Abstract
In Finland, the frequency of isolated cleft palate (CP) is higher than that of isolated cleft lip with or without cleft palate (CL/P). This trend contrasts to that in other European countries but its genetic underpinnings are unknown. We conducted a genome-wide association study in the Finnish population and identified rs570516915, a single nucleotide polymorphism highly enriched in Finns, as strongly associated with CP (P = 5.25 × 10−34, OR = 8.65, 95% CI 6.11–12.25), but not with CL/P (P = 7.2 × 10−5), with genome-wide significance. The risk allele frequency of rs570516915 parallels the regional variation of CP prevalence in Finland, and the association was replicated in independent cohorts of CP cases from Finland (P = 8.82 × 10−28) and Estonia (P = 1.25 × 10−5). The risk allele of rs570516915 alters a conserved binding site for the transcription factor IRF6 within an enhancer (MCS-9.7) upstream of the IRF6 gene and diminishes the enhancer activity. Oral epithelial cells derived from CRISPR-Cas9 edited induced pluripotent stem cells demonstrate that the CP-associated allele of rs570516915 concomitantly decreases the binding of IRF6 and the expression level of IRF6, suggesting impaired IRF6 autoregulation as a molecular mechanism underlying the risk for CP.
Subject terms: Genome-wide association studies, CRISPR-Cas9 genome editing
Here, the authors perform a genome-wide study and identify a genetic variant enriched in the Finnish population that is strongly associated with isolated cleft palate. This finding suggests a genetic basis for the high prevalence of cleft palate in Finland.
Introduction
Non-syndromic orofacial clefts (OFCs) are congenital malformations that affect approximately 1.7 per 1000 newborns globally1. The most common forms of OFCs can be classified as cleft lip only (CL), cleft lip with cleft palate (CLP) or cleft palate only (CP). Epidemiologically, CL and CLP are usually combined (CL/P, i.e., CL with or without CP) because it is unclear whether the cleft palate in individual cases of CLP is secondary to the cleft of the lip which occurs before secondary palate development. The incidence of specific OFCs varies based on the ethnicity and geographic origin of affected families1. In non-Finnish European populations, the prevalence of CL/P is higher than that of CP (0.80 per 1000 births versus 0.55 per 1000 births, respectively), as reported by EUROCAT2,3 for the years 1980–2020. However, Finland has one of the highest rates of non-syndromic (or isolated) CP in the world and the ratio of incidence of CL/P versus CP is reversed, with CP being more common4. Remarkably, among the five Nordic countries where the total incidence of clefts is very similar, Finland stands out for having a higher CP incidence than CL/P; for instance, the CP incidence in Finland is 1.8-fold that in neighboring Sweden4. In Finland the prevalence of births with CL/P over the period 2000–2014 was 1.08 per 1000 births, whereas the prevalence of CP was 1.50 per 1000 births5. Furthermore, there is a significant regional variation in the incidence rate of CP, which increases from the lowest in the south-western-most to highest in the north-eastern-most province of the country5,6. Moreover, 20% of OFC cases in Northern Finland also have a family history of clefts, among whom the highest incidence (77%) was documented for cases with CP7. These epidemiological observations suggest the presence of genetic risk factor(s) that contribute to the exceptionally high incidence and regional variation of CP rate in the Finnish population4.
Previous efforts to detect genetic risk factors that influence risk for CP include six genome-wide association studies (GWAS) which identified 15 loci associated with CP8–13. Candidate genes at these loci are GRHL3, IRF6, CTNNA2, POMGNT2, WHSC1, PARK2, MYC, PTCH1, YAP1, DOCK9, PAX9, DLK1, ISL2/SCAPER, FOXC2/FOXL1 and MAU2. In one study, variants near MLLT3 and SMC2 were associated with increased risk for CP but only if the mother consumed alcohol during the peri-conceptual period, as were variants near TBK1 and ZNF236 but only in the presence of maternal smoking12.
At just one locus, 1p36, the causative variant has been identified and, unusually, it is a coding variant13,14. This variant is a missense single nucleotide polymorphism (SNP) in the GRHL3 gene that disrupts the transactivation function of the transcription factor encoded by this gene. GRHL3 is important for keratinocyte and periderm differentiation15–17, as is IRF618,19, and the IRF6 gene is also associated with CP8,9,11. In zebrafish, Grhl3 acts downstream of Irf6 in periderm differentiation20. Together, these findings imply that the perturbation of a gene regulatory network involving IRF6 and GRHL3, which orchestrates the differentiation of the oral periderm, predisposes individuals to CP. Abnormal periderm differentiation is also proposed to underlie some cases of CL/P21. However, while variants associated with non-syndromic CL/P have been detected near IRF6, none have thus far been reported near GRHL3. Indeed, among hundreds of genes connected to orofacial clefts, IRF6 is extraordinary because rare mutations cause syndromic forms of both CL/P and CP, and in addition, common genetic variants contribute risk for non-syndromic forms of both CL/P and CP.
Here we conducted a GWAS in the Finnish population and report a novel association between isolated CP and the IRF6 gene locus. The risk allele is a low-frequency, non-coding SNP located in MCS-9.7, a known enhancer for the IRF6 gene22. Using in vitro assays, we find evidence that the variant directly affects IRF6 expression through altered binding of IRF6 to this enhancer. This study adds to the short list of common variants whose contribution to etiology of non-syndromic OFCs has been evaluated in molecular detail. To our knowledge, this is the first instance of variants within a single enhancer being linked to the risk of both CP and CL/P.
Results
A low-frequency SNP in the IRF6 enhancer MCS-9.7 is associated with CP
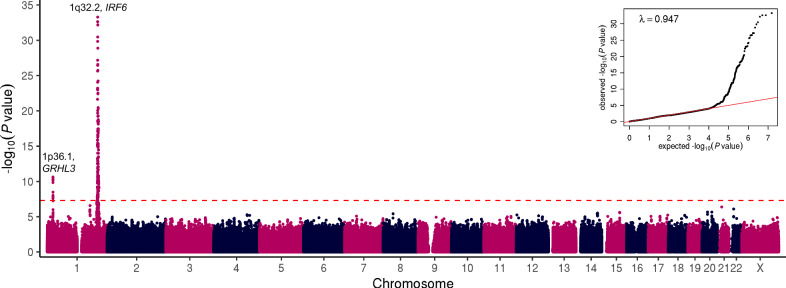
We conducted a GWAS of non-syndromic OFC in the Finnish population using the FinnGen study cohort with a mixture of 355 CL/P and CP cases, and 308,799 ethnicity-matched population controls (Data Freeze [DF] 7). We identified genome-wide significant (P < 5 × 10−8) associations with OFC and two genomic loci located on the long and short arms of chromosome 1 (Supplementary Fig. 1a). Upon dividing the case group into subtypes of clefts, we discovered that the association signals at both loci were driven by the subgroup of 228 cases with CP (Fig. 1). GWAS of 151 cases with CL/P did not yield any genome-wide significant hits (Supplementary Fig. 1b). Due to small sample sizes, we did not conduct independent analyses in the CL and CLP subsets. Variants reaching P values less than 1.0 × 10–4 in all three association analyses are provided in Supplementary Data 1, 2, 3.
Fig. 1. Manhattan and quantile-quantile plots showing GWAS results of 228 cases affected with non-syndromic CP and 308,799 population controls in the FinnGen discovery cohort.
Negative log10 P values (y axis) are plotted for each tested variant against their chromosomal coordinates (x axis) provided in the human genome build GRCh38/hg38. Two-sided P values are obtained from a likelihood ratio test in regression analysis and are not corrected for multiple comparisons. Red dashed line represents the threshold for genome-wide statistical significance (P = 5 × 10–8 or log10(P) = 7.3) after Bonferroni correction for multiple hypothesis testing. A quantile-quantile plot is shown in the inset panel, where the observed ( y axis) negative log10 P values are plotted against the expected (x axis) negative log10 P values under null distribution (red line).
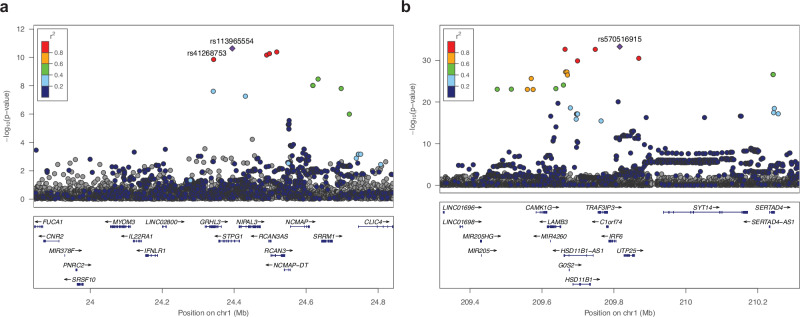
In the GWAS of CP, ten SNPs reached genome-wide significance on chromosome 1p36.1, including the lead SNP rs113965554 (P = 2.32 × 10–11, OR = 2.73) and the previously described etiologic missense variant p.Thr454Met (rs41268753, P = 1.39 × 10–10, OR = 2.66) in the GRHL3 gene (Table 1)13,14. The top five variants with similar P values are in strong linkage disequilibrium (LD) with one another (Fig. 2a). These results indicate that the 1p36.1 signal is likely driven by the GRHL3 missense variant, replicating the association between this common missense SNP and CP in the Finnish population13.
Table 1.
Discovery, replication, and meta-analysis of lead SNPs on chromosome 1p36.1 and 1q32.2 associated with CP in the Finnish and Estonian populations
| SNP | RA | FinnGen Discovery (228 cases, 308,799 controls) | FinnGen Replication (165 cases, 199,935 controls) | Estonian Biobank (71 cases, 198,973 controls) | Meta-analysis | RAF in gnomAD (v3.1.2) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | OR | 95% CI | P value | RAF | OR | 95% CI | P value | RAF | OR | 95% CI | P value | OR | 95% CI | P value | FIN | NFE | All | ||
| rs113965554* (rs41268753) | A (T) | 4.92 (4.89) | 2.73 (2.66) | 2.03–3.66 (1.97–3.58) | 2.32 × 10–11 (1.39 × 10–10) | 4.96 (4.91) | 2.15 (2.17) | 1.47–3.14 (1.49–3.17) | 7.37 × 10-5 (5.64 × 10-5) | 3.21 (3.21) | 2.39 (2.36) | 1.23–4.66 (1.21–4.57) | 0.01 (0.01) | 2.48 (2.45) | 1.99–3.09 (1.96–3.05) | 4.55 × 10–16 (1.95 × 10–15) | 5.19 (5.13) | 3.49 (3.37) | 2.35 (2.24) |
| rs7546104 | A | 24.11 | 1.95 | 1.64–2.33 | 9.53 × 10–14 | 24.40 | 1.92 | 1.56–2.36 | 5.87 × 10–10 | 23.69 | 1.83 | 1.32–2.55 | 0.0003 | 1.92 | 1.70–2.18 | 4.97 × 10–25 | 25.01 | 17.50 | 13.39 |
| rs570516915 | G | 1.20 | 8.65 | 6.11–12.25 | 5.25 × 10–34 | 1.29 | 9.39 | 6.28–14.04 | 8.82 × 10–28 | 0.24 | 13.55 | 4.21–43.63 | 1.25 × 10–5 | 9.14 | 8.89–9.40 | 4.34 × 10–64 | 1.55 | 0.08 | 0.15** |
Two-sided P values are obtained from a likelihood ratio test in regression analysis and are not corrected for multiple comparisons. P values of the meta-analysis are two-sided P values derived from a fixed-effect meta-analysis and were not adjusted for multiple testing. *rs113965554 and rs41268753 (the etiologic missense variant p.Thr454Met in GRHL3) are in nearly complete LD (r2 = 0.99) with each other. **Besides Finnish and non-Finnish Europeans, only 3 alleles were found in African/African Americans in the gnomAD (v3.1.2) database.
RA risk allele, OR odds ratio, CI confidence interval, RAF risk allele frequency (%), FIN Finnish, NFE Non-Finnish European.
Fig. 2. Regional association plots of the chromosome 1 risk loci for non-syndromic CP.
Data are shown for association on chromosome (a) 1p36.1 (GRHL3) and (b) 1q32.2 (IRF6). Negative log10 P values ( y axis) are shown for variants (x axis) within a 1 Mb region centered at the reference SNP. Two-sided P values are obtained from a likelihood ratio test in regression analysis and are not corrected for multiple comparisons. The reference SNP is marked with a purple diamond, and pairwise LD (r 2) between the reference SNP and other variants are indicated by color. The r 2 values were estimated from high-coverage whole-genome sequences of 3775 Finns. Both directly genotyped and imputed SNPs are plotted. Genomic coordinates are shown according to the human genome build GRCh38/hg38.
Most importantly, we observed significant association signals on chromosome 1q32.2, where a total of 240 variants spanning over 7 Mb reached genome-wide significance. Examination of allele frequencies and LD patterns suggested multiple independent signals in the region covering the IRF6 gene (Fig. 2b). One set of variants comprised a 62 kb-long haplotype of strongly correlated variants upstream of IRF6, with the lead SNP rs7546104 reaching a P value of 9.53 × 10–14 (OR = 1.95, Table 1, Supplementary Fig. 2a, b). One of the SNPs within this haplotype, rs72741048 (P = 1.15 × 10–11, Supplementary Fig. 2b), has previously been linked to OFC, including CP, in a large GWAS in the Han Chinese population8.
A second, highly significant set of associated variants in 1q32.2 was defined by the lead SNP rs570516915 (P = 5.25 × 10–34, OR = 8.65, Fig. 2b). This signal is a novel association with CP and the lead SNP changes a highly-conserved nucleotide (Supplementary Fig. 3) in a previously-described cis-regulatory element 9.7 kb upstream of IRF6 transcription start site (i.e., multi-species conserved sequence, 9.7 kb, MCS-9.7)22. The low-frequency risk allele G (1.2%) of rs570516915 appears to have originated on the more prevalent haplotype tagged by the risk allele A (24.1%) of rs7546104. Subsequently, the rs570516915 variant has increased in frequency through the founding bottleneck effect in the Finnish population; occurring 19 times more frequently in Finns compared to non-Finnish Europeans and extremely rare in non-European populations (Table 1). These SNP alleles are in complete disequilibrium (D′ = 1.0) but poorly correlated (r 2 = 0.037) due to their disparate frequencies. After conditioning on rs570516915, the rs7546104 variant showed residual evidence of association exceeding genome-wide significance (ORcond = 1.75, Pcond = 3.98 × 10–10), supporting earlier evidence that rs7546104 tags an independently acting association signal8 (Supplementary Fig. 2c, d). Because of the high D′ value between the two alleles, reciprocal conditioning was not possible. However, the role of rs570516915 in CP susceptibility was already evident by its robust association with large effect size and location in an essential craniofacial gene enhancer. Further analysis of genotype effects on CP risk indicated that the frequency of CP cases rose significantly when the rs570516915 risk allele dosage on the ancestral risk haplotype tagged by rs7546104 was increased to the heterozygous or homozygous state (almost tenfold or fourfold, respectively; Supplementary Data 4). Thus, rs570516915 defines an independent CP risk haplotype that contributes greater, and stronger, risk specifically in the Finnish population.
Furthermore, the risk allele (G) of rs570516915 is linked (D′ = 1.0, r 2 = 0.0034) to the non-risk allele (also G) of the previously described common CL/P risk variant rs642961 in the same enhancer22, suggesting that the association signals are independent of one another. Supporting this conclusion, the rs570516915 variant shows a significant association with CP alone, while rs642961 is strongly associated with CL and modestly associated with CLP22. The risk allele A of rs642961, which has a frequency of 22% in the Finnish population, did not reach statistical significance in the Finnish cohort of CL/P cases evaluated here (P = 0.4), possibly because of the small sample size, and showed nonsignificant negative association with CP (OR = 0.71, 95% CI 0.57–0.89, P = 0.003).
Replication in independent Finnish and Estonian cohorts
To replicate the association signals identified in the CP cohort, we performed GWAS in an independent sample of 165 CP cases and 199,935 populations controls from the FinnGen study (DF8-DF12), and 71 CP cases and 198,973 ethnicity-matched population controls from the Estonian Biobank. The Estonian population is geographically and linguistically relatively close to the Finnish population, and many genetic associations in the Finnish population have been replicated in the Estonian Biobank23. Risk allele of the MCS-9.7 variant rs570516915 also showed genome-wide significant association in the Finnish replication cohort (P = 8.82 × 10–28) and suggestive evidence of association in the Estonian cohort (P = 1.25 × 10–5) with the direction of effect consistent across all three cohorts. In a meta-analysis of the discovery and two replication datasets, the rs570516915 variant yielded a P value of 4.34 × 10–64. Summary statistics of the other two independent signals in the replication cohorts and meta-analysis results are shown in Table 1. Variants reaching P values less than 1.0 × 10–4 in these additional analyses are shown in Supplementary Data 5, 6, 7. Notably, in a small cohort of Estonian CL/P cases (n = 53), none of these variants showed significant association. In summary, the novel association of rs570516915 with CP was confirmed in independent datasets.
Regional correlation of rs570516915 allele frequency with CP prevalence
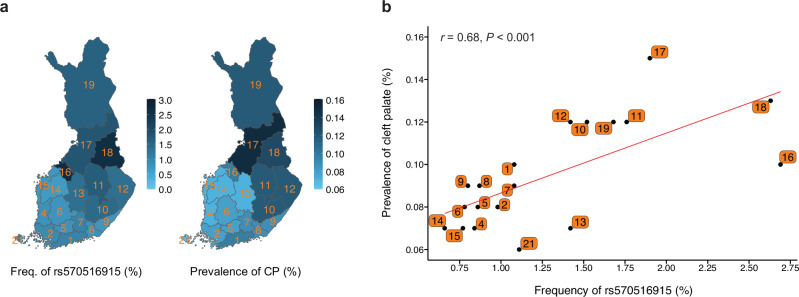
To test whether the distribution of the risk allele tracked with CP prevalence across the 19 administrative regions of Finland, we estimated the distribution of allele frequency using region-level birthplace registry data of 306,678 FinnGen study participants (~5.5% of Finland’s total population). We observed that the risk allele shows higher frequency in the eastern and northern regions of the country, reaching up to 2.7% (Fig. 3a and Supplementary Data 8), where the incidence of CP is also known to be higher compared to the western and southern regions of the country5–7. The risk allele frequency in western and southern regions is lower than the average allele frequency in the entire country (i.e., 1.2%). We next estimated the prevalence of CP in each region of Finland using data from a large, nationwide registry study (FinRegistry, https://www.finregistry.fi). In electronic health records of 5,216,731 individuals, we identified 5162 individuals with ICD codes related to non-syndromic CP. Strikingly, the CP prevalence also showed a geographic distribution that increased from south to north and from west to east (Fig. 3a and Supplementary Data 8). Most importantly, we found a strong and statistically significant correlation (Pearson’s r = 0.68, P < 0.001) between the regional distribution of the risk allele and CP prevalence (Fig. 3b). On the other hand, the frequency of the risk allele of the other two associated SNPs, rs41268753 (p.Thr454Met in GRHL3) and rs7546104 (IRF6 locus), did not vary similarly across the 19 regions in Finland. Therefore, of these three CP-associated SNPs, only rs570516915 appears to contribute to the regional distribution pattern of CP in Finland. This observation also supports our earlier conclusion that the effect of rs570516915 is independent of rs7546104.
Fig. 3. Geographic distribution and correlation of the rs570516915 variant allele frequency with regional prevalence of non-syndromic CP in Finland.
a Distribution of the rs570516915 G allele frequency and CP prevalence in 19 administrative regions of Finland. The variation in allele frequency and CP prevalence across distinct regions are illustrated by the intensity of the blue shading. Regions are labeled by their corresponding codes from Statistics Finland and shown in Supplementary Data 8. b Correlation between the regional allele frequency of rs570516915 and prevalence of CP. Allele frequency was estimated using birthplace data of 306,678 FinnGen study participants. Regional CP prevalences were estimated using nationwide data on the first recorded addresses and CP related diagnosis codes of 5,216,731 individuals (5162 with CP). Red line represents the ordinary least squares regression line. Pearson’s r and P value (two-sided) shown at the top of the plot indicate the strength and statistical significance of correlation. See Supplementary Data 8 for raw numbers.
We also estimated the overall impact of these three variants on CP in the Finnish population by calculating their individual and joint population attributable risks (PAR). Even though rs570516915 has a much lower frequency than rs41268753 and rs7546104 in Finland (4 times and 20 times lower, respectively, Table 1), its PAR (16.2%) is comparable to that of rs41268753 (14.4%) and is nearly half of rs7546104 (27.3%). The latter was calculated using an adjusted odds ratio from the conditional analysis and frequency of the risk allele (22.9%) that is not linked to rs570516915. The reason for the strong PAR by rs570516915 is its large effect size. The large PAR for rs7546104 is likely overestimated since the frequency, effect size and the number of the true causal variant(s) tagged by rs7546104 are unknown. The combined PAR of these three variants on CP is 47.8%.
rs570516915 diminishes the regulatory activity of the IRF6 enhancer MCS-9.7
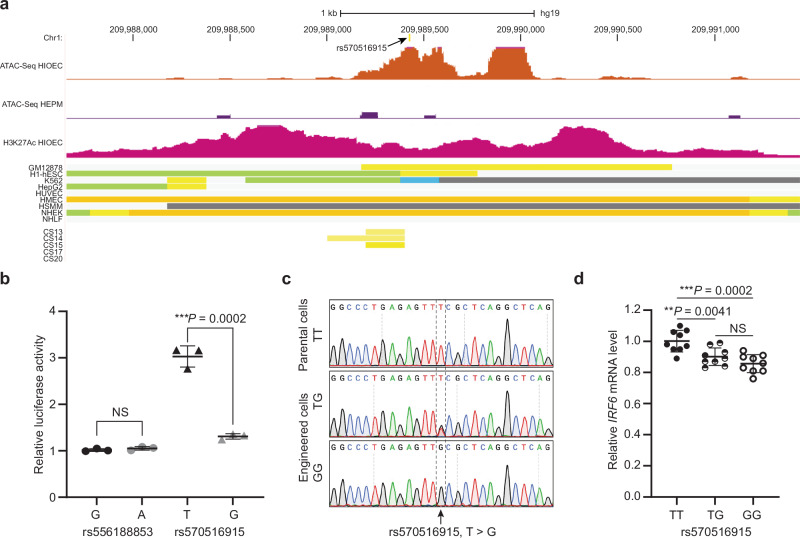
Six SNPs were in strong LD (r 2 > 0.8) with the lead SNP rs570516915. To prioritize these SNPs for possible regulatory variants, we examined their position relative to chromatin marks shown to correlate with active enhancer function including: (i) ATAC-seq peaks, which reveal open chromatin, from the HIOEC oral epithelial cells and the HEPM oral mesenchymal cells24, (ii) H3K27Ac peaks, revealing active enhancers and promoters, from the HIOEC cells24, and (iii) aggregate chromatin modification marks from 9 principal cell types from the ENCODE project25 and from human embryonic facial enhancers datasets26. Of these seven SNPs, the lead SNP and one other fall into regions with chromatin marks consistent with enhancer activity in epithelial cells (HIOEC and NHEK) (Fig. 4a and Supplementary Fig. 4c). As mentioned above, the lead SNP rs570516915 is located in MCS-9.7, a known enhancer for IRF622 in which two deleterious variants have been associated with OFCs previously22,27. The second SNP, rs556188853, is located 58 kb downstream from IRF6. We next engineered 701 bp DNA fragments including the ATAC-seq peak in HIOEC cells containing rs570516915 or rs556188853 and harboring either the risk-associated or non-risk associated allele of the SNP into a luciferase-based reporter vector (Supplementary Methods, Supplementary Fig. 5). We independently transfected these constructs into primary human neonatal epidermal keratinocytes (HEKn). The vector harboring MCS-9.7 with the ancestral, non-risk allele of rs570516915 drove luciferase levels above background. However, the vector harboring the derived risk allele of rs570516915 showed significantly reduced activity relative to the one harboring the non-risk allele (P = 0.0002) (Fig. 4b). By contrast, the vectors harboring the region surrounding rs556188853 did not drive luciferase levels above background, and there was no significant difference in luciferase levels between the vectors harboring the two alleles (Fig. 4b). These results support that the risk allele at rs570516915 is functional by reducing the activity of the MCS-9.7 enhancer in oral keratinocytes.
Fig. 4. The rs570516915 variant reduces enhancer activity of MCS-9.7.
a Browser view of the human genome, GRCh37/hg19, focused on the rs570516915 variant. Top two tracks represent open chromatin peaks detected by ATAC-Seq in HIOEC and HEPM cells, respectively. Next track shows H3K27Ac marks illustrating rs570516915 as a part of enhancer element. Following color coded bars represent chromatin status revealed by ChIP-Seq to various chromatin marks from the ENCODE Project cell lines and facial explants from human embryos at Carnegie stage (CS) 13–20 where orange and yellow bars represent the active and weak enhancer element, respectively; blue, insulator; light green, weak transcribed; gray, Polycomb repressed; light gray, heterochromatin/repetitive; GM12878, B-cell derived cell line; ESC, embryonic stem cells; K562, myelogenous leukemia; HepG2, liver cancer; HUVEC, human umbilical vein endothelial cells; HMEC, human mammary epithelial cells; HSMM, human skeletal muscle myoblasts; NHEK, normal human epidermal keratinocytes; NHLF, normal human lung fibroblasts, CS13-CS20 are facial explants from human embryos. b Scattered dot plot of relative luciferase activity for non-risk and risk alleles of rs556188853 and rs570516915 in HEKn cells. Data are represented as mean values +/− s.d. from three independent experiments. Statistical significance is determined by Student’s t test. P value (two-tailed) is indicated on the plot and NS represents non-significant (P = 0.2452). c Chromatograms illustrating the three genotypes (TT, TG and GG) of rs570516915 in iPSCs generated by CRISPR-Cas9 mediated homology-directed repair. d Scattered dot plot of relative levels of IRF6 mRNA in edited vs parental iOECs assessed by qRT-PCR. Expression levels of IRF6 are normalized against ACTB, GAPDH, HPRT, UBC and CDH1. Data are represented as mean values +/− s.d. from nine replicates of cells harboring each genotype, as indicated in the plot. Statistical significance is determined by Student’s t test (two-tailed). NS represents non-significant (P = 0.1212).
To test the in vivo effect of the risk allele on enhancer activity, we performed a transgenic F0 embryo assay with the MCS-9.7 enhancer fused to the lacZ reporter gene28 (Supplementary Methods). We produced embryos in three experimental batches, and in each batch, we obtained embryos with single or tandem (2 or more) insertions into a safe harbor locus or as random insertions. At E13.5, most embryos carrying the MCS-9.7-lacZ transgene, whether with the risk or non-risk allele, showed staining in the ectoderm of face, torso, limbs, eye and pinna, consistent with previous staining patterns22,29. The intensity of X-gal staining, which showed variation both between and within batches, was assessed by blinded observers in single and tandem insertion embryos separately; random insertion embryos were not evaluated (Supplementary Fig. 6). In three of the sets, non-risk allele carrying embryos were scored on average as having higher X-gal staining intensity than risk-allele carrying embryos (Supplementary Fig. 6 and Supplementary Data 9). However, we did not observe a consistent relationship between genotype and X-gal staining intensity. Therefore, the results based on the transient transgenic embryo assay were inconclusive. Experiments with F1 embryos from multiple transgenic lines may be needed to detect the subtle effects of a common variant on the activity of this enhancer.
We next determined the effect of rs570516915 in its native chromatin context on the expression of IRF6 in embryonic oral epithelial cells derived from induced pluripotent stem cells (iPSCs) (i.e., induced oral epithelial cells, iOECs). Sequencing showed that WTC11 iPSCs were homozygous for the non-risk allele (i.e., TT) of rs570516915. We transfected these cells with appropriate CRISPR-Cas9 reagents to render them heterozygous (i.e., GT) or homozygous (i.e., GG) for the risk allele and isolated three independent clones of both genotypes (Fig. 4c and Supplementary Figs. 7, 8). We subjected iPSCs of each genotype to a differentiation protocol that converts them to embryonic oral epithelium30 (Supplementary Fig. 8). Quantitative RT-PCR and immunostaining revealed induction of multiple epithelial (CDH1, KRT8, KRT18, IRF6 and TP63) and oral (PITX1 and PITX2) markers in iOECs relative to undifferentiated parental iPSCs (Supplementary Figs. 9, 10). Interestingly, IRF6 mRNA levels were lower in cells heterozygous or homozygous for the risk allele compared with the cells homozygous for the non-risk allele (Fig. 4d). These findings provide evidence that the risk allele at rs570516915 directly affects risk for CP by reducing steady state levels of IRF6 mRNA.
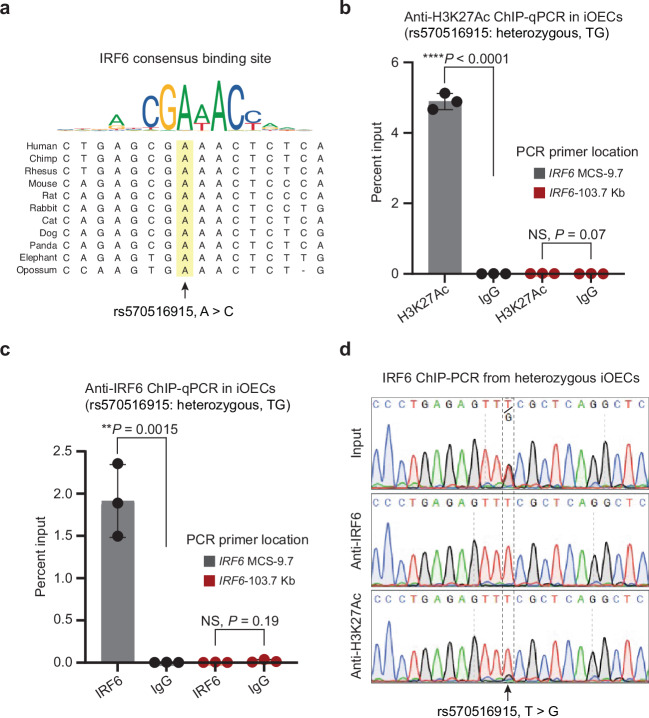
rs570516915 impairs the autoregulatory activity of the IRF6 gene
Motif analysis of the conserved sequences surrounding rs570516915 revealed that the risk-associated allele alters a consensus binding site for IRF6, a transcription factor known to bind DNA (Fig. 5a and Supplementary Fig. 11). Prior in vitro31 and in vivo32 studies showed that IRF6 regulates its own expression, suggesting that the risk-associated allele of rs570516915 impairs IRF6 autoregulation. To test this hypothesis, we performed chromatin immunoprecipitation in iOECs that were heterozygous for the risk and non-risk alleles of rs570516915 with anti-H3K27Ac and with anti-IRF6 antibodies followed by quantitative PCR with primers flanking rs570516915 (ChIP-qPCR), or with primers at a negative control locus lacking evidence for enhancer activity in epithelial cells. Quantitative PCR revealed much higher binding of H3K27Ac and of IRF6 at MCS-9.7 versus at the negative control locus in iOECs (Fig. 5b, c). Interestingly, PCR followed by sequencing of the DNA precipitated by anti-H3K27Ac and by anti-IRF6 revealed that both antibodies preferentially precipitated the non-risk allele of rs570516915 at the MCS9.7 locus (Fig. 5d); the same observation was made in two other biological replicates (Supplementary Fig. 12). In contrast, no allele bias was detected in chromatin precipitated with anti-IRF6 at a heterozygous SNP located beneath a different peak of IRF6 binding that is not within a predicted IRF6 binding site (Supplementary Fig. 13a–c). These data are consistent with a mechanism whereby rs570516915 hinders the binding of IRF6 protein at a critical site in MCS-9.7 to destabilize a positive feedback loop for IRF6 expression that is required for proper palatal development (Supplementary Fig. 14).
Fig. 5. rs570516915 alters an IRF6 binding site and perturbs positive autoregulation of IRF6 expression via the MCS-9.7 enhancer.
a Consensus IRF6 binding motif from the JASPAR database of transcription factor DNA-binding preferences (Matrix ID: PB0036.1) and alignment of the variant site in different species, which shows that the ancestral allele A is completely conserved in mammals and is critical for binding of IRF6. Note that this A corresponds to the T allele of rs570516915 on the opposite DNA strand. Percent input identified by ChIP-qPCR for (b) anti-H3K27Ac and (c) anti-IRF6, respectively, in iOECs heterozygous for rs570516915 using primers specific to the MCS-9.7 enhancer site or, as a negative control, to a region 103.7 kb upstream IRF6 transcription start site that did not harbor active elements identified from ATAC-Seq and H3K27Ac ChIP-Seq in HIOEC or NHEK cells and devoid of predicted IRF6 binding sites. Error bars refer to three ChIP replicates and expressed as mean values +/− s.d. Statistical significance is determined by Student’s t test. P value (two-tailed) is indicated on the plot and NS represents non-significant. d Sequencing chromatograms of anti-IRF6 and anti-H3K27Ac ChIP-PCR products of cells heterozygous for rs570516915.
Discussion
Following multiple waves of settlement after the last glacial period, several genetic founder and bottleneck events have occurred in Finland. Despite the considerable expansion of the population since the early 1800s, regions of genetic isolation have persisted33,34. Consequently, due to strong genetic drift, individuals of Finnish ancestry have numerous deleterious alleles of clinical significance at a relatively high frequency compared to non-Finnish Europeans35. This is well-exemplified by a group of rare monogenic diseases, also known as the Finnish disease heritage, that exists at a higher frequency in Finland than in other Nordic countries36,37, which all share similar living standards, climate, infectious diseases, diet, and cultural habits. Examples of strong regional concentration of polygenic diseases also exist; for instance, schizophrenia, cognitive impairment, and familial hypercholesterolemia are more prevalent in the northeastern region of the Finland than in other regions and show association with regionally enriched genetic variation38,39. It was postulated before the genomics era that the relatively high incidence of CP in Finland compared to other Nordic countries, which all have similar total incidence of clefts as Finland, is likely to result from the distinct components of genetic heritage of Finns4. Here we have shown an association between three independent SNPs and CP in Finland. Their high allele frequency, compared to other populations, and their relatively strong effect size help to account for the high incidence of CP in Finland.
Further, we found evidence that one of the three SNPs, rs570516915, directly elevates risk for CP by perturbing positive regulation by IRF6 of its own expression. Previously, chromatin immunoprecipitation combined with next generation sequencing (ChIP-seq) in primary human keratinocytes identified a 245 bp sequence within the MCS-9.7 enhancer of IRF6 where the IRF6 protein binds31. Furthermore, an in vivo study showed that Irf6 is required for enhancer activity of MCS-9.7 in tissues derived from ectodermal germ layer during lip and palate development in mouse embryos32. Here, we identified an OFC-associated risk variant in a conserved IRF6 binding site within MCS-9.7. The risk allele of this SNP significantly reduced MCS-9.7 enhancer activity in transient luciferase reporter assays in keratinocytes in vitro. As a limitation, we did not see a consistent effect in an F0 transgenic mouse reporter assay, in contrast to a rare Van der Woude syndrome (VWS)-associated variant in MCS-9.7, which had a strong effect in the identical assay27. The differential effects in this assay likely reflect the small and large effects that these two variants have on OFC risk, respectively. Nonetheless, altering rs570516915 within the genome of induced oral epithelial cells was sufficient to significantly decrease binding of IRF6 and expression of IRF6.
These and the published data mentioned above are consistent with a model where autoregulation of IRF6 occurs through direct binding of the IRF6 protein to the conserved IRF6 binding site in MCS-9.7, and that the risk allele of rs570516915 impairs this autoregulation. Nevertheless, more complex scenarios cannot be ruled out. For example, we note that while the conserved IRF6 binding site that contains rs570516915 is located within MCS-9.7, it is located 40 bp outside of the previously described 245 bp region of chromatin bound by IRF631. Similarly, it is located 22 bp from a region of chromatin marked as active enhancer in human embryonic faces collected at Carnegie stages 13-1526 (Supplementary Fig. 15). Thus, indirect mechanisms of autoregulation cannot be excluded.
IRF6 is extraordinary because genetic variation in this gene cause and contribute risk for both syndromic and non-syndromic forms of both CL/P and CP. Remarkably, different variants within the MCS-9.7 enhancer of IRF6 are also associated with this phenotypic spectrum of CL/P, CP, or both. For the syndromic forms, a single nucleotide duplication, chr1:g.209816135dup (GRCh38/hg38), aka 350dupA, was found in one VWS family, where two members had CL/P and one member had CP27. For the non-syndromic forms, the common SNP rs642961 (chr1:209815925), was associated with non-syndromic CL/P22, and now the low-frequency Finnish-enriched SNP rs570516915 (chr1:209816078) described here is associated with non-syndromic CP. All three apparently functional variants (i.e., 350dupA, rs642961, and rs570516915) are located within a 212 bp segment of the MCS-9.7 enhancer (Supplementary Fig. 15). In summary, distinct mutations in the same enhancer predispose individuals to CP and CL/P in both their syndromic and non-syndromic forms.
While MCS-9.7 is active in basal and superficial (periderm) epithelial layers, recapitulating endogenous Irf6 expression patterns29, we favor periderm as the relevant tissue where rs570516915 contributes risk to CP. Both layers are important during fusion of the palate shelves21,40 and during fusion of the lip at the lambdoidal junction41. It is possible that different mutations in MCS-9.7 differentially affect its enhancer activity in basal and periderm layers and thereby differentially influence risk for CP versus CL/P. Our rationale is based on the overlapping genetics and function between IRF6 and GRHL3. Specifically, rs570516915 in IRF6 MCS-9.7 is associated with non-syndromic CP but not with non-syndromic CL/P, which is also true of the common risk variant in GRHL3. Also, while rare variants in both IRF6 and GRHL3 cause VWS, individuals with a mutation in GRHL3 are more likely to have CP than CL/P17. Finally, the primary pathologies in mice that lack either Irf618,19 or Grhl317 are abnormal intraoral epithelial adhesions and failure of periderm formation or function, which hinder development of the palatal shelves and can cause cleft palate40. Precise genome editing at these conserved variant sites in mouse models may reveal the distinct effects of the variants on Irf6 expression in periderm versus basal layers.
In conclusion, the MCS-9.7 regulatory region may represent a mutational hot spot for both rare and common genetic variation that can cause, or contribute risk for, different forms of OFCs potentially by disrupting IRF6 expression in periderm versus in basal layers of oral epithelium.
Methods
FinnGen cohorts
The FinnGen study is a population-based cohort study launched in 2017 as a public-private partnership aiming to identify genotype-phenotype relationships in the Finnish founder population23. Three national and six regional biobanks provide samples to the FinnGen study. Individuals from several previously established prospective epidemiological and disease cohort studies are also included in FinnGen. Participants in FinnGen provided informed consent for biobank research in accordance with the Finnish Biobank Act (law 688/2012). Alternatively, separate research cohorts, collected prior to when the Finnish Biobank Act came into effect (in September 2013) and the start of FinnGen (August 2017), were collected based on study-specific consents and later transferred to the Finnish biobanks after approval by Fimea (Finnish Medicines Agency). Recruitment protocols followed the biobank protocols approved by Fimea. Demographic and clinical data are derived from nationwide electronic health registers. These registers contain records of major health-related events, such as hospitalizations, prescription drug purchases, medical procedures, or deaths with a data collection history spanning over 50 years. This study included 309,154 participants from DF7 as the discovery cohort and 200,100 participants from DF8 through DF12 in the replication cohort. All participants had a Finnish genetic ancestry determined by a principal component (PC) analysis of their genotypes.
FinnGen ethics statement
The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) statement number for the FinnGen study is Nr HUS/990/2017. The FinnGen study is approved by the Finnish Institute for Health and Welfare (THL, permit numbers [PN]: THL/2031/6.02.00/2017, THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019 and THL/1524/5.05.00/2020); the Digital and Population Data Services Agency (PN: VRK43431/2017-3, VRK/6909/2018-3, VRK/4415/2019-3); the Social Insurance Institution (PN: KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, KELA 134/522/2019, KELA 138/522/2019, KELA 2/522/2020, KELA 16/522/2020); Findata (PN: THL/2364/14.02/2020, THL/4055/14.06.00/2020, THL/3433/14.06.00/2020, THL/4432/14.06/2020, THL/5189/14.06/2020, THL/5894/14.06.00/2020, THL/6619/14.06.00/2020, THL/209/14.06.00/2021, THL/688/14.06.00/2021, THL/1284/14.06.00/2021, THL/1965/14.06.00/2021, THL/5546/14.02.00/2020, THL/2658/14.06.00/2021, THL/4235/14.06.00/2021); Statistics Finland (PN: TK-53-1041-17 and TK/143/07.03.00/2020 [earlier TK-53-90-20], TK/1735/07.03.00/2021, TK/3112/07.03.00/2021) and the Finnish Registry for Kidney Diseases (permission/extract from the meeting minutes on July 4, 2019). The Biobank Access Decisions for FinnGen samples and data include: THL Biobank BB2017_55, BB2017_111, BB2018_19, BB_2018_34, BB_2018_67, BB2018_71, BB2019_7, BB2019_8, BB2019_26, BB2020_1, BB2021_65, Finnish Red Cross Blood Service Biobank 7.12.2017, Helsinki Biobank HUS/359/2017, HUS/248/2020, HUS/430/2021 §28, §29, HUS/150/2022 §12, §13, §14, §15, §16, §17, §18, §23, §58, §59, HUS/128/2023 §18, Auria Biobank AB17-5154 and amendment #1 (August 17, 2020) and amendments BB_2021-0140, BB_2021-0156 (August 26, 2021, February 2, 2022), BB_2021-0169, BB_2021-0179, BB_2021-0161, AB20-5926 and amendment #1 (April 23, 2020) and its modifications (September 22, 2021), BB_2022-0262, BB_2022-0256, Biobank Borealis of Northern Finland_2017_1013, 2021_5010, 2021_5010 Amendment, 2021_5018, 2021_5018 Amendment, 2021_5015, 2021_5015 Amendment, 2021_5015 Amendment_2, 2021_5023, 2021_5023 Amendment, 2021_5023 Amendment_2, 2021_5017, 2021_5017 Amendment, 2022_6001, 2022_6001 Amendment, 2022_6006 Amendment, 2022_6006 Amendment, 2022_6006 Amendment_2, BB22-0067, 2022_0262, 2022_0262 Amendment, Biobank of Eastern Finland 1186/2018 and amendment 22§/2020, 53§/2021, 13§/2022, 14§/2022, 15§/2022, 27§/2022, 28§/2022, 29§/2022, 33§/2022, 35§/2022, 36§/2022, 37§/2022, 39§/2022, 7§/2023, 32§/2023, 33§/2023, 34§/2023, 35§/2023, 36§/2023, 37§/2023, 38§/2023, 39§/2023, 40§/2023, 41§/2023, Finnish Clinical Biobank Tampere MH0004 and amendments (21.02.2020 & 06.10.2020), BB2021-0140 8§/2021, 9§/2021, §9/2022, §10/2022, §12/2022, 13§/2022, §20/2022, §21/2022, §22/2022, §23/2022, 28§/2022, 29§/2022, 30§/2022, 31§/2022, 32§/2022, 38§/2022, 40§/2022, 42§/2022, 1§/2023, Central Finland Biobank 1-2017, BB_2021-0161, BB_2021-0169, BB_2021-0179, BB_2021-0170, BB_2022-0256, BB_2022-0262, BB22-0067, decision allowing to continue data processing until August 31, 2024 for projects: BB_2021-0179, BB22-0067,BB_2022-0262, BB_2021-0170, BB_2021-0164, BB_2021-0161, and BB_2021-0169, and Terveystalo Biobank STB 2018001 and amendment August 25, 2020, Finnish Hematological Registry and Clinical Biobank decision June 18, 2021, Arctic biobank P0844: ARC_2021_1001. In addition, for this project ethics approval was given from HUS Helsinki University Hospital (HUS/126/2021-39), and biobank samples were obtained through Fingenious BB_2020-0106, Dnro: HUS/150/2022, 250/2022, from Auria Biobank BB_2020-0106, Biobank Borealis of Northern Finland BB_2022-6002, Biobank of Eastern Finland BB_2020-0106, Dnro: 250/2022, Central Finland Biobank BB_2020-0106, Finnish Clinical Biobank Tampere BB_2020-0106, Helsinki Biobank Dnro: HUS/150/2022, 250/2022, THL Biobank THLBB2022_3.
Estonian cohort
Estonian Biobank is a population-based biobank housed at the Institute of Genomics, University of Tartu. The current cohort size is 200,000 individuals (aged ≥ 18), reflecting the ethnicities, ages, sex, and geographical distribution of the adult Estonian population. All subjects have been recruited by general practitioners and physicians in hospitals and during promotional events. Upon recruitment, all participants completed a questionnaire about their health status, lifestyle, and diet. The Estonian Biobank database is linked with national registries, such as Cancer Registry and Causes of Death Registry, hospital databases, and the national health insurance fund database, which holds treatment and procedure service bills. Diseases and health-related conditions are recorded as International Classification of Diseases (ICD) codes (version 10). These health data are continuously updated through periodical linking to national electronic databases and registries. An in-depth description of Estonian Biobank can be found elsewhere42,43. Ethical approval for this study was obtained from the Research Ethics Review Committee of the University of Tartu. All participants signed informed consent.
Phenotype definitions
Case subjects with non-syndromic forms of OFCs in FinnGen were defined from electronic health records in the Care Register for Health Care of the Finnish Institute for Health and Welfare (THL) and the Causes of Death Register from Statistics Finland, using diagnosis codes of the Finnish version of ICD (versions 10, 9, 8). Codes of versions ICD-10, ICD-9 and ICD-8 were available since 1996, between 1987–1995 and between 1967–1986, respectively. Cases with CP only (discovery cohort n = 228; 157 females and 71 males and replication cohort n = 165; 116 females and 49 males) were defined based on the ICD-10 code Q35, ICD-9 code 7490[ABCE], and ICD-8 code 74900. Cases with CLP (n = 121; 62 females and 59 males) were defined using the ICD-10 code Q37, ICD9 code 7492[ABCDE], and ICD8 codes 74920 and 74924. Cases with CL (n = 54; 28 females and 26 males) were defined using ICD10 code Q36, ICD9 codes 7491[ABCDEFG] and ICD8 codes 74910 and 7491[123]. Syndromic cases with the following ICD-10 codes were excluded: Q75.4 (mandibulofacial dysostosis), Q38.0 (congenital malformations of lips, not elsewhere classified, including VWS), Q86 (congenital malformation syndromes due to known exogenous causes, not elsewhere classified), Q87 (other specified congenital malformation syndromes affecting multiple systems) and Q91 (Edward’s syndrome and Patau’s syndrome). The Estonian cohort consisted of 71 CP cases and 198,973 population controls. Cases with isolated CP (n = 71; 54 females and 17 males) in the Estonian biobank were ascertained using the ICD-10 code Q35 through a questionnaire implemented by the biobank and national health registry-based medical records. The following ICD-10 codes for syndromic cases were excluded from both Estonian cases and controls: Q38.0 (congenital malformations of lips, not elsewhere classified, including VWS), Q38.1 (ankyloglossia), Q38.5 (congenital malformation of palate), Q87 (other specified congenital malformation syndromes affecting multiple systems), and Q91 (Edwards syndrome and Patau syndrome).
Genotyping and imputation
Samples collected and genotyped before the launch of the FinnGen study were genotyped with a mix of Illumina (Illumina) and Affymetrix arrays (Thermo Fisher Scientific). However, recently collected samples were genotyped on two versions of a custom designed Affymetrix array, referred to as FinnGen1 (657,675 markers) and FinnGen2 (664,510 markers), with 655,973 overlapping markers. Array content, sample genotyping and calling, imputation, PC analysis and estimation of population structure and ancestry, and quality control (QC) procedures are described in more detail by Kurki et al.23. Briefly, genotype calls were made with GenCall and zCall algorithms for Illumina, and AxiomGT1 algorithm for Affymetrix data. Data generated on previous reference genome builds were lifted over to build version 38 (GRCh38/hg38), as described in 10.17504/protocols.io.xbhfij6. After basic QC, such as exclusion of samples with missing sex information and those that were mixed up, sample and variant QCs were performed as follows. In sample-wise QC, individuals whose genetic sex mismatched sex provided from registries, with high genotype missingness (>5%), excess heterozygosity (±4 SD) and non-Finnish ancestry were excluded. In variant-wise QC, variants with high missingness (>2%), deviation from Hardy–Weinberg equilibrium (P < 1 × 10–6) and minor allele count (MAC) < 3 were excluded. Samples passing QC were then pre-phased chip-wise with Eagle (v2.3.5)44 using the default parameters, except that the number of conditioning haplotypes was set to 20,000. Finally, genotype imputation was carried out with Beagle 4.1 (version 08Jun17.d8b) by using a Finnish population-specific reference panel built from 3775 high-coverage (25–30×) whole-genome sequences (WGS) in Finns, as described in the following protocol: 10.17504/protocols.io.nmndc5e. Approximately 17 M variants were imputed. Variants with an imputation INFO score <0.8 and minor allele frequency (MAF) < 0.0001 were excluded, resulting in a combination of 15.36 M directly genotyped and imputed variants included in association tests. In total, 309,154 individuals (173,746 females and 135,408 males) passed QC and were included in the discovery GWAS, and 200,100 individuals (113,132 females and 86,968 males) were included in the replication GWAS. Samples from the Estonian Biobank cohort were genotyped at the Core Genotyping Lab of the Institute of Genomics, University of Tartu, using Illumina Infinium Global Screening arrays v1.0, v2.0 and v2.0_EST. Individuals whose sex defined based on chromosome X heterozygosity did not match the sex recorded in phenotype data were excluded from the analysis. Before pre-phasing and genotype imputation, genotyped variants were filtered by call rate <95%, Hardy–Weinberg equilibrium P < 1 × 10−4 (autosomal variants only), and minor allele frequency of 1%. Pre-phasing was performed using Eagle (v2.3.5)44, and imputation was performed using Beagle 5.0 (v.28Sep18.793)45 by using an Estonian population-specific imputation reference panel consisting of 2297 high-coverage (~30×) WGS data from Estonian individuals46.
Association analysis
Association analyses were carried out using the REGENIE program, which uses a two-step, whole-genome regression framework that controls for population stratification and sample relatedness47. We used the code version 2.0.2 of the program which was modified to include dosage-based calculation of allele frequencies in cases and controls. For step 1 LOCO prediction, we included sex as determined from genotypes, age, genotyping batch, and the first 10 PCs as covariates. We used a genotype block size of 1,000. Genetic relatedness in step 1 was calculated using a set of 55,139 LD-pruned, well-imputed, high-quality (INFO > 0.95 and MAF > 0.01) SNPs. LD pruning was performed with PLINK (v2.00a2.3LM)48 with a window size of 1 mb and r 2 = 0.1. Association of the phenotypes with all imputed genetic markers with INFO ≥ 0.8 and MAF ≥ 0.0001 were tested in step 2 using the logistic regression score test. To better control the type I error for rare and low-frequency genetic markers due to severely imbalanced case-control ratios, we used the approximate Firth test for variants with an initial P value of less than 0.01 and computed the standard error based on effect size and likelihood ratio test P value (regenie options --firth --approx --pThresh 0.01 --firth-se). Association testing in the Estonian Biobank cohort data was also performed using REGENIE (v2.2.4), including sex, age and the first 10 PCs as covariates. A minimum MAC of 5 was used when testing variants. Similar to the FinnGen GWAS, the approximate Firth test was applied to control type I error for rare and low-frequency variants due to severely imbalanced case-control ratios, with the following parameters (regenie options --firth --approx --pThresh 0.01 --firth-se). Manhattan plots were generated using the ggplot2 R package49. Regional association plots were generated with the stand-alone version of LocusZoom (v1.3)50, using the Finnish population-specific LD structure estimated in 3,775 whole-genome sequences of Finns. To test genotype distributions, we used BCFtools(v1.18)51 to extract SNP genotypes from VCF files, counted the number of genotypes in R (v4.3.2) and used a chi-squared test for independence of distributions of cases and controls between genotypes.
Meta-analysis
Meta-analysis of the discovery and replication cohort GWAS was performed using the inverse-variance weighted fixed effects model with the METAL software (version 2011-03-25)52.
Conditional analysis
Conditional analysis was performed using the COJO53 approach (--cojo-cond command) implemented in the GCTA software (v1.93.2)54. For this analysis, we used the summary statistics output from the REGENIE GWAS and imputed genotypes from the entire FinnGen cohort for LD estimation within 1 Mb in each direction from the conditioning SNP.
FinRegistry
To assess the geographic distribution of non-syndromic CP prevalence in different regions of Finland, we used data from a nationwide cohort (FinRegistry, https://www.finregistry.fi) that includes 5,216,731 individuals. We assigned each individual to one of the 19 administrative regions based on their first recorded address. CP cases were defined using the same ICD code-based inclusion and exclusion criteria as in FinnGen (see Phenotype definitions). The prevalence was estimated by dividing the number of CP cases in each region by the total number of individuals in that region.
Geographic variation
The geographic distribution of allele frequencies was calculated and plotted using region-level, birthplace data of 306,678 FinnGen study participants obtained from Statistics Finland, with the ggplot2, ggrepel and sf 55 R packages. Spatial polygon data (v3.6) for the Finnish map were downloaded from the database of Global Administrative Areas (https://gadm.org). Correlation between the regional CP prevalence and allele frequency was calculated and plotted using Pearson’s product-moment correlation in base R (v3.6.1).
Population attributable risk
The population attributable risk (PAR) is the proportion of cases that would be prevented if a risk factor were eliminated from the population. Individual and combined PAR of risk variants were calculated as described previously56, using the following formulas:
| 1 |
where p is the population frequency of the risk allele and β is the beta coefficient from the regression analysis, which shows the effect size of the variant on the phenotype and e β expresses odds ratio (OR), and
| 2 |
where the risk variants are assumed to be independent of one another and that their combined effects on disease are multiplicative.
Bioinformatic analysis
SNP overlap with open-chromatin peaks, ChIP-Seq peaks of histone activation marker (H3K27ac) from HIOEC oral epithelial cells and publicly available chromatin modification datasets from 9 principal cell types from the ENCODE Project Consortium25 and from different stages of embryonic facial tissues from the Cotney lab26 were visualized using the UCSC Genome Browser. H3K27Ac ChIP-Seq data from HIOEC oral epithelial cells and ATAC-Seq (Assay for Transposase-Accessible Chromatin using sequencing) data from both the HIOEC oral epithelial cells and the HEPM oral mesenchymal cells were generated previously24. In silico transcription factor binding activities at the risk variant site were determined using HOMER57 and the Transcription factor Affinity Prediction (TRAP) tool58. Position Weight Matrix of the consensus IRF6 binding motif was obtained from the JASPAR database59.
Cell lines
Primary Epidermal Keratinocytes: Normal, Human, Neonatal Foreskin (HEKn) purchased from American Type Culture Collection (ATCC PCS-200-010) were maintained and cultured as per manufacturer’s instruction in dermal cell basal medium (ATCC PCS-200-030) supplemented with keratinocyte growth kit (ATCC PCS-200-040). The human induced pluripotent stem cells (iPSCs), UCSFi001-A ([WTC11], Coriell Institute for Medical Research, Catalog no. GM25256), were maintained in mTeSR1 media (StemCell Technologies) on Matrigel (Corning) coated plates and passaged with Accutase. Cells were incubated at 37 °C in 5% CO2.
Electroporation and dual luciferase reporter assay
Each firefly reporter construct (Supplemental Methods) was co-transfected with a constitutively driven Renilla luciferase plasmid for dual luciferase assays. Briefly, HEKn cells were electroporated using the Amaxa Cell Line Nucleofector Kit V (Lonza, Catalog no. VCA-1003) and the Nucleofector I instrument (Lonza) program T-25. We used a dual-luciferase reporter assay system (Promega, Catalog no. E1980) and FB12 Luminometer (Berthold Detection Systems) to evaluate the luciferase activity following the manufacturer’s instructions. Relative luciferase activity was calculated as the normalized values of the Firefly to the Renilla enzyme activities. Three independent measurements were performed for each transfection group, and three biological replicates were performed. All results were presented as mean ± standard deviation (s.d.). Statistical significance was determined using the Student’s t test (two-tailed).
Transgenic mouse reporter assays
MCS-9.7-lacZ transgenic reporter constructs were generated with the risk and non-risk alleles of rs570516915 on the Finnish haplotype background with a mouse Shh promoter (Supplementary Methods). These constructs were co-injected with the Cas9 protein and gRNA targeting the H11 locus28 into the mouse embryos, and embryos were harvested at E13.5 and processed for X-gal staining as described previously22. Images of transgenic mouse embryos were collected using the Adobe Photoshop Elements 11 software. Plasmid copy number integration for transgenic embryos was estimated by qPCR with probes targeting the mouse Shh promoter region (Mm00560391_cn, Thermo Fisher Catalog no. 4400291). This probe targets both endogenous and plasmid copies. The Tfrc gene was used as a reference locus (Thermo Fisher Catalog no. 4458366). Three non-transgenic mouse DNA samples were used as normalization controls. To calculate an estimated plasmid integration copy number, the 2-ΔΔCt approach was used. Stained embryos were visually assessed and embryos with no staining at all were excluded from the analysis. Embryos were ordered according to the staining intensities in the heads, from the strongest to the weakest, by 14 (2 batches) and 7 (1 batch) independent observers who were blinded to the genotype of the reporter construct28. In each assessment, the embryos were given a score based to their position in the order (1, 2, etc.) and sums of these were used to define a consensus order (Supplementary Fig. 6 and Supplementary Data 9). Mice were housed in the animal facility where their health, food, water, and housing conditions were monitored with daily visual checks by technicians. Mice were housed in bioBUBBLE Clean Rooms, soft-walled enclosures powered by 80–100 air changes per hour of HEPA filtration under Light/Dark Cycle of 12:12 starting at 6AM, at 20.5–24.4 °C, and humidity 30–70%. All animals used in this study were of Mus musculus species and the FVB/NJ strain (7-8 weeks old egg donor females and 2–6 months old stud males) from the Jackson Laboratory, and the BDF-1 strain vasectomized males to generate pseudopregnancies (2 months to 2 years old) and the CD-1 strain recipient females (7−11 weeks old) from Charles River. Transgenic embryos were assayed at embryonic day 13.5. Sex was not considered in the study design as the phenotype of interest does not apply to only one sex. Mouse experiments were performed at the Lawrence Berkeley National Laboratory (LBNL) and were reviewed and approved by the LBNL Animal Welfare and Research Committee.
CRISPR-Cas9 mediated genome editing
Integrated DNA Technologies (IDT) CRISPR-Cas9 guide RNA (gRNA) design checker tool was used to design specific CRISPR RNA (crRNA) harboring the rs570516915 in the seed region to avoid the re-cleavage by CRISPR-Cas9 (Supplementary Fig. 7). A homology directed repair (HDR) template including the desired mutation was designed to have a minimum of 30-35nt long homology arms (Supplementary Fig. 7). Briefly, equimolar concentrations of crRNA and trans-activating crRNA (tracrRNA, IDT: 1072532) were annealed at 95 °C for 5 min followed by cooling at room temperature (RT) to form functional gRNA duplexes. Further, the ribonucleoprotein (RNP) complex was prepared 15 min before transfection by mixing gRNAs (1.5 µM) with Cas9 protein (0.45 µM) (Sigma, Catalog no. CAS9PROT) at RT. RNP complexes together with the repair template were transfected into 1 million WTC11-iPSCs with Amaxa nucleofector (Lonza, Human Stem Cell kit 2, program: A-13) in presence of ROCK inhibitor (Y-27632, Catalog no. 04-0012−10, Stemgent; [10 mM] stock). Individual colonies were hand-picked and plated into 96 well plates. DNA was extracted using Quick Extract DNA extraction solution (Epicenter Catalog no. QE09050). Colonies were screened by PCR and sequenced using primers flanking rs570516915 (Primer sequences are provided in Supplementary Data 10). Three independent clones each of heterozygous (TG), and homozygous risk (GG), and two clones of the homozygous non-risk (TT) genotype were isolated.
In vitro differentiation of iPSCs into embryonic oral epithelial cells
Three independent samples of each genotype (3 clones of the heterozygous genotype, 3 clones of the homozygous risk genotype, and 2 clones plus one sample of the parental WTC-11 cells of the homozygous non-risk genotype) were seeded in triplicate in 12-well plate (50,000 cells per well). Further, the cells were differentiated to embryonic oral epithelial cells using a 10-day differentiation protocol described previously30 (Supplementary Fig. 8). Briefly, on the first day of differentiation stem cell media is replaced with media consisted of EpiCult-C media (StemCell Technologies, Catalog no. 05630) mixed with EpiLife (Thermo, Catalog no. MEPI500CA) at 1:1 ratio, supplemented with 0.1x supplement S7 (Thermo, Catalog no. S0175), 0.1 µM β-mercaptoethanol (Sigma, Catalog no. M7522) and 400 µM smoothened agonist (Selleckchem, Catalog no. S7779). Further, 150pM of bone morphogenic protein-4 (R&D systems, Catalog no. 314-BP-010) is continuously added daily starting from day 3 to day 7 of differentiation. At day 8, the media is supplemented with 1uM of BMP-I inhibitor (LDN-193189) (Tocris, Catalog no. 6053), 5uM of GSK3-Inhibitor (CHIR99021) (Selleckchem, Catalog no. 4423), 500pM epidermal growth factor (R&D systems, Catalog no. 236-EG) and 3.5 μM of Neurotrophin-4 (R&D systems, Catalog no. 268-N4). The cultures were then harvested at day 10 at an oral epithelium stage.
Quantitative RT-PCR on induced embryonic oral epithelial cells
From nine samples of each genotype (Supplementary Fig. 8) RNA was extracted using Quick-DNA/RNA Miniprep kit (Zymo Research) followed by DNase treatment using manufacturer’s instructions (Thermo Fisher Scientific). Reverse transcription was performed using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Real-time PCR was performed using SYBR Green qPCR mix (Bio-Rad) in Bio-Rad CFX Connect Real-Time System. Quantitative RT-PCR reaction for each sample was performed in triplicate. IRF6 expression level was normalized against five genes: four housekeeping genes (ACTB, GAPDH, HPRT, & UBC) and an epithelial marker, CDH1, whose expression is IRF6-independent60 using a robust method61. Data were presented as mean ± s.d. Statistical significance was determined using the Student’s t test (two-tailed). Primer sequences used in the study for qRT-PCR are provided in Supplementary Data 11.
Chromatin immunoprecipitation qPCR
Induced oral epithelial cells (iOECs) derived from iPSC edited to be heterozygous for risk and non-risk alleles of rs570516915 (three heterozygous clones differentiated to oral epithelial cells) were harvested (~3.5 million cells) and fixed in 1% formaldehyde for 15 min at RT. Fixation was stopped with 125 mM glycine for 5 min at RT. Chromatin immunoprecipitation on iOECs was performed using a semi-automated protocol described previously62. Briefly, cells were sheared using PIXUL to obtain genomic DNA fragments averaging 300–600 bp. The sonicated cell lysates were subjected to chromatin immunoprecipitation with a specific antibody (1 μg of anti-H3K27Ac from Millipore-Sigma or 1 μg of Rabbit anti-IRF6 from Dr. Akira Kinoshita, Nagasaki University, as previously described)63. Normal rabbit IgG (1 μg) was used as a negative control antibody. Immuno-precipitated DNA was quantified using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Quantitative PCR was performed with equal mass of DNA (1.25 ng) in triplicate from three ChIP replicates of each antibody and percent input DNA was calculated using a method described previously64. Primer sequences are shown in Supplementary Data 11. Negative control PCR primers were designed to target a sequence 103.7 kb upstream of the IRF6 transcription start site that did not harbor active regulatory elements identified from ATAC-Seq and H3K27Ac ChIP-Seq in HIOEC or NHEK cells and devoid of predicted IRF6 binding sites. Positive control PCR primers were designed within a peak detected in anti-IRF6 ChIP-seq in human keratinocytes31. This region harbors three IRF6 binding sites predicted using JASPAR59 and a SNP (rs34411394) that is heterozygous in WTC-11 iPSCs but that does not lie within any of the predicted IRF6 binding sites. Data were presented as mean ± s.d. Statistical significance was determined using the Student’s t test (two-tailed). GraphPad Prism 10 was used for the statistical data analysis for luciferase reporter assays, RNA expression and ChIP-qPCR experiments.
Immunostaining on iPSCs and induced oral epithelial cells
Human iPSCs and iOECs were fixed in 4% paraformaldehyde (PFA) (EMS, Catalog no. 15710) for 12 min at RT and later washed thrice with 1X PBS for 5 min each on a rocker followed by permeabilization with 0.5% TritonX-100 (Sigma, Catalog no. T9284) at RT on a rocker for 10 min. Later blocking was done for an hour at RT on a rocker with a blocking buffer consisting of 5% goat serum (VWR, Catalog no. 101098-382), 3% BSA (VWR, Catalog no. 9048-46-8), and 0.1% Triton X-100. Then, the cells were incubated overnight in the primary antibodies (PITX2, Santa Cruz, Catalog no. sc-390457, 1:50; ECAD/CDH1, ABclonal, Catalog no. A20798, 1:200) at 4 oC on a rocker. Further, cells were rinsed with 1X PBS thrice for 5 min each and later incubated with corresponding secondary antibodies (Mouse IgG Alexa Flour 488, Thermo Fisher, Catalog no. A-11001, 1:500; Rabbit IgG Alexa Flour 568, Thermo Fisher, Catalog no. A-11036, 1:500) followed by Phalloidin (Phalloidin-iFlour 647 Reagent, abcam, Catalog no. ab176759, 1:500) for an hour at RT on a rocker prepared in the same blocking agent, followed by washing with 1X PBS thrice for 5 min each in the dark. The cells were incubated in autofluorescence quenching solution (Vector Labs, Catalog no. SP-8400) for 5 min at RT under dark conditions on a rocker and rinsed with 1x with PBS. Cells were further incubated in DAPI (Thermo Fisher, Catalog no. D1306) for 10 min then rinsed with 1X PBS, mounted with Vectashield (Vector Labs, Catalog no. H-1700), and imaged using Leica TCS-SPE Confocal microscope. Images were processed with Fiji software distribution of ImageJ v2.9.0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme LCC, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, Novartis Pharma AG, and Boehringer Ingelheim International GmbH. Following biobanks are acknowledged for delivering biobank samples: Auria Biobank, THL Biobank, Helsinki Biobank, Biobank Borealis of Northern Finland, Finnish Clinical Biobank Tampere, Biobank of Eastern Finland, Central Finland Biobank, Finnish Red Cross Blood Service Biobank, Terveystalo Biobank and Arctic Biobank. All Finnish Biobanks are members of BBMRI.fi infrastructure. Finnish Biobank Cooperative–FINBB, is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious® services managed by FINBB. FinRegistry is a collaboration project of the THL and the Data Science Genetic Epidemiology research group at the Institute for Molecular Medicine Finland (FIMM), University of Helsinki. The FinRegistry project has received the following approvals for data access from THL (THL/1776/6.02.00/2019 and subsequent amendments), DVV (VRK/5722/2019-2), Finnish Center for Pension (ETK/SUTI 22003) and Statistics Finland (TK-53-1451-19). The FinRegistry project has received IRB approval from THL (Kokous 7/2019). The FinRegistry project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 945733), starting grant AI-Prevent. Estonian Biobank was supported by research grant (PRG1291) from the Estonian Research Council and the computational work was performed at the High-Performance Computing Center of the University of Tartu. This project was also partially funded by grants from the U.S. National Institutes of Health (NIH): DE027362 (R.A.C.), DE023575 (R.A.C. and B.C.S.), DE033016 (J.M., H. R.-B., and R.A.C.), HG010855 (KB) and CA246503 (KB); and partially funded by grants from HUS Helsinki University Hospital Research Foundation: Y2519SU008 (D.P.R.) and the Finnish Dental Society Apollonia: 20220038 (E.J.). We would like to acknowledge support from the Center of Excellence for Genomics and Translational Medicine (SLTMR16142T/TK142), and the Erasmus+ program of the European Union (2018-1-EL01-KA202-047907; 2018-1-SE01-KA202-039066). W.D.F. acknowledges support from the NIGMS grant R15-GM122030-01 and NIDCR R21-DE033147-01. R.A.C. received support from the Benjamin Hall Endowed Chair in Basic Life Sciences. A.V. was supported by NIH grant R01DE028599 and research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under US Department of Energy Contract DE-AC02-05CH11231, University of California (UC). We thank Dr. Akira Kinoshita (Nagasaki University) for providing the anti-IRF6 antibody and Jennifer Hesson and the Ellison Stem Cell core at ISCRM (University of Washington, Seattle) for assistance with CRISPR gene editing in iPSC. Finally, we would like to thank all participants and investigators of the FinnGen study and the Estonian Biobank.
Author contributions
F.R., P.K., B.C.S., R.A.C. and D.P.R. designed the study. F.R., P.N., P.K., T.N., J.G., A.K., M.Kals., A.G.E. and P.P. analyzed the data. E.J. and A.H. constructed phenotype endpoints. J.K., and M.Kurki carried out genetic data quality control. J.M., A.P.P. and H.R.-B. contributed to genome editing and differentiation of iPSCs. D.M. and K.B. helped with the PIXUL device. W.D.F., K.P. and A.V. performed mouse reporter experiments. A.P., T.E., S.J., A.G. provided resources. F.R., P.N., P.K., B.C.S., R.A.C. and D.P.R. drafted the manuscript. F.R., P.N., P.K., B.C.S., R.A.C. and D.P.R. contributed to the conception of this paper, interpretation of the results, and the revision of the manuscript. All authors critically reviewed the manuscript and approved the final submission.
Peer review
Peer review information
Nature Communications thanks Kerstin Ludwig, Zhaoming Wu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The GWAS data generated in this study have been deposited in the GWAS Catalog under accession codes GCST90448958, GCST90448959 and GCST90448960. Individual-level genotypes and register data from FinnGen participants can be accessed by approved researchers via the Fingenious portal (https://site.fingenious.fi/en) hosted by the Finnish Biobank Cooperative FinBB (https://finbb.fi/en/), in accordance with appropriate permissions. The individual-level data from the Estonian Biobank are available under restricted access administered by the Estonian Genome Center of the University of Tartu in accordance with the regulations of the Estonian Human Genes Research Act; access can be obtained by application at www.biobank.ee. Source data are provided with this paper.
Competing interests
F.R. is a current employee and stockholder of AbbVie, Inc. K.B. is co-founder of Matchstick Technologies, Inc and a co-inventor of PIXUL (US Patents 10809166, 11592366). Andrea Ganna is founder of Real World Genetics Oy. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fedik Rahimov, Pekka Nieminen, Priyanka Kumari.
These authors jointly supervised this work: Brian C. Schutte, Robert A. Cornell, David P. Rice.
Lists of authors and their affiliations appear at the end of the paper.
Contributor Information
Brian C. Schutte, Email: schutteb@msu.edu
Robert A. Cornell, Email: cornellr@uw.edu
David P. Rice, Email: david.rice@helsinki.fi
FinnGen:
Fedik Rahimov, Pekka Nieminen, Juha Karjalainen, Mitja Kurki, Aarno Palotie, Priit Palta, Andrea Ganna, and David P. Rice
Estonian Biobank Research Team:
Tiit Nikopensius, Mart Kals, Abdelrahman G. Elnahas, Tõnu Esko, and Priit Palta
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53634-2.
References
- 1.Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J. & Shaw, W. C. Cleft lip and palate. Lancet374, 1773–1785 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Kinsner-Ovaskainen, A. et al. A sustainable solution for the activities of the European network for surveillance of congenital anomalies: EUROCAT as part of the EU Platform on Rare Diseases Registration. Eur. J. Med. Genet.61, 513–517 (2018). [DOI] [PubMed] [Google Scholar]
- 3.European Surveillance of Congenital Anomalies. European Platform on Rare Disease Registration. URL: https://eu-rd-platform.jrc.ec.europa.eu/eurocat/eurocat-data/prevalence_en (accessed on December 21, 2023).
- 4.Rintala, A. E. Epidemiology of orofacial clefts in Finland: A review. Ann. Plast. Surg.17, 456–459 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Finnish Institute for Health and Welfare. Congenital anomalies 2014. URL: https://urn.fi/URN:NBN:fi-fe2018062626441 Statistical Report 27, (2018).
- 6.Saxen, I. & Lahti, A. Cleft lip and palate in Finland: incidence, secular, seasonal, and geographical variations. Teratology9, 217–223 (1974). [DOI] [PubMed] [Google Scholar]
- 7.Lithovius, R. H., Ylikontiola, L. P., Harila, V. & Sandor, G. K. A descriptive epidemiology study of cleft lip and palate in Northern Finland. Acta Odontol. Scand.72, 372–375 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Huang, L. et al. Genetic factors define CPO and CLO subtypes of nonsyndromic orofacial cleft. PLoS Genet15, e1008357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, M. et al. Genome-wide analyses identify a novel risk locus for nonsyndromic cleft palate. J. Dent. Res.99, 1461–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butali, A. et al. Genomic analyses in African populations identify novel risk loci for cleft palate. Hum. Mol. Genet.28, 1038–1051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu, Y. et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun.8, 14364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaty, T. H. et al. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genet. Epidemiol.35, 469–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslie, E. J. et al. A Genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am. J. Hum. Genet.98, 744–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangold, E. et al. Sequencing the GRHL3 coding region reveals rare truncating mutations and a common susceptibility variant for nonsyndromic cleft palate. Am. J. Hum. Genet.98, 755–762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkin, A. S. et al. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet8, e1002829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashgari, G. et al. Epithelial migration and non-adhesive periderm are required for digit separation during mammalian development. Dev. Cell52, 764–778 e764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyrard-Janvid, M. et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet.94, 23–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson, R. J. et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet.38, 1329–1334 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Ingraham, C. R. et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet.38, 1335–1340 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Garza, G. et al. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol.133, 68–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson, R. J. et al. Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Invest.124, 3891–3900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahimov, F. et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet.40, 1341–1347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature613, 508–518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, H. et al. Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18. Elife9, e51325 (2020). [DOI] [PMC free article] [PubMed]
- 25.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilderman, A., VanOudenhove, J., Kron, J., Noonan, J. P. & Cotney, J. High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep.23, 1581–1597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fakhouri, W. D. et al. An etiologic regulatory mutation in IRF6 with loss- and gain-of-function effects. Hum. Mol. Genet.23, 2711–2720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvon, E. Z. et al. Comprehensive in vivo interrogation reveals phenotypic impact of human enhancer variants. Cell180, 1262–1271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fakhouri, W. D. et al. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev. Dyn.241, 340–349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alghadeer, A. et al. Single-cell census of human tooth development enables generation of human enamel. Dev. Cell58, 2163–2180 e2169 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botti, E. et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. USA108, 13710–13715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metwalli, K. A. et al. Interferon regulatory factor 6 is necessary for salivary glands and pancreas development. J. Dent. Res.97, 226–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltonen, L., Palotie, A. & Lange, K. Use of population isolates for mapping complex traits. Nat. Rev. Genet.1, 182–190 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Kittles, R. A. et al. Dual origins of Finns revealed by Y chromosome haplotype variation. Am. J. Hum. Genet.62, 1171–1179 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim, E. T. et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet.10, e1004494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peltonen, L., Jalanko, A. & Varilo, T. Molecular genetics of the Finnish disease heritage. Hum. Mol. Genet.8, 1913–1923 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Uusimaa, J. et al. The Finnish genetic heritage in 2022 - from diagnosis to translational research. Dis. Model. Mech. 15, dmm049490 (2022). [DOI] [PMC free article] [PubMed]
- 38.Stoll, G. et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci.16, 1228–1237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lahtinen, A. M., Havulinna, A. S., Jula, A., Salomaa, V. & Kontula, K. Prevalence and clinical correlates of familial hypercholesterolemia founder mutations in the general population. Atherosclerosis238, 64–69 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Richardson, R. J., Dixon, J., Jiang, R. & Dixon, M. J. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum. Mol. Genet.18, 2632–2642 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, H., Jones, K. L., Hooper, J. E. & Williams, T. The molecular anatomy of mammalian upper lip and primary palate fusion at single cell resolution. Development146, dev174888 (2019). [DOI] [PMC free article] [PubMed]
- 42.Leitsalu, L., Alavere, H., Tammesoo, M. L., Leego, E. & Metspalu, A. Linking a population biobank with national health registries-the estonian experience. J. Pers. Med.5, 96–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitsalu, L. et al. Cohort profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol.44, 1137–1147 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Loh, P. R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet.48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning, B. L. & Browning, S. R. Genotype imputation with millions of reference samples. Am. J. Hum. Genet.98, 116–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitt, M. et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet.25, 869–876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet.53, 1097–1103 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickham, H. ggplot2: Elegant Graphics for Data Analysis, (Springer-Verlag, New York, 2016).
- 50.Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience10, giab008 (2021). [DOI] [PMC free article] [PubMed]
- 52.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet.44, 369–375 (2012). S361-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet.88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pebesma, E. Simple features for R: standardized support for spatial vector data. R. J.10, 439–446 (2018). [Google Scholar]
- 56.Witte, J. S., Visscher, P. M. & Wray, N. R. The contribution of genetic variants to disease depends on the ruler. Nat. Rev. Genet.15, 765–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas-Chollier, M. et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc.6, 1860–1869 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Rauluseviciute, I. et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 52, D174–D182 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Girousi, E. et al. Lack of IRF6 disrupts human epithelial homeostasis by altering colony morphology, migration pattern, and differentiation potential of keratinocytes. Front. Cell. Dev. Biol.9, 718066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, S. C. et al. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol.37, 761–774 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Bomsztyk, K. et al. PIXUL-ChIP: Integrated high-throughput sample preparation and analytical platform for epigenetic studies. Nucleic Acids Res.47, e69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fakhouri, W. D. et al. Intercellular genetic interaction between Irf6 and Twist1 during craniofacial development. Sci. Rep.7, 7129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Solomon, E. R., Caldwell, K. K. & Allan, A. M. A novel method for the normalization of ChIP-qPCR data. MethodsX8, 101504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The GWAS data generated in this study have been deposited in the GWAS Catalog under accession codes GCST90448958, GCST90448959 and GCST90448960. Individual-level genotypes and register data from FinnGen participants can be accessed by approved researchers via the Fingenious portal (https://site.fingenious.fi/en) hosted by the Finnish Biobank Cooperative FinBB (https://finbb.fi/en/), in accordance with appropriate permissions. The individual-level data from the Estonian Biobank are available under restricted access administered by the Estonian Genome Center of the University of Tartu in accordance with the regulations of the Estonian Human Genes Research Act; access can be obtained by application at www.biobank.ee. Source data are provided with this paper.