Abstract
The severe acute respiratory syndrome (SARS) epidemic of 2003 was responsible for 774 deaths and caused significant economic damage worldwide. Since July 2003, a number of SARS cases have occurred in China, raising the possibility of future epidemics. We describe here a rapid, sensitive, and highly efficient assay for the detection of SARS coronavirus (SARS-CoV) in cultured material and a small number (n = 7) of clinical samples. Using rolling circle amplification (RCA), we were able to achieve sensitive detection levels of SARS-CoV RNA in both solid and liquid phases. The main advantage of RCA is that it can be performed under isothermal conditions with minimal reagents and avoids the generation of false-positive results, a problem that is frequently encountered in PCR-based assays. Furthermore, the RCA technology provides a faster, more sensitive, and economical option to currently available PCR-based methods.
Severe acute respiratory syndrome (SARS) is an emerging disease caused by the novel SARS coronavirus (SARS-CoV) (2, 4, 5, 14). By the end of the SARS epidemic in July 2003, a total of 8,096 SARS cases had been reported from 30 countries, with 774 deaths. Whether future outbreaks of SARS will occur is unknown at present. However, given the recent SARS cases in southern China arising from an unknown source and a number of laboratory-related infections (12), it is important to be prepared for such a possibility. In the absence of a SARS-CoV vaccine or antiviral drugs, the use of strict infection control policies and early diagnosis with rapid, sensitive, and highly specific laboratory methods are essential for the early management of SARS-CoV infection.
Apart from epidemiological linkages, the clinical and radiographic features of the disease are not SARS specific, identifying a need for specific laboratory tests that can confirm SARS-CoV infection early in the course of the illness. Detection of SARS-CoV-specific antibodies is a sensitive and specific but is not possible at clinical presentation (6, 14).
Detection of SARS-CoV by reverse transcription-PCR (RT-PCR) in clinical specimens allows diagnosis in the early stage of the disease. However, in contrast to many other acute respiratory infections, only low levels of SARS-CoV are thought to be present during the early symptomatic phase of infection. On the basis of the results of first-generation RT-PCR assays, SARS-CoV RNA can be detected with a sensitivity of only ca. 30 to 50% in a single respiratory specimen. A higher sensitivity can be achieved if serial samples are collected, particularly during the second week of illness when maximal virus shedding occurs (13, 14). The type of clinical sample (e.g., nasopharyngeal aspirate, throat swabs, stool samples, urine, etc.) also affects the sensitivity of RT-PCR (21).
Recently, the utility of circularizable oligonucleotides, or “padlock probes,” has been demonstrated for the detection of target nucleic acid sequences; this approach shows greater sensitivity than conventional PCR (3, 8, 16). Upon hybridization to a target DNA or RNA sequence, the two ends of the probe become juxtaposed and can be joined by DNA ligase (Fig. 1). The circularized DNA probe then creates an effective template for an exponential, or hyperbranching, rolling-circle amplification (RCA) reaction (Fig. 1), catalyzed by a highly processive DNA polymerase with strand displacement activity. In isothermal conditions, hyperbranching RCA is capable of a 109-fold signal amplification of each circle within 90 min (8). The RCA technique is highly sensitive, and a circularized DNA probe bound to a single target template can be efficiently detected (3). The RCA assay also offers several advantages over other amplification techniques: the ligation requires Watson-Crick base pairing at both ends of the probe hybridize with perfect complementarity, not only permitting the detection of a single-nucleotide polymorphism but also preventing the nonspecific amplification generated by conventional PCR. Circularizable probes can be used for the recognition of both DNA and RNA templates, eliminating the need for RT and creating a uniform assay format for both RNA and DNA detection (11). Single-stranded DNA displaced by the DNA polymerase can be readily bound by primers, thus enabling the reaction to be performed under isothermal conditions and removing the need for a thermocycler. We describe here a simple, scalable assay using RCA technology that allows the rapid, sensitive, and efficient detection of cultured SARS virus in both liquid and solid phases and present preliminary results on a small number of clinical respiratory specimens.
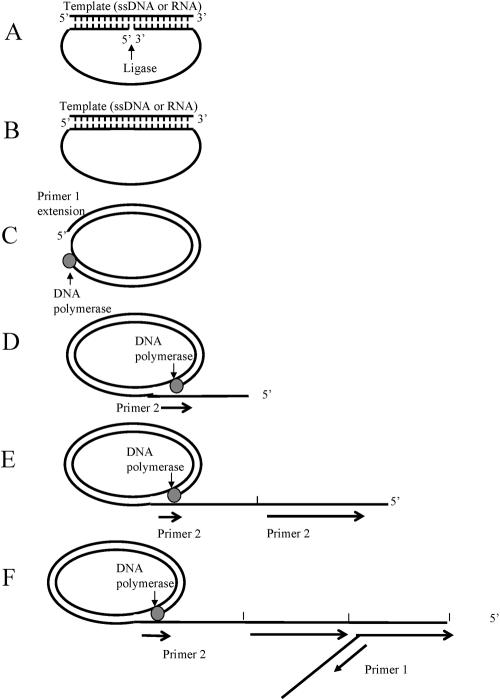
FIG. 1.
Pictorial representation of the RCA method. (A) Padlock probe containing target-complementary segment hybridization to a target DNA or RNA sequence. (B) The padlock probe can be circularized by DNA ligase. (C) Ligated probe and binding of complementary primer (primer 1) for RCA, catalyzed by a strand displacing DNA polymerase. (D) Tandem repeat sequences complementary to the circular probe were generated by RCA. The reverse primer (primer 2) binds to each tandem repeat generated by the rolling circle. (E) Multiple priming events are initiated by primer 2 as the original RCA strand elongates. (F) As these priming events elongate and generate displaced DNA strands, new priming sites for the first primer (P1) are generated. Thus, in the presence of a circular template, the two primers generate a self-propagating, ever-increasing pattern of alternating strand displacement, branching, and DNA fragment release events. As the displaced DNA becomes completely double stranded, it accumulates in fragments of unit length containing one, two, three repeats, etc.
MATERIALS AND METHODS
Culture SARS-CoV and clinical specimens.
In cultured material, SARS CoV (HKU-39849) isolated from a patient with SARS CoV pneumonia was propagated in Vero cells in Dulbecco modified Eagle medium with 10% fetal calf serum. After single passage in Vero cells, viral RNA was extracted by QIAamp virus RNA minikit (Qiagen) according to manufacturer's protocol. SARS-CoV RNA was kindly provided by J. S. M. Peiris at the Department of Microbiology, University of Hong Kong and Queen Mary Hospital, Hong Kong. The sputum samples from seven SARS patients (confirmed by RT-PCR) were collected at day 5 to 13 after the onset of illness. These sputum samples were collected by the National Institute of Virology Beijing, during April and May 2003 by the team of Yunde Hou.
Circularizable probe design.
The oligonucleotide probes used in these experiments were ca. 80 to 90 nucleotides (nt) in length, consisting of two adjacent target complementary sequences (15 to 20 nt) with a spacer region (40 to 50 nt) to facilitate the loop formation and provide a template for RCA primer binding. A total of seven circularizable probes were designed targeting various regions of SARS-CoV (RNA and cDNA templates) (Table 1), carefully selected to ensure that probe binding was SARS-CoV-specific and not sharing similarity to other coronaviruses such as OC43 and 229E strains. The spacer regions were also designed to allow specific RCA primer binding and amplification of the probe-specific signal. To demonstrate a working molecular model and to allow optimization of assay conditions, artificial DNA, or RNA templates were synthesized (Table 1).
TABLE 1.
Circulizable probes with their corresponding artificial templates and RCA primersa
| Probe and primer | Probe and primer sequence |
|---|---|
| Open circle probe A: OCP A, 90 nt | 5′-(P)GTCCTCCATTCTGGTTATTGTCAGTAGTACGCTGATATTCGTGTCAAGATACGCGTAATGACCTTAGTTACTTTGGCCTTGCCCCATTGC-3′ |
| Open circle probe B: OCP B, 83 nt | 5′-(P)CAAGACGTAATGACTGTTCAGATTCTTGCATGGTCACACGTCGTTCTAGTACGCTTCTAGTACGCTTTAAACCATGGCGTCGA-3′ |
| Open circle probe B2: OCP B2, 84 nt | 5′-(P)GTTGTCTGATATCACACATTGAGTACGCTGATATTCGTGTCAAGATACGCGTAATGACCTTAGTTACTCAACTACGAATAGGA-3′ |
| Open circle probe: OCP C, 83 nt | 5′-(P)GTTGTCTGATATCACACATTGATTCTTGCATGGTCACACGTCGTTCTAGTACGCTTCTAGTACGCTTTCAACTACGAATAGGA-3′ |
| Open circle probe: OCP C2, 83 nt | 5′-(P)CAAGACGTAATGACTGTTCAGAGTACGCTGATATTCGTGTCAAGATACGCGTAATGACCTTAGTTACTAAACCATGGCGTCGA-3′ |
| Open circle probe: OCP E, 80 nt | 5′-(P)GACGCCATGGTTTATACTATTCTTGCATGGTCACACGTCGTTCTAGTACGCTTCTAGTACGCTTTCAGTCATTACGTCTT-3′ |
| Open circle probe: OCP F, 85 nt | 5′-(P)CAACTCCTATTCGTAGTTGAATTCTTGCATGGTCACACGTCGTTCTAGTACGCTTCTAGTACGCTTTAATGTGTGATATCAGA-3′ |
| Artificial template for OCP A (28171-28215) | 5′-CAACTGACAATAACCAGAATGGAGGACGCAATGGGGCAAGGCCAA-3′ |
| Artificial RNA template for OCP B and B2 (1966-2006) | 5′-UCUGAACAGUCAUUACGUCUUGUCGACGCCAUGGUUUAUACU-3′ |
| Artificial RNA template for OCP C and C2 (14735-14768) | 5′-ACAAUGUGUGAUAUCAGACAACUCCUAUUCGUAGU-3′ |
| Artificial template for OCP-E (2016-1957) | 5′-GCAGGTCTGAAGTATAAACCATGGCGTCGACAAGACGTAATGACTGTTCAGAAATACCATC-3′ |
| Artificial template for OCP F (14785-14716) | 5′-TTTATCAACAACTTCAACTACGAATAGGAGTTGTCTGATATCACACATTGTTGGCAGATTATAACGATAA-3′ |
| Artificial RNA template with poly(A) for OCP C and C2 | 5′-CAAUGUGUGAUAUCAGACAACUCCUAUUCGUAGUUGAAAAAAAAAAAAAAAAAAAAAAAAAA-3′ |
| Primer 1 for OCP B, C, E, and F | 5′-AACGACGTGTGACCATGCAAGAAT-3′ |
| Primer 2 for OCP B, C, E, and F | 5′-CTAGTACGCTTCTAGTACGCTTT-3′ |
| Primer 1 for OCP A, B2, and C2 | 5′-TATCTTGACACGAATATCAGCGT-3′ |
| Primer 2 for OCP A, B2, and C2 | 5′-CGTAATGACCTTAGTTACT-3′ |
The underlined regions of the probe sequences are template-binding sites. The genomic locations of the artificial DNA and RNA template are also listed in the table as compared to SARS coronavirus BJ01 (AY278488).
Molecular model of RCA in the liquid phase.
Liquid-phase RCA was performed in two steps. The first was a ligation reaction, wherein the probe specifically binds the target template and becomes circularized by DNA ligase. Probe (20 pmol) was hybridized with 1011 artificial single-stranded DNA template molecules and circularized by incubation with 20 U of T4 DNA ligase (New England Biolabs) in 1× reaction buffer at 37°C for 30 min in a 20-μl reaction volume. Ligations using artificial RNA templates were performed by using 40 U of T4 DNA ligase with 1× reaction buffer containing10 mM MgCl2 and 10 μM ATP in a total volume of 20 μl. Separate control reactions were also performed with human genomic DNA as a template. Five units of ampligase (a thermostable DNA ligase for double-stranded DNA [dsDNA]; Astral Scientific, Caringbah, Australia) were mixed with human genomic DNA (1 μg) and circularizable probe (20 pmol). Genomic DNA was denatured at 95°C for 5 min, and a ligation step then performed at 45°C for 25 min.
After ligation and circularization of the probe, a second step was performed to amplify the circular probe by RCA by using DNA polymerase with strand-displacing activity. Two DNA polymerases were chosen to perform the RCA: Vent (exo-) DNA polymerase and Bst DNA polymerase (New England Biolabs). RCA reactions were performed in a 50-μl volume containing Bst DNA polymerase (8 U) or Vent DNA polymerase (20 U), 400 μM deoxynucleoside triphosphate mix, 40 pmol of each RCA primer, and 5 μl of the original ligation mix. Circularized probe signals were amplified by incubation at 65°C for 90 min and visualized on a 1.5% agarose gel under UV illumination.
To optimize the reaction and further increase amplification efficiency, we modified the reaction mix by adding bacteriophage T4 Gene 32 protein (Roche Applied Science), a single-stranded DNA-binding protein. Gene 32 protein can destabilize dsDNA helices (10) and has been used to enhance PCR (7). During RCA reactions with 500 copies of artificial template, T4 gene 32 protein was added to the reaction in decreasing concentrations (5 μg to 0.5 ng/reaction). In separate experiments, 1 to 10% dimethyl sulfoxide (DMSO) was added to RCA reactions. DMSO can destabilize dsDNA helices while stabilizing polymerase-template complexes (20), thus further increasing the efficiency of the reaction.
Molecular model of RCA in the solid phase.
Excess probe molecules carried over from the ligation step may bind RCA primers and form short dsDNA molecules, a process that may increase background signals and hamper quantitative analysis. To circumvent this, a solid phase RCA assay was designed by using magnetic beads coated with oligo(dT) (Dynal mRNA DIRECT Kit, Dynal Biotech, Oslo, Norway) which bind mRNA molecules and provide a solid platform for the RCA reaction. Artificial RNA templates with a 3′-poly(A) tail were used to assess the sensitivity of this assay. Serially diluted artificial RNA templates were incubated with 40 μl of oligo(dT)- coated magnetic beads and 40 pmol of probe at room temperature for 10 min. The artificial RNA templates and probe complexed with beads were extracted by magnetic separation according to the manufacturer's protocol. Probe bound to artificial RNA templates was circularized by incubation with 40 U of T4 DNA ligase in 1× buffer containing 10 mM MgCl2 and 10 μM ATP at 37°C for 30 min. The RCA reaction was performed as described above in the liquid phase by using Bst DNA polymerase.
Detection of SARS-CoV RNA target.
After the specificity and sensitivity of RCA was confirmed by molecular modeling, the assay was then tested on SARS-CoV RNA samples derived from cultured material and sputum samples of SARS patients. In solid-phase RCA, SARS-CoV RNA was extracted by incubation of specimens with 500 μl of lysis/binding buffer (supplied by the kit) for 10 min at room temperature, followed by the addition of oligo(dT)-coated magnetic beads (40 μl) to bind to RNA molecules. The RNA-bonded beads were washed four times with the wash buffer provided in the kit at room temperature and separated by using a magnet to remove possible inhibitors. Then 40 pmol of probe was added and mixed with bead-bonded viral RNA molecules. The ligation of probe and viral RNA was performed by incubation with 40 U of T4 ligase in 1× reaction buffer (10 mM MgCl2 and 10 μM ATP) at 37°C for 30 min. After the ligation, a second washing step was carried out with the wash buffer provided with the kit to remove the unbound probe, and the circularized probe, viral RNA, and magnetic beads complex were separated by using a magnet. RCA reactions were then performed as described above.
RESULTS
Circularizable probe design and amplification of probe signal using RCA.
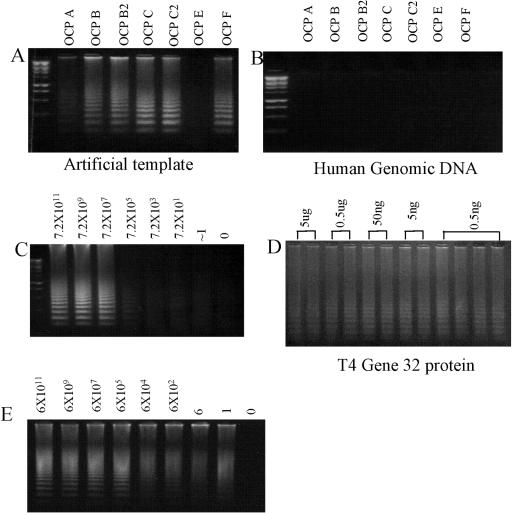
Seven circularizable probes were designed to target various RNA and cDNA regions of SARS-CoV (Table 1). To demonstrate a working molecular model, complementary artificial DNA or RNA templates were also synthesized. Initial testing of the seven probes and their corresponding artificial templates indicated that six of seven probes readily bound to the template and probe signals could be easily detected (Fig. 2A). One probe did not generate a signal from its corresponding template, a finding that could be due to poor quality of the probe and/or secondary structure(s) in the template/probe. When human DNA samples were used as nonspecific template for specificity tests, no signals were observed, indicating highly specific binding for all six probes (Fig. 2B).
FIG. 2.
(A) RCA detection using 20 pmol of circularizable probe and 1011 artificial DNA and RNA template. Six of seven probes were shown to detect their corresponding artificial template (Table 1), the negative signal (OCP-E) could be due to the quality of the probe and/or secondary structure of the probe and template. (B) Specificity testing with 1 μg of human genomic DNA as a nonspecific template. No probe signal can be detected. (C) Liquid-phase RCA assay performed on serially diluted artificial template. Based on the optical density and measurements of stock oligonucleotides, the detection can reach the single-copy level. However, the signal was weak at low target template numbers. (D) Effect of gene 32 protein on liquid-phase RCA. Using 500 copies of artificial template and T4 gene 32 protein with decreasing concentrations from 5 μg to 0.5 ng/reaction indicated an minor enhancement of signal at a concentration of 0.5 ng. (E) Solid-phase RCA assay performed on serially diluted artificial SARS template. The solid-phase assay was able to detect serial diluted templates at the single-copy level.
RCA in the liquid phase.
To assess the sensitivity of the technique, RCA was performed on serial dilutions of the target template (artificial DNA and RNA templates), and a sensitivity of detection close to the single template level was achieved. However, the signal derived was relatively weak in many cases when the target template copy number was below 100 (Fig. 2C). Several strategies have been indicated to improve signal strength in RCA, including the addition of DMSO or T4 gene 32 protein (7, 20, 22) to destabilize dsDNA helices and stabilize polymerase-template complexes. Tests with 500 copies of artificial template and T4 gene 32 protein with decreasing concentrations from 5 μg to 0.5 ng/reaction indicated a minor enhancement of signal at a concentration of 0.5 ng (Fig. 2D). In a separate experiment, 1 to 10% DMSO added to the RCA reaction by using 500 copies of artificial template gave no significant signal enhancement (data not shown). More tests are required to validate optimal conditions for use of these enhancers to maximize signal strength. In addition to weak signal strength at low copy numbers, unbound probe molecules carried over from the ligation mix sometimes resulted in smear-like background during the liquid-phase RCA assay. The presence of background signal makes liquid-phase RCA detection of SARS-CoV less suitable for possible quantitative applications.
RCA in the solid phase.
To circumvent these problems and prevent the formation of background signals, we developed a variant “solid-phase” RCA assay using magnetic beads coated with oligo(dT). The poly(A) signal located at the 3′ end of the SARS-CoV RNA binds to the oligo(dT) molecules on the beads, facilitating the rapid isolation of RNA and further providing a solid-phase medium for the RCA reaction. A molecular model was also established by using artificial RNA templates with poly(A) sequences located at the 3′ end. Serial dilutions of template used in RCA proved that the solid-phase method could detect close to a single artificial copy with a strong signal (Fig. 2E).
Detection of SARS-CoV RNA by RCA.
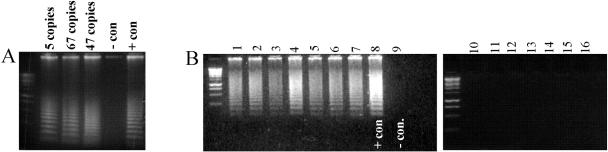
The molecular models we developed demonstrated the sensitivity of RCA for SARS-CoV detection, particularly in the solid phase. Testing the SARS-CoV RNA samples derived from cultured material with known copy numbers clearly indicated that as few as five copies of SARS viral genome (defined by using a RealArt HPA Coronavirus kit; RealArt, Hamburg, Germany) could be easily detected by RCA in the solid phase (Fig. 3A). Testing of SARS-CoV from patient samples was also performed. In total, all seven RNA samples derived from sputum of SARS-positive patients for SARS-CoV by RT-PCR showed a strong positive signal by RCA (Fig. 3B). Control samples of healthy individuals showed no signal in solid-phase RCA experiments.
FIG. 3.
(A) Solid-phase RCA detection of SARS-CoV RNA from cultured material. Based on copy numbers determined by the commercially available RT-PCR kit, the RCA test was easily successful in detecting under five SARS-CoV RNA copies. (B) Solid-phase RCA detection of SARS-CoV RNA from positive sputum samples (lanes 1 to 7) generated strong signals. The positive control (line 8) in this test derived from cultured SARS virus, whereas RCA performed on uninfected human RNA samples showed no signals (lanes 10 to 16).
DISCUSSION
This report describes an efficient method for the sensitive detection of SARS-CoV genome using RCA in both liquid and solid phases.
Given the occasional reappearance of SARS after the 2003 epidemic ended, the development of diagnostic assays for early, rapid, and reliable detection of SARS-CoV should remain a high priority. At present, the RT-PCR-based assays are the most widely used initial diagnostic tests. However, the sensitivity of SARS-CoV RT-PCR has been estimated to be as low as 30%, depending on the type of specimens tested and the timing of collection relative to the onset of symptoms (13, 14). One recent report has described an increase in sensitivity for the detection of the virus, benefiting earlier diagnosis (15). Despite the obtainable magnitude of amplification, these methods are time-consuming (e.g., RNA extraction, RT, and real-time PCR) and require a high-precision thermocycler, along with expensive reagents. In addition, cross-contamination and carryover, which lead to false-positive results, remains a continued problem. RCA offers a distinct advantage because it is based on the amplification of signal rather than target amplification.
The availability of ligases that accurately distinguish DNA sequences has made oligonucleotide ligation assays increasingly popular in clinical diagnostics (1). DNA ligases have been isolated from a number of organisms and characterized for their ability to discriminate between matched and mismatched substrates (9, 17, 18, 19). The ATP-dependent T4 DNA ligase can join DNA oligonucleotides on RNA targets, although this reaction is quite inefficient at ATP concentrations commonly used for DNA-templated DNA ligation. Recently, however, reaction conditions have been defined that allow efficient RNA-template DNA ligation by the T4 DNA ligase (11), extending the use of padlock probes to RNA targets.
Although RCA has been shown to be highly sensitive and able to detect a single template (3, 8, 16), PCR remains the method of choice in both research and clinical diagnostics for sensitive detection of nucleic acids. However, in recognition of the promising features and considerable advantages of RCA and similar methods, padlock probes are becoming more widely adopted. PCR involves exponential target amplification, thereby increasing the risk of amplicon cross-contamination. In contrast, RCA specifically amplifies the probe signal avoiding the target template accumulation over time, minimizing the risk of contamination and making it more attractive for high-throughput analysis. Moreover, RCA can be performed in isothermal conditions, a factor that is useful where access to scientific equipment is minimal.
In the present study, we have successfully adopted the RCA technique for the detection of SARS-CoV from artificial and patient-derived material and have clearly demonstrated the advantages of this sensitive and specific diagnostic test. In molecular modeling with artificial DNA templates, we demonstrated that this technique is capable detecting down to the single template level. These results demonstrate a considerable potential of this technique in future clinical diagnostic testing of SARS-CoV, although the enhancement of signals in the liquid phase by DMSO and T4 gene 32 protein at low copy numbers of the template may require further optimization. In addition, the direct attachment to RNA molecules by circular probes can eliminate the RT step required by many RNA studies, providing a useful mechanism for the sensitive and accurate detection and identification of RNA sequences. The recent development of efficient RNA-templated DNA ligation (11), coupled with the powerful signal amplification generated by RCA down to the detection of a single circular probe (3), has facilitated the design of a sensitive assay for SARS-CoV RNA detection. More than 80% of the RNA template present can be recognized by DNA probes (11) and successfully ligated by T4 DNA ligase, making RCA detection of RNA template greater in sensitivity than conventional RT-PCR technology.
The relatively abundant unbound probe remaining in the ligation reaction during liquid-phase RCA can in some cases generate a background signal interfering with signal detection. This may be of concern, particularly in quantitative applications using fluorescently labeled primer at low copy numbers. However, the application of a solid-phase assay using oligo(dT)-coated magnetic beads provides a promising solution to this problem. This solid-phase format has the added advantage of rapid RNA extraction through the binding of oligo(dT) molecules on the beads, with poly(A) signals located at 3′ end of the mRNAs. Hybridization between the target RNA and complementary probe binding region results in the formation of a helix with four turns (10 nt per turn, 20 nt long in each complementary region), allowing a ligation reaction to create a padlock structure. This interaction can withstand stringent washing, allowing the removal of unbound probe molecules. The solid-phase RCA assay demonstrated close to single artificial RNA template detection in molecular modeling with stronger signal, and five copies of SARS-CoV RNA were easily detectable. This clearly demonstrates the bright future of RCA in the diagnosis of SARS-CoV and indeed of other diseases. Testing conducted on patient samples further highlighted the usefulness of this assay as a future diagnostic test for SARS-CoV. It remains unclear why solid-phase RCA can generate a stronger signal than the liquid phase, but the reduction of background signal by solid-phase alone confirms it as an attractive option for ultrasensitive detection. Nonetheless, it is important to reiterate that we could successfully achieve low-copy detection in both solid- and liquid-phase RCA. Our data also showed that the RCA reaction is most efficient when catalyzed by thermostable strand-displacing enzymes such as Bst DNA polymerase. Additional work will be required to develop the ideal conditions for RCA in SARS-CoV detection, although the reaction sensitivity and signal strength already compares very favorably with PCR. Future studies are required for the development of a quantitative assay. Studies performed by Faruqi et al. (3) clearly demonstrated the quantitative potential of RCA, showing it to be capable of accurately detecting numbers of circularizable probe bound to genomic DNA. We predict that the inherent simplicity of this method will allow RCA to be used in a wide variety of clinical applications.
Acknowledgments
This study was supported by grant from NHMRC grant (264435) to B.W. B.W. is supported by postdoctoral fellowship U2000 (University of Sydney).
REFERENCES
- 1.Dequeker, E., and J. J. Cassiman. 2000. Genetic testing and quality control in diagnostic laboratories. Nat. Genet. 25:259-260. [DOI] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Faruqi, F. A., S. Hosono, M. D. Driscoll, F. B. Dean, O. Alsmadi, R. Bandaru, G. Kumar, B. Grimwade, Q. Zong, Z. Sun, Y. Du, S. Kingsmore, T. Knott, and R. S. Lasken. 2001. High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics 2:4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 5.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, G., X. Chen, and A. Xu. 2003. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 349:508-509. [DOI] [PubMed] [Google Scholar]
- 7.Liu, Q., and S. S. Sommer. 1998. Subcycling-PCR for multiplex long-distance amplification of regions with high and low GC content: application to the inversion hotspot in the factor VIII gene. BioTechniques 25:1022-1028. [DOI] [PubMed] [Google Scholar]
- 8.Lizardi, P. M., X. H. Huang, Z. G. Zhu, P. Bray-Ward, D. C. Thomas, and D. C. Ward. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19:225-232. [DOI] [PubMed] [Google Scholar]
- 9.Luo, J., D. E. Bergstrom, and F. Barany. 1996. Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res. 24:3071-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris, C. F., N. K. Sinha, and B. M. Alberts. 1975. Reconstruction of bacteriophage T4 DNA replication apparatus from purified components: rolling circle replication following de novo chain initiation on a singlestranded circular DNA template. Proc. Natl. Acad. Sci. USA 72:4800-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson, M., D. O. Antson, G. Barbany, and U. Landegren. 2001. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 29:578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parry, J. 2004. Breaches of safety regulations are probable cause of recent SARS outbreak, W.H.O. says. BMJ 328:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peiris, J., C. Chu, V. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, K. Y. Yuen, et al. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, and SARS study group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon, L. L. M., K. H. Chan, O. K. Wong, W. C. Yam, K. Y. Yuen, Y. Guan, Y. M. Lo, and J. S. Peiris. 2003. Early diagnosis of SARS coronavirus infection by real-time RT-PCR. J. Clin. Virol. 28:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweitzer, B., and S. Kingsmore. 2001. Combining nucleic acid amplification and detection. Curr. Opin. Biotechnol. 12:21-27. [DOI] [PubMed] [Google Scholar]
- 17.Shuman, S. 1995. Vaccinia virus DNA ligase: specificity, fidelity, and inhibition. Biochemistry 34:16138-16147. [DOI] [PubMed] [Google Scholar]
- 18.Sriskanda, V., and S. Shuman. 1998. Specificity and fidelity of strand joining by Chlorella virus DNA ligase. Nucleic Acids Res. 26:3536-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong, J., W. Cao, and F. Barany. 1999. Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res. 27:788-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varadaraj, K., and D. M. Skinner. 1994. Denaturants or cosolvents improve the specificity of PCR amplification of a G+C-rich DNA using genetically engineered DNA polymerases. Gene 140:1-5. [DOI] [PubMed] [Google Scholar]
- 21.Yam, W. C., K. H. Chan, L. L. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. Peiris. 2003. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, D. Y., M. Brandwein, T. Hsuih, and H. B. Li. 2001. Ramification amplification: a novel isothermal DNA amplification method. Mol. Diagn. 6:141-150. [DOI] [PubMed] [Google Scholar]