Abstract
Uropathogenic Escherichia coli is the most common cause of urinary tract infection (UTI). Cystitis in women is by far the most common UTI; pyelonephritis in both sexes and prostatitis in men are more severe but are less frequent complaints. The ability of E. coli to cause UTI is associated with specific virulence determinants, some of which are encoded on pathogenicity islands (PAI). One such PAI (PAI IICFT073), of the prototypical uropathogenic E. coli strain CFT073, contains 116 open reading frames, including iron-regulated genes, carbohydrate biosynthetic genes, the serine protease autotransporter picU, a two-partner secretion system, a type I secretion system, mobility genes, and a large number of hypothetical genes. To determine the association of PAI IICFT073 with UTI, PCR was used to examine the prevalence of the five virulence-associated loci among the ECOR collection and a collection of E. coli isolated from patients with cystitis, pyelonephritis, prostatitis, or septicemia. All PAI IICFT073 loci were found to be more prevalent among the B2 phylogenetic group than any other group within the ECOR collection and among invasive prostatitis strains than were cystitis or pyelonephritis strains. These data support the theory that clinical isolates causing prostatitis are more virulent than those producing cystitis or pyelonephritis in women.
Urinary tract infections (UTIs) continue to be among the most common extraintestinal diseases. In the United States, UTIs account for 8.7 million annual physician and 2.3 million hospital visits (49). Cystitis in women is by far the most common UTI; pyelonephritis in both sexes and prostatitis in men are more severe but less frequent complaints. Several bacterial species may cause UTIs, but Escherichia coli is by far the most common cause (14), accounting for 50% of all nosocomial UTIs and 90% of infections among ambulatory patients.
It is believed that uropathogenic E. coli (UPEC) are adapted to colonizing the urinary tract. The liberation of specific virulence factors may aid attachment to host mucosal tissue, allow evasion of immune defenses, and promote invasion of the normally sterile urinary tract and tissues (14, 15). Among these factors, adhesins, capsule, aerobactin, toxins, and proteases have been described (1, 15, 36, 46). Analysis of the prevalence of virulence factors among commensal E. coli and those causing different UTIs has indicated a greater virulence potential of the disease-causing strains (18, 33, 46). Moreover, virulence determinants appear to be more prevalent among strains causing invasive disease (46). Interestingly, half of all UPEC isolates possess none, or only one, of the virulence factors characterized thus far. As such, it is reasonable to assume other, as-yet-uncharacterized, bacterial factors may be important in the pathogenesis of UTI (33).
The genome sequence of the UPEC strain CFT073 has been determined, and this has allowed the identification of potential virulence genes (54). Several of the genes associated with the acquisition and development of UTIs are encoded on pathogenicity islands (PAIs), e.g., hemolysin and P fimbriae (3). By definition, PAIs contain genes that are associated with virulence and are absent from avirulent or less virulent strains of the same species. Multiple PAIs varying in size and gene complement have been described in UPEC isolates (2, 13, 34). Recently, we described the existence within uropathogenic E. coli strains of the serine protease autotransporter PicU (40). In silico analysis of the genomic context of the gene encoding PicU revealed that, like other virulence factors, it was located on a PAI. The 100-kb PAI contains 116 open reading frames (ORFs) which, in addition to PicU, encode a type I protein secretion system, a member of the two-partner protein secretion system (TPSS), iron-sequestering proteins, proteins involved in carbohydrate metabolism, insertion elements, and ORFs of unknown function. The PAI encoding PicU demonstrates homology with PAI IICFT073, a 71.6-kb PAI previously characterized in E. coli CFT073 (43). In silico investigations of the previously described PAI IICFT073 revealed that it was incorrectly assembled from three distinct regions of the E. coli CFT073 complete genome sequence. Here we describe the correct genetic organization of PAI IICFT073 and investigate the prevalence of these genes among populations of E. coli including pathogenic E. coli causing cystitis and pyelonephritis in women, prostatitis in men, and septicemia in both sexes.
MATERIALS AND METHODS
Bioinformatic analysis of the E. coli CFT073 genome. To characterize the genomic context of the gene encoding the PicU autotransporter, the E. coli CFT073 genome sequence was compared to other E. coli and Shigella strains by using coliBASE (http://colibase.bham.ac.uk), an online database for E. coli comparative genomics (6). This contains all of the complete E. coli, Shigella, and Salmonella genome sequences, together with preliminary data from a number of sequencing projects currently in progress at The Wellcome Trust Sanger Institute. The database contains precalculated genome alignments performed by using MUMmer and PROmer (6, 10) and provides user-friendly tools to display pairwise comparisons between equivalent regions of different strains to highlight chromosomal insertions, deletions, and rearrangements.
Bacterial strains and growth conditions.
Ninety-four E. coli strains causing cystitis or pyelonephritis in women or prostatitis in men were obtained from J. Ruiz (Barcelona, Spain). These strains were previously characterized for prevalence of nine uropathogenic virulence factors (46). The ECOR collection, a phylogenetically characterized collection of E. coli strains representing the four major phylogenetic groups (A, B1, B2, and D) (38), was obtained from M. J. Pallen (University of Birmingham, Birmingham, United Kingdom). A collection of 43 consecutive E. coli blood culture isolates were obtained from the clinical laboratories at the Queen Elizabeth Hospital in Birmingham in 2003. All strains were grown overnight at 37°C on LB agar plates to check purity. Overnight LB broth cultures were then used for DNA preparations by using the DNeasy tissue kit (Qiagen, Crawley, United Kingdom), according to the manufacturer's instructions. The prototypical UPEC strain CFT073 was kindly provided by H. L. T. Mobley.
Phylogenetic grouping of the blood culture isolates was performed by using the method described by Clermont et al. (7). Briefly, DNA from all strains was amplified with primers for chuA, positive samples were subsequently amplified with primers corresponding to yjaA, whereas negative samples were amplified with primers corresponding to tspE4C2 as follows: samples positive for yjaA represent the B2 cluster, samples negative for yjaA represent the D group, samples positive for tspE4C2 represent the B1 cluster, and samples negative for tspE4C2 represent the A group. Primers are listed in Table 1.
TABLE 1.
PCR primers used in this study
| Target (orientation)a | Primer sequence (5′-3′) | Genomic positionb |
|---|---|---|
| PAI (F) | AGCGGTAGTTCAGTCGGTTAGAATA | 248556-248580 |
| PAI (R) | TTTTATGATAAGCGTCATCTGTTCC | 349062-349086 |
| fbp (F) | TTGCTCGTCGGGCTGAAAAAGTTGT | 278466-278490 |
| fbp (R) | TTGCACCATCCACTATCACCATCGA | 279008-278984 |
| efu (F) | TATCGTGTTTATTCCCGCTATGG | 302752-302774 |
| efu (R) | CCCCTCGGAGAAAACCAGATATT | 303203-303181 |
| etp (F) | CCACCATCACGGCAGGACAACTCAC | 315712-315736 |
| etp (R) | CCCGGTTTTCCAGTCGTTCAGCCTC | 317178-317154 |
| picU (F) | TCAGGCCGGTAAGAACAGCAAAAT | 327844-327821 |
| picU (R) | ACGGTAAGAGTGTGGATGGCGGAGTC | 327473-327498 |
| tos (F) | ATAGCATCACTGGCCACCCGTTTCC | 338629-338605 |
| tos (F) | CAGTTTTTGAGGTTACGGTTGACGG | 343750-343774 |
| chuA (F) | GACGAACCAACGGTCAGGAT | |
| chuA (R) | TGCCGCCAGTACCAAAGACA | |
| ypaA (F) | TGAAGTGTCAGGAGACGCTG | |
| ypaA (R) | ATGGAGAATGCGTTCCTCAAC | |
| tspE4C2 | GAGTAATGTCGGGGCATTCA | |
| tspE4C2 | CGCGCCAACAAAGTATTACG |
F, forward; R, reverse.
Relative to the UPEC strain CFT073 genome (53).
PCR detection of potential virulence genes.
The gene-specific PCR primers used in the present study were designed against the PAI nucleotide sequence of UPEC strain CFT073 (54). Primers are listed in Table 1. Genomic DNA preparations were tested for these genes in 50-μl PCR mixtures containing 15 pmol of each of the forward and reverse primers, 10 nmol of each deoxynucleoside triphosphate, 1 U of Taq DNA polymerase (Invitrogen, Paisley, United Kingdom), and 2 mM MgCl2 in 1× PCR buffer (Invitrogen). The PCR conditions were as follows: initial incubation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30s, and extension at 72°C for 45 s. A final extension step of 72°C for 10 min was also included. Products were analyzed by electrophoresis through 0.8% (wt/vol) gels with ethidium bromide as visualization agent. The sizes of amplicons were determined by comparison to the 1-kb plus DNA ladder (Invitrogen). E. coli K-12 (MG1655) and UPEC E. coli CFT073 were used as negative and positive controls, respectively, in all assays. Negative reactions were retested with annealing at 45 and 50°C to allow for small variations in primer-site sequence between strains. The prevalence data was analyzed online (http://www.matforsk.no/ola/fisher.htm) by using a two-tailed Fisher exact test to test for statistical significance.
RESULTS
Correct structure and gene content of PAI IICFT073.
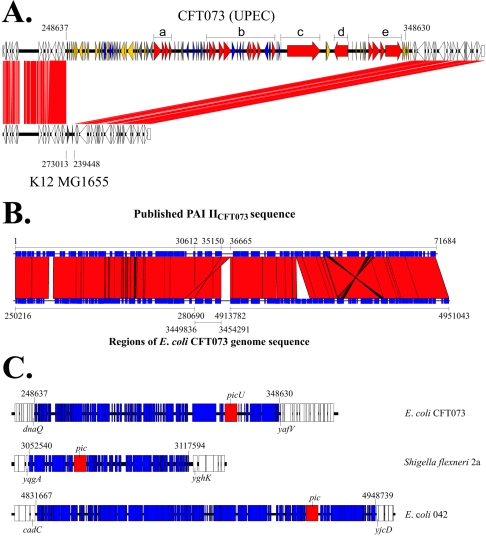
We previously identified a mucinase termed PicU in the UPEC strain CFT073 (40). PicU is a serine protease autotransporter of the Enterobacteriaceae (SPATE) demonstrating a high level of homology (95% identity) with the previously described PAI-encoded Pic mucinase of enteroaggregative E. coli and S. flexneri 2a (21). Analysis of the E. coli CFT073 genome sequence indicated that picU was present on a 100-kb PAI inserted between the E. coli K-12 genes dnaQ and yafV (see Fig. 1A). This region of the K-12 genome includes two genes, yafT and yafU, which are absent from E. coli CFT073, and the aspV tRNA gene, which is present in both genomes. Comparison of E. coli O157:H7, S. flexneri 2a, and EAEC 042 with E. coli K-12 demonstrates that each strain possesses a PAI inserted between dnaQ and yafV. These inserts are dissimilar to each other (data not shown) differing in size (36.0, 57.6, and 31.3 kb, respectively) and gene content, suggesting that the dnaQ-yafV region represents a hotspot for insertions into the backbone sequence.
FIG. 1.
Genetic organization of the PAI IICFT073. (A) Comparison of the E. coli CFT073 PAI IICFT073 and flanking sequences with the equivalent region of E. coli K-12 MG1655. ORFs common to E. coli K-12 and E. coli CFT073 are white. Other ORFs are indicated as follows: ORFs with no homology to anything in the GenBank databases are gray; ORFs with homology to genes associated with IS elements, phage, and mobility genes are yellow; ORFs with homology to genes of unknown function are blue; and ORFs associated with putative virulence determinants are red. Regions that may be associated with virulence are designated by thick black lines above the PAI and represent the fbp iron-sequestering locus (a), the efu sugar-metabolizing loci (b), the etp two-partner secretion system (c), the picU autotransporter (d), and the tos type I secretion system (e). (B) Comparison of the E. coli CFT073 PAI IICFT073 from the complete genome (54) with that of the previously published by Rasko et al. (43). The earlier sequence appears to be comprised of three distinct regions of the E. coli CFT073 genome. The numbers on the bottom represent the position of the different DNA fragments within the E. coli CFT073 genome. (C) Comparison of PAI IICFT073 from E. coli CFT073 with the pic-containing islands of S. flexneri and E. coli 042. The islands are divergent in size, gene complement, organization and chromosomal location. Figures represent the point of insertion into the chromosome of each strain.
Analyses of the nucleotide sequence from the correct PAI IICFT073 revealed the presence of 116 ORFs (c0253 to c0368). Homology searches indicated a close relationship between this PicU-encoding PAI and a 71.6-kb PAI previously characterized in E. coli CFT073 and designated PAI IICFT073 (43). Although both islands share the same first 48 ORFs (representing only 30% of the 100-kb island described here), the remainder of the island is significantly different. Further analysis of the 71.6-kb island revealed it to be a mosaic, comprised of two distinct regions of the E. coli CFT073 genome separated by an inverted repeat of 1,515 bp (Fig. 1B), suggesting the sequence of this putative 71.6-kb island was incorrectly assembled in the first instance. We have retained the PAI IICFT073 nomenclature for the corrected island.
Twenty-four ORFs from the correct PAI IICFT073 were associated with IS, phage and mobility genes, and 67 encoded hypothetical proteins of unknown function. The remainder of the genes, including picU, were associated with virulence and are listed in Table 2. As mentioned above, Pic was characterized previously in EAEC 042 and S. flexneri 2a (21). PAI IICFT073 is distinct in size, organization, and gene composition from the pic-containing islands of the S. flexneri 2a and E. coli 042 (Fig. 1C). Furthermore, the islands are inserted in different regions in each strain, suggesting that pic and picU are mobile elements and can move independently of the PAI.
TABLE 2.
PAI IICFT073 virulence-associated loci
| Locus | GenPept IDa | Position | Product | Homology |
|---|---|---|---|---|
| Fbp locus | ||||
| c0294 | AAN78782.1 | 274013‥276178 | FbpA TonB-dependent receptor | Putative TonB-dependent receptor Erwinia carotovora (CAG76463) |
| c0295 | AAN78783.1 | 276186‥277178 | FbpB periplasmic siderophore-binding protein | Putative iron-chelating periplasmic binding protein Erwinia carotovora (CAG76462) |
| c0296 | AAN78784.1 | 277197‥278255 | FbpD ABC-type Fe3+ siderophore transport permease | Putative ABC transport protein Salmonella enterica serovar Typhimurium LT2 (AAL19708) |
| c0297 | AAN78785.1 | 278252‥279019 | FbpC ABC-type Fe3+ siderophores transport ATPase components | Putative Fe3+ siderophore transport ATPase component Salmonella enterica serovar Typhimurium LT2 (AAL19709) |
| Efu locus | ||||
| c0318 | AAN78806.1 | Complement (289106‥289744) | EfuA sugar-phosphate isomerase | Putative sugar-phosphate isomerase Xanthomonas axonopodis (AAM35062) |
| c0319 | AAN78807.1 | 289986‥291158 | EfuB oligogalacturonide lyase | Putative oligogalacturonate lyase Erwinia carotovora (CAG75329) |
| c0321 | AAN78809.1 | 291188‥291988 | EfuC gluconate 5-dehydrogenase | Putative 5-keto-d-gluconate 5-reductase Streptococcus pyogenes (AAM79055) |
| c0322 | AAN78810.1 | 292732‥294246 | EfuD oligogalacturonide transporter | Putative oligogalacturonide transporter Erwinia carotovora (CAG73730) |
| c0323 | AAN78811.1 | 294176‥296482 | EfuE exopolygalacturonate lyase | Putative exopolygalacturonate lyase Erwinia carotovora (CAG77405) |
| c0330 | AAN78818.1 | Complement (300643‥301428) | EfuF deoxyribose operon repressor | Hypothetical protein Escherichia coli 536 (CAE85174) |
| c0331 | AAN78819.1 | 301731‥302651 | EfuG ribokinase | Hypothetical protein Escherichia coli 536 (CAE85173) |
| c0332 | AAN78820.1 | 302679‥303995 | EfuH l-fucose permease | Hypothetical protein Escherichia coli 536 (CAE85172) |
| c0333 | AAN78821.1 | 304007‥305020 | EfuI cytoplasmic protein | Hypothetical protein Escherichia coli 536 (CAE85171) |
| c0334 | AAN78822.1 | Complement (305943‥307199) | EfuJ sugar-specific permease | Unknown protein Escherichia coli AL862 (AAK27335) |
| c0335 | AAN78823.1 | Complement (307212‥307499) | EfuK sugar phosphotransferase component II B | Unknown protein Escherichia coli AL862 (AAK27336) |
| c0336 | AAN78824.1 | Complement (307515‥307958) | EfuL PTS system, mannitol (cryptic)-specific IIA component | Unknown protein Escherichia coli AL862 (AAK27337) |
| Eth locus | ||||
| c0342 | 310860‥312131 | EthB two-partner secretion system membrane exporter | Hypothetical protein Escherichia coli 536 (CAD42056) | |
| c0343 | 312310‥312684 | EthB two-partner secretion system membrane exporter | Hypothetical protein Escherichia coli 536 (CAD42056) | |
| c0344 | 312321‥312632 | EthB two-partner secretion system membrane exporter | Hypothetical protein Escherichia coli 536 (CAD42056) | |
| c0345 | AAN78830.1 | 312645‥322295 | EthA two-partner secretion system exoprotein | Hypothetical protein Escherichia coli 536 (CAD42055) |
| PicU locus | ||||
| c0350 | AAN78833.1 | Complement (326209‥330324) | PicU serine protease precursor | Pic serine protease precursor Shigella flexneri 2a (AAD23953) |
| Tos locus | ||||
| c0360 | AAN78841.1 | 336046‥337293 | TosC TolC-like outer membrane protein | Putative outer membrane protein Ralstonia eutropha (ZP_00170441) |
| c0361 | AAN78842.1 | 337318‥339465 | TosB HlyB-like cytoplasmic membrane export protein | Putative ABC-type exporter Ralstonia eutropha (ZP_00170444) |
| c0362 | AAN78843.1 | 339511‥340773 | TosD HlyD-like membrane spanning export protein | Putative HlyD family secretion protein Salmonella enterica serovar Typhimurium LT2 (AAL21580) |
| c0363 | AAN78844.1 | 340972‥345804 | TosA RTX family exoprotein A gene | Unknown protein Synechocystis sp. strain PCC 6803 (NP_442018) |
ID, identification code.
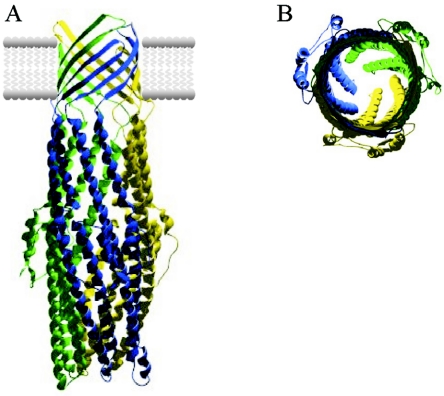
Including picU, PAI IICFT073 contains three loci that are associated with protein secretion. The adjacent locus (c0360 to c0363) encodes a type I protein secretion system, designated the tos locus (for “type one secretion”). In silico analyses demonstrate that TosC encodes a TolC-like outer membrane protein, TosB encodes an ABC-transporter protein of the HlyB family, TosD encodes a protein of the HlyD family of membrane fusion proteins, and TosA encodes a 164-kDa repetitive protein similar to hemagglutinins and hemolysins. In BLAST searches against finished and unfinished microbial genomes, TosC, TosB, and TosD were found to be most similar to hypothetical proteins encoded by a cluster of three ORFs in the unfinished Dichelobacter nodosus VCS1703A genome (28, 56, and 53% identity, respectively), whereas the best TosA homologue was found in Ralstonia solanacearum (24% identity) (47). Among homologues of known function, TosC is most similar to those required for protein secretion (as opposed to cation or drug efflux), suggesting that TosA may be the cognate substrate for this putative export system. With analogy to the haemolysin TolC-HlyBD system (17, 28, 52, 53), TosC would form a trimeric 12-stranded β-barrel pore in the outer membrane connected to an elongated α-helical trans-periplasmic channel, which would transiently dock with the TosD/TosB translocase complex at the inner membrane to facilitate the secretion of TosA from the cytoplasm. A multiple alignment of TosC with other TolC homologues indicates that a notable sequence gap in TosC falls within the extracellular loop linking α-strands 4 and 5 in the α barrel. Although this would produce a loop several residues shorter than the equivalent loop in other TolC homologues (28, 29), it appears from the three-dimensional model of TosC that α-strands 4 and 5 are of sufficient length to span the hydrophobic core of the outer membrane lipid bilayer (Fig. 2).
FIG. 2.
(A) Homology model of the TosC trimer. The individual protomers are colored blue, yellow, and green. A side view of the TosC trimer embedded in the bacterial outer membrane is shown. (B) Top view of the TosC trimer in the same orientation as in panel A demonstrating the periplasmic channel formed by the TosC trimer. The TosC homology model was generated by using the Swissmodel server (www.expasy.org/swissmod/) in alignment mode with the TolC crystal structure (1EK9) as a template. The TosC-TolC pairwise alignment on which the model is based was manually edited to ensure the integrity of conserved secondary structures and residues according to the multiple alignment of 115 TolC homologues from the COG1538 group.
The third protein secretion locus resembles a TPSS (designated etp for “E. coli two-partner secretion”), which normally consists of two genes encoding a secreted effector molecule and a transmembrane pore-forming domain (25). The etpA gene (c0345) encodes a putative effector molecule of 328.4-kDa representing the largest protein encoded by E. coli CFT073 and which shows homology with the HecA colonization factor of Erwinia chrysanthemi (30% identity over 1,712 amino acids) and other large proteins secreted via the TPSS (45). The initial annotation indicated that EtpB, the pore-forming domain, was nonfunctional being represented by three pseudogenes (c0342 to c0344). However, closer inspection revealed c0342 and c0343 constitute one ORF. Furthermore, scrutiny of c0343 and c0344 suggested that they might represent a single ORF and that a small sequencing error introduced an in-frame stop codon (Amber) into the sequence in place of a tryptophan residue (TGG→TAG). However, we resequenced this region and confirmed the presence of an in-frame Amber stop codon. An Amber stop codon is relatively infrequent in E. coli (30); this, combined with the fact that this is an in-frame termination signal, suggests that this termination signal may be suppressed, with one of a number of potential amino acids being inserted at this position. Furthermore, etpB of UPEC strain 536 is an intact ORF, suggesting that this is a functional locus (13) and not a pseudogene; however, empirical evidence is needed to clarify this.
A further locus (c0294 to c0297) designated fbp (for “ferric binding protein”) may be involved in the acquisition of iron. In silico analyses revealed that FbpA belongs to the TonB-dependent family of outer membrane receptor proteins mostly associated with iron transport, whereas FbpB, FbpD, and FbpC belong to the ferric enterobactin family of iron sequestering proteins, possessing homology to the FepB, FepD, and FepC proteins, respectively.
The remaining virulence-associated locus comprises genes associated with sugar metabolism (c0318 to c0336). These loci appear to be involved with the ability to degrade the specific carbohydrates fucose and pectin, and we have designated this the efu locus (for “E. coli fucose metabolism”).
Distribution of PAI IICFT073 genes among the ECOR collection.
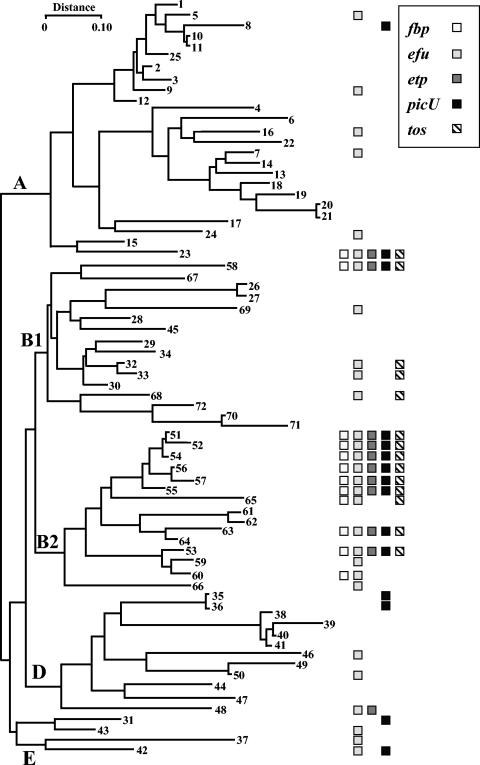
To determine the phylogenetic distribution of PAI IICFT073, we surveyed the well-defined ECOR collection, which is richly varied in terms of phylogeny and zoological and geographical strain origins (38). Using primers corresponding to dnaQ and yafV, which flank either side of the aspV tRNA site, the presence of an appropriately sized PCR product could be detected in amplifications with E. coli K-12 genomic DNA. In contrast, and as expected, no product could be detected in reactions with E. coli CFT073 DNA. The failure of the PCR indicated the presence of PAI IICFT073. Similar reactions with genomic DNA preparations from all members of the ECOR collection yielded negative reactions comparable to those of control E. coli CFT073, indicating that every strain within the ECOR collection possessed an insert in this region and suggesting that the aspV tRNA site is a hotspot for recombination.
The prevalence of the five PAI IICFT073-encoded virulence-associated loci among the ECOR collection is shown in Fig. 3. The fbp locus was found to have low prevalence among non-B2 ECOR strains (3.5%) and a significantly higher prevalence among the B2 group (66.7%; P < 0.00000003). The genes for the carbohydrate metabolism locus (efu) were found at a significantly higher frequency in the B2 phylogenetic locus (80%) than the non-B2 group (29.8%; P < 0.0006), even though this locus demonstrated the widest distribution among the non-B2 isolates. The two-partner secretion system locus (etp) was also found more frequently in the B2 ECOR group (53.3%) than in non-B2 strains (7.0%; P < 0.0002). The picU gene was found in 7.0% (4 of 57) non-B2 ECOR strains, which was significantly less than among the B2 group (53.3%; P < 0.0008). The presence of the tos locus also occurred at a higher frequency in the B2 cluster (60%) than in the non B2 groups (8.8%; P < 0.00007). Interestingly, the etp and tos loci are found only in strains in which the efu locus was detected. None of the remaining loci demonstrated a strict concomitant presence.
FIG. 3.
Distribution of PAI IICFT073 virulence associated loci among the ECOR collection. A phylogenetic tree of the ECOR isolates shows the distribution of the five virulence-associated loci, where each loci is represented by a shaded box as indicated in the figure. The number of the ECOR isolate is given in boldface, and each of the major phylogenetic branches are indicated. The complete complement of virulence associated loci is preferentially associated with the B2 phylogentic cluster. Adapted from reference 23 with permission.
Only two non-B2 strains were positive for the full complement of the PAI IICFT073 virulence associated loci, namely, ECOR23, an ECOR group A strain from a healthy elephant and ECOR58, an ECOR group B1 strain from a healthy lion (38). To ensure that these positive results did not arise through contamination of the original stocks with a B2 isolate, the phylogenetic grouping was determined for each strain by using the method described by Clermont et al. (7). These experiments confirmed that ECOR23 belonged to group A and ECOR58 to the B1 group, indicating that PAI IICFT073 had been acquired by horizontal transfer.
Prevalence of PAI IICFT073 genes among UTI isolates.
To determine whether PAI IICFT073 was associated with a particular UTI, the presence of the five virulence-associated loci were detected by PCR in a collection of pathogenic E. coli strains isolated from patients with cystitis, pyelonephritis, and prostatitis. The results are shown in Fig. 4.
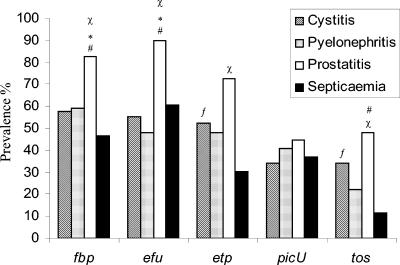
FIG. 4.
Prevalence of PAI IICFT073 virulence associated loci in E. coli clinical isolates. The fbp, efu, etp, picU, and tos loci were detected by PCR in clinical isolates of E. coli. Prevalence is indicated as a percentage of the total population of strains associated with each clinical syndrome. ✽, A statistically significant difference between prostatitis and cystitis;, #, a statistically significant difference between prostatitis and pyelonephritis; χ, a statistically significant difference between prostatitis and septicemia isolates; ƒ, a statistically significant difference between cystitis and septicemia isolates.
When the cystitis and pyelonephritis strains were analyzed, no statistically significant difference was observed between the distribution of the fbp locus (57.9% versus 59.3%), the efu locus (55.3% versus 48.1%), the etp locus (52.6% versus 48.1%) the picU locus (34.2% versus 40.7%), or the tos locus (34.2% versus 22.2%). Similarly, no statistically significant difference was observed between the distribution of the etp and picU loci when prostatitis isolates were compared to cystitis strains (72.4% versus 52.6% and 44.8% versus 34.2%, respectively) or pyelonephritis strains (72.4% versus 48.1% and 44.8% versus 40.7%, respectively). In contrast, the iron-binding locus fbp demonstrates a higher prevalence in prostatitis strains compared to cystitis (82.8% versus 57.9%, P < 0.03) and pyelonephritis (82.8% versus 59.3%, P < 0.05). In a comparable fashion, the distribution of efu, the sugar metabolizing locus, in prostatitis isolates, is significantly higher compared to cystitis (89.7% versus 55.3%, P < 0.003) and pyelonephritis (89.7% versus 48.1%, P < 0.001). Finally, a negative correlation was observed between the distribution of the tos locus in prostatitis strains and pyelonephritis isolates (22.2% versus 48.3%, P < 0.05) but not between prostatitis and cystitis isolates (48.3% versus 34.2%).
In all cases the presence of the tos locus was found concurrently with the fbp and efu loci and, with the exception of one strain, with the picU gene. The distributions of the hly, fim, aer, cnf1, pap, sat, and prs loci have previously been described for this collection of UTI isolates (46). Comparison of the current data with the previous data did not reveal a strict concomitant correlation between the presence of any of the previously investigated loci and the data generated for the present study.
Prevalence of PAI IICFT073 genes among E. coli septicemia isolates.
The prototypical uropathogenic E. coli isolate CFT073 was isolated from the blood of a woman suffering from pyelonephritis, indicating that it had the capacity to invade the bloodstream (35). To investigate whether PAI IICFT073 contributed to the ability of E. coli to invade the bloodstream, the distribution of the five virulence-associated loci in 43 strains of E. coli isolated from patients with septicemia was examined by PCR (Fig. 4). There were no statistically significant differences between the prevalences of any of the PAI IICFT073 loci in the septicemia and pyelonephritis isolates. Similarly, there was no statistically significant difference between the distribution of the fbp, efu, and picU loci in the septicemia isolates compared to the cystitis isolates, and no difference was observed in the distribution of the picU gene when septicemia isolates were compared to the prostatitis strains. Interestingly, the tos and etp loci demonstrated a statistically lower prevalence in the septicemia isolates than in cystitis isolates (P < 0.02 and P < 0.05, respectively).
Unexpectedly, the prevalence of the efu, tos, etp, and fbp loci was significantly lower in the septicemia isolates than in the prostatitis isolates (P < 0.05, P < 0.0009, P < 0.0007, and P < 0.002, respectively). To determine whether the distribution was altered by a population of septicemia isolates that was not representative of the B2 phylogenetic cluster, the phylogenetic grouping was determined for each isolate by using the method described by Clermont et al. This analysis revealed that the collection of septicemia isolates was composed of 26 strains from the B2 phylogenetic group, 4 strains from the B1 group, 7 strains from the D group, and 3 strains from the A group. Comparison of the B2 septicemia isolates with the ECOR B2 isolates revealed no statistically significant difference in the prevalence of efu, picU, etp, or fbp. Interestingly, the tos locus was significantly underrepresented in the blood-borne isolates when the two B2 groups were compared (60% versus 19.2%, P < 0.02). Comparison of the distribution of the PAI IICFT073 loci among the nonB2 ECOR collection and the nonB2 septicemia isolates revealed no statistical difference between the prevalence of the efu, tos, etp, or fbp loci. Surprisingly, the nonB2 septicemia isolates demonstrated a higher prevalence of picU than the nonB2 ECOR collection (58.9% versus 10.5%, P < 0.0001) and the B2 septicemia isolates (58.9% versus 23.1%, P < 0.0008).
DISCUSSION
The number of proteins identified in gram-negative bacteria that are secreted via the autotransporter secretion pathway (11, 12) has grown almost exponentially since the initial description of the secretion of gonococcal immunoglobulin A1 protease (42). Recently, we described the existence of several autotransporters in UPEC strain CFT073, including the SPATE protein PicU (40). Further investigation revealed PicU was encoded on a 100-kb PAI inserted at the aspV tRNA locus. Like other PAIs, PAI IICFT073 possesses several loci, in addition to the picU locus, which may be involved in virulence. Among these are a member of the two-partner secretion system (etp), a type I secretion system (tos), iron acquisition genes (fbp), and a carbohydrate metabolism locus (efu). Interestingly, the genetic content of the PAI encoding PicU is homologous (30% identical) to a PAI (PAI IICFT073) that was reported as being inserted in the chromosomal backbone of E. coli in a similar position (43). The reason for the discrepancy between the complete genome sequence and that published by Rasko et al. (43) is unclear but could be due to several reasons, including a recombination event, concatamerization of inserts in the cosmid clones used to assemble the sequence of Rasko et al. (43), or misassembly of the sequenced regions. Nevertheless, we have retained the PAI IICFT073 nomenclature for the PicU-encoding island.
E. coli is a highly clonal species represented by four major phylogentic groups (A, B1, B2, and D) (26). Most extra-intestinal pathogenic E. coli strains, including those with the most robust virulence factor repertoires and those that are most able to infect noncompromised hosts, derive from phylogenetic group B2 with group D representing the second largest group contributing to the extraintestinal clones (26, 27, 55). In contrast, diarrheagenic pathotypes occur almost exclusively in the A, B1, D, and ungrouped phylogenetic groups (26). The full complement of virulence associated loci was found in eight of the 15 B2 isolates, however six of these were clustered together, suggesting that acquisition of PAI IICFT073 occurred late in the divergence of the B2 cluster and that other strains have acquired the island by horizontal gene transfer, a hypothesis supported by the observation of the intact PAI in ECOR23 and ECOR58, group A and B1 strains, respectively. However, an alternative hypothesis is that PAI IICFT073 was acquired early in the B2 cluster and that various genes were lost by mutational attrition. This hypothesis is supported by the presence of the full complement of genes on two additional branches of the B2 cluster. Nevertheless, it is clear that the island and its constituent loci are mainly associated with the B2 cluster, which is representative of extraintestinal infectious organisms, and commensal rectal isolates.
A question that remains unanswered is whether the more invasive extraintestinal strains, such as those which cause pyelonephritis, prostatitis, or septicemia, are simply more virulent than their counterpart cystitis isolates or whether they are separate pathogens with specialized repertoires of virulence factors that have evolved to allow colonization of their specific niches. Previous investigations by many different groups working with many different pathogens have demonstrated that the acquisition of iron is often essential for full virulence (8, 16, 44). In addition, many PAIs possess iron acquisition mechanisms (4, 5, 31). The locus encoding the FbpA-D iron siderophore system was previously shown to be associated with UPEC and to be more represented in these pathogens than in intestinal isolates of E. coli (43). However, in the same study Rasko et al. suggested that the presence of this locus was more strongly correlated with survival in the bladder because of the presence of the locus in a significantly greater number of cystitis isolates (43). In contrast, we found no difference between the presence of the fbp locus in pyelonephritis and cystitis strains. However, prostatitis isolates showed a significantly greater prevalence of the fbp locus than cystitis or pyelonephritis isolates, indicating the greater virulence of these strains. The existence of this locus and additional iron acquisition loci suggests that E. coli CFT073 may be adept at scavenging this important nutrient.
The presence of the sugar-metabolizing locus (efu) within the PAI has no obvious connection with a role in pathogenesis of E. coli CFT073. However, recent signature-tagged mutagenesis analyses of Klebsiella pneumoniae identified mutants in three separate genes in the fucose-metabolizing locus that were unable to colonize the intestine of mice (32). Furthermore, in Bacteroides thetaiotaomicron, a commensal intestinal isolate, expression of the fucose metabolic pathway is coordinated with the production of fucosylated glycans in enterocytes that are hypothesized to act a receptors for this organism (24). Interestingly, the presence in certain women of fucosylated structures on the surface of uroepithelial cells appears to have a protective effect from UTI (48). The extraordinary virulence of E. coli CFT073 may be due in part to the ability to degrade these fucosylated structures. Although a role for the efu locus specifically in the pathogenesis of UTI appears unlikely due to its wide distribution among all phylogenetic groups of E. coli, and an equal distribution among cystitis, pyelonephritis, and septicemia strains, it is interesting that three of the loci (c0334 to c0336) are among the top 15 genes upregulated during growth of bacteria in urine (50).
A range of different adhesive factors have been described for extraintestinal pathogenic E. coli, including the type 1, P, and S fimbrial systems (37). Several groups have demonstrated that possession of these different adhesive systems contributes to the ability of UPEC to infect different niches, e.g., expression of P fimbriae contributes to an ascending UTI leading to pyelonephritis (19). Unlike the fimbrial systems, the etpBA locus encodes a putative adhesin secreted via a two-partner secretion system in which EtpA represents the secreted protein and EtpB the transmembrane pore that facilitates secretion of EtpA (25). EtpA demonstrates a high level of similarity to HecA of Erwinia chrysanthemi and filamentous hemagglutinin of Bordetella pertussis; both proteins have been shown to be important in the pathogenesis of disease by mediating adherence and bacterial cell aggregation (45, 51), suggesting that EtpA might lead to colonization of specific niche within the urinary tract. Indeed, the presence of the etp locus was observed at a statistically higher frequency among prostatitis strains compared to cystitis and pyelonephritis. Interestingly, filamentous hemagglutinin is a component of the acellular pertussis vaccines currently in use (41), suggesting that this homologous protein might offer potential for therapeutic uses.
Members of the SPATE family of autotransporters are proteins from E. coli and Shigella spp. that have been widely implicated in virulence (22). Previous investigations in our lab demonstrated that the SPATE protein PicU is a multifunctional enzyme which, like its enteric counterpart (Pic), can digest mucin (21, 40). Since the urinary tract is lined with mucin, PicU may play a role in breaching this protective layer to allow onset of disease. Indeed, previous data have shown that disruption of the urinary tract mucus layer prior to bacterial challenge increases the level of colonization and the severity of disease (9). Previously, we found picU to be present in 22.5% of UTI isolates compared to only 12% of rectal isolates with no statistical difference between cystitis and pyelonephritis (40), whereas in the current study we found no difference between cystitis, pyelonephritis, prostatitis, or septicemia isolates. In contrast, Heimer et al. (20) detected picU in a significantly higher number of pyelonephritis strains than cystitis isolates; however, the reasons for the disparate results is not clear. Perhaps the most interesting observation regarding picU is its distribution among the septicemia isolates. Although the prevalence of picU among the B2 phylogenetic groups in the ECOR and septicemia isolates is similar, among non-B2 groups it demonstrated a higher prevalence in the septicemia isolates, a finding which could be explained if picU contributed to the ability of bacteria to invade the bloodstream or to survive within the bloodstream. In this respect Hbp, a homologue of PicU, has been demonstrated to recruit iron through the degradation of hemoglobin, suggesting that PicU might have the same ability (39). The ability of PicU or Pic to scavenge iron in a similar fashion has yet to be investigated.
Pfam and SMART domain searches clearly show that TosB, TosD, and TosC are members of the HlyB, HlyD, and TolC protein families, respectively. Although TosC shares much lower identity with its respective homologues than TosB and TosD, this is a typical feature of all TolC-like proteins (28). Importantly, TosC shares the stabilizing ring of aromatic residues at the base of the α-barrel and the majority of conserved proline residues in the interdomain region between the α-barrel and the periplasmic helices (Fig. 2). In general, the closest homologues of TosB and TosD are encoded in clusters in association with genes encoding TosA and TosC homologues, although the order is not always conserved with E. coli CFT073. Given the close association of tosA with tosCDB, it appears likely, but not certain, that the 164-kDa product of this gene is secreted in a type I-dependent fashion via the TosBDC secretion apparatus. TosA is a highly repetitive protein, with several glycine- and leucine-rich repeats that appear to be similar to the calcium-binding repeats found in the RTX family of type I-secreted proteins. Classical RTX toxins, such as HlyA, share low-level identity over the first 500 to 600 residues, followed by a discrete region of calcium-binding nonamers and a C-terminal secretion domain (28). In contrast, TosA and its close homologues have no significant similarity to the N-terminal domain of RTX toxins and are not found in operons encoding HlyC homologs (the acyl modification enzyme required for activation of RTX toxins); therefore, they represent a distinct family of large repetitive proteins. The presence of a single transmembrane domain in the N-terminal region of TosA suggests that, postsecretion, the molecule may be tethered to the outer membrane and act as an adhesin. Interestingly, although orthologous clusters are found throughout the Enterobacteriaceae, including the majority of Salmonella species, no function has yet been ascribed to a TosA homologue. However, the tos locus is present at a low frequency in pyelonephritis and septicemia isolates compared to cystitis, suggesting that expression of this adhesin may inhibit the ability of bacteria to invade renal tissue and the bloodstream but may contribute to bladder colonization.
In summary, we have demonstrated that the presence of PAI IICFT073 is associated with extraintestinal group B2 isolates of E. coli. Furthermore, PAI IICFT073 is more prevalent in strains causing prostatitis than cystitis or pyelonephritis, and the presence of the fbp, efu, etp, and picU loci does not contribute to ascending urinary tract infections, whereas the presence of the TosA adhesin may inhibit the ability of E. coli strains to ascend the urinary tract and/or invade the bloodstream. Although fbp, efu, etp, and tos do not contribute to the ability of E. coli to invade the bloodstream, the presence of picU may enhance the ability of E. coli from groups A, B1, D, and E to survive or invade the bloodstream. The full contribution of these loci to prostatitis is currently under investigation.
Acknowledgments
We thank Guy Plunkett III for access to the prepublication sequence of E. coli CFT073, and Harry Mobley for providing the E. coli CFT073 strain, and Chengjie Liu for technical assistance. The data from several ongoing E. coli genome-sequencing projects were made available by the Wellcome Trust Sanger Institute and the E. coli Genome Project (http://www.genome.wisc.edu/). Preliminary sequence data for the D. nodosus genome was obtained from the Institute for Genomic Research (http://www.tigr.org/), whose funding was provided by the U.S. Department of Agriculture.
This study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) grants 81/D14955 and 81/P14130 to I.R.H. S.A.B. is supported by an MRC (United Kingdom) fellowship.
REFERENCES
- 1.Bauer, R. J., L. Zhang, B. Foxman, A. Siitonen, M. E. Jantunen, H. Saxen, and C. F. Marrs. 2002. Molecular epidemiology of three putative virulence genes for Escherichia coli urinary tract infection—usp, iha, and iroN (E. coli). J. Infect. Dis. 185:1521-1524. [DOI] [PubMed] [Google Scholar]
- 2.Bingen-Bidois, M., O. Clermont, S. Bonacorsi, M. Terki, N. Brahimi, C. Loukil, D. Barraud, and E. Bingen. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126:189-195. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 5.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri, R. R., A. M. Khan, and M. J. Pallen. 2004. coliBASE: an online database for Escherichia coli, Shigella, and Salmonella comparative genomics. Nucleic Acids Res. 32(database issue):D296-D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelissen, C. N. 2003. Transferrin-iron uptake by gram-negative bacteria. Front. Biosci. 8:836-847. [DOI] [PubMed] [Google Scholar]
- 9.Cornish, J., J. P. Lecamwasam, G. Harrison, M. A. Vanderwee, and T. E. Miller. 1988. Host defence mechanisms in the bladder. II. Disruption of the layer of mucus. Br. J. Exp. Pathol. 69:759-770. [PMC free article] [PubMed] [Google Scholar]
- 10.Delcher, A. L., A. Phillippy, J. Carlton, and S. L. Salzberg. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvaux, M., N. J. Parham, and I. R. Henderson. 2004. The autotransporter secretion system. Res. Microbiol. 155:53-60. [DOI] [PubMed] [Google Scholar]
- 12.Desvaux, M., N. J. Parham, and I. R. Henderson. 2004. Type V protein secretion: simplicity gone awry? Curr. Issues Mol. Biol. 6:111-124. [PubMed] [Google Scholar]
- 13.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands [PAI I(536) to PAI IV(536)] of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg, M., and R. Welch. 1996. Virulence determinants of uropathogenic Escherichia coli, p. 135-174. In H. Mobley and J. Warren (ed.), Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, D.C.
- 15.Emody, L., M. Kerenyi, and G. Nagy. 2003. Virulence factors of uropathogenic Escherichia coli. Int. J. Antimicrob. Agents 22(Suppl. 2):29-33. [DOI] [PubMed] [Google Scholar]
- 16.Faraldo-Gomez, J. D., and M. S. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell. Biol. 4:105-116. [DOI] [PubMed] [Google Scholar]
- 17.Gentschev, I., G. Dietrich, and W. Goebel. 2002. The Escherichia coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 10:39-45. [DOI] [PubMed] [Google Scholar]
- 18.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanisch, F. G., J. Hacker, and H. Schroten. 1993. Specificity of S fimbriae on recombinant Escherichia coli: preferential binding to gangliosides expressing NeuGc…a(2-3)Gal and NeuAc…a(2-8)NeuAc. Infect. Immun. 61:2108-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heimer, S. R., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect. Immun. 72:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper, L. V., P. G. Falk, and J. I. Gordon. 2000. Analyzing the molecular foundations of commensalism in the mouse intestine. Curr. Opin. Microbiol. 3:79-85. [DOI] [PubMed] [Google Scholar]
- 25.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. 2002. Evolution of pathogenic Escherichia coli, p. 55-77. In M. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, London, England.
- 27.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 28.Koronakis, V. 2003. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 555:66-71. [DOI] [PubMed] [Google Scholar]
- 29.Koronakis, V., J. Li, E. Koronakis, and K. Stauffer. 1997. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol. Microbiol. 23:617-626. [DOI] [PubMed] [Google Scholar]
- 30.Lacadena, J., A. Martinez del Pozo, J. M. Mancheno, M. Gasset, M. Onaderra, and J. G. Gavilanes. 1995. Escherichia coli JA221 can suppress the UAG stop signal. Lett. Appl. Microbiol. 21:96-98. [DOI] [PubMed] [Google Scholar]
- 31.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroncle, N., D. Balestrino, C. Rich, and C. Forestier. 2002. Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 70:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marrs, C. F., L. Zhang, P. Tallman, S. D. Manning, P. Somsel, P. Raz, R. Colodner, M. E. Jantunen, A. Siitonen, H. Saxen, and B. Foxman. 2002. Variations in 10 putative uropathogen virulence genes among urinary, faecal and peri-urethral Escherichia coli. J. Med. Microbiol. 51:138-142. [DOI] [PubMed] [Google Scholar]
- 34.Middendorf, B., B. Hochhut, K. Leipold, U. Dobrindt, G. Blum-Oehler, and J. Hacker. 2004. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin, M. D., and W. J. Hopkins. 2002. Identification of virulence genes in uropathogenic Escherichia coli by multiplex polymerase chain reaction and their association with infectivity in mice. Urology 60:537-541. [DOI] [PubMed] [Google Scholar]
- 37.Mulvey, M. A. 2002. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 4:257-271. [DOI] [PubMed] [Google Scholar]
- 38.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto, B. R., S. J. van Dooren, J. H. Nuijens, J. Luirink, and B. Oudega. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parham, N. J., U. Srinivasan, M. Desvaux, B. Foxman, C. F. Marrs, and I. R. Henderson. 2004. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol. Lett. 230:73-83. [DOI] [PubMed] [Google Scholar]
- 41.Podda, A., E. C. De Luca, B. Contu, R. Furlan, A. Maida, A. Moiraghi, D. Stramare, L. Titone, F. Uxa, F. Di Pisa, et al. 1994. Comparative study of a whole-cell pertussis vaccine and a recombinant acellular pertussis vaccine. J. Pediatr. 124:921-926. [DOI] [PubMed] [Google Scholar]
- 42.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 43.Rasko, D. A., J. A. Phillips, X. Li, and H. L. Mobley. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J. Infect. Dis. 184:1041-1049. [DOI] [PubMed] [Google Scholar]
- 44.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 100:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz, J., K. Simon, J. P. Horcajada, M. Velasco, M. Barranco, G. Roig, A. Moreno-Martinez, J. A. Martinez, T. Jimenez de Anta, J. Mensa, and J. Vila. 2002. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J. Clin. Microbiol. 40:4445-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 48.Schaeffer, A. J., N. Rajan, Q. Cao, B. E. Anderson, D. L. Pruden, J. Sensibar, and J. L. Duncan. 2001. Host pathogenesis in urinary tract infections. Int. J. Antimicrob. Agents 17:245-251. [DOI] [PubMed] [Google Scholar]
- 49.Schappert, S. M. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States, 1997. Vital Health Stat. 13:i-iv, 1-39. [PubMed] [Google Scholar]
- 50.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuomanen, E., A. Weiss, R. Rich, F. Zak, and O. Zak. 1985. Filamentous hemagglutinin and pertussis toxin promote adherence of Bordetella pertussis to cilia. Dev. Biol. Stand. 61:197-204. [PubMed] [Google Scholar]
- 52.Welch, R. A. 1991. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 53.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 257:85-111. [DOI] [PubMed] [Google Scholar]
- 54.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]