Abstract
Acne is a prevalent inflammatory disease in dermatology, and its pathogenesis may be associated with inflammation, immunity, and other mechanisms. It commonly manifests in young individuals and frequently imposes a heavy economic, physical, and psychological burden on patients. Gut microbes and blood metabolites, as significant immune and inflammatory regulators in the body, have been hypothesized to form the “neurocutaneous axis.” Nonetheless, the precise causal relationships among the gut microbes, circulating blood metabolites, and acne development have yet to be elucidated. This study employed bidirectional two-sample Mendelian randomization (MR) to probe the causal impacts of 412 distinct gut microbes and 249 blood metabolites on acne. Single nucleotide polymorphisms (SNPs), which are closely associated with gut microbes and blood metabolites, were utilized as instrumental variables. This approach was taken to discern whether these elements serve as pathogenic or protective factors in relation to acne. Furthermore, a mediation analysis encompassing gut microbes, blood metabolites, and acne was conducted to explore potential correlations between gut microbes and blood metabolites, as well as their cumulative effects on acne. This was done to substantiate the notion of causality. Bidirectional two-sample MR analysis revealed 8 gut bacteria, 6 bacterial metabolic abundance pathways determined by birdshot, and 8 blood metabolites significantly associated with acne. The mediation MR analysis revealed 2 potential causal relationships, namely, Bifidobacterium-DHA-Acne and Bifidobacterium-Degree of Unsaturation-Acne. This study identified gut microbes and blood metabolites that are causally associated with acne. A potential causal relationship between gut microbes and blood metabolites was obtained via mediation analysis. These insights pave the way for the identification of new targets and the formulation of innovative approaches for the prevention and treatment of acne.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-78603-z.
Keywords: Gut microbes, Blood metabolites, Acne, Mendelian randomization, Mediation analysis
Subject terms: Computational biology and bioinformatics, Skin diseases, Genetics research
Introduction
Gut microbes are associated with a variety of diseases, including diabetes, obesity, nonalcoholic fatty liver disease, and rosacea1,2. The parallels and interconnections between gut microbes and the skin have been recognized for a substantial duration. It has been suggested that the gut and skin share physiological and functional similarities, as both are populated by a vast array of microbes and serve a role similar to a “barrier”. The concept of the “gut‒skin axis” was introduced as far back as the previous century3,4. The linkage between the two might be ascribed to blood metabolites that are generated or modulated by gut microbes5.
Acne is a prevalent and frequently encountered dermatologic condition, with a global prevalence of approximately 9.4%. It is the eighth most prevalent disease worldwide6. Research has indicated that the development of this condition may be associated with genetic factors, hormone levels, medication use, consumption of dairy products, a high-fat diet, and infection with Propionibacterium acnes7. In light of the recent association between gut microbes and skin conditions, gut microbes are anticipated to emerge as a novel focus for the prevention and management of acne. The presence of acne has been linked to dietary patterns8,9, and dietary composition can impact the diversity of gut microbes.
Blood metabolites, either produced or influenced by gut microbes, are connected with the mammalian target of the rapamycin (mTOR) signaling pathway. The mTOR signaling pathway has been demonstrated to affect the composition of the gut flora and, consequently, the progression of acne10. In instances of acne and gut microbial imbalance, an increase in the expression of substance P and a subsequent amplification of substance P-containing nerves have been noted. Substance P triggers an increase in the levels of proinflammatory mediators, including interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α), which have also been implicated in the pathogenesis of acne10.
Mendelian randomization (MR) is an analytical approach rooted in genetics. It leverages genetic variants associated with a potential risk factor as proxies to infer causal relationships regarding the influence of the exposure on the outcome. MR essentially mimics the randomization process, thereby increasing the likelihood of reducing the effect of additional confounding factors. This addresses the constraints of randomized controlled clinical trials and enables the drawing of causal connections between potential risk factors and outcomes. Using MR, we scrutinized the causal link between gut microbes, blood metabolites, and acne.
Materials and methods
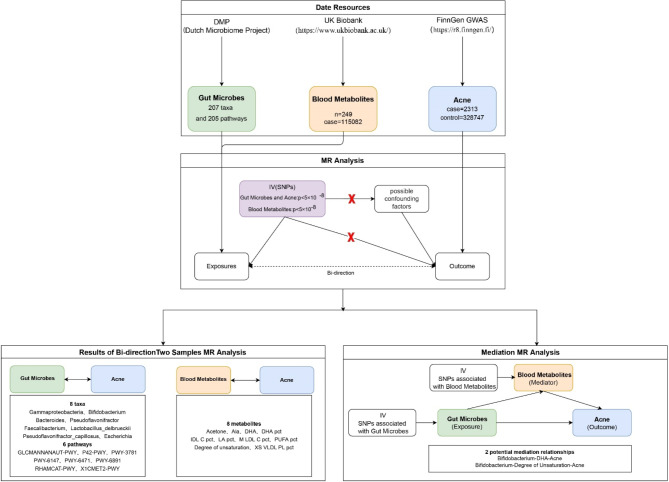
This study utilized gut microbes and blood metabolites as exposure variables, with acne as an outcome. The study design is depicted in Fig. 1. Initially, we conducted two bidirectional two-sample MR analyses, using gut microbes and blood metabolites as exposure factors, to investigate and validate their causal relationships with acne. Subsequently, mediation MR analyses were conducted on gut microbes, blood metabolites, and acne via two-step MR and multivariable MR, with the aim of identifying potential causal relationships.
Fig. 1.
Flow chart of the study.
Study design
GWAS database
Pooled data on gut microbes, blood metabolites, and acne were acquired from the database of previous genome‒wide association studies (GWASs). The data on gut microbes were acquired from the most recent extensive cohort study conducted by the Dutch Microbiome Project (DMP). The study examined the abundance of gut microbes and their functional pathways in a sample of 8208 individuals from the Netherlands11, which included a three-generation cohort of 2756 families, 99.5% of whom were of Dutch–European descent. Lopera-Maya et al. conducted shotgun metagenomic sequencing on 7738 individuals from the DMP and acquired GWAS data on gut microbes for the study. Our analysis incorporated an extensive array of 207 taxa, spanning 5 phyla, 10 classes, 13 orders, 26 families, 48 genera, and 105 species. Additionally, we included 205 distinct metabolic pathways in our study framework12. The data for 249 blood metabolites were collected from the UK Biobank GWAS of 115,082 individuals of European descent13. The data for the outcome phenotype, acne, were sourced from the FinnGen GWAS results, comprising 2,313 acne patients and 328,747 controls. The corresponding phenotype code was “L12_ACNE”. Acne is classified by the ICD-10 code L70.000.
Instrumental variable (IV) selection
The genetic instrumental variables of microbes and blood metabolites that were utilized underwent GWAS, with a P value threshold of < 1 × 10− 5) and an effect allele frequency (EAF) > 0.01. Setting different p values as cutoff values results in the screening of different numbers of independent variables (IVs), which in turn produces different MR results. In this study, we set the p value threshold for the blood metabolite and acne data at p < 5 × 10− 8, aligning with the approach used in other studies14,15, to increase the depth of information for the study. However, when analyzing the gut microbe data, we used a threshold of p < 5 × 10− 8 to screen the IVs and found that the number of SNPs was either too low or that there was a lack of significant SNPs available. To optimize the amount of genetic variance accounted for by genetic predictors, and in synergy with other extant studies involving gut microbes16, we set the threshold at p < 1 × 10− 5 to screen for IVs.
Drawing from the reference panel of the European population in the 1000 Genomes Project, we established a threshold for linkage disequilibrium at r2 < 0.001 within a span of ± 100 kb around the genetic variation. Following this, we implemented a clumping procedure to prune the collection of significant SNPs. When no single nucleotide polymorphisms (SNPs) were accessible in the exposure and results, data with a linkage disequilibrium coefficient (r2) of 0.8 or higher were utilized as a proxy. The SNPs included in the study all presented effect allele frequencies exceeding 0.01. Additionally, SNPs with an F statistic < 10 were omitted to mitigate the potential influence of weak instrumental bias17.
Statistical analysis and sensitivity analysis
Initially, a bidirectional two-sample Mendelian randomization (MR) analysis was executed to determine the causal relationship between gut microbes and acne, as well as between blood metabolites and acne. The inverse variance weighted (IVW) method was adopted as the primary strategy for causality estimation. Moreover, a composite of five methods, including weighted median, MR Egger, simple mode, weighted mode, and the Wald ratio, was used for result evaluation. A statistically significant association in our analysis was denoted by P < 0.05. The MR Egger’s intercept and MR-PRESSO methods were applied to evaluate heterogeneity and multiplicity, respectively.
Following this, within the framework of mediated Mendelian randomization analysis, we used the inverse variance weighted (IVW) method as the primary technique to explore potential mediated causality. In addition, the weighted median and MR‒Egger methods were employed to evaluate the outcomes. We leveraged the Cochran Q test to assess the heterogeneity and multiplicity of the results, while the MR-PRESSO method was applied to scrutinize their sensitivity.
The obtained results were adjusted for the false discovery rate (FDR). Ultimately, only gut microbes and blood metabolites with a significance level of p < 0.05, a sensitivity (SNP > 3), absence of heterogeneity and pleiotropy (heterogeneity_Q p > 0.05, P_intercept p > 0.05), and no evidence of reverse causality were included in the study.
Mediation analysis is a sophisticated statistical method designed to explore the intermediary role of a variable within the causal relationship between two other variables. The mediation MR analysis can establish a pathway from exposure factors to outcome phenotypes, which involves scrutinizing the mediators to shed light on the potential mechanisms through which exposure factors impact outcome phenotypes. For example, if a causal relationship exists between gut microbes, blood metabolites, and acne and if gut microbes also have a causal relationship with blood metabolites, it forms a triangular relationship15.
In our study, we designated gut microbes as the exposure variable, blood metabolites as the mediator, and acne as the outcome variable. We employed Mendelian randomization analyses focused on mediation to uncover potential causal pathways linking gut microbes and blood metabolites with the incidence of acne. Initially, univariate MR analyses were conducted by individually comparing specific gut microbes as exposures with selected blood metabolites as the outcome. The analysis involved a comparison of selected blood metabolites as exposures with selected gut microbes as outcomes, leading to a reversal of the initial findings. The proportion of mediating effects was calculated via the two-step MR method, which employs the following equation: 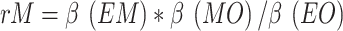 18, 19). Where rM represents the mediating effect ratio of blood metabolites, β(EM) denotes the MR effect value of exposure on the mediator, β(MO) signifies the MR effect value of the mediator on the outcome, β(EM)*β(MO) indicates the “indirect” effect via the mediator, and β(EO) represents the total effect of exposure on the outcome. The multivariable MR method employs the equation
18, 19). Where rM represents the mediating effect ratio of blood metabolites, β(EM) denotes the MR effect value of exposure on the mediator, β(MO) signifies the MR effect value of the mediator on the outcome, β(EM)*β(MO) indicates the “indirect” effect via the mediator, and β(EO) represents the total effect of exposure on the outcome. The multivariable MR method employs the equation 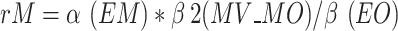 . Where rM represents the mediator effect ratio for blood metabolites, α(EM) denotes the MR effect value of exposure on the mediator, β2(MV_MO) signifies the mediator’s adjusted effect value on the outcome via multivariate Mendelian randomization, and β(EO) indicates the total effect value of exposure on the outcome. Furthermore, given that mediators may be influenced by various exposures, a multivariate MR was conducted to ascertain the primary exposures, and the two approaches were cross-validated. The aforementioned analyses were conducted via the R packages TwoSampleMR20,21, RMediation, and ggplot2.
. Where rM represents the mediator effect ratio for blood metabolites, α(EM) denotes the MR effect value of exposure on the mediator, β2(MV_MO) signifies the mediator’s adjusted effect value on the outcome via multivariate Mendelian randomization, and β(EO) indicates the total effect value of exposure on the outcome. Furthermore, given that mediators may be influenced by various exposures, a multivariate MR was conducted to ascertain the primary exposures, and the two approaches were cross-validated. The aforementioned analyses were conducted via the R packages TwoSampleMR20,21, RMediation, and ggplot2.
Results
MR Analysis results of gut microbes (bidirectional)
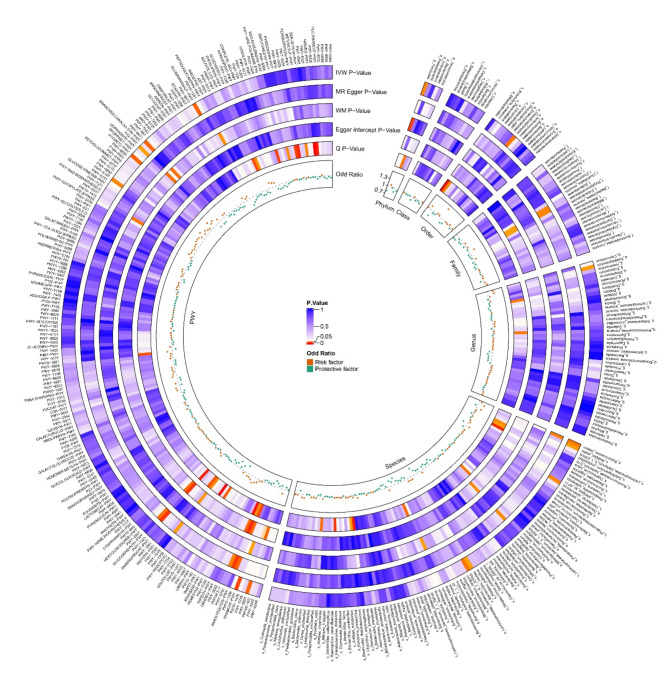
Through two-sample MR analysis of gut microbes and acne, we identified eight bacteria that are significantly associated with acne via the inverse variance weighted (IVW) method. Additionally, we found that eight bacterial metabolic abundance pathways, which were determined via the birdshot method, were significantly associated with acne. These 16 bacteria and bacterial metabolic abundance pathways are significantly associated with acne, and other negative results can be visualized in the circo heatmap in Fig. 2.
Fig. 2.
Circo Heatmap showing the causal effects of gut microbes and bacterial metabolic pathways on acne. The prefixes p_, c_, o_, f_, g_, s_ and PWY represent Phylum, Class, Order, Family, Genus, Species, and Pathway, respectively. IVW, inverse-variance weighted; WM, weighted median.
The bacteria included c_Gammaproteobacteria, g_Bifidobacterium, g_Bacteroides, g_Pseudoflavonifractor, g_Faecalibacterium, s_Lactobacillus_delbrueckii, s_Pseudoflavonifractor_capillosus, and s_Escherichia. The bacterial metabolic abundance pathways include GLCMANNANAUT-PWY, P42-PWY, PWY-3781, PWY-6147, PWY-6471, PWY-6891, RHAMCAT-PWY, and X1CMET2-PWY. Among the positive findings, PWY-3781 was excluded because of a low nSNP of 2, which prevented the calculation of the Egger intercept value and p value. Additionally, PWY-6891 was excluded because horizontal pleiotropy was identified during sensitivity analysis. The remaining 14 bacteria and bacterial metabolic abundance pathways are displayed in the scatterplots in Fig. 3.
Fig. 3.
Scatterplots showing the causal effects of gut microbes and bacterial metabolic pathways on acne.
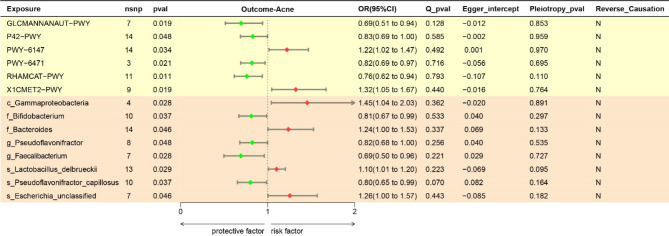
As illustrated in the forest plots in Fig. 4, we used the IVW method and found that the genera g_Bifidobacterium, g_Pseudoflavonifractor, and g_Faecalibacterium and the species s_Pseudoflavonifractor _capillosus, GLCMANNAUT-PWY, P42-PWY, PWY-6471, and RHAMCAT-PWY had protective effects against acne pathogens; the remaining bacteria and bacterial abundance pathways were associated with negative pathogenic effects. C_Gammaproteobacteria was found to be associated with the highest risk of acne (OR = 1.45, 95% CI 1.04–2.03, P = 0.028).
Fig. 4.
Forest plots showing the causal effects of gut microbes and bacterial metabolic pathways on acne. The results of OR and 95%CI in this table were obtained by the method of inverse–variance weighted. OR, odds ratio; CI, confidence interval.
The reverse MR analysis of gut microbes and acne did not reveal any evidence of reverse causality, as indicated by p values exceeding 0.05 for all gut microbes (pathways) via the IVW method. Two-sample MR analysis from gut microbes to acne revealed a total of 6 positive results for gut microbe pathways and 8 for gut bacteria. The specific findings are depicted in Figs. 2 and 3, and 4.
MR Analysis results of blood metabolites (bidirectional)
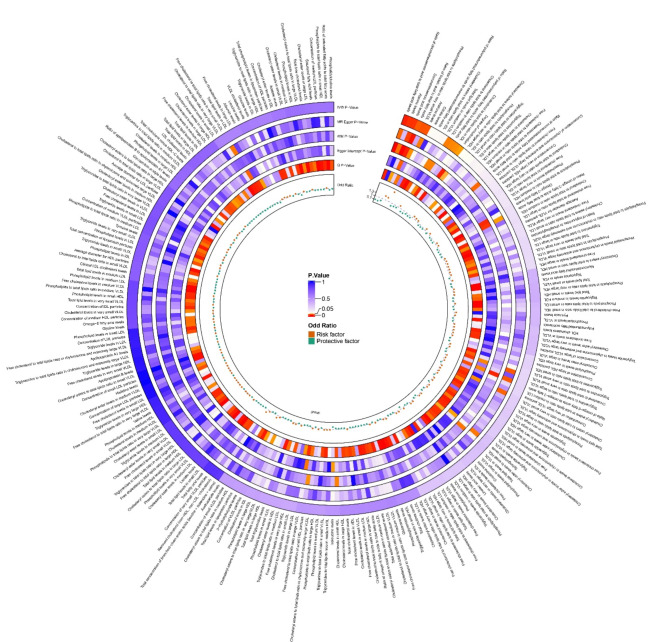
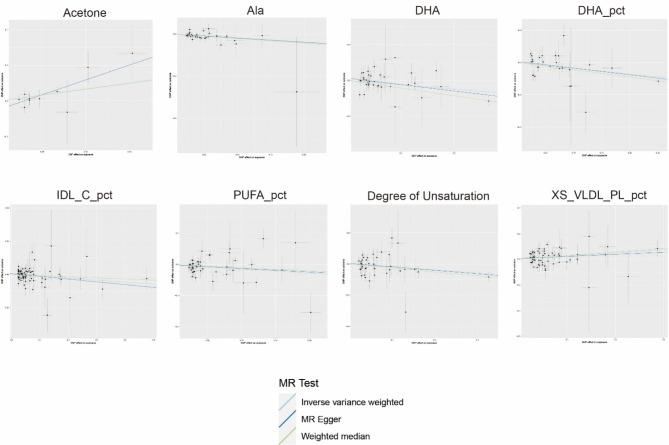
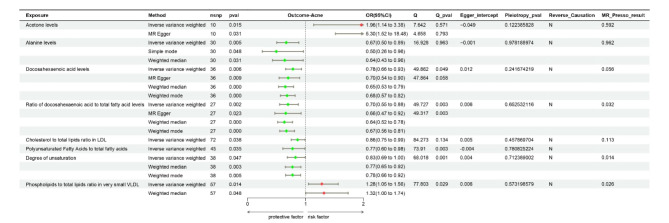
Through the application of two-sample MR analysis of blood metabolites and acne, the results of a total of 249 blood metabolites can be seen in the circo heatmap in Fig. 5. We discovered 10 blood metabolites that are significantly associated with acne, including acetone levels, alanine levels (Ala), docosahexaenoic acid levels (DHA), the ratio of docosahexaenoic acid to total fatty acid levels (DHA pct), the cholesterol to total lipid ratio in IDL (IDL C pct), the ratio of linoleic acid to total fatty acids (LA pct), the cholesterol to total lipid ratio in medium LDL (M LDL C pct), the ratio of polyunsaturated fatty acids to total fatty acids (PUFA pct), the degree of unsaturation, and the phospholipid to total lipid ratio in very small VLDL (XS VLDL PL pct). The M LDL C pct was excluded from the analysis because of its observed inverse causal relationship with acne in the reverse MR (P value = 0.048, 95% CI = 0.947–0.100, Beta = -0.027, OR = 0.973). LA pct sensitivity analysis revealed a P intercept value of less than 0.05, indicating horizontal pleiotropy, and as a result, it was rounded. Following sensitivity analysis calibration, both the DHA and PUFA pct values exhibited heterogeneity, which was deemed acceptable after MR-Presso correction, leading to their retention in the study. Consequently, a total of 8 blood metabolites were found to be significantly associated with acne. Acetone levels and the phospholipid-to-total lipid ratio in very small VLDL were identified as risk factors for acne development. Specifically, acetone levels had the most significant effect on the risk of acne development (OR = 5.3, 95% CI 1.52–18.48, P = 0.031 by MR Egger’s method). The specific findings are depicted in Figs. 5 and 6, and 7.
Fig. 5.
Circo Heatmap showing the causal effects of blood metabolites on acne. VLDL, very low density lipoprotein; LDL, low density lipoprotein; HDL, high density lipoprotein; IDL, intermediate density lipoprotein.
Fig. 6.
Scatterplots showing the causal effects of blood metabolites on acne. The Suffixes pct represent percentage. Ala, alanine; DHA, docosahexaenoic acid; IDL_C, intermediate-density lipoprotein cholesterol; PUFA, polyunsaturated fatty acid; XS_VLDL_PL, extra small very low density lipoprotein particle levels.
Fig. 7.
Forest plots showing the causal effects of blood metabolites and acne.
Bidirectional mediation analysis
To explore the possible mediated causal relationships among gut microbes, blood metabolites, and acne, we executed a mediated MR analysis focusing on the gut microbe–blood metabolite–acne pathway. We carried out a two-sample-mediated MR analysis, with gut microbes as the exposure and blood metabolites as the outcome, which yielded seven significant discoveries. These included GLCMANNANANAUT-PWY and DHA pct, RHAMCAT-PWY and M LDL C pct, g_Bifidobacterium and DHA, g_Bifidobacterium and degree of unsaturation, g_Bacteroides and LA pct, g_Bacteroides and PUFA pct, g_Faecalibacterium and LA pct. The LA pct was excluded from the two results containing LA pct because of pleiotropy in the two-sample MR with acne (p_intercept = 0.019). The inclusion of M_LDL_C_pct was deemed inappropriate because of the presence of reverse causality in the two-sample Mendelian randomization analysis with acne. Consequently, the result incorporating M_LDL_C_pct was excluded from the analysis. A total of four significant positive results were obtained, as shown in Table 1. No evidence of reverse causality was detected in the reverse two-sample MR analysis with blood metabolites as the exposure and gut microbes as the outcome. This served as the foundation for the mediated MR analysis.
Table 1.
Two-samples mendelian randomization analysis of exposure(gut microbes) and mediator(blood metabolites).
| Exposure | Outcome | Method | nSNP | Beta | OR | 95%CI | P value |
|---|---|---|---|---|---|---|---|
| GLCMANNANAUT-PWY | DHA pct | IVW | 6 | -0.052 | 0.949 | 0.911–0.989 | 0.012 |
| g_Bifidobacterium | DHA | IVW | 10 | 0.055 | 1.057 | 1.000-1.117 | 0.048 |
| g_Bifidobacterium | Degree of Unsaturation | IVW | 10 | 0.059 | 1.061 | 1.006–1.118 | 0.028 |
| g_Bacteroides | PUFA pct | IVW | 13 | -0.034 | 0.967 | 0.938–0.996 | 0.026 |
This study investigated the potential mediating effects of gut microbes (pathways) on acne through blood metabolites via two-step MR and multivariable MR. The two-step method identified a total of seven potential gut microbe-blood metabolite-acne mediating effects. However, GLCMANNANANAUT-PWY with DHA pct, RHAMCAT-PWY with M LDL C pct, g_Bacteroides with LA pct, and g_Faecalibacterium with LA pct were excluded because of discrepancies between the positive and negative directions of the beta effect value in the two-step method and the total effect value. The g_Bacteroides with PUFA pct was excluded because of the potential for a masking effect. Using the multivariable MR method, a total of four potential mediating relationships were identified, of which GLCMANNANANAUT-PWY and DHA pct and g_Bacteroides and PUFA pct were excluded because they produced mediating effect ratios of less than 5%. Consequently, this study successfully identified two significant gut microbes and metabolites that mediate the effects of acne through a two-step and multivariable MR approach. Specifically, these factors included Bifidobacterium-DHA (with a two-step MR mediating effect ratio of 5.53% and a multivariable MR mediating effect ratio of 8.42%) and Bifidobacterium-Degree of Unsaturation (with a two-step MR mediating effect ratio of 5.88% and a multivariable MR mediating effect ratio of 6.65%). The findings are presented in Fig. 8; Table 2.
Fig. 8.
Gut microbes mediate the mediating effect and mediating effect ratio of metabolites on acne(two-step method). The causal effect of Bifidobacterium on acne was found to be mediated by DHA and the Degree of Unsaturation. β(EM) and P(EM) represent the beta and P values obtained by MR analysis of exposure and mediator. β(MO) and P(MO) represent the beta and P values obtained by MR analysis of the mediator and outcome. β(EO) represents the beta and P values obtained by MR analysis of the exposure and outcome.
Table 2.
Gut microbes mediate the mediating effect and mediating effect ratio of metabolites on acne(multivariable MR).
| Exposure | Mediator | Outcome | β1 | β2 | β(EO) | α*(EM) | mediating effect | mediating effect ratio |
|---|---|---|---|---|---|---|---|---|
| g_Bifidobacterium | DHA | Acne | -0.195 | -0.317 | -0.208 | 0.055 | -0.018 | 8.42% |
| g_Bifidobacterium | Degree of Unsaturation | Acne | -0.211 | -0.235 | -0.208 | 0.059 | -0.014 | 6.65% |
Discussion
While acne typically resolves on its own, it is widespread among adolescents and can take several years to diminish. Additionally, acne may cause permanent scarring or post-inflammatory hyperpigmentation, leading to heavy physical and psychological burdens for patients22–24. Problems with current acne treatments, such as drug resistance, have been identified over the years25. Consequently, new ideas and methods for managing acne need to be explored, with a particular emphasis on early intervention. Although the concept of the gut‒skin axis has been postulated for a substantial length of time and has consistently been backed by various studies, the exact mechanism linking acne with gut microbes remains unclear. Building upon similarities with other autoimmune-related inflammatory skin conditions, such as psoriasis, rosacea, and atopic dermatitis4,26,27, and in accordance with previous research on the gut–skin axis, it seems reasonable to suggest that the link between acne development and gut microbes could originate from the influence that alterations in gut microbes exert on blood metabolites. These metabolites are implicated in the body’s inflammatory immune response10, potentially exerting either positive or negative effects on acne.
In this study, the most recent GWAS genetic data for gut microbes and acne were utilized to reduce the impact of acquired and external confounding factors. MR analysis was utilized to scrutinize the causal associations between gut microbes and blood metabolites in the context of acne. The results pointed to the existence of 14 and 8 metabolites within the gut microbial pathways that may conceivably play a causal role in the onset of acne. Two potential mediating causal relationships were identified through mediation MR analysis.
In two-sample MR studies examining the relationship between gut microbes and acne, it is important to consider that each outcome within the gut microbe abundance pathway comprises various components, including multiple gut bacteria, metabolites, and metabolic pathways. These components may be influenced by numerous confounding factors. Therefore, the discussion focus specifically on the impact of eight clearly defined gut bacteria, especially g_Bifidobacterium, on the pathogenesis of acne.
g_Bifidobacterium is recognized as a probiotic for humans and has been demonstrated in several studies to have the ability to inhibit pathogenic microorganisms both in vitro and in vivo28. Lee’s research indicated that Bifidobacterium not only curtailed the proliferation of Propionibacterium acnes but also impeded the progression of the inflammatory response29. Additionally, Rahmayani et al. investigated the variations in IL-10 levels in 33 acne patients who received oral administration of a probiotic blend containing Bifidobacterium for a period of 30 days. Notable differences were noted in the serum IL-10 levels before and after the oral consumption of a combination of probiotics. Research conducted by Fabio Rinaldi and colleagues revealed that the oral intake of a probiotic mixture comprising Bifidobacterium shortum, Lactobacillus casei, and plant extracts led to an increase in the levels of the anti-inflammatory cytokine IL-10 in patients. This increase has a beneficial impact on the treatment of acne30. Compared with those in the baseline period, notable decreases in oil secretion, flaking scores, mean porphyrin counts, and Propionibacterium acnes and Staphylococcus aureus levels were observed in patients who consumed dietary supplements31. Therefore, the oral intake of probiotics, including Bifidobacterium, might offer a potential therapeutic avenue for future acne prevention or treatment. G_Faecalibacterium and s_Pseudoflavonifractor capillosus are associated with the synthesis of short-chain fatty acids (SCFAs), crucial metabolic byproducts of gut microbes. A previous study revealed that g_Faecalibacterium, s_Pseudoflavonifractor capillosus, and the SCFAs produced by these bacteria have positive effects on immunomodulation, intestinal barrier function, and regulation of the intestinal microbiota32,33. While the current research lacks animal and clinical studies exploring its relationship with acne, this area could serve as a focal point for future investigations.
The presence of g_Bacteroides, s_Lactobacillus delbrueckii, and s_Escherichia has been pinpointed as potential risk factors potentially contributing to the emergence of acne. However, further research is needed to elucidate their potential association with acne.
For blood metabolites, we focus on two positive results: DHA and the degree of unsaturation. The degree of unsaturation cannot be explicitly used to characterize which substances. Therefore, it is not discussed here.
DHA, a member of the Omega-3 family of polyunsaturated fatty acids, has been demonstrated to reduce the levels of IGF-1. This particular hormone has been implicated in sebum production and the obstruction of hair follicles34. Omega-3 FAs also inhibit the synthesis of inflammatory leukotriene B4, thereby reducing the occurrence of inflammatory acne lesions35. A study carried out in Shanghai reported lower concentrations of Omega-3 fatty acids (FAs) in the erythrocytes of male patients with acne than in those of a control group. Specifically, the study highlighted a marked reduction in docosahexaenoic acid (DHA) levels36. FA levels in erythrocytes are indicative of the body’s long-term dietary intake37. Consequently, the reduced concentration of DHA in the red blood cells of individuals with acne may be linked to their dietary habits. Clinical research has indicated that enhancing a diet with omega-3 fatty acids, specifically those rich in DHA, leads to notable improvements in both the severity and appearance of acne-related skin lesions and symptoms38. In line with the results of our current research, it can be inferred that DHA acts as a protective factor in mitigating the onset of acne.
During the mediation MR analyses, two potential gut microbe-metabolite-acne mediation relationships were identified: Bifidobacterium-DHA-Acne and Bifidobacterium-Degree of Unsaturation-Acne. Interactions between gut microbes and blood metabolites may have an impact on the host’s metabolism39. The incorporation of feed that contains Bifidobacterium into the diets of mice led to a notable increase in the levels of fatty acids, including DHA, within the liver and brain tissues40. Concurrently, a decrease in the levels of proinflammatory cytokines, namely, tumor necrosis factor-alpha (TNF-α) and interferon-gamma (IFN-γ), has been observed41. In vitro intervention studies have demonstrated that DHA suppresses the growth of Escherichia and enhances the growth of Glomerobacterium, thereby facilitating metabolic benefits. It is also plausible that DHA could decrease endotoxin and inflammation levels by interfering with gut microbes, thereby increasing the concentration of propionic acid in SCFAs, which are key metabolic products synthesized by gut microbes42. SCFAs engage with G protein-coupled receptor 43 (GPR43), facilitating a decrease in certain inflammatory responses in vivo. Moreover, the interplay between SCFAs and GPR43 has a substantial effect on modulating inflammatory processes43. In our present study, no potential reverse causality was found between DHA and Bifidobacterium. Consequently, further comprehensive studies are necessary to explore the underlying mechanisms and associations involved. It is plausible that Bifidobacterium, which functions as a probiotic, could modulate the levels of metabolites, such as SCFAs, through its involvement in the breakdown of dietary fibers. This, in turn, may impact blood metabolites such as DHA, potentially exerting an anti-inflammatory influence and contributing to the amelioration of acne. This finding elucidates the reason for the milder manifestation of acne symptoms in individuals who consume a greater quantity of fresh fruits and vegetables than in those who follow a high-fat diet. This phenomenon can be attributed to the increased intake of fresh fruits and vegetables, which are rich in dietary fiber. This leads to elevated levels of beneficial SCFAs produced by gut microbes44,45, subsequently influencing downstream metabolites such as DHA. The latter is known for its protective effects against acne. High-fat diets have been linked to a decrease in the diversity of the gut microbiota and an increase in the levels of endotoxins. Such dietary patterns can compromise the integrity and barrier function of the intestinal epithelium, lead to thinning of the mucosal layer in the gut, and increase the concentrations of proinflammatory cytokines46. Hyperglycemia and hyperinsulinemia resulting from a high-sugar diet induce an increase in IGF-1 and a decrease in IGFBP-3. This process may stimulate excessive proliferation of keratin-producing cells and increase androgen levels47, which in turn can lead to the overactivity of sebaceous glands and the subsequent onset of acne. The degree of unsaturation in the other mediator results is not thoroughly discussed due to the lack of clarity regarding the unsaturated substances. We hypothesized that it may be unsaturated fatty acids, etc. We expect that further comprehensive studies can elucidate this phenomenon.
The present study has several limitations. The results included a portion of the gut microbe and blood metabolite data, which were specifically related to gut microbe metabolic pathways and metabolite ratios. These findings are subject to potential confounding factors, which complicates the full elucidation of their association with acne. The GWAS dataset employed in this research comprised exclusively samples from European ancestry, and sample data from ethnic populations in other regions, such as Asia, were lacking. The presumed causal relationships among gut microbes, blood metabolites, and acne could be confounded by factors such as race, dietary patterns, and comorbidities. Moreover, there is a dearth of validation in meticulously planned, extensive-sample, multicenter clinical randomized controlled trials, and the majority of the existing experimental studies have been conducted in animals or in vitro intervention trials. Further clinical human trials are imperative in the future to substantiate causality and explore potential novel therapies for probiotic or metabolite interventions in acne.
Conclusion
In conclusion, our two-sample MR analysis identified six gut microbe pathways, eight gut microbes, and eight blood metabolites that are causally associated with acne. This study also revealed two putative causal links between gut microbes, blood metabolites, and acne. These identified gut microbes and blood metabolites could serve as innovative targets or biomarkers for the prophylaxis, detection, and management of acne, presenting promising avenues for future investigations into the etiology of this skin condition.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We express our gratitude to the Dutch Microbiome Project, UK Biobank, and FinnGen databases for making their GWAS data publicly and freely accessible. We are also grateful to all the subjects who kindly provided samples, and to the associated researchers for their prior efforts and commitment.
Author contributions
XM, HX, ZZy, and JHy designed the study. ZZy, JHy, and GQr analysed dates. LH, WKb collected dates. All authors contributed to the drafting and revision of the manuscript. QYs made critical revisions to this manuscript. XM, as the study’s guarantor, had complete access to all the data involved in the research and assumes responsibility for the integrity of the data and the precision of the data analysis. All authors have reviewed and consented to the final version of the manuscript for publication.
Funding
This study was funded by the State Administration of Traditional Chinese Medicine through the Second National Famous Traditional Chinese Medicine Doctor’s (TCM) Workshop Development Project (Grant No. Guozhongyongyong Renjiaofa [2022] No. 5).
Data availability
The GWAS dataset employed in this research was sourced from publicly accessible GWAS repositories on the internet, including the UK Biobank (:搜索 --- : Search (ox.ac.uk)) and FinnGen GWAS results (https://r8.finngen.fi/). The study titled “FinnGen: Unique genetic insights from combining isolated population and national health register data” provides further details (doi: 10.1101/2022.03.03.22271360).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuesi Qin, Email: qinyuesi@126.com.
Min Xiao, Email: xiaomin@cdutcm.edu.cn.
References
- 1.Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol.19 (1), 55–71 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Hou, K. et al. Microbiota in health and diseases. Signal. Transduct. Target. Therapy. 7 (1), 135 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee, H. J. & Kim, M. Skin barrier function and the Microbiome. Int. J. Mol. Sci. ;23(21). (2022). [DOI] [PMC free article] [PubMed]
- 4.Mahmud, M. R. et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. 14 (1), 2096995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill, C. A., Monteleone, G., McLaughlin, J. T. & Paus, R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. BioEssays: News Reviews Mol. Cell. Dev. Biology. 38 (11), 1167–1176 (2016). [DOI] [PubMed] [Google Scholar]
- 6.VosT et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the global burden of Disease Study 2010. Lancet (London England). 380 (9859), 2163–2196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams, H. C., Dellavalle, R. P. & Garner, S. Acne Vulgaris. Lancet (London England). 379 (9813), 361–372 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Dall’Oglio, F., Nasca, M. R., Fiorentini, F. & Micali, G. Diet and acne: review of the evidence from 2009 to 2020. Int. J. Dermatol.60 (6), 672–685 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Baldwin, H. & Tan, J. Effects of Diet on Acne and its response to treatment. Am. J. Clin. Dermatol.22 (1), 55–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salem, I., Ramser, A., Isham, N. & Ghannoum, M. A. The gut Microbiome as a Major Regulator of the gut-skin Axis. Front. Microbiol.9, 1459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature. 604 (7907), 732–739 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Lopera-Maya, E. A. et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat. Genet.54 (2), 143–151 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Richardson, T. G. et al. Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol.20 (2), e3001547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Q. et al. Dissecting Causal relationships between Gut Microbiota, Blood metabolites, and stroke: a mendelian randomization study. J. Stroke. 25 (3), 350–360 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan, H. et al. Systematic mendelian randomization study of the effect of gut microbiome and plasma metabolome on severe COVID-19. Front. Immunol.14, 1211612 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanna, S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet.51 (4), 600–605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess, S. & Thompson, S. G. Avoiding bias from weak instruments in mendelian randomization studies. Int. J. Epidemiol.40 (3), 755–764 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Chen, L. et al. Systematic mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat. Commun.13 (1), 6143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter, A. R. et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur. J. Epidemiol.36 (5), 465–478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess, S. & Thompson, S. G. Interpreting findings from mendelian randomization using the MR-Egger method. Eur. J. Epidemiol.32 (5), 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife ;7. (2018). [DOI] [PMC free article] [PubMed]
- 22.Bhate, K. & Williams, H. J. T. B. Epidemiol. acne Vulgaris ;168(3):474–485. (2013). [DOI] [PubMed] [Google Scholar]
- 23.Thiboutot, D. et al. Practical management of acne for clinicians: An international consensus from the Global Alliance to Improve Outcomes in Acne. ;78:S1-S23.e1. (2018). [DOI] [PubMed]
- 24.Webster, G. F. The pathophysiology of acne. Cutis. 76 (2 Suppl), 4–7 (2005). [PubMed] [Google Scholar]
- 25.Moon, S. H. et al. Antibiotic resistance of microbial strains isolated from Korean acne patients. J. Dermatol.39 (10), 833–837 (2012). [DOI] [PubMed] [Google Scholar]
- 26.De Pessemier, B. et al. Gut-skin Axis: current knowledge of the interrelationship between Microbial Dysbiosis and skin conditions. Microorganisms ;9(2). (2021). [DOI] [PMC free article] [PubMed]
- 27.Navarro-López, V. et al. Probiotics in the therapeutic Arsenal of dermatologists. Microorganisms ;9(7). (2021). [DOI] [PMC free article] [PubMed]
- 28.Servin, A. L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev.28 (4), 405–440 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Lee, D. K. et al. In vitro evaluation of antibacterial activities and anti-inflammatory effects of Bifidobacterium spp. addressing acne vulgaris. Arch. Pharm. Res.35 (6), 1065–1071 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Rahmayani, T., Putra, I. B. & Jusuf, N. K. The effect of oral probiotic on the Interleukin-10 serum levels of Acne Vulgaris. Open. Access. Macedonian J. Med. Sci.7 (19), 3249–3252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinaldi, F. et al. Facial acne: a Randomized, Double-Blind, placebo-controlled study on the clinical efficacy of a Symbiotic Dietary supplement. Dermatology Therapy. 12 (2), 577–589 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barcenilla, A. et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol.66 (4), 1654–1661 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlier, J. P. et al. Proposal to unify Clostridium orbiscindens Winter 1991 and Eubacterium plautii (Séguin. Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. International journal of systematic and evolutionary microbiology. 2010;60(Pt 3):585 – 90. (1928). [DOI] [PubMed]
- 34.Logan, A. C. Omega-3 fatty acids and acne. Arch. Dermatol.139 (7), 941–942 (2003). author reply 2–3. [DOI] [PubMed] [Google Scholar]
- 35.Simopoulos, A. P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Experimental biology and medicine. (Maywood NJ). 233 (6), 674–688 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Cao, K. et al. Fatty acid profiling in Facial Sebum and erythrocytes from adult patients with moderate acne. Front. Physiol.13, 921866 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arab, L. Biomarkers of fat and fatty acid intake. J. Nutr.133 (3), 925s–32s (2003). [DOI] [PubMed] [Google Scholar]
- 38.Jung, J. Y. et al. Effect of dietary supplementation with omega-3 fatty acid and gamma-linolenic acid on acne vulgaris: a randomised, double-blind, controlled trial. Acta dermato-venereologica. 94 (5), 521–525 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Canfora, E. E., Meex, R. C. R., Venema, K. & Blaak, E. E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Reviews Endocrinol.15 (5), 261–273 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Wall, R. et al. Impact of administered bifidobacterium on murine host fatty acid composition. Lipids. 45 (5), 429–436 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Wall, R. et al. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am. J. Clin. Nutr.89 (5), 1393–1401 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Zhuang, P. et al. Eicosapentaenoic and docosahexaenoic acids attenuate hyperglycemia through the microbiome-gut-organs axis in db/db mice. Microbiome. 9 (1), 185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 461 (7268), 1282–1286 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macfarlane, S. & Macfarlane, G. T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc.62 (1), 67–72 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Simpson, H. L. & Campbell, B. J. Review article: dietary fibre-microbiota interactions. Aliment. Pharmacol. Ther.42 (2), 158–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreno, B. et al. Understanding innate immunity and inflammation in acne: implications for management. J. Eur. Acad. Dermatology Venereology: JEADV. 29 (Suppl 4), 3–11 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Matsui, M. S. Update on diet and acne. Cutis. 104 (1), 11–13 (2019). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS dataset employed in this research was sourced from publicly accessible GWAS repositories on the internet, including the UK Biobank (:搜索 --- : Search (ox.ac.uk)) and FinnGen GWAS results (https://r8.finngen.fi/). The study titled “FinnGen: Unique genetic insights from combining isolated population and national health register data” provides further details (doi: 10.1101/2022.03.03.22271360).