Abstract
Hepatitis delta virus (HDV), in association with hepatitis B virus, is responsible for severe acute and chronic hepatitis. Treatment of the infection relies on the long-term administration of high doses of alpha interferon (IFN), and the treatment efficiency is monitored by the detection of anti-HDV immunoglobulin M and HDV genome in serum. Like the case for other chronic viral infections, HDV genome quantification in serum should be useful for the follow-up of infected patients. The aims of this study were to develop a quantitative assay for the detection of any type of HDV in serum and to evaluate the benefit of HDV RNA quantification for the follow-up of chronically infected patients receiving IFN. A real-time reverse transcription-PCR assay was developed to quantify the HDV RNA load in serum. Its efficacy was evaluated with 160 serum samples, 76 of which were collected from 11 chronically infected patients who were treated with pegylated IFN. The assay was sensitive (100 copies/ml of serum) and efficient for all HDV types, including type 3 and the recently described types 5, 6, and 7. The viral load determinations for treated patients allowed us to identify different profiles of virological responses to IFN therapy with more accuracy than that attainable with the qualitative approach. In conclusion, we have developed a quantitative HDV RNA assay for serum which is adapted to the follow-up of antiviral treatment for patients infected with any HDV type. The assay will help us to understand the natural history of HDV infection and to define guidelines for the management of chronic delta hepatitis.
Hepatitis delta virus (HDV) is a 36-nm viral particle which depends on the hepatitis B virus (HBV) for virion assembly and propagation (16). The HDV particle consists of a circular single-stranded RNA genome of 1,672 to 1,697 nucleotides (30, 38) which assembles with two viral proteins, sHD and LHD, to form a ribonucleoprotein. During the course of viral replication, the ribonucleoprotein buds through the hepatocyte endoplasmic reticulum membrane and acquires a lipid envelope in which the hepatitis B virus surface antigens are embedded. Extensive intramolecular complementarity of the HDV genome leads to the formation of a pseudo-double-stranded RNA structure (38). Sequence analyses of numerous isolates have led to the classification of HDV into at least seven distinct clades (or types) with different geographic distributions (30). Briefly, HDV type 1 viruses (genotype I, hereafter named HDV-1) are found in most areas, types 2 and 4 (genotypes IIA and IIB, referred to as HDV-2 and HDV-4, respectively) are located in Far Eastern areas, type 3 (genotype III, referred to as HDV-3) is found in the northern part of South America, and types 5, 6, and 7 (HDV-5, HDV-6, and HDV-7, respectively) are found in Western and Central Africa (5, 19, 30, 35, 40).
HDV infection can cause severe liver disease, with fulminant hepatitis occurring at least 10 times more often than for HBV infection alone and with a chronicity rate reaching 70 to 90% in cases of superinfection (10, 16, 32). In many cases, chronic delta hepatitis evolves to cirrhosis (60 to 70%) and hepatocellular carcinoma (13, 34), and the current treatment efficacy is disappointing (11, 33). Indeed, after 12 months of treatment with alpha interferon (IFN-α) at a high dose (27 MU weekly) (12), only about 50% of patients respond positively, but 40% of responders relapse during the 6 months following the end of therapy (11). Neither lamivudine, a nucleosidic analog inhibitor of the HBV DNA polymerase (23, 39), nor ribavirin, acyclovir, or famciclovir is effective at controlling HDV replication (1, 14, 42). The diagnosis of HDV infection usually relies on the detection of specific anti-HDV antibodies, with the presence of anti-HDV immunoglobulin M (IgM) reflecting ongoing viral replication. However, the serological approach for the detection of virus replication lacks sensitivity (8, 25). The HDV antigen is rarely detected in the serum, except in cases of acute infections (prior to the antibody response) or chronic infections in severely immunosuppressed patients (17). Therefore, HDV replication is most efficiently evaluated by the detection of HDV RNA in serum by the use of homemade qualitative reverse transcription-PCR (RT-PCR) assays, as no commercial test is available (21). Several semiquantitative methods have been proposed to evaluate HDV RNA levels in the serum, but they remain imprecise and difficult to handle with routine laboratory procedures (4, 7, 21).
Recently, Yamashiro et al. developed a real-time RT-PCR assay to quantify HDV RNA in serum, and they suggested a possible correlation between the viral load and the clinical stage of liver disease (41). This test, which is sensitive and simple to perform, was designed to detect HDV-2 and HDV-4 viruses. However, these HDV types are mostly located in Far Eastern regions of the world and are not commonly detected in European countries. For the present study, a consensus real-time RT-PCR assay was developed to quantify HDV RNAs of all known types in serum. The assay was sensitive enough to detect 100 HDV RNA copies per ml of serum. It was used to quantify viral loads in sequential sera collected from patients with chronic delta hepatitis during the course of IFN treatment. The results revealed different patterns of viral load reduction among the patients, indicating that the treatment efficacy might be monitored more accurately with an RT-PCR-based quantitative approach than with a qualitative RT-PCR approach. This assay is proposed for the follow-up of chronically infected patients with the aim of defining guidelines for standardized treatments.
MATERIALS AND METHODS
Patients and samples.
Eighty-four blood samples (numbered 1 to 84) were used for validation of the assay. HDV-negative samples (no. 1 to 26) were tested to evaluate the specificity of the assay. Among these samples, 5 were positive for the HBV surface antigen and HBV DNA (no. 1 to 5) and 16 were negative for the HBV surface antigen but positive for either hepatitis C virus (HCV) RNA (no. 6 to 11), hepatitis A virus (HAV) IgM (no. 12 to 15), human immunodeficiency virus (HIV) RNA (viral load, >10,000 copies/ml; no. 16 to 20), or hepatitis E virus RNA (no. 21). The remaining five samples were negative for HCV RNA, HAV IgM, and HIV RNA. Samples 41 to 79, which according to Radjef et al. (30) corresponded to HDV-1 (no. 41 to 63), HDV-2 (no. 64), HDV-5 (no. 65 to 70), HDV-6 (no. 71 to 74), and HDV-7 (no. 75 to 79), were tested with both a qualitative RT-PCR assay designed to amplify the R0 region of the HDV genome (nucleotides 885 to 1285) (20, 30) and the new quantitative real-time RT-PCR assay. HDV-3 samples (no. 80 to 84) were available from a collection of dried blood spot specimens on filter paper collected by means of fingertip pricks from infected individuals in the Amazonia region (T. Hanslik and G. de Catheu, unpublished). The filter papers were air dried and stored at room temperature until use. For the assay, a 7-mm disk was punched out of the center of each dried blood spot, soaked in 500 μl of a 9‰ NaCl solution, and stored at −20°C. RNAs were extracted from 250-μl eluates by use of a QIAamp MinElute virus vacuum (QIAGEN, Courtabœuf, France) according to the manufacturer's recommendations.
Eleven patients, labeled A to K (76 samples), who had been treated or were being treated with IFN (or its pegylated form, PEG-IFN) for chronic HDV infection and for whom sequential serum samples had been kept frozen at −80°C, were selected for retrospective HDV RNA quantification. These samples had previously been collected for routine diagnostic purposes and tested with the qualitative assay cited above. The age, sex, HDV type, and HIV status of each patient are presented in Table 1.
TABLE 1.
Clinical features of patients selected for retrospective sequential quantification of HDV RNA in blood samples
| Patient | Sexa | Age | HDV typeb | HIV statusc | No. of samples tested |
|---|---|---|---|---|---|
| A | M | 54 | 1 | NEG | 7 |
| B | M | 40 | 1 | NEG | 4 |
| C | F | 45 | 1 | NEG | 8 |
| D | M | 37 | 1 | NEG | 6 |
| E | M | 52 | 1 | NEG | 4 |
| F | M | 44 | 1 | NEG | 11 |
| G | M | 42 | 1 | NEG | 4 |
| H | M | 37 | 1 | POS | 9 |
| I | M | 43 | 1 | POS | 4 |
| J | M | 54 | 1 | POS | 10 |
| K | M | 40 | 5 | POS | 9 |
M, male; F, female.
HDV types were determined by sequencing the R0 region (or by restriction fragment length polymorphism analysis for patients A, C to E, and G).
NEG, negative HIV serology; POS, positive HIV serology.
Aliquots containing 40 or 4,000 HDV RNA copies derived from the total liver RNA from a woodchuck (woodchuck 5) infected with the HDV-1 prototype (9, 29) were used as controls.
Real-time PCR quantification in serum samples.
HDV RNAs were extracted from 250 μl of serum or plasma by use of a QIAamp MinElute virus vacuum (QIAGEN, Courtabœuf, France). cDNAs were synthesized as previously described (20) and were purified with Montage PCR centrifugal filter devices (Millipore, Molsheim, France).
The primers and probe were designed to take into account the genetic variability of HDVs (30). The forward primer was selected to target the ribozyme region of the genome, and the reverse primer targeted region I of the antigenome ribozyme (28). The probe, which hybridized to the same region as the reverse primer, was designed to anneal to the antigenomic sequence to avoid base pairing with the reverse primer. The names and sequences of the primers and probe were as follows: Delta-F (forward primer), 5′-GCATGGTCCCAGCCTCC-3′; Delta-R (reverse primer), 5′-TCTTCGGGTCGGCATGG-3′; and Delta-P (probe), 5′-FAM-ATGCCCAGGTCGGAC-MGB-3′. Because of the existence of one mismatch with the sequences of HDV-3 genomes, the following second direct primer was specifically designed for the amplification of HDV-3 isolates: T3-Delta-F, 5′-GCATGGCCCCAGCCTCC-3′.
Real-time PCRs were performed by using the TaqMan Universal PCR master mix (Applied Biosystems, Courtabœuf, France). Purified cDNA (10 μl) was added to a 40-μl PCR mixture containing 25 μl of TaqMan Universal PCR master mix, 300 μmol/liter of each primer, and 200 μmol/liter of fluorogenic probe. The reaction consisted of one initiating step of 2 min at 50°C, followed by 10 min at 95°C and then 45 cycles of amplification including 15 s at 95°C and 1 min at 60°C. The reactions, data acquisition, and analyses were performed with the ABI PRISM 7000 sequence detection system (Applied Biosystems, Courtabœuf, France).
The plasmid pCRII-dFr45-R′1 (30), containing one copy of the R′1 region of the HDV genome (nucleotides 305 to 1,161 according to Wang et al. [38]), was used as a standard for HDV cDNA quantification. After EcoRI (Biolabs, St. Quentin en Yvelines, France) digestion of the plasmid, the R′1 region was purified from an agarose gel by the use of QIAEX II (QIAGEN, Courtabœuf, France). The concentration of purified R′1 DNA was determined with a multichannel spectrophotometer, and the corresponding copy number was calculated.
Qualitative RT-PCR for HDV RNA detection in serum samples.
HDV RNA detection in serum samples was performed as previously described (20). HDV type determination was performed with the amplified R0 region of the genome (nucleotides 885 to 1285), using either a two-step restriction fragment length polymorphism procedure (E. Gordien, N. Radjef, M. Tamby, P. Dény) or a previously described phylogenetic analysis of the sequence (30).
Statistical analysis.
The Mann-Whitney nonparametric U test (StatView 4.02) was used for statistical comparisons. Differences were considered significant at P values of ≤0.05.
RESULTS
Specificity, sensitivity, and reproducibility of real-time PCR assay for detection and quantification of HDV RNA in serum.
The HDV R′1 fragment was used as a standard to quantify HDV RNA. Tenfold serial dilutions ranging from 10 to 107 copies were tested in triplicate, with the mean cycle threshold (CT) values plotted against the copy number to establish a standard curve. The correlation coefficient was repeatedly >0.997 and the slope was −3.5. The amplification efficiency, calculated as [10(−1/slope) − 1] × 100, was 93%. Dilutions corresponding to inputs of 1 and 10 copies per reaction were repeatedly detected (results not shown). Thus, according to the dilution factors used during the RNA extraction and RT procedures, the sensitivity of the assay to detect HDV RNA in clinical samples was 100 copies/ml of serum and the linearity of quantification ranged from 103 to 109 copies/ml.
The total liver RNA from woodchuck 5 infected with the HDV-1 prototype (9, 29) was subjected to the assay for HDV RNA quantification. Dilutions were then performed to obtain 10-μl aliquots containing 40 and 4,000 copies. These aliquots were stored at −80°C and used as low- and medium-level controls for the assay. A run was considered valid when the low control value was between 30 and 50 copies/10 μl and the medium control value was between 3,200 and 5,000 copies/10 μl (corresponding to a coefficient of variation [CV] of 30%).
The assay specificity was ascertained by the negative RT-PCR results obtained after 45 cycles of amplification for all 26 control samples with negative HDV serology (results not shown).
The intra-assay reproducibility was evaluated by two approaches. First, eight replicates of 10-fold standard dilutions ranging from 10 to 107 copies per reaction were tested in the same experiment. The CVs of the CT values ranged from 0.4% to 2.6% and are reported in Table 2. Second, three replicates of seven serum samples (no. 27 to 33) were independently subjected to RNA extraction, cDNA synthesis, and real-time PCR in the same experiment. The mean viral load of each sample ranged from 7,000 to 2,130,000 copies/ml, and the CV ranged from 1.8% to 25% (Table 2). For estimation of the interassay variability, RNAs were extracted and amplified from four serum samples (no. 34 to 37) in triplicate (three different operators on three distinct days). The mean viral load of each sample ranged from 1,700 to 220,000 copies/ml, and the CV ranged from 3.3% to 25.4% (Table 2).
TABLE 2.
Intra- and interassay reproducibility
| Amt of sample (for standards) or sample no. | Mean CT value | Mean viral load (copy/ml)d | % CV of CT | % CV of viral loadd |
|---|---|---|---|---|
| Standards (intraassay)a | ||||
| 10 copies/10 μl | 38.74 | NA | 2.60 | NA |
| 102 copies/10 μl | 32.75 | NA | 1.60 | NA |
| 103 copies/10 μl | 29.66 | NA | 1.40 | NA |
| 104 copies/10 μl | 26.04 | NA | 0.50 | NA |
| 105 copies/10 μl | 22.57 | NA | 0.70 | NA |
| 106 copies/10 μl | 19.21 | NA | 0.40 | NA |
| 107 copies/10 μl | 16.92 | NA | 0.60 | NA |
| Intraassay samplesb | ||||
| 27 | 20.95 | 2.13 × 106 | 0.25 | 2.70 |
| 28 | 23.17 | 4.70 × 105 | 1.89 | 7.40 |
| 29 | 24.13 | 3.20 × 105 | 0.18 | 1.80 |
| 30 | 24.36 | 2.10 × 105 | 2.48 | 7.20 |
| 31 | 26.49 | 7.50 × 104 | 0.57 | 6.70 |
| 32 | 29.38 | 1.30 × 104 | 1.63 | 25.00 |
| 33 | 30.41 | 7.00 × 103 | 0.34 | 7.10 |
| Interassay samplesc | ||||
| 34 | 24.69 | 2.20 × 105 | 3.03 | 25.40 |
| 35 | 25.46 | 1.30 × 105 | 1.66 | 24.10 |
| 36 | 26.50 | 8.80 × 104 | 3.54 | 3.30 |
| 37 | 32.61 | 1.70 × 103 | 0.88 | 17.30 |
Intraassay reproducibility was evaluated with eight replicates of 10-fold standard dilutions.
Intraassay reproducibility was evaluated with three replicates of serum samples (27 to 33) subjected independently to extraction.
Interassay reproducibility was evaluated with three replicates of serum samples (34 to 37).
NA, not applicable.
The real-time PCR assay and the qualitative assay were compared in terms of sensitivity. Three cDNAs obtained from three distinct serum samples (no. 38 to 40) were diluted to determine the end-point dilution value for each assay. Purified cDNAs were diluted from 1/2 to 1/10,000, and each dilution was tested in parallel with both the qualitative and quantitative assays. With the qualitative assay, the last detectable dilution after ethidium bromide staining of the agarose gel was 1/100 for sample 38 and 1/1,000 for samples 39 and 40. With the real-time PCR assay, the end-point dilution values were 1/500, 1/1,000, and 1/5,000 for samples 38, 39, and 40, respectively, indicating that the qualitative and quantitative assays displayed the same range of sensitivity (results not shown).
HDV RNA quantification for blood samples.
The quality of each assay was monitored with different external controls. Negative controls consisting of two aliquots of water (one subjected to the entire experimental process, including the extraction step, and one subjected to real-time PCR only) and one HDV-negative serum were tested in each assay in order to detect RNA or DNA (plasmid) contamination. Low- and medium-viral-load controls consisting of total HDV-1-infected woodchuck liver RNA were used to monitor the quantification reproducibility of the assay. Finally, each run included a serum sample which had been quantified in a previous run. The assay was considered valid if the interassay CV of this sample remained beneath 30%.
The ability of the assay to quantify HDV RNAs of different types was evaluated with a panel of 39 serum samples, containing either type 1 viruses (n = 23, samples 41 to 63) or viruses of type 2, 5, 6, or 7 (n = 16, samples 64 to 79). For the 23 HDV-1 samples, the median copy number was 195,000 (5.29 log10), with values ranging from 200 (2.3 log10) to 98,600,000 (7.99 log10) copies/ml of serum (viral loads of <1,000 copies/ml were extrapolated from the standard curve). For serum samples containing HDV-2 (n = 1), HDV-5 (n = 6), HDV-6 (n = 4), and HDV-7 (n = 5), the median viral load was 130,000 (5.11 log10), with values ranging from 200 (2.3 log10) to 100,000,000 (8 log10) copies/ml of serum. Thus, the viral load values obtained for HDV-1 viruses and viruses of other types were not significantly different, indicating that the assay could equally detect and quantify HDV genomes of any of these types.
The ability of the assay to detect HDV-3 RNA in samples 80 to 84 was evaluated separately, as these RNA samples had been extracted from the elution products of dried blood spots (which did not allow a quantitative approach). These samples had been found to be positive for HDV RNA with the qualitative assay (although only a faint signal was detected on the ethidium bromide-stained agarose gel). Samples 80 and 81 were found to be positive with the real-time RT-PCR assay using either Delta-F or T3Delta-F as a forward primer. For samples 82 and 83, HDV RNA was detected with primer T3Delta-F only, and sample 84 remained negative under both conditions (Table 3). However, one could expect that viral RNAs obtained directly from serum might give better results.
TABLE 3.
HDV-3 RT-PCR results
| Sample no. | Qualitative RT-PCR resulta | Real-time RT-PCR CT valueb with indicated primer
|
|
|---|---|---|---|
| Delta-F | T3-Delta-F | ||
| 80 | ++ | 44.55 | 35.38 |
| 81 | + | 39.54 | 32.84 |
| 82 | ± | >45 | 41.28 |
| 83 | ± | >45 | 36.56 |
| 84 | ± | >45 | >45 |
Visual appreciation of the ethidium bromide signal intensity: ±, faintly visible band; +, band of low intensity; ++, band of medium intensity.
CT values of >45 indicate a negative result.
HDV RNA quantification for chronically infected patients.
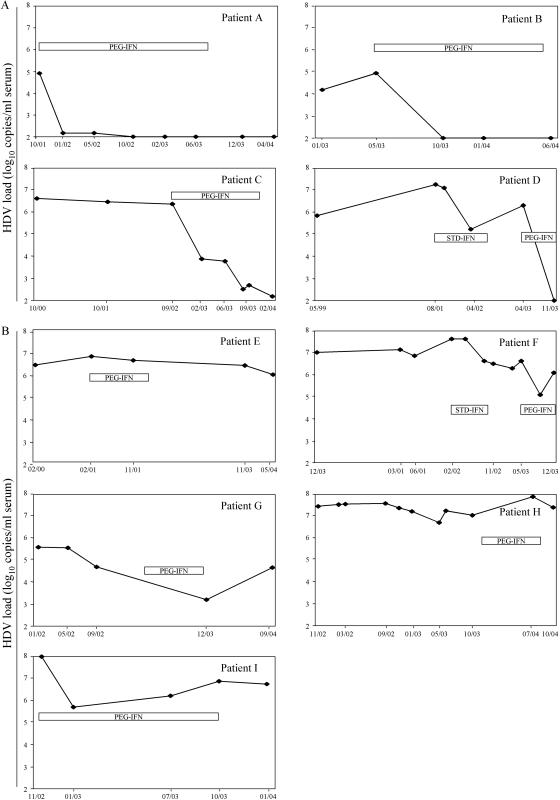
HDV viral loads were retrospectively determined by use of the real-time RT-PCR assay with a collection of serial samples that had been previously tested in the laboratory with the qualitative RT-PCR assay. Eleven patients who had received or were currently receiving IFN for chronic delta hepatitis were considered for this evaluation. Their viral loads, expressed as log10 copy numbers per ml of serum, are reported in Fig. 1.
FIG. 1.
Evolution of HDV load in chronically infected patients treated with IFN. (A) Viral load kinetics suggesting a virological response to PEG-IFN. Patients A and B seemed to respond to the treatment and to negate their viral loads. Patients C and D had detectable viral loads at the end of the treatment, with decreases of >5 log10 copies/ml of serum. (B) Viral load kinetics suggesting no virological response to PEG-IFN. Viral load decreases did not exceed 2.5 log10 copies/ml during the course of the treatment. In the cases of patients E, F, H, and I, the initial viral load was >6 log10 copies/ml.
Two groups of patients could be distinguished on the basis of the HDV viral load evolution. For the first group (Fig. 1A), a reduction of at least 5 log10 copies per ml was observed during the course of the treatment, suggesting a favorable virological response to IFN therapy. For patients A and B, HDV RNA was undetectable with both the qualitative and the quantitative assays at the end of the treatment. Patient C, who had received PEG-IFN for 1 year, presented a viral load which was dramatically reduced but still detectable 2 months after the end of the treatment. Patient D, who had not responded to previous IFN therapy, had a decrease of 5 log10 copies per ml after 6 months of PEG-IFN. This could indicate the initiation of a virological response. In these two cases (patients C and D), the usual qualitative assay had remained positive, thus providing no indication of the viral load decrease. For the second group of patients (Fig. 1B), the reduction of the viral load upon PEG-IFN therapy did not exceed 2.5 log10 copies/ml. Patients E, F, H, and I presented viral loads of >6.5 log10 copies/ml at the initiation of therapy, and patients H and I were coinfected with HIV. Interestingly, and despite the small number of patients, the pretherapeutic viral load level appeared to be significantly higher for the second group of patients than for the first group (P = 0.05; Mann-Whitney U test).
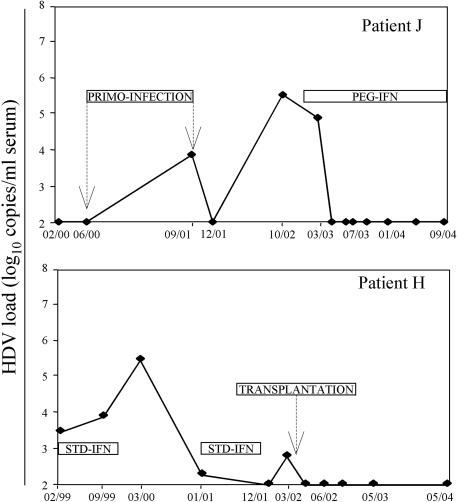
Patient J (Fig. 2), who was chronically infected with HBV, had been negative for HDV markers until a superinfection with HDV occurred between June 2000 and September 2001. In December 2001, viral RNAs became undetectable, suggesting a spontaneous virus neutralization. However, in October 2002, the viral load was raised up to 5.5 log10 copies/ml. A treatment with PEG-IFN was then initiated in February 2003, leading to a rapid reduction of the viral load, which has remained undetectable ever since, with both quantitative and qualitative assays. Patient K, who was unsuccessfully treated with IFN in 1999, developed a hepatic carcinoma in 2001 while undergoing a new treatment. This patient underwent hepatic transplantation in May 2002, and his viral load has remained undetectable since.
FIG. 2.
Evolution of HDV load in infected patients. Patient J, who was coinfected with HBV and HIV, experienced an HDV superinfection. Patient K had a liver transplantation after two treatment failures.
DISCUSSION
HDV is associated with HBV infection in approximately 5% of HBV surface antigen carriers worldwide. A chronic HBV-HDV mixed infection frequently leads to cirrhosis, and the mortality rate for mixed infections is higher than that for chronic HBV infections alone (34). The treatment of chronic hepatitis D has made very little progress, although the long-term benefit of high doses of IFN seems to be confirmed (12). Like the case for chronic hepatitis C (26), the use of the pegylated form of IFN might improve the treatment efficacy, but this has not yet been evaluated in the case of HDV. To date, there is no consensus for the treatment of chronic hepatitis D. Efficacy is usually evaluated with biochemical, histological, and virological parameters, such as specific anti-HDV IgM or HDV RNA. The detection of HDV RNA in serum or plasma is performed by the use of homemade qualitative RT-PCR techniques and reflects viral replication (8, 31, 36). During the last decade, the management of various chronic viral infections has become more and more dependent on quantitative molecular approaches. In the case of HCV, the viral load is now used as a predictive marker of treatment efficacy. For HDV, the severity of the infection and the uncertainty of the treatment outcome are incentives for the development of reliable HDV RNA quantification assays to monitor treatment efficacy.
Real-time PCR based on TaqMan technology provides an accurate and sensitive means of quantifying viral genomes, with the major advantage of avoiding post-PCR handling that can be a source of DNA carryover. Several studies have reported the benefit of this technique for the quantification of viral genomes in blood samples (15, 18, 37). The large dynamic range (10 to 107 copies) of the technique makes it particularly attractive for HDV RNA quantification. Indeed, on the one hand, large viral loads are expected to be detected in the case of immunodeficiency or acute infection (as observed after direct liver inoculation in chimpanzees [2]), and on the other hand, very small amounts of RNA need to be detected during the follow-up of patients under treatment.
The development of an accurate and sensitive test for HDV RNA quantification in blood samples raises several technical problems. First, the “rod-like” structure of HDV RNA, which is due to intramolecular base pairing of >70% of the sequence (22, 38), is likely to impair cDNA synthesis and therefore PCR efficiency. Second, the genetic variability of the virus (5, 19, 30, 35, 40) requires the design of primers and probes to target the most conserved regions of the genome (27). Indeed, the divergence between HDV types has been shown to be as high as 37% over the entire nucleotide sequence of the genome (30). As expected (3, 6), a comparative analysis of HDV sequences of different types indicated that the most conserved regions of the genome are located within the ribozymes (30). The primers and probe were thus designed for these regions, and despite the strong secondary structures in the primer annealing region, the PCR efficiency was repeatedly over 93%. The quantitative real-time RT-PCR assay that was developed in our laboratory aimed to replace the qualitative assay as a routine diagnosis procedure. This was made possible because of the sensitivity of the assay and its ability to detect HDV RNAs of all known types, thus avoiding false-negative results due to a low copy number or sequence variability. HDV-4 isolates were not available for our study, but sequence alignments indicate that our assay should work for the detection and quantification of HDV-4 genomes. In the case of HDV-3 sequences, the existence of a mismatch in the forward primer led us to propose the use of a specific forward primer when an HDV-3 infection is suspected based on the clinical data, the geographical origin of the patient, or the presence of specific anti-HDV IgM with no detectable HDV RNA. Another problem concerns the standardization of the assay and its comparison to other existing techniques. To date, there is no available international standard or control to calibrate a quantitative assay for HDV. Moreover, apart from the assay described by Yamashiro et al. (41), which was specifically designed to quantify HDV-2 and HDV-4 isolates (such isolates are only very occasionally involved in HDV infections among European and African patients in our environment, who are usually infected with HDV-1, -5, -6, or -7), there is no reliable test available for HDV RNA quantification in serum. Taken together, our results indicate that the real-time RT-PCR assay reported here is sensitive enough to replace the qualitative assay for the detection of HDV RNA in serum (samples for which the real-time RT-PCR assay detected 100 to 1,000 copies/ml were considered positive even when their values were below the quantification level) and reproducible enough to follow viral load kinetics in infected patients, with the quantification reproducibility being monitored by the use of external controls.
The clinical impact of HDV RNA quantification in serum remains to be fully established, but a recent study already indicated the possibility of an association between the HDV RNA load and liver damage (41). In the present study, 11 chronically infected patients for whom sequential serum samples were available were evaluated in terms of the HDV viral load. Two patterns of virological response to IFN were distinguished according to the viral load reduction during treatment. Whether the qualification (based on the HDV viral load) of “responder” or “nonresponder” to treatment is appropriate would need further evaluation, since treatment failure could be due to a relapse after the end of therapy. Other factors such as the alaine amino-transferase level may need to be taken into account. However, our results suggest that HDV RNA quantification might help with monitoring the treatment duration. Indeed, in some cases, a viral load decrease might take a long time (as in the case of patient C), and some patients might need to be treated for >1 year, as previously described (24). In the case of patient D, a significant decrease in the HDV RNA load was observed during the course of PEG-IFN therapy, although the patient had remained resistant to a prior 1-year treatment with IFN. This might indicate more efficacy of PEG-IFN for the treatment of chronic delta hepatitis. A quantitative RT-PCR assay will be useful to evaluate this possibility. Patients E to I maintained high viral loads during PEG-IFN therapy, indicating that other factors might interfere with the treatment efficacy. For example, the initial viral loads were higher for the nonresponder group than for the responders (P = 0.05), and this could indicate that the baseline viral load might predict treatment efficacy and could be taken into account during the management of treatment in terms of dose and duration.
In summary, we have developed a sensitive assay to quantify HDV RNA in plasma or serum. Most importantly, the assay is consensual in that it performs equally across all known HDV types. Indeed, one should keep in mind that the HDV genome may diverge up to 37% at the nucleotide sequence level, and such variability is usually an impediment to the development of a universally sensitive assay. Our procedure overcomes this difficulty. The medical impact of HDV RNA quantification in serum remains to be established, and studies involving a larger number of patients are currently under way to address this point. However, this assay should help with the management of chronically infected patients, with specifying their evolutionary profiles before, during, and after treatment, and with the study of the natural history of HDV infection. It could also be used in large-scale prospective studies to define treatment guidelines and to evaluate the efficacy of new drugs.
Acknowledgments
We thank Corinne Castelnau, Michel Beaugrand, Jean-Claude Trinchet, Dominique Roulot, and Philippe Podevin for providing clinical information. We are grateful to Camille Sureau and Antoine Garbarg-Chenon for helpful discussions and critical readings of the manuscript. We thank Patrice Allard for technical advice and Catherine Aslo, Tiana Ralaimhihoatra, and Samira Mendil for excellent technical assistance.
This work was supported by the Assistance-Publique-Hôpitaux de Paris fund Progrès Médical et Innovation Thérapeutique and by the Ministère de la Santé (Laboratoire associé au Centre National de Références des hépatites B et C).
REFERENCES
- 1.Berk, L., R. A. de Man, C. Housset, P. Berthelot, and S. W. Schalm. 1991. Alpha lymphoblastoid interferon and acyclovir for chronic hepatitis delta. Prog. Clin. Biol. Res. 364:411-420. [PubMed] [Google Scholar]
- 2.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branch, A. D., J. A. Polaskova, and D. R. Schreiber. 1995. Tm studies of a tertiary structure from the human hepatitis delta agent which functions in vitro as a ribozyme control element. Nucleic Acids Res. 23:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cariani, E., A. Ravaggi, M. Puoti, G. Mantero, A. Albertini, and D. Primi. 1992. Evaluation of hepatitis delta virus RNA levels during interferon therapy by analysis of polymerase chain reaction products with a nonradioisotopic hybridization assay. Hepatology 15:685-689. [DOI] [PubMed] [Google Scholar]
- 5.Casey, J. L., T. L. Brown, E. J. Colan, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 90:9016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, Y. C., M. F. Chang, I. Gust, and M. M. Lai. 1990. Sequence conservation and divergence of hepatitis delta virus RNA. Virology 178:384-392. [DOI] [PubMed] [Google Scholar]
- 7.Deny, P., G. Fattovich, F. Le Gal, G. Giustina, C. Lecot, G. Morsica, H. Poinsot, A. Alberti, and C. Brechot. 1994. Polymerase-chain-reaction-based semi-quantification of hepatitis D viraemia in patients treated with high doses of alpha 2b interferon. Res. Virol. 145:287-295. [DOI] [PubMed] [Google Scholar]
- 8.Deny, P., C. Lecot, V. Jeantils, L. Ovaguimian, A. Krivitzky, and C. Brechot. 1993. Polymerase chain reaction-based detection of hepatitis D virus genome in patients infected with human immunodeficiency virus. J. Med. Virol. 39:214-218. [DOI] [PubMed] [Google Scholar]
- 9.Deny, P., A. L. Zignego, N. Rascalou, A. Ponzetto, P. Tiollais, and C. Brechot. 1991. Nucleotide sequence analysis of three different hepatitis delta viruses isolated from a woodchuck and humans. J. Gen. Virol. 72:735-739. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P. 2003. Delta hepatitis: an update. J. Hepatol. 39:S212-S219. [DOI] [PubMed] [Google Scholar]
- 11.Farci, P., A. Mandas, A. Coiana, M. E. Lai, V. Desmet, P. Van Eyken, Y. Gibo, L. Caruso, S. Scaccabarozzi, D. Criscuolo, J. C. Ryff, and A. Balestrieri. 1994. Treatment of chronic hepatitis D with interferon alfa-2a. N. Engl. J. Med. 330:88-94. [DOI] [PubMed] [Google Scholar]
- 12.Farci, P., T. Roskams, L. Chessa, G. Peddis, A. P. Mazzoleni, R. Scioscia, G. Serra, M. E. Lai, M. Loy, L. Caruso, V. Desmet, R. H. Purcell, and A. Balestrieri. 2004. Long-term benefit of interferon alpha therapy of chronic hepatitis D: regression of advanced hepatic fibrosis. Gastroenterology 126:1740-1749. [DOI] [PubMed] [Google Scholar]
- 13.Fattovich, G., G. Giustina, E. Christensen, M. Pantalena, I. Zagni, G. Realdi, and S. W. Schalm. 2000. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut 46:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garripoli, A., V. Di Marco, R. Cozzolongo, C. Costa, A. Smedile, A. Fabiano, F. Bonino, M. Rizzetto, G. Verme, A. Craxi, et al. 1994. Ribavirin treatment for chronic hepatitis D: a pilot study. Liver 14:154-157. [DOI] [PubMed] [Google Scholar]
- 15.Gault, E., Y. Michel, A. Dehee, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerin, J. L., J. L. Casey, and R. H. Purcell. 2002. Hepatitis delta virus, p. 169-182. In F. B. Hollinger, R. H. Purcell, and J. L. Gerin (ed.), Viral hepatitis. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 17.Goudeau, A., F. Dubois, and P. Leturcq. 1987. Delta hepatitis. Presse Med. 16:2117-2122. [PubMed] [Google Scholar]
- 18.Gourlain, K., D. Salmon, E. Gault, C. Leport, C. Katlama, S. Matheron, D. Costagliola, M. C. Mazeron, and A. M. Fillet. 2003. Quantitation of cytomegalovirus (CMV) DNA by real-time PCR for occurrence of CMV disease in HIV-infected patients receiving highly active antiretroviral therapy. J. Med. Virol. 69:401-407. [DOI] [PubMed] [Google Scholar]
- 19.Imazeki, F., M. Omata, and M. Ohto. 1991. Complete nucleotide sequence of hepatitis delta virus RNA in Japan. Nucleic Acids Res. 19:5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivaniushina, V., N. Radjef, M. Alexeeva, E. Gault, S. Semenov, M. Salhi, O. Kiselev, and P. Deny. 2001. Hepatitis delta virus genotypes I and II cocirculate in an endemic area of Yakutia, Russia. J. Gen. Virol. 82:2709-2718. [DOI] [PubMed] [Google Scholar]
- 21.Jardi, R., M. Buti, M. Cotrina, F. Rodriguez, H. Allende, R. Esteban, and J. Guardia. 1995. Determination of hepatitis delta virus RNA by polymerase chain reaction in acute and chronic delta infection. Hepatology 21:25-29. [PubMed] [Google Scholar]
- 22.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (delta) virus possesses a circular RNA. Nature 323:558-560. [DOI] [PubMed] [Google Scholar]
- 23.Lau, D. T., E. Doo, Y. Park, D. E. Kleiner, P. Schmid, M. C. Kuhns, and J. H. Hoofnagle. 1999. Lamivudine for chronic delta hepatitis. Hepatology 30:546-549. [DOI] [PubMed] [Google Scholar]
- 24.Lau, D. T., D. E. Kleiner, Y. Park, A. M. Di Bisceglie, and J. H. Hoofnagle. 1999. Resolution of chronic delta hepatitis after 12 years of interferon alfa therapy. Gastroenterology 117:1229-1233. [DOI] [PubMed] [Google Scholar]
- 25.Madejon, A., T. Cotonat, J. Bartolome, I. Castillo, and V. Carreno. 1994. Treatment of chronic hepatitis D virus infection with low and high doses of interferon-alpha 2a: utility of polymerase chain reaction in monitoring antiviral response. Hepatology 19:1331-1336. [PubMed] [Google Scholar]
- 26.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 27.Modahl, L. E., and M. M. Lai. 2000. Hepatitis delta virus: the molecular basis of laboratory diagnosis. Crit. Rev. Clin. Lab. Sci. 37:45-92. [DOI] [PubMed] [Google Scholar]
- 28.Perrotta, A. T., and M. D. Been. 1991. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature 350:434-436. [DOI] [PubMed] [Google Scholar]
- 29.Ponzetto, A., P. J. Cote, H. Popper, B. Hoyer, W. T. London, E. Ford, B. Ferruccio, R. Purcell, and J. Gerin. 1984. Transmission of the hepatitis B virus-associated delta agent to the Eastern woodchuck. Proc. Natl. Acad. Sci. USA 81:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radjef, N., E. Gordien, V. Ivaniushina, E. Gault, P. Anais, T. Drugan, J. C. Trinchet, D. Roulot, M. Tamby, M. C. Milinkovitch, and P. Deny. 2004. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a Deltavirus genus of at least seven major clades. J. Virol. 78:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasshofer, R., M. Buti, R. Esteban, R. Jardi, and M. Roggendorf. 1988. Demonstration of hepatitis D virus RNA in patients with chronic hepatitis. J. Infect. Dis. 157:191-195. [DOI] [PubMed] [Google Scholar]
- 32.Rizzetto, M., G. Verme, S. Recchia, F. Bonino, P. Farci, S. Arico, R. Calzia, A. Picciotto, M. Colombo, and H. Popper. 1983. Chronic hepatitis in carriers of hepatitis B surface antigen, with intrahepatic expression of the delta antigen. An active and progressive disease unresponsive to immunosuppressive treatment. Ann. Intern. Med. 98:437-441. [DOI] [PubMed] [Google Scholar]
- 33.Rosina, F., C. Pintus, C. Meschievitz, and M. Rizzetto. 1991. A randomized controlled trial of a 12-month course of recombinant human interferon-alpha in chronic delta (type D) hepatitis: a multicenter Italian study. Hepatology 13:1052-1056. [PubMed] [Google Scholar]
- 34.Saracco, G., F. Rosina, M. R. Brunetto, P. Amoroso, F. Caredda, P. Farci, P. Piantino, F. Bonino, and M. Rizzetto. 1987. Rapidly progressive HBsAg-positive hepatitis in Italy. The role of hepatitis delta virus infection. J. Hepatol. 5:274-281. [DOI] [PubMed] [Google Scholar]
- 35.Shakil, A. O., S. Hadziyannis, J. H. Hoofnagle, A. M. Di Bisceglie, J. L. Gerin, and J. L. Casey. 1997. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology 234:160-167. [DOI] [PubMed] [Google Scholar]
- 36.Smedile, A., M. Rizzetto, K. Denniston, F. Bonino, F. Wells, G. Verme, F. Consolo, B. Hoyer, R. H. Purcell, and J. L. Gerin. 1986. Type D hepatitis: the clinical significance of hepatitis D virus RNA in serum as detected by a hybridization-based assay. Hepatology 6:1297-1302. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 38.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 39.Wolters, L. M., A. B. van Nunen, P. Honkoop, A. C. Vossen, H. G. Niesters, P. E. Zondervan, and R. A. de Man. 2000. Lamivudine-high dose interferon combination therapy for chronic hepatitis B patients co-infected with the hepatitis D virus. J. Viral Hepat. 7:428-434. [DOI] [PubMed] [Google Scholar]
- 40.Wu, J. C., T. Y. Chiang, and I. J. Sheen. 1998. Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 79:1105-1113. [DOI] [PubMed] [Google Scholar]
- 41.Yamashiro, T., K. Nagayama, N. Enomoto, H. Watanabe, T. Miyagi, H. Nakasone, H. Sakugawa, and M. Watanabe. 2004. Quantification of the level of hepatitis delta virus RNA in serum, by real-time polymerase chain reaction, and its possible correlation with the clinical stage of liver disease. J. Infect. Dis. 189:1151-1157. [DOI] [PubMed] [Google Scholar]
- 42.Yurdaydin, C., H. Bozkaya, S. Gurel, H. L. Tillmann, N. Aslan, A. Okcu-Heper, E. Erden, K. Yalcin, N. Iliman, O. Uzunalimoglu, M. P. Manns, and A. M. Bozdayi. 2002. Famciclovir treatment of chronic delta hepatitis. J. Hepatol. 37:266-271. [DOI] [PubMed] [Google Scholar]