Abstract
We identified two homozygous truncating variants in GON4L [NM_001282860.2:c.62_63del, p.(Gln21Argfs*12) and c.5517+1G>A] in two unrelated families who presented prenatal-onset growth impairment, microcephaly, characteristic face, situs inversus, and developmental delay. The frameshift variant is predicted to invoke nonsense-mediated mRNA decay of all five known GON4L isoforms resulting in the complete loss of GON4L function. The splice site variant located at a region specific to the longer isoforms; therefore, defects of long GON4L isoforms may explain the phenotypes observed in the three patients. Knockdown of Gon4l in rat PC12 cells suppressed neurite outgrowth in vitro. gon4lb knockdown and knockout zebrafish successfully recapitulated the patients’ phenotypes including craniofacial abnormalities. We also observed situs inversus in gon4lb-knockout zebrafish embryo. To our knowledge, the relationship between craniofacial abnormalities or situs inversus and gon4lb has not been reported before. Thus, our data provide evidence that GON4L is involved in craniofacial and left-right patterning during development.
Subject terms: Neurodevelopmental disorders, Neurological disorders
Introduction
GON4L (MIM*610393) at 1q22 encodes GON4-like protein (GON4L), which localizes in the nucleus and is a putative transcriptional regulator associated with cell division, proliferation and differentiation1–10. At least five GON4L isoforms are registered in RefSeq. Three long isoforms encode three functional domains: paired amphipathic helix 1 (PAH1), PAH2, and Myb-like domains. The PAH domain mediates protein–protein interaction11,12, and the Myb-like domain is similar to the SANT [switching-defective protein 3 (Swi3), adaptor 2 (Ada2), nuclear receptor co-repressor (N-CoR), transcription factor (TF)IIIB] domain, which is related to chromatin-remodeling and chromatin accessibility13–15. In addition, GON4L forms complexes with the transcriptional regulators, YY1, SIN3A, and HDAC116.
Ubiquitously expression of GON4L in adult human tissues was shown by Northern blotting and RT-PCR2. GON4L is evolutionarily highly conserved and is involved in several developmental pathways in multiple animal models. The gon-4 gene in Caenorhabditis elegans is involved in gonadogenesis17. In Drosophila, the Gon4l homolog, mute, is a component of the histone locus body, potentially regulating muscle cell differentiation by controlling histone expression6. In zebrafish, the GON4L homolog, gon4lb, plays critical roles in primitive hematopoiesis, notochord boundary formation, tail formation, and anteroposterior axis establishment during gastrulation, and homozygous zebrafish mutants (udu/gon4lbtu24, udu/gon4lbvu66, and udu/gon4lsq1 arising from nonsense variants) showed short body axis, bent-down tail, small head and eyes, lack of blood circulation, and increased apoptosis1,3,4,8. In mice, Gon4l is involved in heart and hematopoietic cell development5,9,18 and homozygous Gon4l null mice are embryonic lethal18. Furthermore, in Fleckvieh cattle, a homozygous frameshift variant of GON4L (ENSBTAT00000027126:c.4287delC) is associated with low birth weight, small size, and craniofacial abnormalities (brachygnathia inferior, elongated narrow head and structural deformities of the muzzle), spinal distortions, excessive skin, and a disproportionately large head19.
In humans, GON4L has not been established as a disease-related gene. Some monoallelic GON4L variants (missense, synonymous, and canonical splice site variants) have been reported in individuals with autism spectrum disorder20–24, developmental disorder20, bipolar disorder25, and hydrocephalus26. In addition, one family with a homozygous GON4L splice site variant, NM_001282860.2: c.5517+1G>A, show syndromic intellectual disability, but the clinical details were not well described27. Therefore, to our knowledge, human GON4L-related disorders have not been fully described.
Results
Clinical features
Patient 1 was the first child of a consanguineous couple (first cousins) in Family 1 (Figs. 1a and 2a–d). There was no relevant family history. He was born at term after an uneventful pregnancy with a birth weight of 2000 g (−2.8 SD). Small head circumference (not recorded) and left microphthalmia were noted at birth. Ultrasound of the eyes showed normal right eye axial length (22 mm) while the axial length of the left eye was 15 mm. In addition, opaque lens, vitreous floaters, and membranes extending to the retina showing vitreoretinal adhesion and thickened choroid were documented. Further ophthalmological examination showed bilateral corneal opacity not affecting the vision on the right side, but the left side showed anterior segment dysgenesis. Visual evoked potential and electroretinography at the age of 1 year showed a normal functioning right eye. Echocardiography showed a small ventricular septal defect, an atrial septal defect, and tricuspid regurgitation. Abdominal ultrasound and electroencephalography showed normal results. A TORCH examination was negative.
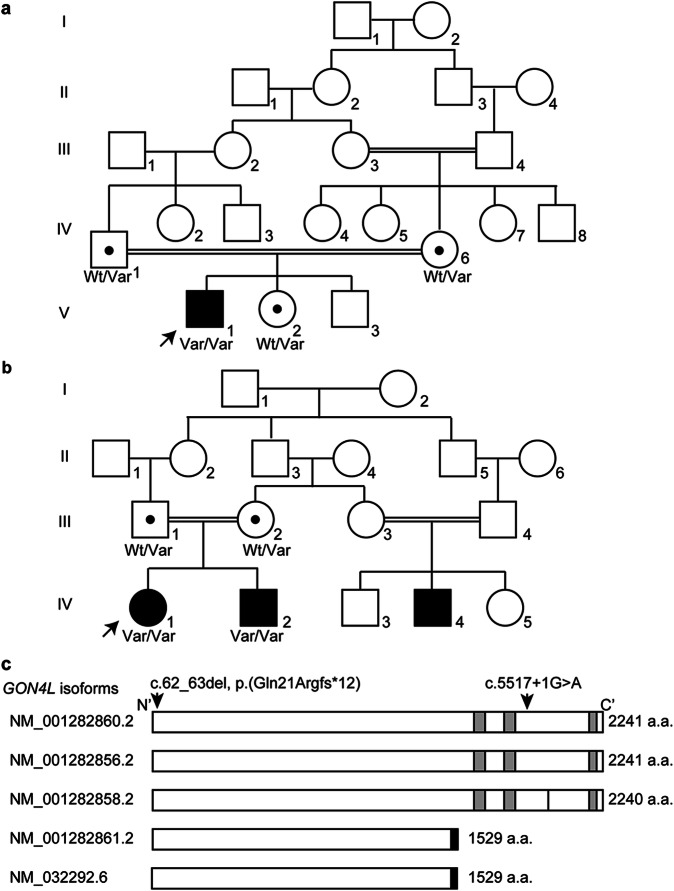
Fig. 1. Genetic analysis of two families with homozygous GON4L variants.
Pedigree of Family 1 (a) and Family 2 (b). Wt wild-type, Var variant allele. Black dots indicate a GON4L variant carrier. Arrows indicate probands. c Human GON4L isoforms with the previously reported canonical splice variant (c.5517+1G>A) which is also identified in Family 2 and the frameshift variant identified in Family 1 [c.62_63del, p.(Gln21Argfs12*)] in NM_001282860.2 are indicated. The black bar in NM_001282858.2 indicates the lack of p.Ala1958. The short isoforms (NM_001282861.2 and NM_032292.6) have the same amino acid sequence from the first methionine to p.Gln1490 as the long isoforms, and the black boxes indicate the unique regions of the short isoforms. The gray boxes indicate paired amphipathic helix 1 (PAH1, 1624–1696 amino acids), PAH2, (1706–1777 amino acids), and Myb-like (2148–2201 amino acids) domains from N- to C-terminus, respectively, based on UniProtKB (Q3T8J9).
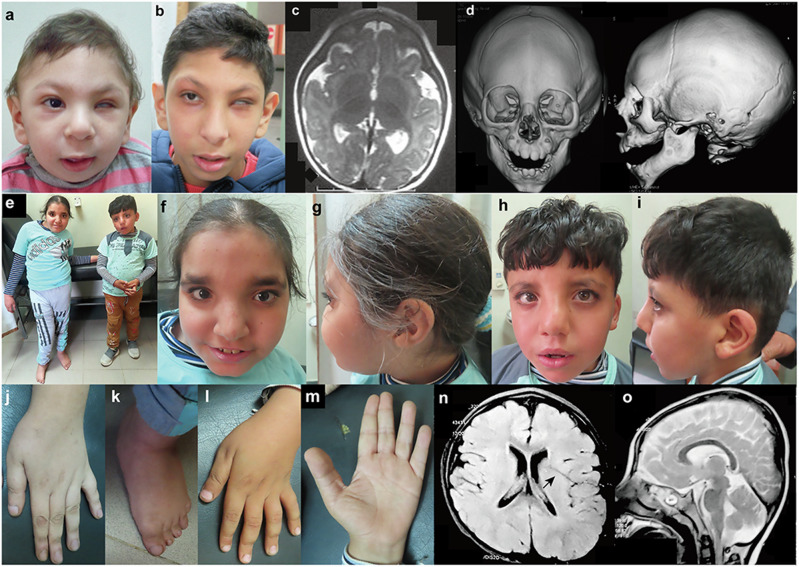
Fig. 2. Clinical features of the patients homozygous for GON4L variants.
Photographs of the face of Patient 1 at the age of 10 months (a) and 9 years and 6 months (b). He showed left microphthalmia. c Brain MRI of Patient 1 at the age of 5 months showed simplified gyral pattern. d Three-dimensional cranial CT images of Patient 1 at 10 months indicated metopic craniosynostosis. e Full picture of Patient 2 (left) and 3 (right). f, g Front and side of the face of Patient 2 at the age of 13 years old. She showed asymmetric face, a high forehead, thick eyebrows, upward slanting palpebral fissures, strabismus, broad nasal root, a broad nose with a beaked tip, deviated nasal septum, a prominent left cheek, short philtrum, thin upper lip, everted lower vermillion, broad chin, and low-set ears with folded helix. h, i Front and side of the face of Patient 3 at the age of 8 years. He shows dysmorphic features similar to Patient 2. Long fingers (j) and deviated feet with pes planus (k) of Patient 2. l, m Dorsum and palma of the hand with incomplete single transverse crease of Patient 3. Brain MRI of Patient 3 at the age of 6 [Axial T1- (n) and Sagittal T2-weighted (o)]. Deep Sylvian fissure is indicated by an arrow.
At the age of 10 months, his weight was 7 kg (−2 SD), length was 68 cm (−2 SD), and head circumference was 35 cm (−6.6 SD). His early developmental milestones showed mild delay (sitting at 9 months, standing with support at 14 months, and walking independently at 20 months). He had a history of repeated infection in the first years of life. At the age of 2 years, his parents noticed delayed speech, although a hearing test and auditory brain response were normal. Three-word sentences were not achieved until the age of 6 years, although the words were not pronounced clearly. He had a monotonous speech pattern (with a nasal tone and similar to that in some individuals with palatal anomalies). At 7 years of age, his weight was 18 kg (−1.6 SD), height was 120 cm (mean), and head circumference was 41 cm (−8.3 SD). At this age, the visual acuity of his right eye was normal. His muscle tone and reflexes were normal. No history of seizures or neurological regression was recognized. He showed hyperactivity with impulsivity but he had a very pleasant and friendly personality and was very cooperative with people. His intelligence quotient (IQ) was 65 at the age of 7 years and 55 at the age of 12 years, according to the Wechsler Preschool and Primary Scale of Intelligence. He was enrolled in regular school but performed poorly in reading, writing, and arithmetic tasks. He could read and write only a few words and counted with difficulty. At the age of 5 months, a brain MRI showed a simplified gyral pattern (Fig. 2c), hypogenesis of the corpus callosum, and a slightly small-sized pons. A cranial three-dimensional CT scan at the age of 10 months showed metopic craniosynostosis (Fig. 2d). Chromosomal G-banding showed a normal male karyotype (46,XY).
In Family 2, two affected siblings were born to first cousin healthy parents from Upper Egypt (IV-1 and IV-2 in Figs. 1b and 2e–o). History of a similarly affected male cousin with developmental delay and situs inversus totalis was recorded in the family pedigree, but we were not able to examine him (IV-4 in Fig. 1b). Patient 2 (IV-1 in Fig. 1b), a proband of family 2, is a 13-year-old female who was born after uneventful vaginal delivery. Her birth weight was 2.3 kg (−2.1 SD), length 45 cm (−2 SD), and head circumference 33 cm (−1.2 SD). Her Apgar score was 8/10 (1 min/5 min) and she did not require special neonatal care. Bilateral talipes equinovarus was noted. Several plaster casts were applied to the feet during the first year of life for talipes correction, which produced gradual improvement. At 4 months of age, she experienced febrile seizures (atonic and generalized tonic-clonic seizures) and a severe respiratory infection, and was admitted to an intensive care unit for 4 days. After this episode, the febrile seizures did not recur. Her developmental milestones were delayed: sitting at 18 months, standing without support at 2 years, and walking independently and speaking a meaningful word at 3 years. At the age of 7 years, she was continent. On examination, she was cooperative, understood and obeyed orders, reacted with her surroundings, talked in short sentences with dysarthric speech, and had an abnormal gait. At the age of 13 years her weight was 39 kg (−1 SD), height 117 cm (−5.7 SD), and head circumference 51 cm (−2 SD). Scoliosis was noted and the left lower limb was 5 cm longer than the right one. She had specific asymmetric facial features; a long face, a high forehead, thick eyebrows, upward slanting palpebral fissures, strabismus, broad nasal root, a broad nose with a beaked tip, deviated nasal septum, a prominent left cheek, short philtrum, thin upper lip, everted lower vermillion, broad chin, and low-set ears with folded helix (Fig. 2f, g). The extremities showed mild hyperextensibility of interphalangeal joints, long fingers (Fig. 2j), bilateral single transverse and increased palmar creases, and deviated feet with pes planus (Fig. 2k). In addition, echocardiography, abdominal ultrasonography, and chest and abdominal CT documented situs inversus totalis with normal heart structure. She showed hypotonia, normal reflexes, and good coordination. Her IQ was 62 on the Wechsler Intelligence Scale for Children at the age of 13 years. Brain MRI showed mild dilated lateral ventricles, prominent cortical sulci, a minimal high signal of white matter around the occipital horn, thin corpus callosum, and a relatively small vermis. Spine X-rays and chest CT documented scoliosis with Cobb’s angle of 35 degrees (moderate). Pubertal assessment showed breast B2, pubic hair P2, axillary hair A1 by Tanner stage, and at 13 years of age her menarche had not occurred.
Patient 3 (IV-2 in Fig. 1b) is the younger brother of Patient 2, and is now 8 years old. He was delivered by Caesarian section with no specific perinatal events. At birth, his weight was 2.5 kg (−1.7 SD), length 46 cm (−1.5 SD), and head circumference 33 cm (−1.2 SD). He also showed developmental delay, but with better achievement than his sister; sitting at 12 months, standing without support at 18 months, and walking independently and speaking a meaningful word at 2 years of age. He was continent at 5 years of age with infrequent nocturnal enuresis. On examination at 8 years, he was cooperative, understood orders, talked in short sentences with some unclear words, and had a normal gait. His weight was 26.8 kg (−0.5 SD), height 102 cm (−5 SD), and head circumference 49 cm (−2.7 SD). His dysmorphic features were similar to those of his sister (Fig. 2h, i). He also presented dextrocardia and situs inversus totalis with normal heart structure, and calcific hepatic foci with mild hepatomegaly. He showed mild hyperextensibility of interphalangeal joints, incomplete single transverse palmar crease on both hands (Fig. 2l, m), pes planus, and bilateral descended testicles. At the age of 8 years, neurological assessment showed hypotonia and normal reflexes and his IQ was 65 on the Wechsler Intelligence Scale for Children. No history of seizures was recorded. Brain MRI at the age of 6 years showed mild cortical simplified gyration, deep Sylvian fissure, thin corpus callosum, and small vermis (Fig. 2n, o). Blood examination including liver enzymes, metabolic screening, organic acid profile, electroencephalography, fundus, auditory brainstem response, and karyotype were normal in both Family 2 patients.
Genetic analysis
To identify the genetic cause of the patients’ conditions, we performed whole exome sequencing. For Patient 1, we did not identify any strong candidate variants in genes known to be associated with human diseases. However, we identified a homozygous variant in GON4L [NM_001282860.2: c.62_63del, p.(Gln21Argfs*12), dbSNP ID: rs755827429], which was confirmed by Sanger sequencing (Supplementary Fig. 1). His parents and sibling were heterozygous for this variant, but presented no obvious phenotype (Fig. 1a). At least five GON4L isoforms are archived in the RefSeq database. Two long isoforms are 2241 amino acids (NM_001282860.2 and NM_001282856.2) and a third long isoform lacks p. Ala1958 of the other two long forms and is 2240 amino acids (NM001282858.2). Two short isoforms have the same amino acid sequence from the first methionine to p.Gln1490 as the long isoforms and have unique C-terminal regions of 39 amino acids (Fig. 1c and Supplementary Fig. 2). The frameshift variant, c.62_63del, p.(Gln21Argfs*12), identified in Patient 1 is located in a region common to all five isoforms (in exon 2 out of 32 coding exons in NM_001282860.2) (Fig. 1c). All isoforms with this variant are predicted to be subject to nonsense-mediated mRNA decay, which would result in complete loss of GON4L function. In addition, this variant was rare in gnomAD, with a minor allele frequency (MAF) = 0.00004377, and no record of homozygosity. We also identified a homozygous GON4L canonical splice site variant (c.5517+1G>A) in two affected individuals in an unrelated pedigree (Family 2, Fig. 1b). Sanger sequencing confirmed the familial segregation of the variant (Supplementary Fig. 3). This variant is located in a region specific for longer isoforms (NM_001282860.2, NM_001282856.2, NM_012812858.2) (Fig. 1c), and c.5517 is located in exon 27 of 32 coding exons (NM_001282860.2). If this variant causes a frameshift, the mRNA would be subject to nonsense-mediated mRNA decay resulting in no protein production. The clinical similarities among these three patients (Table 1 and Supplementary Table 1) indicate that the longer isoforms are important in human development. Both variants were located within the absence of heterozygosity regions: Chr1: 154,112,167–155,934,683 (1.8 Mb) in Family 1 and Chr1: 52,281,228–171,076,768 (18.8 Mb) in Family 2.
Table 1.
Comparison of the clinical characteristics of diseases caused by pathogenic variants in GON4L, YY1, and SIN3A
| Gene | GON4L | YY1 | SIN3A | |||
|---|---|---|---|---|---|---|
| Patient ID or disease name | Patient 1 | Patient 2 | Patient 3 | Sum | Gabriele-de Vries syndrome | Witteveen–Kolk syndrome |
| MIM phenotype # | N.A. | 617557 | 613406 | |||
| Inheritance mode | AR | AD | AD | |||
| Sex | Male | Female | Male | |||
| Intrauterine growth retardation | + | + | + | 3/3 | + | + |
| Short stature | + | + | + | 3/3 | + | + |
| Microcephaly | + | + | + | 3/3 | − | + |
| Facial asymmetrya | − | + | + | 2/3 | + | + |
| Broad/high forehead | + | + | + | 3/3 | + | + |
| Long face | + | + | + | 3/3 | − | + |
| Downslanted palpebral fissures | + | + | + | 3/3 | + | + |
| Strabismus | N.A. | + | + | 2/2 | + | + |
| Large ears | + | − | − | 1/3 | − | + |
| Thick lower lip | + | − | − | 1/3 | + | + |
| Pointed chin | + | − | + | 2/3 | + | + |
| Cardiac abnormalities | + | + | + | 3/3 | + | + |
| Cryptorchidism | − | N.A. | − | 0/2 | + | + |
| Extremity abnormalities | − | + | − | 1/3 | + | + |
| Developmental delay | + | + | + | 3/3 | + | + |
| Intellectual disability | + | + | + | 3/3 | + | + |
| Speech delay | + | + | + | 3/3 | + | + |
| Autistic features | + | − | − | 1/3 | + | + |
| Feeding problems | − | − | − | 0/3 | + | + |
| Brain structure abnormalities | + | + | + | 3/3 | + | + |
| Cortical abnormalities | + | + | + | 3/3 | + | + |
| White matter abnormalities | + | + | + | 3/3 | + | + |
| Hypoplastic/thin corpus callosum | + | + | + | 3/3 | + | + |
Patient 1. The left eye shows microphthalmia and only the right eye has normal vision. Therefore, the presence of strabismus cannot be determined. Patient 2. She is female.
AD autosomal dominant inheritance, AR autosomal recessive inheritance. N.A. not applicable.
aExcept for eyes.
Human GON4L expression
To further explore GON4L expression in humans, we analyzed multiple human tissues at fetal and adult stages using TaqMan assays with two probes common for short and long isoforms, and found GON4L to be ubiquitously expressed, including in the fetal brain (Supplementary Fig. 4a, b). Additionally, the long isoforms specific GON4L expressed ubiquitously including the fetal brain (Supplementary Fig. 4c), supporting that the longer isoforms are crucial in human brain development.
Neurite outgrowth in PC12 cells
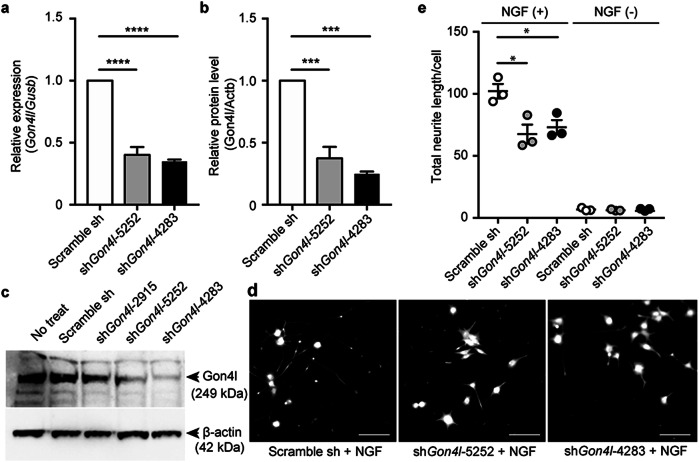
To analyze the impact of GON4L on nerve development, we generated Gon4l-knockdown PC12 cells. PC12 cells treated with nerve growth factor (NGF) have been widely used as a neuronal differentiation model28,29. We designed three short hairpin RNA (shRNA) sequences for Gon4l knockdown: shGon4l_2915/5252/4283, two of which, shGon4l_5252 and shGon4l_4283, lowered Gon4l mRNA levels and GON4L protein levels efficiently (Fig. 3a–c). Gon4l knockdown did not influence PC12 cell growth (Supplementary Fig. 5). To analyze the effects of Gon4l knockdown on neural development, neurite length after NGF treatment was measured. Neurites of PC12 cells were elongated by NGF treatment, whereas neurite lengths of Gon4l-knockdown cells were shorter than those of control cells (Fig. 3d, e).
Fig. 3. Gon4l knockdown in PC12 cells and neurite outgrowth.
Knockdown efficiencies of Gon4l mRNA (a) and GON4L protein (b, c) levels in PC12 cells. mRNA levels were normalized against Gusb, and protein levels were normalized against ACTB. Data are shown as the mean ± standard error of the mean (SEM) from three independent experiments. d, e Neurite outgrowth assay in Gon4l-knockdown PC12 cells in response to NGF treatment. Representative images (d) and total neurite length in one cell (e) are shown. Dots indicate means of each independent experiment (n = 3), and the bars represent means and SEM of three independent experiments. *p ≤ 0.05, ***p ≤ 0.001, ****p ≤ 0.0001 using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test.
Zebrafish gon4lb-knockdown phenotype and rescue by human GON4L mRNA
We next examined the morphological effects of GON4L knockdown in zebrafish. In zebrafish, two orthologous genes of GON4L are known: gon4la (XM_003200603.5, XP_003200651.2) on chromosome 19 and gon4lb (NM_001201535.1, NP_001188464.1) on chromosome 16 (in the NCBI database). Neither encoded protein shows high homology with human GON4L, but only gon4lb is “validated” in RefSeq (the status of gonl4a is only “model”) at present (accessed March 5, 2024) and shows a more similar protein sequence alignment using Clustal W (https://www.genome.jp/tools-bin/clustalw) (Supplementary Fig. 6). In addition, zebrafish phenotypes have been well studied in gon4lb previously1,3,4,8, so we chose gon4lb as a GON4L ortholog. We generated gon4lb-knockdown zebrafish using an MO, and reconfirmed the decreased eye size, head size, and body length at 50 hpf, and increased apoptosis in the central nervous system as previously reported1,3,4,8 (Supplementary Figs. 7 and 8).
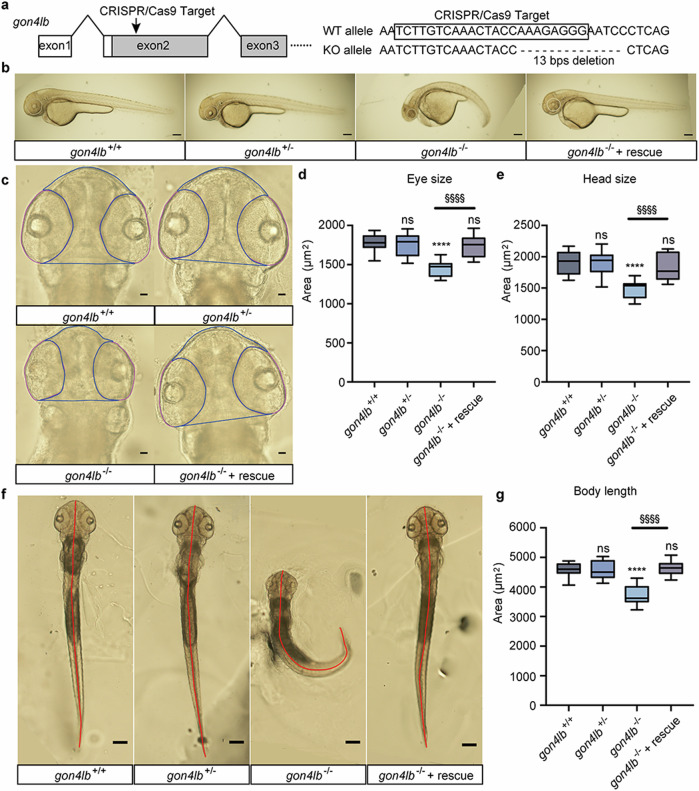
gon4lb-knockout zebrafish phenotype and rescue by human GON4L mRNA
We created gon4lb-knockout zebrafish using the CRISPR/Cas9 system and successfully obtained a knockout germline allele, a 13 bp deletion in exon 2, that leads to a premature stop codon (Fig. 4a and Supplementary Fig. 9). A lateral view of gon4lb-knockout embryo morphology is shown in Fig. 4b. Consistent with our MO knockdown experiments and previous reports1,3,4,8, functional impairment of gon4lb resulted in significant reduction of three measured parameters: eye size (19%), head size (22%), and body length (19%) compared with those of gon4lb+/+ embryos (Fig. 4c–g). Moreover, in comparison to gon4lb+/+ embryos, gon4lb+/− embryos did not exhibit notable alterations in the size of eye, head, or body length. Rescue experiments conducted on gon4lb-knockout embryos demonstrated that injection of human GON4L mRNA was capable of rescuing phenotypic abnormalities, recovering eye size (Fig. 4c, d), head size (Fig. 4c, e), and body length (Fig. 4f, g), thereby reconfirming the essential role of gon4lb in head, eye, and body axis development as reported previuosly1,3,4,8. Similarly, akin to the results of MO knockdown, Acridine Orange staining revealed that impaired gon4lb function also caused a significant increase in the number of positive cells in the head region corresponding to dead cells, whereas gon4lb+/− embryos did not exhibit a significant change in the number of positive cells in the head region compared to gon4lb+/+ embryos (Supplementary Fig. 10). Rescuing gon4lb-knockout zebrafish showed that the number of positive cells in the head region was recovered (Supplementary Fig. 10). Therefore, the loss-of-function gon4lb induced cell death in developing brain of zebrafish, as previously reported1,3,4,8.
Fig. 4. Establishment of gon4lb-null zebrafish and rescue using human GON4L mRNA.
a Target design to knockout gon4lb using CRISPR/Cas9. A crRNA targeted exon 2 of gon4lb and resulted in a 13 bp deletion that generated a premature termination codon to knockout. b Representative images of 50-hpf zebrafish embryos from the gon4lb-knockout line: gon4lb+/+, gon4lb+/−, gon4lb−/−, and gon4lb−/− injected with human GON4L mRNA (rescue). All images are lateral views, with the anterior surface to the left. Scale bar: 200 µm. c Representative images of eye size and head size of 50-hpf zebrafish embryos from the gon4lb-knockout line. All images are dorsal views, with the anterior aspect at the top. The blue line delineates the contour of the head, and the pink line delineates the contour of the eye. Scale bar: 50 µm. Quantitative data showing eye (d) and head size (e) of 50-hpf zebrafish embryos: gon4lb+/+ (n = 12), gon4lb+/− (n = 12), gon4lb−/− (n = 10), and gon4lb−/− with rescue (n = 15). f Representative images of the body length of 50-hpf zebrafish embryos. All images are dorsal views, with the anterior aspect at the top. The red line indicates the position for measuring the body axis. Scale bar: 200 µm. g Quantitative data showing the body length of 50-hpf zebrafish embryos. Sample numbers are as follows: gon4lb+/+ (n = 30), gon4lb+/− (n = 30), gon4lb−/− (n = 31), and gon4lb−/− embryos with rescue (n = 17). Data are shown as the mean ± SEM; ****p ≤ 0.0001, §§§§p ≤ 0.0001 using one-way ANOVA with post hoc Tukey’s test.
gon4lb-knockout and knockdown zebrafish exhibits craniofacial abnormalities
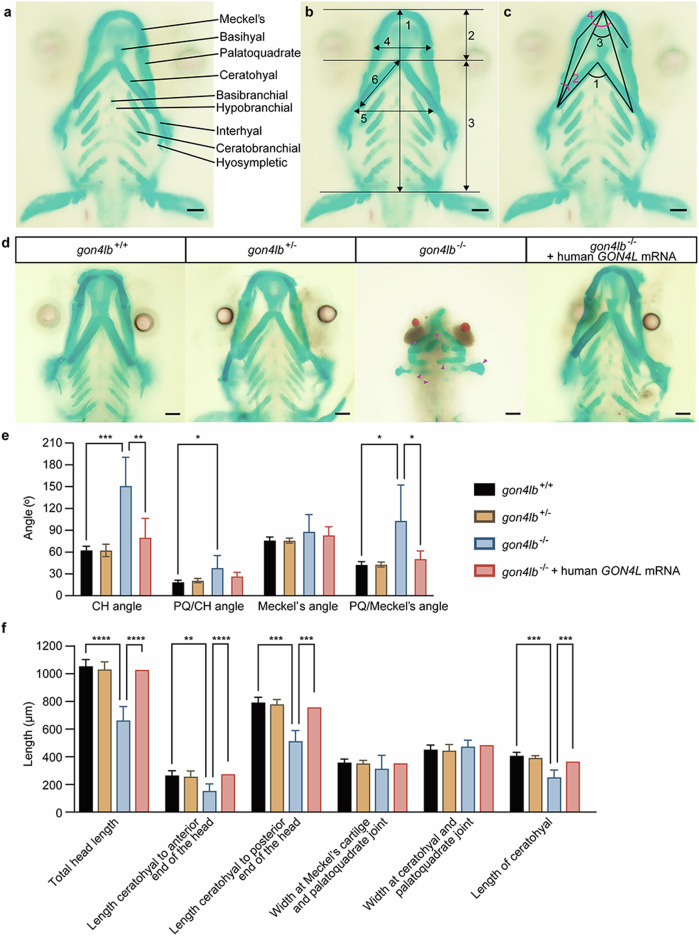
We performed Alcian blue/Alizarin Red bone staining on gon4lb-knockdown and knockout zebrafish larvae at 5 dpf. In knockout model (Fig. 5), we found that in 78.6% of gon4lb−/− larvae, although ventral cartilage was formed, craniofacial cartilage abnormalities were observed. These included a smaller and narrower Meckel’s cartilage, ectopic palatoquadrate cartilage, and a lack of ceratobranchial cartilage in gon4lb−/− deformities, similar to gon4lb MO knockdown zebrafish larvae, and the ceratohyal angle expanded to nearly 180° (Fig. 5d). Compared with gon4lb+/+ larvae, three out of four craniofacial cartilage angle parameters significantly increased in gon4lb−/− larvae (Fig. 5e), and four out of six craniofacial cartilage length parameters were significantly decreased (Fig. 5f). Subsequent rescue experiments were conducted on gon4lb-knockout embryos. In 5 dpf gon4lb−/− larvae injected with human GON4L mRNA, there was no lack of ceratobranchial cartilage, no morphological abnormalities of Meckel’s cartilage and palatoquadrate cartilage, and no abnormalities of the ceratohyal angle (Fig. 5d). Compared with gon4lb+/+ larvae, all four significantly decreased craniofacial cartilage length parameters in gon4lb−/− larvae were rescued, and two out of three significantly increased craniofacial cartilage angle parameters were also rescued (Fig. 5e, f). These results also indicate that the craniofacial cartilage developmental abnormalities in 5-dpf larvae are caused by the loss of gon4lb function. The similar phenotypes were observed in knockdown zebrafish (Supplementary Fig. 11).
Fig. 5. gon4lb knockout causes abnormal craniofacial development.
a Craniofacial bone structure of zebrafish larvae at 5 dpf. b Measurement parameters of craniofacial bone length in wild-type zebrafish larvae: (1) Total length of the head, (2) length from the ceratohyal cartilage to the anterior end of the head, (3) length from the ceratohyal cartilage to the posterior end of the head, (4) width of the Meckel’s cartilage and palatoquadrate joint, (5) width of the ceratohyal cartilage and palatoquadrate joint, and (6) length of the ceratohyal cartilage. c Four angle parameters for craniofacial cartilage: (1) the angle between ceratohyal (CH) cartilages (CH Angle), (2) the angle between palatoquadrate (PQ) cartilage and ceratohyal cartilage (PQ/CH Angle), (3) the angle between Meckel’s cartilages (Meckel’s Angle), and (4) the angle between palatoquadrate cartilage and Meckel’s cartilage (PQ/Meckel’s Angle). All images are ventral views with the anterior at the top. Scale bars: 100 µm. d Representative images of the craniofacial cartilage of 5 dpf zebrafish larvae stained with Alcian blue and Alizarin Red (10 embryos each). The magenta-colored arrows in the panel of gon4lb−/− represent the abnormal craniofacial cartilages. All images are ventral views with the anterior at the top. Scale bar: 100 µm. e Quantitative data showing the angles of four different mineralized craniofacial cartilage elements 5 dpf zebrafish larvae. f Quantitative data showing the lengths of six different mineralized craniofacial cartilage elements in 5 dpf zebrafish larvae. Data are represented as mean ± SEM; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001 using two-way ANOVA with Tukey’s multiple comparisons test (α = 0.05). CH ceratohyal, PQ palatoquadrate.
Situs inversus in gon4lb-knockout zebrafish
Since two of three patients showed situs inversus totalis, we examined the heart positioning in 48 hpf gon4lb mutant zebrafish embryos. Ectopic heart positioning on the right side was more frequently observed in gon4lb−/− (82%) compared to gon4lb+/+ (15%) with the statistical significance of p < 0.01 (Supplementary Fig. 12). We also examined the liver positioning in 5 dpf gon4lb mutant embryos. Normally, the left lobe of the liver (as observed from the left lateral view) was significantly larger than the right lobe (from the right lateral view) (Supplementary Fig. 13). Abnormal liver positioning was more frequently observed in gon4lb−/− embryos (37%) compared to gon4lb+/+ (5%) with the statistical significance of <0.001.
Discussion
In this study, we identified two homozygous truncating GON4L variants [NM_001282860.2: c.62_63del, p.(Gln21Argfs*12) and c.5517+1G>A] in three patients from two families with prenatal-onset growth impairment, developmental delay, mild intellectual disability, speech impairment, progressive and disproportionate microcephaly, facial asymmetry, congenital heart anomaly, and brain structure abnormalities (Fig. 2 and Table 1). In addition, the probability of being loss-of-function intolerant (pLI) score of GON4L was 0.95 in gnomAD v2.1.1. The most prevalent loss-of-function variant in GON4L [chr1-155823547-TTCTTC-T (GRCh37), NM_001282860.2:c.20_24del, p.(Arg7Asnfs*10)] in gnomAD was very rare (MAF = 0.000366) and compatible with being causative of rare recessive diseases. Parents of Patients 1 and 2 and Patient 1’s sister are heterozygotes with no symptoms including autism spectrum disorder, developmental disorder, bipolar disorder, and hydrocephalus which were observed in individuals with heterozygous GON4L variant20–26; therefore, it is likely that biallelic GON4L truncating variants cause a human phenotype. In other words, monoallelic GON4L variants are less likely to cause a human phenotype considering their carrier status.
At least five GON4L isoforms are known. The frameshift variant is predicted to make all five GON4L isoforms subject to nonsense-mediated mRNA decay, which would lead to complete loss of GON4L function. According to the GTEx portal (https://gtexportal.org/home), long isoforms of GON4L seems the canonical ones (Supplementary Figs. 4 and 14). The canonical splice site variant (c.5517+1G>A) affects only longer isoforms (NM_001282860.2, NM_001282858.2, and NM_001282856.2, Fig. 1c); however, this variant results in a similar phenotype to the null allele variant affecting the all isoforms. There is one previous report of a family who has an identical canonical splice site variant (c.5517+1G>A) and a missense (p.Asp1522Gly) as homozygote27. These individuals also have intellectual disability, facial dysmorphism, and malformed ears, but no detailed clinical information was reported27. The identical variant in another family supports disruption of the longer isoforms as being responsible for the phenotype.
Our in vitro study showed that Gon4l knockdown affects neurite outgrowth in PC12 cells, indicating that GON4L plays an important role in neurogenesis. Furthermore, we successfully recapitulated some of the proband’s phenotypic features (growth impairment, microphthalmia, microcephaly, and brain structure abnormalities) in knockdown and knockout zebrafish. In addition, we observed situs inversus in mutant zebrafish (right-sided heart displacement and liver positional anomalies) as observed in our patients, which has not been reported before. So, our study highlighted a potential key role for gon4l in the left-right pattern. Overall, our data indicate that a biallelic GON4L loss-of-function variant can explain the patient’s phenotype.
GON4L forms complexes with the transcriptional regulators, YY1, SIN3A, and HDAC116. Among them, pathogenic variants in YY1 (MIM*600013, NM_003403.5) and SIN3A (MIM*607776, NM_001145358.2) cause monogenic diseases. YY1 is a ubiquitous transcription factor involved in embryogenesis, differentiation, replication, and cell proliferation30. YY1 haploinsufficiency causes Gabriele-de Vries syndrome (MIM #617557), which is characterized by intrauterine growth retardation, psychomotor developmental delay, intellectual disability, skeletal dysplasia, and various congenital abnormalities involving the face, brain, eye, heart, kidney, and genitourinary system31–33. SIN3A is a transcriptional corepressor34 and heterozygous SIN3A variants cause Witteveen–Kolk syndrome (MIM# 613406) through haploinsufficiency35. This syndrome is characterized by developmental delay, intellectual disability, facial dysmorphism, and short stature. It is noteworthy that pre- and postnatal growth impairment, facial asymmetry, broad/high forehead, downslanted palpebral fissures, strabismus, thick lower lip, pointed chin, cardiac abnormalities, developmental delay, intellectual disability, speech delay, autistic features, and brain structure abnormalities are observed in these three conditions (Table 1 and Supplementary Table 1)31,35–38.
Heterozygous GON4L variants have been reported in cases with mild neurodevelopmental defects involving autism, autism spectrum disorder, developmental disorder, bipolar disorder, and hydrocephalus20–25. All parents in both families and a sister of Patient 1 are heterozygous for the same GON4L truncating variants; however, they do not present any of the patients’ symptoms. Of note, the pLI score of GON4L in gnomAD v.2.1.1 is 0.95; therefore, it is possible that a monoallelic loss-of-function GON4L variant can cause the disease via haploinsufficiency with incomplete penetrance. The number of cases is very limited, and it remains unclear whether heterozygous loss of GON4L function can cause human disease. Further genetic investigation and clinical evaluation of individuals carrying mono- and bi-allelic GON4L variants are needed to clarify this point.
Our experiments using gon4lb-knockout and knockdown zebrafish revealed distinct morphological and size abnormalities in the craniofacial cartilage of zebrafish larvae. These abnormalities included reductions in Meckel’s cartilage dimensions, palatoquadrate cartilage misalignment, ceratobranchial cartilage deficiencies, and an increase in the angle of ceratohyal cartilage. This implies that these anomalies result from the loss of gon4lb function. Moreover, human GON4L mRNA was able to rescue craniofacial cartilage phenotypic abnormalities in zebrafish larvae, suggesting that the human GON4L can functionally substitute for the zebrafish gon4lb. These results underscore the pivotal and conserved role of GON4L/gon4lb in craniofacial cartilage development.
We observed increased levels of apoptosis in the brains of gon4lb-knockdown and -knockout zebrafish, as previously reported1,3,4,8. Interestingly, the mechanism by which gon4l variants produce a phenotype differs among organs; for example, cardiomyocyte deficiency in gon4l/udu−/− zebrafish is caused by reduced proliferation rather than TP53-dependent apoptosis9, while erythropoiesis in udu(gon4lb)-loss-of-function zebrafish is caused by aberrant cell cycle control and increased TP53-mediated apoptosis3. B cell deficiencies in Gon4l-deficient mice are also caused by abnormal cell cycle progression and increased TP53-mediated apoptosis7. Our results in the brain using the zebrafish model were consistent with the data on hematopoietic inhibition associated with increased apoptosis. Here, we again observed growth impairment, microcephaly, and microphthalmia using our zebrafish model, which enabled intense investigation of craniofacial cartilage development for the first time; therefore, further studies exploring the phenotypes of additional patients with biallelic GON4L variants are required to uncover the exact pathophysiology of loss of GON4L function.
Methods
DNA preparation and genetic analysis
Clinical information and a peripheral blood sample were collected from all participants after written informed consent was obtained. For pediatric patients, written informed consent to participate in this study and publish this study was obtained from their parents. Written informed consent to participate in this study and publish this study was obtained from the parents as well. Genomic DNA was isolated from peripheral blood using the QuickGene-610L system (Fujifilm, Tokyo, Japan) for Family 1 and the Qiagen Blood DNA Kit (Qiagen, Hilden, Germany) for Family 2. Trio-based exome sequencing for Family 1 (Fig. 1a) was performed by Takara Bio (Kusatsu, Japan) using the Sure Select Human All Exon kit V4 (Agilent Technologies, Santa Clara, CA) and the NovaSeq 2000 system (Illumina, San Diego, CA). Exome sequencing for only the proband of Family 2 (VI-1 in Fig. 1b) was performed using the SureSelect Human All Exome 50 Mb Kit (Agilent, Santa Clara, CA) and the HiSeq2000 system (Illumina). The obtained sequences were aligned to the UCSC human genome assembly, version GRCh37/hg19, and variants were called by the GATK pipeline. Candidate variants were selected using publicly available control databases [esp6500 (http://evs.gs.washington.edu/EVS/), ExAC (https://exac.broadinstitute.org/), gnomAD (https://gnomad.broadinstitute.org/), Human Genetic Variation Database (HGVD)] and in-house controls (n = 575 in Family 1, n > 1500 in Family 2). The candidate variants were validated by Sanger sequencing. Absence of heterozygosity regions were confirmed by AutozygosityMapper39 for Family 1 and AutoMap40 for the proband of Family 2 using vcf files from exome sequencing. This study was approved by the Institutional Review Board of the Yokohama City University School of Medicine and the Medical Research Ethics Committee of the National Research Centre, Egypt. We have complied with all relevant ethical regulations including the Declaration of Helsinki. Parents of the pediatric patients provided informed consent for the publication of the photographs in Fig. 2.
Determination of human GON4L expression
Quantitative PCR (qPCR) was performed using adult and fetal stage cDNA libraries purchased from Clontech Laboratories (Mountain View, CA). TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, Waltham, MA) with pre-designed TaqMan probes for human GON4L (Hs00228595_m1 and Hs01685033_mH for long and short isoforms, and Hs00250331_m1 specific for long isoform, Thermo Fisher Scientific) and the human housekeeping gene beta-actin (ACTB, Thermo Fisher Scientific) were used for mRNA quantification. PCR was performed on a Rotor-Gene Q (Qiagen) and expression levels were normalized against ACTB. Data were processed by the 2−ΔΔCt method, and relative quantities are shown.
Gon4l knockdown in PC12 cells
PC12 cells were purchased from the RIKEN cell bank (Tsukuba, Japan). The cells were maintained in low glucose Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO) and 10% horse serum (BioWest, Nuaillé, France). Short hairpin RNAs (shRNAs) for Gon4l were subcloned into the lentiviral vector, pLKO.1-TRC41 [a gift from David Root (Addgene plasmid # 10878; http://n2t.net/addgene:10878; RRID:Addgene_10878)]. The target-specific shRNA sequences were: shGon4l_2915, GCTCCCAACAACGTGATTAAA; shGon4l_5252, CAGCATATCCCTGGCAAATAT; shGon4l_4283, CCATCGAAGGATTTATTAAAC. Lentiviruses were generated in HEK293T cells by co-transfecting the three plasmids, lentiviral vector (pLKO-shControl or pLKO-shGon4l), psPAX2 and pCMV-VSV-G-Rev (all purchased from RIKEN Bioresource Center, Tsukuba, Japan), using XtremeGENE 9 (Merck, Darmstadt, Germany). At 48 h post-transfection, virus-containing supernatants were collected for infection. For viral transduction, the lentiviruses were incubated with PC12 cells. At 72 h post-infection, cells were selected with 3 µg/ml puromycin.
Western blotting
Cells were lysed for 30 min in ice-cold lysis buffer [50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EDTA, 0.5% NP40, 10% glycerol] containing proteinase inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). After centrifugation at 15,000 × g and 4 °C for 20 min, supernatants were collected. The lysates were incubated with NuPAGE LDS sample buffer (Thermo Fisher Scientific) containing 5% 2-mercaptoethanol at 70 °C for 10 min. Proteins were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked with 5% non-fat milk in Tris-buffered saline plus 0.1% Tween 20. Immunoblotting was performed with a rabbit anti-GON4L antibody (MBS9609854, 1:1000; MyBioSource, San Diego, CA), a mouse anti-β-actin antibody (ab6276, 1:5000; Abcam, Cambridge, UK) and a horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG Goat antibody (1:10,000; Jackson ImmunoResearch Laboratories, West Grove, PA) and detected with a ChemiDoc Touch imaging system (BioRad, Hercules, CA). All blots derive from the same experiment and they were processed in parallel. Uncropped representative blots were shown in Supplementary Fig. 15.
Nerve growth factor treatment and neurite outgrowth assay
PC12 cells (1 × 103 cells/well) were seeded into 24-well plates coated with Atelocollagen/Native Collagen Acidic Solutions (KOKEN, Tokyo, Japan) and cultured overnight. The cells were then treated with 100 ng/ml nerve growth factor (NGF 2.5S, Alomone Labs, Jerusalem, Israel) and the medium changed every 48–72 h. On the 6th day, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS)(-) for 20 min. Permeabilization was performed with 0.1% TritonX/PBS for 10 min and blocking was performed with 5% goat serum/PBS for 30 min. Samples were incubated with anti-Neurofilament heavy polypeptide antibody (EPR20020, 1:100; Abcam) overnight and subsequently incubated with Alexa Fluor 488 goat anti-rabbit IgG (1:1000; Invitrogen, Carlsbad, CA). The nuclei were stained with DAPI solution (DOJINDO, Kumamoto, Japan). The cells were visualized and analyzed with an IN Cell Analyzer 6000 (Cytiva, Marlborough, MA).
Quantitative PCR
RNA samples were isolated using RNeasy Mini Kit (Qiagen) and subjected to reverse transcription using PrimeScript RT Master Mix (Takara Bio). qPCR was performed on the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). TBgreen Premix ExTaqII (Takara Bio) was used for quantification. Data were processed by the 2−ΔΔCt method, and quantities relative to the shControl were calculated. The primer sequences were: Gon4l forward, 5′-CATGGAGGATGGTGGACTCT-3′; reverse, 5′-GTACGTCATCGTCCCTTGCT-3′; Gusb forward, 5′-CCGTGGAACAGGGAATGAG-3′; reverse, 5′-CTCAGGTGTTGTCATCGTCA-3′.
Zebrafish strains
All zebrafish were cared for and experimental procedures were conducted following protocols approved by the Institutional Animal Care and Use Committee of Waseda University. The RIKEN Wako (RW) WT zebrafish strain was obtained from the Zebrafish National BioResource Center, Japan. Experimental embryos were obtained via light-induced spawning. Zebrafish embryos were maintained at a temperature of 28.0 ± 0.5 °C. Zebrafish were housed in an aquaneering semi-recirculating housing system with a 12-h light and 12-h dark photoperiod cycle. pH and conductivity levels were maintained at ~7.4 and 1100 µS, respectively. To prevent pigmentation, 0.003% 1-phenyl-2-thiourea was added to the fish water within 24 h of fertilization.
Antisense morpholino oligonucleotides and mRNA injections
Antisense morpholino oligonucleotides (MOs) were designed and obtained from Gene Tools (Philomath, OR). The MO sequences were: gon4lb, 5′-TAACACTACACTCACCACCCCTTTT-3′; Standard control, 5′-CCTCTTACCTCAGTTACAATTTATA-3′. The MOs were injected into 1–2 cell stage zebrafish embryos at a concentration of 4 µg/µL. For rescue experiments, human GON4L mRNA was synthesized using the mMessage mMachine Kit (Thermo Fisher Scientific) and co-injected with gon4lb MO into embryos at a concentration of 300 ng/µL.
Immunohistochemistry
Immunohistochemistry was performed on whole-mount zebrafish embryos at 50 hpf. The embryos were anesthetized using a 0.0016 M tricaine solution (Sigma-Aldrich, St. Louis, MO) and euthanized, followed by fixation in 4% paraformaldehyde. Subsequently, the embryos were washed sequentially with 0.1% PBST (PBS containing 0.1% Tween 20), 150 mM Tris-HCl buffer (pH 9.0), and acetone. After blocking with 10% blocking reagent (diluted in 0.1% PBST) for 1 h at room temperature, mouse anti-HuC/HuD (RRID:AB_221448, 1:100; Thermo Fisher Scientific) and Histofine Simple Stain MAX PO (M) (Nichirei, Tokyo, Japan) were used for staining. Staining was visualized by incubating the samples with a 3,3-diaminobenzidine chromogen (Fujifilm) for 1 h.
Anatomical measurement of the head and eye
Zebrafish embryos at 50 hpf were anesthetized by exposure to a 0.0016 M tricaine solution (Sigma-Aldrich) and immobilized in a 1.5% low melting point agarose matrix. The specimens were examined by light microscopy and acquired photographs were subjected to image analysis using ImageJ software. Assessment of the dorsal portion of the eye and head areas was performed in a blinded fashion. The boundaries of eyes and heads were demarcated by red and green contours.
Establishment of gon4lb-knockout zebrafish
gon4lb, a zebrafish ortholog of human GON4L, was knocked out in zebrafish using the CRISPR/Cas9 system. A target-specific CRISPR RNA (crRNA) sequence, 5′-TCTTGTCAAACTACCAAAGAGGG-3′, was designed to target exon 2 of gon4lb (NM_001201535). Pre-four-cell stage embryos, typically around the 1–2-cell stage, were microinjected with ∼1 nL of a solution containing CRISPR/Cas9 components, including 500 ng/µl Cas9 endonuclease, 25 ng/µl crRNA (Fasmac, Atsugi, Japan), 100 ng/µl tracrRNA (Fasmac), and phenol red. To verify the success of the injection, 200 ng/µL mCherry mRNA was also injected into the embryos. After injection, the embryos were raised in 28 °C fish water, and mCherry fluorescence was observed at 28 hpf. Some mCherry-positive embryos were raised to 50 hpf, and DNA was then extracted for T7 endonuclease assays and genomic sequencing, while others were raised to adulthood to establish the mutant line. F0 zebrafish carrying mutations were then bred with wild-type fish to produce F1 offspring, which were subsequently screened for mutations. Through this process, a small deletion at the targeted site, resulting in a premature stop codon, was generated.
Variant detection in knockout zebrafish
Adult zebrafish ~2 months old were subjected to genotyping. The caudal fin of each fish was excised and incubated in lysis buffer containing proteinase K (Fujifilm) at 55 °C overnight. Following DNA purification, the target region was amplified by PCR using primers specific for the gon4lb gene. The forward primer was 5′-GCTCACTGGGCTTGGGTAAA-3′, and the reverse primer was 5′-TGGAGTAGAGGACTGGATAC-3′. The amplified PCR product was then cloned into the T-Vector pMD20 (Takara Bio) and transformed into Escherichia coli (Toyobo, Tokyo, Japan). Plasmids from selected E. coli colonies were isolated and sequenced using either M13 Forward or M13 Reverse primers (Eurofins, Tokyo, Japan).
To identify descendants carrying the same variant identified in gon4lb F1 fish, PCR was conducted using specific primers. The forward primer for wild-type gon4lb was 5′-CAAACTACCAAAGAGGGAAT-3′, and the forward primer for the variant was 5′-CTTGTCAAACTACCAAAGTA-3′. The reverse primer for both wild-type and variant alleles was 5′-GTCACTGTCATCTTCAACAT-3′ (Fasmac).
Acridine Orange staining
To quantify the extent of cell death, zebrafish embryos at 50 hpf were immersed in E3 medium supplemented with 10 µg/mL Acridine Orange dye (Sigma) at 28.5 °C and shielded from light for 60 min. After multiple rinses using E3 medium, embryos were anesthetized using a 0.0016 M tricaine solution (Sigma) and embedded in 1.5% low melting point agarose. Finally, fluorescence images were acquired by confocal microscopy (FV1000, Olympus, Toko, Japan) using UNPlanFL 20× (NA = 0.50) and LUMPlanFLN 40× (NA = 0.80) water immersion objectives.
Cartilage and bone staining
A modified acid-free staining protocol was employed to visualize cartilage and bone in 5-day post-fertilization (dpf) zebrafish larvae, as reported previously42. The larvae were fixed in 4% paraformaldehyde for 2 h and then washed with water and 80% ethanol. To stain cartilage, the larvae were incubated in an Alcian blue solution (Fujifilm) containing 40 mM MgCl2 for 1.5 h. Subsequently, the larvae were washed again in ethanol and subjected to bleaching using a peroxide solution consisting of 0.8% KOH, 1.1% H2O2, and 0.2% Triton, followed by two wash steps, the first in 0.2% Triton and the second in a saturated sodium tetraborate solution. Next, the larvae were treated with a trypsin solution (1 mg/ml) for 12 min, followed by another wash with 0.2% Triton. For bone staining, the larvae were immersed in a 0.003% Alizarin Red solution (Nacalai Tesque) overnight, cleared using a graded series of glycerol concentrations (25%, 50%, and 75%), and stored in 100% glycerol for imaging.
Microscopy and imaging
Light microscopy was conducted using a BX51 microscope (Olympus) equipped with a UPlanApo 10× objective (NA = 0.40). Images were captured with a DP704 digital camera (Olympus). Fluorescence microscopy was performed using a FV1000 confocal laser scanning microscope (Olympus) with UNPlanFL 20× (NA = 0.50) and LUMPlanFLN 40× (NA = 0.80) water immersion objectives. The acquired images were processed using either ImageJ, Adobe Illustrator, or Adobe Photoshop software.
Statistical analysis for zebrafish measurements
For all experiments, images that contained unclassifiable features were excluded during the counting process. Each independent experiment was repeated two to three times. The presented data are expressed as the mean ± standard deviation (SD) or the mean ± standard error of the mean (SEM). The statistical significance between pairs of groups was assessed using either Student’s unpaired t-test or the Mann–Whitney test, depending on the normality of the data distribution. In cases where multiple comparisons were necessary, a one-way or two-way analysis of variance (ANOVA) was used, followed by Tukey’s test to determine the statistical significance of the differences in frequencies between groups. p values were calculated using GraphPad Prism and are indicated as: ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, and ns, indicating a lack of statistical significance.
Observational methods for anatomical positioning in zebrafish embryos and larvae
To accurately observe the heart position in zebrafish embryos at 48 hpf and liver positioning in zebrafish larvae at 5 dpf, we initially selected embryos and larvae from the population. At specific developmental time points, the zebrafish embryos and larvae were anesthetized using a 0.0016 M solution of tricaine (Sigma-Aldrich). Post-anesthesia, the samples were immediately fixed in 4% paraformaldehyde. Following fixation, samples were transferred to 100% glycerol for preservation to prevent desiccation and damage. Prior to microscopic examination, the fixed samples were embedded in 1.5% low melting point agarose. The specimens were observed using a light microscope BX51 (Olympus), equipped with a UPlanSApo 20× objective lens (NA = 0.40, Olympus). All images captured via light microscopy were subsequently subjected to image analysis using ImageJ software to quantify the anatomical changes in the positions of the heart and liver.
Supplementary information
Acknowledgements
We thank the affected individuals and their families for participating in this study. We also thank Ms. Sayaka Sugimoto and Ms. Kaori Takabe from Yokohama City University Graduate School of Medicine and Ms. Miwa Nakano from the National Center for Global Health and Medicine for their technical assistance. We also thank Dr. Atsuo Kawahara at the Laboratory for Developmental Biology, Center for Medical Education and Sciences, Graduate School of Medical Science, University of Yamanashi for his helpful advice for revising zebrafish study. We are also grateful to Jeremy Allen, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was supported by AMED under grant numbers JP24ek0109674, JP24ek0109760, JP24ek0109617, JP24ek0109648, and JP24ek0109677 (N.Ma.), and JP21ek0109484 (T.O.); JSPS KAKENHI under grant numbers JP19H03621 and 22H03047 (N.Mi.), and JP24K02230 (N.Ma), the Takeda Science Foundation (N.Ma.), the NCGM Intramural Research Fund under grant numbers 21A1011 and 23A1014 (N.Mi.), and the Science and Technology Development Fund (STDF), Academy of Science Research and Technology, Egypt (grant number: 33650) (M.S.A.H.).
Author contributions
T.O., N.Ma., and N.Mi. contributed to the conception and design of the study; S.L., S.T., G.M.H.A., M.Y.I., M.S.Z., M.S.A., A.M.S.S., E.K., A.F., R.F., T.O., and N.Mi. contributed to the acquisition and analysis of data; S.T., T.O., N.Ma., and N.Mi. contributed to drafting and critically reviewing the text and preparing the figures.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The linked genotype and phenotype data were registered as MGS000085 and MGS000086 in MGeND (https://mgend.ncgm.go.jp/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Simo Li, Sanami Takada.
Contributor Information
Toshio Ohshima, Email: ohshima@waseda.jp.
Naomichi Matsumoto, Email: naomat@yokohama-cu.ac.jp.
Noriko Miyake, Email: nomiyake@ri.ncgm.go.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-024-00437-5.
References
- 1.Hammerschmidt, M. et al. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development123, 143–151 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Kuryshev, V. Y. et al. An anthropoid-specific segmental duplication on human chromosome 1q22. Genomics88, 143–151 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Liu, Y. et al. The zebrafish udu gene encodes a novel nuclear factor and is essential for primitive erythroid cell development. Blood110, 99–106 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Lim, C. H., Chong, S. W. & Jiang, Y. J. Udu deficiency activates DNA damage checkpoint. Mol. Biol. Cell20, 4183–4193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu, P. et al. The Justy mutation identifies Gon4-like as a gene that is essential for B lymphopoiesis. J. Exp. Med.207, 1359–1367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulchand, S., Menon, S. D., George, S. E. & Chia, W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci.123, 2697–2707 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Barr, J. Y., Goodfellow, R. X., Colgan, D. F. & Colgan, J. D. Early B cell progenitors deficient for GON4L fail to differentiate due to a block in mitotic cell division. J. Immunol.198, 3978–3988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams, M. L. K. et al. Gon4l regulates notochord boundary formation and cell polarity underlying axis extension by repressing adhesion genes. Nat. Commun.9, 1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budine, T. E. et al. Gon4l/Udu regulates cardiomyocyte proliferation and maintenance of ventricular chamber identity during zebrafish development. Dev. Biol.462, 223–234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai, S. M., Chu, K. C. & Jiang, Y. J. Newly identified Gon4l/Udu-interacting proteins implicate novel functions. Sci. Rep.10, 14213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spronk, C. A. et al. The Mad1-Sin3B interaction involves a novel helical fold. Nat. Struct. Biol.7, 1100–1104 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Wang, H., Clark, I., Nicholson, P. R., Herskowitz, I. & Stillman, D. J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol.10, 5927–5936 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer, L. A. et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell10, 935–942 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Boyer, L. A., Latek, R. R. & Peterson, C. L. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol.5, 158–163 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Aasland, R., Stewart, A. F. & Gibson, T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci.21, 87–88 (1996). [PubMed] [Google Scholar]
- 16.Lu, P. et al. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J. Biol. Chem.286, 18311–18319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, L., Santa Anna-Arriola, S., Hodgkin, J. & Kimble, J. gon-4, a cell lineage regulator required for gonadogenesis in Caenorhabditis elegans. Dev. Biol.228, 350–362 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Colgan, D. F., Goodfellow, R. X. & Colgan, J. D. The transcriptional regulator GON4L is required for viability and hematopoiesis in mice. Exp. Hematol.98, 25–35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzenbacher, H. et al. A frameshift mutation in GON4L is associated with proportionate dwarfism in Fleckvieh cattle. Genet. Sel. Evol.48, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner, T. N. et al. Sex-based analysis of de novo variants in neurodevelopmental disorders. Am. J. Hum. Genet.105, 1274–1285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosmicki, J. A. et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet.49, 504–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, E. T. et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci.20, 1217–1224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koire, A. et al. A method to delineate de novo missense variants across pathways prioritizes genes linked to autism. Sci. Transl. Med.1310.1126/scitranslmed.abc1739 (2021). [DOI] [PMC free article] [PubMed]
- 25.Nishioka, M. et al. Systematic analysis of exonic germline and postzygotic de novo mutations in bipolar disorder. Nat. Commun.12, 3750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, S. C. et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat. Med.26, 1754–1765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Najmabadi, H. et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature478, 57–63 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Das, K. P., Freudenrich, T. M. & Mundy, W. R. Assessment of PC12 cell differentiation and neurite growth: a comparison of morphological and neurochemical measures. Neurotoxicol. Teratol.26, 397–406 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Wiatrak, B., Kubis-Kubiak, A., Piwowar, A. & Barg, E. PC12 cell line: cell types, coating of culture vessels, differentiation and other culture conditions. Cells910.3390/cells9040958 (2020). [DOI] [PMC free article] [PubMed]
- 30.Verheul, T. C. J., van Hijfte, L., Perenthaler, E. & Barakat, T. S. The why of YY1: mechanisms of transcriptional regulation by Yin Yang 1. Front. Cell Dev. Biol.8, 592164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriele, M. et al. YY1 haploinsufficiency causes an intellectual disability syndrome featuring transcriptional and chromatin dysfunction. Am. J. Hum. Genet.100, 907–925 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales-Rosado, J. A., Kaiwar, C., Smith, B. E., Klee, E. W. & Dhamija, R. A case of YY1-associated syndromic learning disability or Gabriele-de Vries syndrome with myasthenia gravis. Am. J. Med. Genet. A176, 2846–2849 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Zurkirchen, L. et al. Yin Yang 1 sustains biosynthetic demands during brain development in a stage-specific manner. Nat. Commun.10, 2192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grzenda, A., Lomberk, G., Zhang, J. S. & Urrutia, R. Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta1789, 443–450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witteveen, J. S. et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet.48, 877–887 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Koruga, N. et al. First reported case of Gabriele-de Vries syndrome with spinal dysraphism. Children1010.3390/children10040623 (2023). [DOI] [PMC free article] [PubMed]
- 37.Ercoskun, P. & Yuce Kahraman, C. Witteveen-Kolk syndrome: the first patient from Turkey. Am. J. Med. Genet. A185, 617–619 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Coenen-van der Spek, J. et al. DNA methylation episignature for Witteveen-Kolk syndrome due to SIN3A haploinsufficiency. Genet. Med.25, 63–75 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Steinhaus, R., Boschann, F., Vogel, M., Fischer-Zirnsak, B. & Seelow, D. AutozygosityMapper: identification of disease-mutations in consanguineous families. Nucleic Acids Res.50, W83–W89 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinodoz, M. et al. AutoMap is a high performance homozygosity mapping tool using next-generation sequencing data. Nat. Commun.12, 518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell124, 1283–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Walker, M. B. & Kimmel, C. B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem.82, 23–28 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The linked genotype and phenotype data were registered as MGS000085 and MGS000086 in MGeND (https://mgend.ncgm.go.jp/).