5′-ACCGCTGGAATACAAATCAG | 748-1011 | M20169 |
| 5′-TGGGCTGTCTCTGGTTATTC | |||
| PB2 | 5′-CTGTTAAGGGACAATGAAGC | 57-301 | M20163 |
| 5′-CACATTTGGCCTTTGGTTCC |
Six additional oligo primers were also used for sequencing the NA gene fragment: 5′-ATTGTAGTATCC CCCTGGTT, 5′-ACTCCGATATATGTCCATTC, 5′-TTCCACATGGCAGCTTGGAG, 5′-TCTTAAGATTCGAGAGGGCC, 5′-TTATCTCTACAGGCACATTC, and 5′-GTACCACCTTCCAATCTTGG.
RESULTS
Characterization of influenza B viruses by sequencing HA1 gene fragments.
Based on HA1 sequences, we found that 96.2% of the specimens belonged to the B/Victoria/2/87 lineage, which were further subgrouped into B/Hong Kong/330/01-like (27.7%) and B/Hong Kong/1351/02-like (72.3%) viruses. Of the 105 viruses examined, only a small percentage of the viruses (3.8%) was found to belong to the B/Yamagata/16/88 lineage (Table 3, group 1). Among these, significant antigenic drift was present in the deduced amino acid sequences of the HA1 protein compared to those of the reference strain B/Victoria/504/00. A total of 17 amino acid substitutions were identified, and 6 of them occurred at positions 29, 88, 162, 179, 183, and 184, respectively. These substitutions were identical to those found in the HA1 protein of B/Hong Kong/330/01. In contrast, the HA1 drift in the B/Hong Kong/330/01-like viruses and B/Hong Kong1351/02-like viruses was minimal (Table 3, group 2) or sporadic (Table 3, group 3), respectively. However, all the isolates contained the same amino acid substitution at position 197.
TABLE 3.
| Group, lineage, and strain or isolate | Amino acid at indicated position | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 (n = 4),c B/Yamagata/16/ 88 lineage | 29 | 40 | 48 | 56 | 75 | 88 | 116 | 126 | 131 | 162 | 168 | 172 | 179 | 180 | 183 | 184 | 197 |
| Reference strain: B/Victoria/504/00 | A | H | K | T | I | K | K | N | P | R | T | P | H | I | K | E | D |
| SA10 | V | Y | R | D | T | R | N | D | - | K | N | S | Y | V | E | G | N |
| SA40 | V | - | R | D | T | R | N | D | L | K | N | - | Y | - | E | G | N |
| SA43 | V | - | R | D | T | R | N | D | L | K | N | - | Y | - | E | G | N |
| SA96 | V | - | R | D | T | R | N | D | L | K | N | - | Y | - | E | G | N |
| B/Hong Kong/330/01 | V | - | - | K | - | R | R | - | - | K | - | S | Y | - | E | G | S |
| Group 2 (n = 28), B/Victoria/2/87 lineage | 34 | 197 | |||||||||||||||
| Reference strain: B/Hong Kong/330/01 | T | S | |||||||||||||||
| SA115 | M | N | |||||||||||||||
| 27 others | - | N | |||||||||||||||
| Group 3 (n = 73), B/Victoria/2/87 lineage | 15 | 37 | 56 | 90 | 118 | 121 | 144 | 154 | 165 | 190 | 197 | 199 | |||||
| Reference strain: B/Hong Kong/1351/02 | V | T | K | V | R | T | P | A | N | V | K | T | |||||
| SA11 | M | - | - | - | - | - | - | - | - | - | N | - | |||||
| SA30, SA31, SA88, SA94 | - | I | - | - | - | - | - | - | - | - | N | - | |||||
| SA9, SA19 | - | - | E | - | - | - | - | - | - | - | N | - | |||||
| SA15 | - | - | E | I | - | - | - | - | - | - | N | A | |||||
| SA21, SA28, SA90, SA91 | - | - | - | - | - | - | L | - | - | - | N | - | |||||
| SA27 | - | - | - | - | - | - | - | S | - | - | N | - | |||||
| SA92 | - | - | - | - | K | - | - | - | - | - | N | - | |||||
| SA84 | - | - | - | - | - | A | - | - | - | - | N | - | |||||
| SA53 | - | - | - | - | - | - | - | - | K | G | N | - | |||||
| SA62, SA63 | - | - | - | - | - | - | - | - | Y | - | N | - | |||||
| 55 others | - | - | - | - | - | - | - | - | - | - | N | - | |||||
Amino acid residues of reference sequences are shown in boldface. Positions are listed for each group.
-, amino acid residues identical with the sequences of respective reference strain.
Italicized residues are identical to those of B/Hong Kong/330/01.
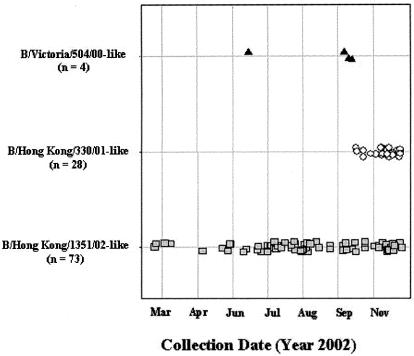
The time distribution of our specimens is shown in Fig. 1. It is interesting to note that the specimens containing the B/Hong Kong/330/01-like viruses were clustered at the end of the collection period during October and November of 2002. Since B/Hong Kong/330/01 was circulating in previous influenza seasons, we further investigated whether there was any change in its genetic compositions.
FIG. 1.
Time distribution of influenza B specimens. The viruses were classified as B/Victoria/504/00-like (triangles), B/Hong Kong/330/01-like (circles), or B/Hong Kong/1351/02-like (squares) based on the HA1 sequences.
Real-time PCR assay of the NA gene of influenza B viruses.
To determine the origin of the NA gene of the B viruses, a real-time RT-PCR assay was employed. The specificity of the primers-probes was tested using two reference virus strains: B/Hong Kong/330/01 and B/Hong Kong/1351/02. The results indicated that the assay is highly specific in distinguishing the two influenza B strains (data not shown). Among the clinical specimens, we unexpectedly found that the B/Hong Kong/330/01-like viruses also contained the NA gene of B/Hong Kong/1351/02.
Sequence analysis of the six internal gene fragments.
To further confirm the PCR results of the NA gene, we selectively amplified and sequenced a 1.4-kb NA gene fragment (corresponding to amino acids 1 to 436) from four new variants of B/Hong Kong/330/01-like viruses. Multiple alignment analysis not only confirmed these sequences were B/Hong Kong/1351/02-like phylogenetically (data not shown) but also revealed five amino acid substitutions in the deduced NA amino acid sequence at positions 35 (Asn to Gly), 46 (Thr to Ile), 248 (Val to Ile), 395 (Ala to Thr), and 416 (Lys to Arg) compared to those of B/Hong Kong/1351/02. Three of them, 35Gly, 46Ile, and 416Arg, are found in all four new B/Hong Kong/330/01 variants but not in any sequence from the specimens that are B/Hong Kong/1351/02-like based on HA1 sequences.
In addition, we analyzed the six internal gene fragments NS, NP, M, PA, PB1, and PB2 of the new B/Hong Kong/330 variants by RT-PCR and sequencing and then compared the resulting sequences with those of B/Hong Kong/330/01 and B/Hong Kong/1351/02. Phylogenetic analysis of the sequences of the six internal genes showed that four of them, M, NS, PA, and PB2, were B/Hong Kong/1351/02 in origin, whereas NP and PB1 retained B/Hong Kong/330/01-like origin (Table 4).
TABLE 4.
| Isolate | Gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HA | NA | NS | NP | M | PA | PB1 | PB2 | |
| SA1 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | Not done | Not done | Not done |
| SA7 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 | B/HK1351 |
| SA46 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 |
| SA47 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 | Not done | Not done | Not done |
| SA51 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 | Not done | Not done | Not done |
| SA65 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 | B/HK1351 | B/HK330 | B/HK1351 |
Based on the phylogenetic analysis of sequences of respective gene segments.
The boldface letters indicate a different origin from that of the HA gene.
DISCUSSION
Surveillance and monitoring antigenic changes in circulating influenza viruses provide important information for annual selection of vaccine strains. Molecular methods such as real-time PCR and sequencing are rapid and sensitive methods for characterizing the genetic composition and classification of the viruses. In addition, identifying changes in the codons of the HA1 domain of the hemagglutinin gene, especially those under positive selection, can facilitate the prediction of future lineages (2). In this study, we analyzed 105 wild-type influenza B viruses collected in 2002 from southeast Asia, specifically the Philippines and Thailand. Two lineages of influenza B viruses were detected by sequencing the HA1 gene fragment. The B/Victoria/2/87-like viruses, including B/Hong Kong/330/01-like and B/Hong Kong/1351/02-like viruses, dominated the southeast Asia region in 2002. A small percentage of B/Yamagata/16/88-like viruses were also found, and these viruses exhibited more extensive antigenic drift compared to B/Victoria/504/00 (Table 3). Among these, the deduced amino acid sequence of SA10 (group 1 virus, Table 3) is very similar to that of isolate B/jhb/77/01 from South Africa (1), except at positions 88, 172, and 197, and it is nearly identical to that of WV5, an isolate from Europe in the 2001-2002 season (4), except at position 172. Within the B/Victoria/2/87 group, we found a substitution of amino acid 197 in all the HA1 amino acid sequences of B/Hong Kong-like viruses. Previously, we reported this change (4) and showed that a new N-glycosylation site (NET) was created as a result of the substitution. Nakagawa et al. also found a new glycosylation site in the HA of influenza B isolates in Japan during 2002-2003 (11) and demonstrated that the amino acid sequence 197 to 199 determined reactivity to certain monoclonal antibodies. In our study, all but one B/Hong Kong-like HA1 sequence exhibited NET at amino acids 197 to 199.
Genetic reassortment has recently been a major feature in the evolution of both influenza A and B viruses (7). Cocirculation of multiple subtypes and lineages of influenza viruses can allow genetic reassortment to occur during mixed infections. Shaw et al. first reported the reappearance and global spread of variants of B/Victoria/2/87 lineage viruses, represented by the B/Hong Kong/330/01 strain in the 2000-2001 season and B/Hong Kong/1351/02 strain in the 2001-2002 season (13). We confirmed these findings in our previous report (4). In this study, we identified a new variant of influenza B virus that has the combined features of B/Hong Kong/330/01 and B/Hong Kong/1351/02: the HA, NP, and PB1 genes are B/Hong Kong/330/01 in origin, while the NA, NS, M, PA, and PB2 genes are B/Hong Kong/1351/02 in origin, which could arise as a result of reassortment between B/Hong Kong/330/01 and B/Hong Kong/1351/02 viruses. Often not distinguished by HAI tests, B/Hong Kong/1351/02 can be genetically distinguished from B/Hong Kong/330/01 by (1) the absence of three signature amino acids at positions 116, 121, and 164 in the HA1 gene fragment (13) and (2) the presence of the B/Yamagata/16/88 NA gene instead of the B/Victoria/2/87 NA gene. In addition to B/Hong Kong/1351/02, a group of viruses isolated in 2001-2002 also carry similar features, such as B/India/772/01 and B/New York/1/02 (16). Such reassortants may have certain selective advantages, since B/Hong Kong/1351/02-like viruses appear to have quickly become globally dominant (4, 5).
In 2003, Matzuzaki et al. isolated a B/Victoria/2/87-like virus that had acquired the NA, NS, M, and PA gene segments from a Shiga/T30/98-like virus (9). In this study, we determined the origin of the six internal genes of new influenza B variants and showed that the NS, M, PA, and PB2 gene segments belong to the B/Hong Kong/1351/02 clade. The fact that these viruses appeared late in 2002 suggests a continuous evolution of influenza B viruses through antigenic drift and reassortment.
Nucleotide sequence accession numbers.
GenBank accession numbers for nucleotide sequences of the HA gene are B/Victoria/2/87, M22943; B/Yamagata/16/88, M36105; B/Victoria/504/00, ISDN20057; B/Hong Kong/330/01, AF532549; B/Hong Kong/1351/02, AF532545. Accession numbers for the NA gene sequences are B/Hong Kong/330/01, AY139066; B/Hong Kong/1351/02, AY139074. Clinical isolates are AY880074 to AY880178 (HA sequences for 105 specimens); AY880066 to AY880073 (NA sequences of SA1, -7, -32, -46, -47, -51, -59, and -65, respectively). Partial nucleotide sequences for NP, NS, M, PA, PB1, and PB2 genes of selected specimens are available upon request.
Acknowledgments
We thank Giuseppe Palladino for providing wild-type influenza reference strains and Melissa Colella, Fenglan Li, and Tammy La for technical assistance.
REFERENCES
- 1.Besselaar, T. G., L. Botha, J. M. McAnerney, and B. D. Schoub. 2004. Phylogenetic studies of influenza B viruses isolated in southern Africa: 1998-2001. Virus Res. 103:61-66. [DOI] [PubMed] [Google Scholar]
- 2.Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, S. M., R. Vainionpaa, P. Zhao, F. Li, A. Hu, B. Forrest, and R. Rappaport. 2004. Detection of influenza B in clinical specimens: comparison of high throughput RT-PCR and culture confirmation. Virus Res. 103:85-90. [DOI] [PubMed] [Google Scholar]
- 4.Chi, X. S., T. V. Bolar, P. Zhao, R. Rappaport, and S. M. Cheng. 2003. Co-circulation and evolution of two lineages of influenza B viruses in Europe and Israel in the 2001-2002 season. J. Clin. Microbiol. 41:5770-5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi, X. S., A. Hu, R. Rappaport, and S. M. Cheng. 2004. Molecular characterization of the hemagglutinin and neuraminidase genes of influenza B circulating worldwide in the 2001-2002 season, p. 704-707. In Y. Kawaoka (ed.), Options for the control of influenza V. Elsevier, Amsterdam, The Netherlands.
- 6.Hu, A., M. Colella, J. S. Tam, R. Rappaport, and S. M. Cheng. 2003. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real-time PCR. J. Clin. Microbiol. 41:149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin, Y. P., V. Gregory, M. Bennett, and A. Hay. 2004. Recent changes among human influenza viruses. Virus Res. 103:47-52. [DOI] [PubMed] [Google Scholar]
- 8.Lindstrom, S. E., Y. Hiromoto, H. Nishimura, T. Saito, R. Nerome, and K. Nerome. 1999. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J. Virol. 73:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzaki, Y., K. Sugawara, E. Takashita, Y. Muraki, S. Hongo, N. Katsushima, K. Mizuta, and H. Nishimura. 2004. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J. Med. Virol. 74:132-140. [DOI] [PubMed] [Google Scholar]
- 10.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa, N., R. Kubota, A. Maeda, and Y. Okuno. 2004. Influenza B virus Victoria group with a new glycosylation site was epidemic in Japan in the 2002-2003 season. J. Clin. Microbiol. 42:3295-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rota, P. A., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 13.Shaw, M. W., X. Xu, Y. Li, S. Normand, R. T. Ueki, G. Y. Kunimoto, H. Hall, A. Klimov, N. J. Cox, and K. Subbarao. 2002. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology 303:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Wagner, R., M. Matrosovich, and H. D. Klenk. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12:159-166. [DOI] [PubMed] [Google Scholar]
- 15.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 16.Xu, X., S. E. Lindstrom, M. W. Shaw, C. B. Smith, H. E. Hall, B. A. Mungall, K. Subbarao, N. J. Cox, and A. Klimov. 2004. Reassortment and evolution of current human influenza A and B viruses. Virus Res. 103:55-60. [DOI] [PubMed] [Google Scholar]
- 17.Xu, X., J. Shaw, C. B. Smith, N. J. Cox, and A. I. Klimov. 2001. Multiple lineages co-circulation and genetic reassortment of the neuraminidase and hemagglutinin genes within influenza viruses of the same type/subtype. Elsevier Science, New York, N.Y.
- 18.Yamashita, M., M. Krystal, W. M. Fitch, and P. Palese. 1988. Influenza B virus evolution: cocirculating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 163:112-122. [DOI] [PubMed] [Google Scholar]