Abstract
Bacterial contamination remains one of the major risks associated with blood product transfusion. The kinetics of bacterial growth in red blood cell concentrates (RBCC) is different than otherwise due to storage at 4°C, conditions in which most bacteria do not survive. Psychrophilic bacteria such as Yersinia enterocolitica, however, can proliferate from a very low level of contamination to clinically significant levels at 4°C and are known to cause severe transfusion-related infections. A screening method allowing the early detection of very low levels of bacteria in RBCC would improve transfusion safety. The Scansystem method has been previously described for detection of bacteria in platelet concentrates. We present here a modification of the system for detection of low levels of bacteria in RBCC. The Scansystem RBC kit protocol requires three steps, i.e., the agglutination and selective removal of RBCs, a labeling stage during which bacteria are labeled with a DNA-specific fluorophore, and finally recovery of bacteria on the surface of a black membrane for analysis using the Scansystem. The entire procedure from sampling to result can be completed in 90 min. Both gram-negative and gram-positive bacteria in RBCC are detected with a higher sensitivity than with currently available culture-based methods. The Scansystem RBC kit is shown to be sensitive enough to identify low-level bacterial contamination in a single unit tested in a pool of up to 20 RBCC samples (detection limit of between 1 and 10 CFU/ml depending on the bacterial strain). The method therefore lends itself to incorporation into high-sample-throughput screening programs.
Due to the availability of highly sensitive virus screening kits, in donated blood products bacterial contamination, now the greatest risk for patients receiving a transfusion, is found 50 to 250 times more frequently than viral infection such as human immunodeficiency virus, hepatitis virus, or human T-cell lymphotropic virus infection. Transfusion-related bacterial infections are usually associated with a rapid onset of sepsis with high mortality rates (4, 16). Platelet concentrate preparations are known to carry a greater risk of bacterial infections, as they are required to be stored at 20 to 24°C (3, 6, 7, 10). With the significantly higher number of red blood cell concentrates (RBCC) transfused, however, the incidence of transfusion-associated bacterial contamination is similar (5, 11, 18, 28). In the French Bacthem case-control study, 41 bacterium-related transfusion reactions were reported during a 2-year period, including 25 following transfusion with RBCC, with 4 fatalities, and 16 following platelet transfusion, with 2 fatalities (23).
Currently, the examination of the RBCC bag for evidence of hemolysis just prior to transfusion is the only screening method carried out to check for bacterial contamination (12). However, visual inspection is subjective, and hemolysis also occurs in noncontaminated units nearing the end of their shelf life. Identification of low levels of bacteria and therefore hemolysis, although potentially clinically significant, would also be missed.
In this article we present a highly sensitive method for RBCC screening for bacterial contamination by solid-phase cytometry. The system, which was described previously for detection of bacteria in platelet concentrates (25), includes an erythrocyte removal step based on the induction of red cell agglutination. In the subsequent bacterial labeling of the erythrocyte-depleted sample, the membranes of gram-positive and gram-negative bacteria are permeabilized, allowing the entry and DNA-specific binding of a fluorophore marker. The sample is then filtered through a 0.4-μm black membrane, which retains the bacteria on its surface before being transferred to the Scansystem analyzer. The bacteria, particulate debris, and residual leukocytes are discriminated by the analyzer software based on their specific fluorescences, sizes, and shapes. The method described here is sensitive enough to enable the detection of very low levels of bacterial contamination in a single RBCC test sample when screened at a dilution ratio equivalent to that of a pool of 20 RBCC samples.
RBCC samples spiked with five different bacterial strains associated with transfusion reactions (16, 23), at spiking concentrations ranging from 1 to 100 CFU/ml, were analyzed over a period of 36 days with the Scansystem and detection sensitivity compared to the reference method of plate culture. The Scansystem RBC kit was shown to be able to detect very low levels of dividing as well as quiescent bacteria in packed RBC units stored at 4°C.
MATERIALS AND METHODS
Propagation of bacteria.
All bacterial strains used in this study originated from the bacterial collection of the Pasteur Institute, Paris; the strains were Staphylococcus epidermidis ATCC 12228, Staphylococcus aureus ATCC 6538, Bacillus cereus ATCC 7064, Klebsiella oxytoca ATCC 13182, and Yersinia enterocolitica CIP 101776. Following overnight culture (16 to 18 h) in Trypticase soy broth, the culture turbidity was adjusted to match a 0.5 McFarland standard and dilutions were performed in phosphate-buffered saline (PBS), pH 7.4. In all experiments, bacterial spiking concentrations were confirmed by culture on Mueller-Hinton plates or by esterase labeling (13).
Blood products.
RBCC were obtained from the French blood transfusion services, Etablissement Français du Sang (EFS). RBCC were prepared according to established French regulatory guidelines (Journal Officiel Français, 28 May 2003, no. 123, p. 9109). All experiments were carried out on RBCC between 1 and 42 days of collection under sterile conditions.
Scansystem RBC kit.
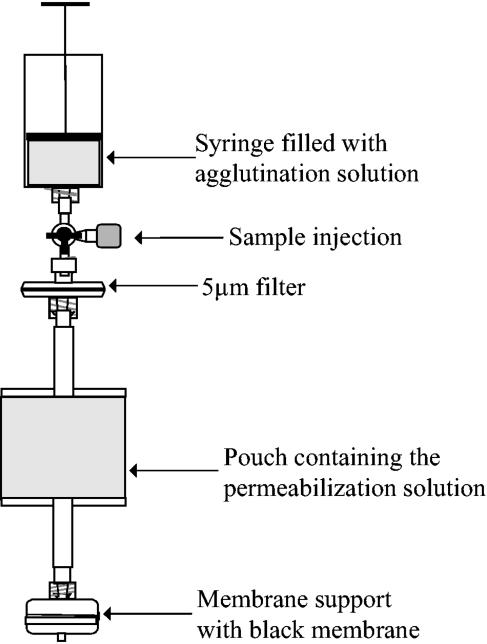
The Scansystem RBC kit is a closed device (Fig. 1) prefilled with 9 ml agglutination solution (polyethylene glycol 35000, 10 g/liter; lectin, 6.66 mg/liter; Picogreen, 0.17 ml/liter; in PBS) in a 20-ml syringe and 3 ml labeling solution (polyethylenimine, 20 mg/liter; EDTA, 4.34 g/liter; nisin, 18.7 mg/liter; N-octyl-β-d-glucopyranoside, 5.83 g/liter; chlorhexidine-diacetate, 116.7 mg/liter; prepared in PBS) in a 15-ml pouch, separated by a 5-μm filter (Pall Inc.). A 0.4-μm black membrane is held in a plastic membrane support (Millipore) with connectors for attaching to a vacuum pump for filtration. Any bacteria present in the sample are retained on the surface of the membrane, which is then transferred to the Scansystem (described below) for analysis.
FIG. 1.
Diagram of the design of the Scansystem red blood cell kit (Hemosystem, France).
Five milliliters of the RBCC was drawn into the kit syringe and mixed with the agglutination solution before being suspended on a dedicated support for a 45-min incubation period at room temperature. The agglutinated erythrocytes precipitate cleanly, and by depressing the syringe plunger, 8 ml of the supernatant was forced through the 5-μm filter (Pall Inc.), removing any residual red cells. The resulting RBC-depleted sample was incubated for 45 min at room temperature with 3 ml labeling reagent. The sample was then filtered through a 0.4-μm black membrane (Whatman), which retains the bacteria on its surface. The black membrane was transferred to the Scansystem solid-phase cytometer, and the number of labeled bacteria was determined (25).
Determination of the bacterial detection sensitivity of the RBC kit.
RBCC were spiked with the panel of gram-negative and gram-positive bacteria at 1, 5, 10, 50, and 100 CFU/ml. Eight milliliters of the spiked sample was incubated overnight (18 h) in 15-ml sterile polystyrene tubes at 37°C. Five milliliters of the spiked sample was then tested separately (single test), or 250 μl was mixed with 4.75 ml sterile RBCC to give a dilution equivalent to that of a pool of 20 samples. The noncontaminated (sterile) negative control RBCC was incubated at 37°C for the same period as the spiked test sample, and sterility was confirmed by plate culture.
Detection of bacterial growth in RBCC.
Four-day-old RBCC units were spiked with the different bacterial strains at 10 CFU/ml at day 0 and stored at 4°C. Each bacterial strain was inoculated into two RBCC units. At days 0, 4, 7, 11, 14, 18, 21, 25, 28, 32, and 38, a 5-ml sample was removed under sterile conditions and analyzed in single tests with the RBC kit. One hundred microliters of each sample at each time point was also filtered on a gridded membrane which was cultured on Mueller-Hinton petri dishes for bacterial enumeration. At days 14 and 28, an 8-ml sample was removed and incubated in a sterile 15-ml polystyrene tube for 18 h at 37°C. These samples were then diluted with sterile RBCC to give a dilution ratio equivalent to that of a pool of 20 samples and analyzed with the RBC kit. Samples from two sterile RBCC units were tested at each time point as negative controls.
Scansystem solid-phase cytometry.
The solid-phase scanning cytometry method has been described previously (19). In brief, (i) the black membrane is scanned by an argon laser (wavelength of excitation light, 488 nm) in the analyzer module; (ii) after membrane scanning, which takes 3 min, all fluorescent signals detected are analyzed and discriminated by the software based on their size, shape, and fluorescence intensity, and the result is displayed on the computer screen as a map showing the number of discriminated spots and their position on the membrane; and (iii) the membrane is transferred to an epifluorescence microscope for computer-assisted visual confirmation. In the experiments described here, 25 spots were chosen at random on the computer map and confirmed as being either microorganisms or particles.
RESULTS
Determination of RBC kit sensitivity.
The RBC kit sensitivity was determined with single samples of RBCC spiked with different strains of either gram-positive or gram-negative bacteria.
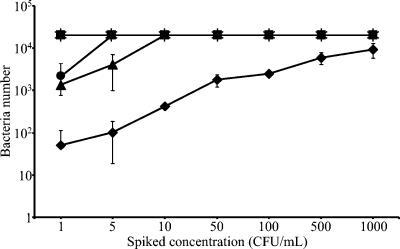
The Scansystem RBC kit allowed the detection of all bacterial strains at concentrations down to 1 CFU/ml (Fig. 2). At the lowest concentration, a mean of 50 bacteria per membrane was detected for the S. epidermidis strain, which is the slowest-growing strain, while 1,346 and 2,149 bacteria were detected for S. aureus and Y. enterocolitica, respectively. The number of detected bacteria increased with increased spiking concentrations for all the strains except K. oxytoca and B. cereus, for which the Scansystem gave a reading of more than 20,000 bacteria per membrane for all the concentrations tested, including 1 CFU/ml. Accurate bacterial enumeration for samples with excessively high levels of bacterial contamination was not possible; when bacterial concentrations exceeded 20,000 per sample, the analyzer detected a uniform coverage of the membrane and gave a “high-baseline warning” message. All sterile RBCC control samples tested (n = 10) gave negative results by both plate culture and the Scansystem RBC kit.
FIG. 2.
Sensitivity of the Scansystem red blood cell kit with single samples. S. epidermidis (♦), S. aureus (▴), B. cereus (▪), K. oxytoca (✻), and Y. enterocolitica (•) were spiked at concentrations ranging from 1 to 1,000 CFU/ml in RBCC. After 18 h of incubation at 37°C, samples were processed with the Scansystem RBC kit and bacteria were detected and counted with the Scansystem analyzer. The mean number of detected bacteria and the standard deviation are reported.
Screening sensitivity in testing a pool of 20 samples.
Single RBCC samples were spiked with the different bacterial strains before being mixed with sterile RBCC at a dilution ratio equivalent to 1 contaminated sample with 19 noncontaminated samples. The sensitivity of bacterial detection was determined with spiking concentrations ranging from 1 to 10 CFU/ml. Each experiment was performed between 4 and 18 times.
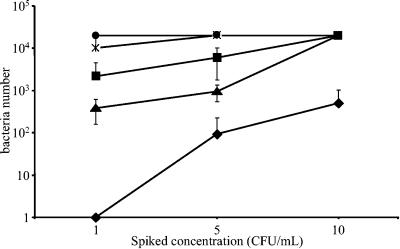
With the exception of S. epidermidis, bacteria were detected in all tests with spiking concentrations of 1 CFU/ml and above (Fig. 3). S. epidermidis was negative for the 1-CFU/ml test and positive in 16 out of 18 tests with 5 CFU/ml. Bacteria were detected in all pooled sample tests spiked with the five different bacterial strains at 10 CFU/ml. Similar results were obtained for Streptococcus agalactiae, Serratia marcescens, Acinetobacter baumannii, and Escherichia coli detected from 1 CFU/ml (data not shown). No bacteria were detected in the sterile RBCC negative controls (n = 10).
FIG. 3.
Sensitivity of the Scansystem red blood cell kit with diluted samples equal to a pool of 20 samples. S. epidermidis (♦), S. aureus (▴), B. cereus (▪), K. oxytoca (✻), and Y. enterocolitica (•) were spiked at concentrations ranging from 1 to 10 CFU/ml in RBCC. After 18 h of incubation at 37°C, the contaminated sample was mixed with a volume of noncontaminated RBCC sample to represent the pooling of a total of 20 samples. An aliquot was then processed with the Scansystem RBC kit. Bacteria detected were counted with the Scansystem analyzer. The mean number of detected bacteria and the standard deviation are reported.
Bacterial growth in RBCC stored at 4°C.
The Scansystem RBC kit was used to measure bacterial growth at 4°C after a spiking at 10 CFU/ml at set time points from 0 to 32 days. Of the two RBCC units spiked, there was no growth in one bag with K. oxytoca and no growth in either bag with S. epidermidis, S. aureus, or B. cereus. The absence of bacterial growth was confirmed by plate culture.
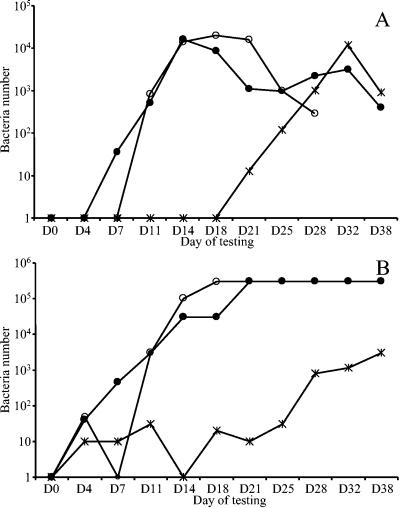
Bacteria were detected by the Scansystem RBC kit in the second bag spiked with K. oxytoca from day 21 to 38 (Fig. 4A). Plate cultures showed constant concentrations of K. oxytoca, ranging from 0 to 20 CFU/ml in samples taken from the same unit between days 0 and 21 and increasing from day 25 (Fig. 4B). Bacteria were consistently detectable by the kit in tests on samples with bacterial spiking concentrations down to 10 CFU/ml. Bacteria in the two bags contaminated with Y. enterocolitica were detected by the RBC kit from days 7 and 11, respectively. This corresponded to concentrations of as low as 450 CFU/ml as enumerated on the corresponding plates. The experiment could not be continued with the second bag contaminated with Y. enterocolitica after day 28, as the RBCs were completely lysed. Bacterial concentrations for Y. enterocolitica and K. oxytoca were shown to decrease in all bags from day 18 to 21 and from day 32, respectively. No bacterial growth was demonstrated in any of the negative control samples.
FIG. 4.
Detection of bacterial growth in single RBCC samples. S. epidermidis, S. aureus, B. cereus, K. oxytoca, and Y. enterocolitica were inoculated in RBCC bags at day 0, and their growth was analyzed during 38 days with the Scansystem method (A) and compared to that of plate cultures (B). Growth was observed only in one bag for K. oxytoca (✻) and in two bags for Y. enterocolitica (• and ○). The number of bacteria detected either with the Scansystem RBC kit or with plates is reported.
Detection of bacteria in RBCC samples at 14 and 28 days postspiking.
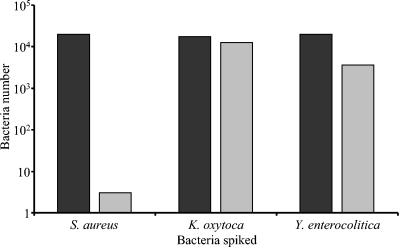
At days 14 and 28 postspiking, an 8-ml sample was removed and incubated at 37°C for 18 h, and this sample was then diluted with sterile RBCC to represent the equivalent dilution of a pool of 20 samples and tested by the Scansystem RBC kit. As indicated previously (Fig. 4), bacteria were observed only on plates from samples taken from bags spiked with Y. enterocolitica on day 14 and from those spiked with Y. enterocolitica and K. oxytoca on day 28. Bacteria were detected by the Scansystem RBC kits for both bacterial strains on both day 14 and 18 samples (Fig. 5). S. aureus-spiked samples were also found to be positive by the RBC kit but showed no growth on plates. S. epidermidis- and B. cereus-spiked samples showed no growth on culture plates and no presence of bacteria by the RBC kit. For the three bacterial strains detected by either method, the number of bacteria recorded was lower on day 28 than on day 14.
FIG. 5.
Detection of bacterial growth in sample dilutions representing pooled RBCC samples 14 and 28 days after spiking. The growth of spiked bacteria was analyzed after 14 days (blacks bars) or 28 days (grey bars) of refrigerated storage. After an 18-h incubation step at 37°C, the contaminated sample was mixed with the equivalent of 19 noncontaminated samples and the pool was analyzed with the Scansystem RBC kit. The number of bacteria detected is reported for the three strains that displayed growth; S. epidermidis and B. cereus were not observed.
DISCUSSION
There is currently no system available for the screening of RBCC for bacterial detection except visual inspection of the bags prior to transfusion. This method, however, is subjective and inadequately sensitive. Even with a visual reference, the method has been described as biased, inaccurate, and misleading (12). Automated culture systems which were developed for testing whole blood samples in hospital settings (9, 20) and are used for platelet concentrate screening (8, 17, 21) have not yet been developed for routine RBCC screening.
Although storage of RBCC at a low temperature (2 to 8°C) inhibits most bacterial growth, they are still at risk of contamination with psychrophilic bacteria such as Yersinia enterocolitica, which is now the most common cause of transfusion-associated bacterial contamination (5, 11, 18, 22, 28). The presence of these bacteria in blood products has often been associated with asymptomatic bacteremia and gastrointestinal illness of the donor (11, 28). Other bacterial strains observed in severe transfusion reactions include S. epidermidis (16) and S. aureus and B. cereus (23), which are thought to be the result of inadequate skin disinfection or transient bacteremia of the donor at the time of donation.
The 4°C storage of RBCC strongly influences the reliability of being able to detect bacteria in contaminated units because the lag phase before the bacteria start to proliferate can be very long; even psychrophilic bacteria remain in a nondividing state for up to 10 days (29). The possibility of screening the donated unit for bacterial contamination immediately after collection is not practical, however, as the level of contaminating bacteria in the unit is, in the majority of cases, too low to ensure representation and detection in the test sample volume removed. The time between unit collection and removal of test sample and the volume of the sample for screening are critical to the sensitivity of the bacterial screening method. The longer the unit is left before screening, the greater the chance that any dividing bacteria present will reach a level adequate for reliable detection. On the other hand, the fresher the unit offered for transfusion, the greater the viability of the erythrocytes and benefit to the patient.
One solution to this problem would be to screen every unit of RBCC immediately prior to transfusion. This would reduce the risk of the transfusion of RBCC which had been stored for 2 to 3 weeks and which had clinically significant levels of bacteria that had missed detection in early screening. The other possibility would be to increase the reliability of the early bacterial screening methods by incorporating a period of incubation of the test sample to allow any bacteria present to have grown sufficiently to increase the probability of detection. In line with these two possibilities, the Scansystem RBC kit was developed for direct single-sample testing or incorporation into multiple-sample screening programs with a sample incubation phase. After testing of different 37°C incubation periods (data not shown), 18 h was shown to be sufficient for even slow-growing bacteria inoculated at between 1 and 10 CFU/ml to reach concentrations detectable by our analyzer.
The use of a solid-phase cytometer has already been described as a highly sensitive bacterial screening system for various biologic products, including platelet concentrates (2, 19, 25; P. Morel, M. Dechaseaux, X. Bertrand, C. Naegelen, and D. Talon, Transfusion 43:SP13, 2003). When adapting the method for use with RBCC, a lysis step to remove red blood cells was tested; however, the free hemoglobin generated too high a background of fluorescence. The solution to the problem of RBC removal is based on the ability to induce their agglutination (1, 24, 26). In a 20-min incubation period in the agglutination solution, more than 99% of the RBCs compacted into a pellet. Residual RBCs could then be removed by filtration and the cell free supernatant analyzed by the solid-phase scanning cytometry system (25). Very low levels of bacteria can be concentrated from a large test sample volume by filtration, giving a significant advantage over plate culture methods. The large sample volume, 18-h incubation phase, and high sensitivity of the Scansystem enables the reliable screening of pools of up to 20 samples in a single test, making the system suitable for incorporation into high-sample-throughput screening programs. No false-positive results were found during these experiments. The sensitivity for single samples was as low as 1 CFU/ml, which is comparable to detection limits achieved with automated culture techniques on blood samples (14, 15). In pooled samples (incubated for 18 h), the sensitivity was also as low as 1 CFU/ml for eight out of the nine strains tested. The exception was S. epidermidis, which was positive in all tests down to 10 CFU/ml.
The sensitivity of detection of bacteria with the Scansystem was quite similar to that of the reference method of petri dish cultures. The Scansystem allowed early detection of bacteria for strains that were able to grow in refrigerated conditions, such as Y. enterocolitica and K. oxytoca. The growth kinetics were found to be comparable to those previously reported in the literature (29).
The test time points of 14 days and 28 days for the pooled-sample experiments were chosen to represent, for Y. enterocolitica, an active phase of growth and the beginning of the plateau, respectively (27). In addition to these two fast-growing strains, the system also allowed the detection of S. aureus (<10 CFU/ml), which was not detectable by plate culture. The lower bacterial numbers detected at day 28 than at day 14 were in agreement with the reported growth curves.
The Scansystem method for bacterial detection is qualitative and therefore difficult to compare directly to the quantitative plate culture method. However, in blood product screening, the presence of bacteria and not their absolute number determines the exclusion of the RBCC from transfusion. The identification of RBCC units with low bacterial contamination levels would prevent life-threatening transfusion-associated bacterial contamination in immunocompromised patients. The Scansystem is furthermore able to detect dead or quiescent as well as actively dividing bacteria, protecting the recipient from the effects of the bacterial toxins.
In conclusion, the Scansystem RBC kit process is rapid, simple, and sensitive. It allows detection of bacteria in single units within 24 h following RBCC preparation and can equally be used to screen units at any time during the 42 days of storage to just prior to transfusion, requiring only 90 min. The incorporation of an incubation period increases the system sensitivity to 1 CFU/ml to allow the testing of pooled samples in high-throughput programs, offering a significant advantage over standard culture methods. The routine use of this screening system could prevent RBCC transfusion-transmitted bacterial contamination and the associated clinical outcomes, including shock, severe sepsis, and patient mortality.
Acknowledgments
This work was supported by Hemosystem, Marseille.
We thank C. Lafontaine and N. Goncalves for critical reading of the manuscript and useful comments.
REFERENCES
- 1.Baskurt, O. K., M. Bor-Kucukatay, O. Yalcin, H. J. Meiselman, and J. K. Armstrong. 2000. Standard aggregating media to test the “aggregability” of rat red blood cells. Clin. Hemorheol. Microcirc. 22:161-166. [PubMed] [Google Scholar]
- 2.Bauters, T. G. M., D. Swinne, V. Stove, and H. J. Nelis. 2003. Detection of single cells of Cryptococcus neoformans in clinical samples by solid-phase cytometry. J. Clin. Microbiol. 41:1736-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blajchman, M. A., and M. Goldman. 2001. Bacterial contamination of platelet concentrates: incidence, significance, and prevention. Semin. Hematol. 38:20-26. [DOI] [PubMed] [Google Scholar]
- 4.Blajchman, M. A. 2002. Incidence and significance of the bacterial contamination of blood components. Dev. Biol. 108:59-67. [PubMed] [Google Scholar]
- 5.Boulton, F. E., S. T. Chapman, and T. H. Walsh. 1998. Fatal reaction to transfusion of red-cell concentrate contaminated with Serratia liquefaciens. Transfus. Med. 8:15-18. [DOI] [PubMed] [Google Scholar]
- 6.Brecher, M. E., P. V. Holland, A. A. Pineda, G. E. Tegtmeier, and R. Yomtovian. 2000. Growth of bacteria in inoculated platelets: implications for bacteria detection and the extension of platelet storage. Transfusion 40:1308-1312. [DOI] [PubMed] [Google Scholar]
- 7.Brecher, M. E., N. Means, C. S. Jere, D. Heath, S. Rothenberg, and L. C. Stutzman. 2001. Evaluation of an automated culture system for detecting bacterial contamination of platelets: an analysis with 15 contaminating organisms. Transfusion 41:477-482. [DOI] [PubMed] [Google Scholar]
- 8.Brecher, M. E., S. N. Hay, and S. J. Rothenberg. 2004. Validation of BacT/ALERT plastic culture bottles for use in testing of whole-blood-derived leukoreduced platelet-rich-plasma-derived platelets. Transfusion 44:1174-1178. [DOI] [PubMed] [Google Scholar]
- 9.Fontanals, D., I. Sanfeliu, I. Pons, D. Mariscal, and M. Torra. 1998. Evaluation of the BacT/Alert and VITAL blood culture systems for the diagnosis of bacteremia. Clin. Microbiol. Infect. 4:88-93. [DOI] [PubMed] [Google Scholar]
- 10.Heal, J. M., S. Singal, E. Sardisco, and T. Mayer. 1986. Bacterial proliferation in platelet concentrates. Transfusion 26:388-390. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, J., D. Jamaer, J. Vandeven, M. Wouters, C. Vermylen, and J. Vandepitte. 1989. Yersinia enterocolitica in donor blood: a case report and review. J. Clin. Microbiol. 27:1119-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janatpour, K. A., T. G. Paglieroni, V. L. Crocker, D. J. DuBois, and P. V. Holland. 2004. Visual assessment of hemolysis in red blood cell units and segments can be deceptive. Transfusion 44:984-989. [DOI] [PubMed] [Google Scholar]
- 13.Jones, D. L., M. A. Brailsford, and J.-L. Drocourt. 1999. Solid-phase, laser-scanning cytometry: a new two-hour method for the enumeration of microorganisms in pharmaceutical water. Pharmacop. Forum 25:7626-7645. [Google Scholar]
- 14.Kellogg, J. A., J. P. Manzella, and J. H. McConville. 1984. Clinical laboratory comparison of the 10-ml Isolator blood culture system with BACTEC radiometric blood culture media. J. Clin. Microbiol. 20:618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellogg, J. A., J. P. Manzella, and D. A. Bankert. 2000. Frequency of low-level bacteremia in children from birth to fifteen years of age. J. Clin. Microbiol. 38:2181-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehnert, M. J., V. R. Roth, N. R. Haley, K. R. Gregory, K. V. Elder, G. B. Schreiber, M. J. Arduino, S. C. Holt, L. A. Carson, S. N. Banerjee, and W. R. Jarvis. 2002. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 41:1493-1499. [DOI] [PubMed] [Google Scholar]
- 17.Macauley, A., A. Chandrasekar, G. Geddis, K. G. Morris, and W. M. McClelland. 2003. Operational feasibility of routine bacterial monitoring of platelets. Transfus. Med. 13:189-195. [DOI] [PubMed] [Google Scholar]
- 18.McDonald, C. P., J. A. Barbara, P. E. Hewitt, S. Hartley, P. Telfer, R. Gale, E. James, and H. G. Prentice. 1996. Yersinia enterocolitica transmission from a red cell unit 34 days old. Transfus. Med. 6:61-63. [DOI] [PubMed] [Google Scholar]
- 19.Mignon-Godefroy, K., J.-G. Guillet, and C. Butor. 1997. Solid phase cytometry for detection of rare events. Cytometry 27:336-344. [PubMed] [Google Scholar]
- 20.Mirrett, S., L. B. Reller, C. A. Petti, C. W. Woods, B. Vazirani, R. Sivadas, and M. P. Weinstein. 2003. Controlled clinical comparison of BacT/ALERT standard aerobic medium with BACTEC standard aerobic medium for culturing blood. J. Clin. Microbiol. 41:2391-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munksgaard, L., L. Albjerg, S. T. Lillevang, B. Gahrn-Hansen, and J. Georgsen. 2004. Detection of bacterial contamination of platelet components: six years' experience with the BacT/ALERT system. Transfusion 44:1166-1173. [DOI] [PubMed] [Google Scholar]
- 22.Orozova, P., N. Markova, and T. Radoucheva. 2001. Properties of Yersinia enterocolitica and Yersinia pseudotuberculosis in red blood cell concentrate of different ABO groups during 30-day storage at 4 degrees C. Clin. Microbiol. Infect. 7:358-361. [DOI] [PubMed] [Google Scholar]
- 23.Perez, P., L. R. Salmi, G. Follea, J. L. Schmit, B. de Barbeyrac, P. Sudre, R. Salamon, the BACTHEM Group, and the French Haemovigilance Network. 2001. Determinants of transfusion-associated bacterial contamination: results of the French BACTHEM case-control study. Transfusion 41:862-872. [DOI] [PubMed] [Google Scholar]
- 24.Rao, U. J., P. R. Ramasarma, D. R. Rao, and K. V. Prasad. 1994. Detection of lectin activity on Western blots using erythrocytes. Electrophoresis 15:907-910. [DOI] [PubMed] [Google Scholar]
- 25.Ribault, S., K. Harper, L. Grave, C. Lafontaine, P. Nannini, A. Raimondo, and I. B. Faure. 2004. Rapid screening method for detection of bacteria in platelet concentrates. J. Clin. Microbiol. 42:1903-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sewchand, L. S., and D. Bruckschwaiger. 1980. Observed differences in dextran and polyvinylpyrrolidone as rouleaux-inducing agents. Can. J. Physiol. Pharmacol. 58:271-274. [DOI] [PubMed] [Google Scholar]
- 27.Siblini, L., B. Lafeuillade, A. Ros, J. C. Le Petit, and B. Pozzetto. 2002. Reduction of Yersinia enterocolitica load in deliberately inoculated blood: the effects of blood prestorage temperature and WBC filtration. Transfusion 42:422-427. [DOI] [PubMed] [Google Scholar]
- 28.Strobel, E., J. Heesemann, G. Mayer, J. Peters, S. Muller-Weihrich, and P. Emmerling. 2000. Bacteriological and serological findings in a further case of transfusion-mediated Yersinia enterocolitica sepsis. J. Clin. Microbiol. 38:2788-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zavizion, B., D. Serebryanik, I. Serebryanik, J. Chapman, and A. Purmal. 2003. Prevention of Yersinia enterocolitica, Pseudomonas fluorescens, and Pseudomonas putida outgrowth in deliberately inoculated blood by a novel pathogen-reduction process. Transfusion 43:135-142. [DOI] [PubMed] [Google Scholar]