Abstract
A gram-negative bacillus, SMC-8986T, which was isolated from the purulent exudate of an epidermal cyst but could not be identified by a conventional microbiologic method, was characterized by a variety of phenotypic and genotypic analyses. Sequences of the 16S rRNA gene revealed that this bacterium belongs to the genus Bordetella but diverged distinctly from previously described Bordetella species. Analyses of cellular fatty acid composition and performance of biochemical tests confirmed that this bacterium is distinct from other Bordetella species. Furthermore, the results of comparative sequence analyses of two protein-coding genes (risA and ompA) also showed that this strain represents a new species within the genus Bordetella. Based on the evaluated phenotypic and genotypic characteristics, it is proposed that SMC-8986T should be classified as a new species, namely Bordetella ansorpii sp. nov.
The genus Bordetella now consists of eight species, including three classical species, Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica (4, 13). B. pertussis and B. parapertussis are strict human pathogens causing the respiratory tract infection called whooping cough (6). Even though B. bronchiseptica is a commensal of the respiratory tract in many animals, it also infrequently causes respiratory tract infections in humans (3). Bordetella hinzii, mainly a colonizer of the respiratory tract of poultry, has been found in immunocompromised humans (11) and was recently reported as a causative agent of fatal septicemia (7). Bordetella holmesii and Bordetella trematum exclusively infect humans. B. holmesii has been found repeatedly in blood of young adults and often in sputum (10, 14). B. trematum causes ear and wound infections (12). Bordetella avium, a pathogen of birds, causes coryza or rhinotracheitis in poultry, but it has never been found in humans. Lastly, Bordetella petrii, which was identified very recently, is a unique member of the genus Bordetella isolated from the environment and capable of anaerobic growth (13).
In this paper, we report a novel Bordetella species isolated from the purulent exudate of an epidermal cyst. This bacterium could not be identified by a conventional method. Comparative 16S rRNA sequence analysis showed that it belongs to the genus Bordetella, but it does not correspond to any previously characterized species. Thus, we suggest a new species name, B. ansorpii, for this microorganism based on phenotypic and genotypic characteristics.
Case report.
A 19-year-old female was admitted for anticancer chemotherapy. She had received chemotherapy due to rhabdomyosarcoma of the nasal cavity and right orbit since 1 year prior to admission. On admission, a 2-cm, soft, tender, and erythematous mass was detected on her right posterior neck. Her body temperature was normal. The leukocyte count was 5,310/mm3 (normal range, 3,200 to 9,000/mm3) with segmented neutrophils at 72% (normal, 40 to 74%) and lymphocytes at 18% (normal, 20 to 50%); the hemoglobin level was 10.2 g/dl (normal, 11.2 to 14.8 g/dl); the erythrocyte sedimentation rate was 21 mm/h (normal, 0 to 27 mm/h); and the C-reactive protein level was 0.07 mg/dl (0 to 0.3 mg/dl). Amoxicillin-clavulanate was given under the impression of the infected mass. As no improvement was observed despite antibiotic therapy for 3 days, the mass was biopsied and the drainage was examined by microscope and cultured. The pathology revealed an infected epidermal cyst. The purulent exudate of an epidermal cyst was cultured on blood and MacConkey agar plates and incubated at 37°C. Gram staining of the colonies on agar plates demonstrated gram-negative bacilli. However, the isolated bacterium could not be identified by conventional automated methods such as VITEK (bioMérieux, Hazelwood, Mo.) and MicroScan (Dade-Microscan, Sacramento, Calif.). The infected mass had improved with amoxicillin-clavulanate after incision and drainage. She received another cycle of chemotherapy uneventfully.
The strain SMC-8986T, a gram-negative bacillus, grew on both blood and MacConkey agar at 37°C. The VITEK GNI+ card (gram-negative identification card; bioMérieux, Hazelwood, Mo.) and the API 20NE (bioMérieux, Hazelwood, Mo.) were used for identification according to the recommendations of the manufacturer. SMC-8986T was positive only for citrate utilization in repeated tests with the VITEK GNI+ card. It was identified as B. avium with an accuracy of 82% by repeated tests with API 20NE. Briefly, it was positive for gelatin hydrolysis and adipate, malate, citrate, and phenylacetate assimilation, while negative for oxidase, reduction of nitrates to nitrites, indole production, acidification, arginine dihydrolase, urease, β-glucosidase hydrolysis, β-galactosidase, and assimilation of glucose, arabinose, mannose, mannitol, N-acetyl-glucosamine, maltose, gluconate, and caprate (Table 1). The organism was motile on LB swarming agar (0.8% NaCl, 0.4% agar [wt/vol]), which distinguishes it from B. holmesii (Table 1).
TABLE 1.
Comparison of phenotypic characteristics of Bordetella species, including strain SMC-8986T
| Characteristic | Result for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SMC-8986T | B. pertussis | B. parapertussis | B. bronchiseptica | B. avium | B. hinzii | B. holmesii | B. trematum | B. petrii | |
| Growth on: | |||||||||
| Blood agar | + | − | + | + | + | + | + | + | + |
| MacConkey agar | + | − | ± | + | + | + | + | + | + |
| Oxidase | − | + | − | + | + | + | − | − | + |
| Nitrate reduction | − | − | − | + | − | − | − | ± | − |
| Urease production | − | − | + | + | − | ± | − | − | − |
| Motility | + | − | − | + | + | + | − | + | − |
Analysis of cellular fatty acid (CFA) composition was performed for SMC-8986T, B. avium ATCC 35086T, and B. brochiseptica ATCC 4601T, using a Hewlett Packard 6890A gas chromatograph and the MIDI aerobe method (Chem Station ver. 4.02) at MicroID (Seoul, Korea). The CFA profiles determined in this study were compared with those of several reports from the literature (7, 12, 14). CFA analysis results are shown in Table 2. The cellular fatty acid profile of strain SMC-8968T was compared with those of other Bordetella species and was found to have a similar predominance of C16:0 (32.4%). However, the overall CFA composition of SMC-8986T, which included C16:1ω7c (17.6%), C18:1ω7c (12.9%), and C17:0cyclo (9.6%), did not correspond to any previously described Bordetella species (Table 2). The lower composition rate of C17:0cyclo is the significant difference between strain SMC-8986T and other Bordetella species. The G+C content of strain SMC-8986T, which was determined by thermal denaturation (5), was 63.8 mol%. The G+C contents of other Bordetella species ranged from 60 to 69 mol% (11-14).
TABLE 2.
Comparison of fatty acid composition of Bordetella species
| Fatty acid | Proportion (%) of fatty acid in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SCM-8986T | B. aviuma | B. bronchisepticaa | B. hinziib | B. parapertussisb | B. pertussisb | B. holmesiib | B. trematumb | |
| C12:0aldehyde | 1.3 | 0.47 | 0.48 | 0.4 | ||||
| C12:02OH | 1.1 | 2.16 | 2.08 | 2.34 | 2.9 | 2.7 | ||
| C14:0 | 2.4 | 0.99 | 4.83 | 0.61 | 6 | 5 | 1.1 | |
| C14:02OH | 4.9 | 2.86 | 3.38 | 3.4 | 4.6 | |||
| C16:1iso | 6.7 | 7.33 | 6.49 | NAc | NA | NA | 7.9 | 10.0 |
| C16:1ω7c | 17.6 | 4.31 | 11.0 | 3.80 | 6 | 40 | NA | NA |
| C16:0 | 32.4 | 39.13 | 42.23 | 34.53 | 40 | 32 | 41.5 | 37.5 |
| C17:0cyclo | 9.6 | 29.52 | 26.27 | 33.24 | 35 | 34.2 | 31.6 | |
| C16:03OH | 1.1 | 0.35 | 0.23 | 0.48 | NA | NA | NA | NA |
| C17:0 | 0.92 | 0.93 | 0.84 | 3 | NA | |||
| C18:1ω7c | 12.9 | 0.67 | 1.42 | 1.65 | NA | |||
| C18:0 | 1.9 | 4.39 | 2.80 | 7.60 | 5 | 8 | 4.9 | 4.2 |
| C19:0cycloω8c | 4.6 | 0.19 | 0.19 | 1.26 | NA | NA | NA | NA |
For genotypic characterization, genomic DNA of SMC8986T, B. avium ATCC 35086T, and B. bronchiseptica ATCC 4617T was extracted from bacterial colonies by a simple boiling-lysis method (1). Briefly, colonies were suspended in lysis buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% Triton X-100), and were incubated at 80°C for 10 min. The mixture was then centrifuged for a moment, and the aqueous phase was used as a template for PCR. The 16S rRNA was amplified with universal primers 16S-F3 (5′-CAGGCCTAACACATGCAAGT-3′) and 16S-R3 (5′-GGGCGGWGTGTACAAGGC-3′) (15). Template DNA and 50 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Daejon, Korea) (8). The reaction mixture was then subjected to 35 cycles for amplification. Each cycle consisted of 30 s at 95°C for denaturation, 30 s at 58°C, and 1 min at 72°C for extension, followed by final extension at 72°C for 5 min. Amplified PCR product was purified for sequencing using a PCR purification kit (CoreOne, Seoul, Korea). The purified PCR product was sequenced directly using the same primers of PCR amplification and another primer, 16S-F5 (5′-TATTGGGCGTAAAGCGAGCGC-3′), which was designed by us. DNA sequences were determined with an ABI prism Rhodomine terminator cycle sequencing kit (PE Biosystems, Foster City, CA) and an ABI 3710 automated sequence (PE Biosystems, Foster City, CA). The determined 16S rRNA nucleotide sequences of SMC-8986T, B. avium ATCC 35086T, and B. brochiseptica ATCC 4617T (1,424 bp, 1,422 bp, and 1,422 bp, respectively) were used for phylogenetic comparison.
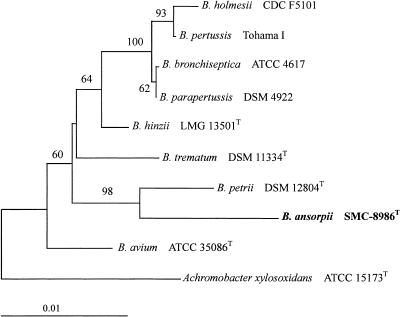
Table 3 presents sequence dissimilarities among 16S rRNA sequences of Bordetella species and strain SMC-8986T, which were analyzed with the MegAlign program in DNASTAR (DNASTAR, Madison, WI). The 16S rRNA gene sequence of SMC-8986T was compared to published or determined sequences of other Bordetella species, and dissimilarities ranged from 1.7% to 2.6%. Considering the range of dissimilarities (0.1% to 1.9%) among the characterized Bordetella species, except SMC-8986T, and the common difference limit in species definition (1.0%) (2), SMC-8986T is regarded as a new Bordetella species. A phylogenetic tree reconstructed by the method of neighbor joining (9) also suggests a phylogenetic relationship indicating that SMC-8986T is a member of the genus Bordetella but is distinct from other Bordetella species (Fig. 1). With respect to the phylogenetic relationships, strain SMC-8986T was closely related to B. petrii, which was supported robustly by the bootstrap value (98%).
TABLE 3.
Similarities and dissimilarities among 16S rRNA sequences of Bordetella speciesa
| Species | % Similarity or dissimilarity to:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| B. holmesii | B. pertussis | B. bronchiseptica | B. parapertussis | B. hinzii | B. trematum | B. avium | B. petrii | SMC-8986T | |
| B. holmesii | 99.8 | 99.5 | 99.6 | 99.0 | 98.4 | 98.4 | 98.1 | 97.4 | |
| B. pertussis | 0.2 | 99.8 | 99.8 | 99.2 | 98.6 | 98.4 | 98.4 | 97.5 | |
| B. bronchiseptica | 0.5 | 0.2 | 99.9 | 99.3 | 98.6 | 98.6 | 98.4 | 97.6 | |
| B. parapertussis | 0.4 | 0.2 | 0.1 | 99.5 | 98.7 | 98.7 | 98.4 | 97.6 | |
| B. hinzii | 1.0 | 0.8 | 0.7 | 0.5 | 99.1 | 99.3 | 98.2 | 97.7 | |
| B. trematum | 1.6 | 1.4 | 1.4 | 1.3 | 0.9 | 99.0 | 98.1 | 97.7 | |
| B. avium | 1.6 | 1.6 | 1.4 | 1.3 | 0.7 | 1.0 | 98.0 | 97.8 | |
| B. petrii | 1.9 | 1.6 | 1.6 | 1.6 | 1.8 | 1.9 | 2.0 | 98.3 | |
| SMC-8986T | 2.6 | 2.5 | 2.4 | 2.4 | 2.3 | 2.3 | 2.2 | 1.7 | |
The strain number of each species is represented in Fig. 1.
FIG. 1.
Phylogenetic relationships of B. ansorpii sp. nov. and other Bordetella species inferred from 16S rRNA sequences, which were aligned using the multiple alignment program Clustal X. This tree was reconstructed by the neighbor-joining method. Achromobacter xylosoxidans ATCC 15173T was used as an outgroup. Numbers at branching nodes are percentages of 1,000 bootstrap replications. Only values greater than 50% are indicated. The scale bar represents one substitution per 100 nucleotides.
In addition to 16S rRNA, the homologues of risA and ompA genes of B. avium were partially amplified and sequenced using the same primers as von Wintzingerode et al. (13). Determined risA and ompA sequences of SMC-8986T, B. avium ATCC 35086T, and B. brochiseptica ATCC 4617T were aligned with those of other Bordetella species retrieved from GenBank, respectively. risA and ompA sequences of SMC-8986T showed 11.2 to 11.6% and 17.2 to 26.3% divergences from those of B. pertussis, B. parapertussis, B. bronchiseptica, B. avium, and B. petrii, respectively. These results also confirm that strain SMC8986T is a distinct species from other Bordetella species, although a few Bordetella species could not be included in this analysis.
Thus, in light of the complementary nature of all the results described above, in terms of biochemical tests, cellular fatty acid composition, and molecular genetic analysis, a new species of the genus Bordetella, B. ansorpii, is proposed for strain SMC-8986T isolated from the purulent exudate of an epidermal cyst.
Description of Bordetella ansorpii sp. nov.
The species name, ansorpii, stands for ANSORP, Asian Network for Surveillance of Resistant Pathogens.
It is a gram-negative bacillus. It grows on both blood agar and MacConkey agar. It is negative for indole production, oxidase, urease, arginine dihydrolase, esculinase, gelatinase, β-galactosidase, nitrate reduction, and assimilation of glucose, mannose, mannitol, N-acetyl-glucosamine, malonate, gluconate, and caprate but positive for citrate, adipate, malate, and phenyacetate utilization, gelatinase activity, and motility. It has the cellular fatty acids 16:0, 16:1ω7c, 18:1ω7c, and 17:0cyclo as the major fatty acid components. It was isolated from the purulent exudate of an epidermal cyst, but its pathogenic significance remains unknown so far. It has a G+C content of 63.8 mol%. The type strain of B. ansorpii is strain SMC-8986T, which has been deposited at ABB (Asian Bacterial Bank, Seoul, Korea).
Nucleotide sequence accession number.
The sequences of 16S rRNA, risA, and ompA of strain SMC-8986T have been deposited in the GenBank database under accession numbers AY594190 to AY594192.
Acknowledgments
We thank Mi Young Lee and Na Young Kim for technical assistance.
This study was partly supported by the SBRI (Samsung Biomedical Research Institute) (grant C-A4-316-1).
REFERENCES
- 1.Baek, J. Y., K. S. Ko, W. S. Oh, S.-I. Jung, Y. S. Kim, H.-H. Chang, H. Lee, S. W. Kim, K. R. Peck, N. Y. Lee, and J.-H. Song. 2004. Unique variations of pbp2b sequences in penicillin-nonsusceptible Streptococcus pneumoniae isolates from Korea. J. Clin. Microbiol. 42:1746-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microb. Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 5.Goodfellow, M. 1985. Chemical methods in bacterial systematics, p. 67-93. Academic Press, London, United Kingdom.
- 6.Hoppe, J. E. 1999. Bordetella, p. 614-624. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 7.Kattar, M. M., J. F. Chavez, A. P. Limaye, S. L. Rassoulian-Barrett, S. L. Yarfitz, L. C. Carlson, Y. Houze, S. Swanzy, B. L. Wood, and B. T. Cookson. 2000. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J. Clin. Microbiol. 38:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko, K. S., H. K. Lee, M.-Y. Park, K.-H. Lee, Y.-J. Yun, S.-Y. Woo, H. Miyamoto, and Y.-H. Kook. 2002. Application of RNA polymerase β-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J. Clin. Microbiol. 40:2653-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 10.Tang, Y. W., M. K. Hopkins, C. P. Kolbert, P. A. Hartley, P. J. Severance, and D. H. Persing. 1998. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin. Infect. Dis. 26:389-392. [DOI] [PubMed] [Google Scholar]
- 11.Vandamme, P., J. Hommez, M. Vancanneyt, M. Monsieurs, B. Hoste, B. Cookson, C. H. Wirsing von König, K. Kerster, and P. J. Blackall. 1995. Bordetella hinzii sp. nov., isolated from poultry and humans. Int. J. Syst. Bacteriol. 45:37-45. [DOI] [PubMed] [Google Scholar]
- 12.Vandamme, P., M. Heyndrickx, M. Vancanneyt, B. Hoste, P. de Vos, E. Falsen, K. Kerster, and K.-H. Hinz. 1996. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Rüger and Tan 1983. Int. J. Syst. Bacteriol. 46:849-858. [DOI] [PubMed] [Google Scholar]
- 13.von Wintzingerode, F., A. Schattke, R. A. Siddiqui, U. Rösick, U. B. Göbel, and R. Gross. 2001. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51:1257-1265. [DOI] [PubMed] [Google Scholar]
- 14.Weyant, R. S., D. G. Hollis, R. E. Weaver, M. F. M. Amin, A. G. Steigerwalt, S. P. O'Connor, A. M. Whitney, M. I. Daneshvar, C. W. Moss, and D. J. Brenner. 1995. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]