Abstract
Despite their indisputable importance in neuroblastoma (NB) pathology, knowledge of the bases of NB plasticity and heterogeneity remains incomplete. They may be rooted in developmental trajectories of their lineage of origin, the sympatho-adrenal neural crest. We find that implanting human NB cells in the neural crest of the avian embryo allows recapitulating the metastatic sequence until bone marrow involvement. Using deep single cell RNA sequencing, we characterize transcriptome states of NB cells and their dynamics over time and space, and compare them to those of fetal sympatho-adrenal tissues and patient tumors and bone marrow samples. Here we report remarkable transcriptomic proximities restricted to an early sympathetic neuroblast branch that co-exist with phenotypical adaptations over disease progression and recapitulate intratumor and interpatient heterogeneity. Combining avian and patient datasets, we identify a list of genes upregulated during bone marrow involvement and associated with growth dependency, validating the relevance of our multimodal approach.
Subject terms: Paediatric cancer, Metastasis, Embryonal neoplasms, CNS cancer, Cancer genomics
The roots of neuroblastoma plasticity and heterogeneity remain poorly understood. Here, the authors characterise the transcriptional states of neuroblastoma cells and their changes throughout development using avian embryo models, single-cell RNA-sequencing, and patient samples; they find transcriptomic changes related to early sympatho-adrenal development.
Introduction
Pediatric cancers are highly heterogeneous diseases as reflected by their emergence from a range of developing tissues. Half of them originate from embryonic cell lineages, thus at stages of organ formation and maturation, when many lineage differentiation processes are underway1,2. Neuroblastoma (NB) is the most common extracranial solid tumor in childhood and derives from the embryonic neural crest committed to generate the cell populations of the sympatho-adrenal (SA) peripheral nervous system3. This specific origin contributes to the formation of NB primary tumors within the adrenal gland or all along the sympathetic ganglionic chain. High-risk NBs represent 50% of cases and are typically associated with a wide metastatic profile detected at diagnosis, in particular in the bone marrow, suggesting early and potent dissemination4. This clinical presentation considerably limits our understanding of NB dissemination mechanisms and also constrains the development of therapeutic interventions targeting the metastatic foci.
Recent studies formally timed NB initiation as early as during the first trimester of pregnancy and emergence of metastatic clones before diagnosis, pointing at an occurrence at early steps of SA lineage development5,6. Although still being explored, the developmental complexity of the SA lineage is thought to ground the NB high interpatient and intratumor heterogeneity7–9. In particular, at least two types of NB cell states were depicted: cells having neuronal features and associated transcriptomic landscape known as ADR- or NOR-type, and non-neuronal cells with mesenchymal traits, referred to as MES-type cells10,11. Interconversion between these two states reveals high NB cell plasticity potential, and is suspected to be rooted in lineage-related developmental transitions12,13.
The developmental trajectories of the SA lineage have been deeply investigated, in particular with single cell RNA Sequencing (scRNA-seq) strategies14–17. Both in mice and in humans, these studies revealed that two successive neural crest-derived progenitor states contribute to SA cell contingents: (1) the first one undergoing an early ventromedial migration, referred to as Sympathetic Neuron Progenitors Cells (SNPCs), and (2) the second one associated with a later ventrolateral axon-bound migration, assigned to the Schwann Cell Precursors (SCPs)7,8. SNPCs were found to generate neuroblasts of the sympathetic ganglia and, at least in mice, a fraction of adrenal neuroblasts. SCPs generate a major fraction of chromaffin cells and neuroblasts of the fetal adrenal gland both in mice and in human species. The full mapping of SA lineage transcriptome trajectories is complexified by the series of hub and transitory intermediate cell states that may be fast and hard to capture by time-fixed analyses7,18.
Attempts to precisely trace NB cell of origin by aligning NB tumor scRNA-seq profiles on the human SA physiological atlas identified several types of transcriptomic similarities between NB and neuroblastic “NOR” features or proximal transient states, but above all, highlighted lineage-related heterogeneity and plasticity of NB transcriptomic states14–17,19,20. Recent scRNA-seq analysis reported that NB cells from paired primary tumour/ bone marrow metastasis samples share common states that align on the sympathoadrenal differentiation axis21. Nevertheless, many questions remain unanswered as to when these particular states emerge and how plastic they are from the moment the metastatic process is initiated. Indeed, correlating the dynamics of NB cell physical progression from the primary tumor to the metastatic sites with that of their transcriptomic states has been limited by the lack of suited experimental models, and the scarcity of NB patient samples harvested both at the primary tumor and metastatic sites. Consequently, it is unclear whether the dynamics of NB transcriptomic states across disease progression remains governed by the developmental rules of the SA lineage and what adaptations to the different microenvironments of the primary tumor sites, metastatic routes and secondary foci NB cells undergo.
To address these challenges, we took advantage of an embryonic avian model of NB that we previously set up consisting in engrafting human NB cells at embryonic day 2 (E2) within the avian pre-migratory sympatho-adrenal neural crest22–24. Our cellular and molecular characterization of this model showed that NB cells exposed to such embryonic microenvironment, mimicking that of the disease emergence, migrate along a stereotypic ventral path under the action of environmental cues. This migration drives them towards the primordia of sympathetic ganglia (SG) and adrenal medulla (ADR) in which they stop to form highly cohesive primary tumors at E422–24. From E6 to E9, we observed by light sheet microscopy and confocal microscopy that NB cells take advantage of physiological sympathetic signals to escape from these tumors, forming clusters disseminating at distance along the dorsal aorta and the peripheral nerves22,23. In this work, using this model, we conduct a longitudinal study of the dynamics of NB transcriptome states, from the primary tumor formation, to the dissemination step and the bone marrow involvement. By generating high depth scRNA-seq data, we investigate NB transcriptomic and clonal evolution across disease progression in light of the SA lineage of origin. We show that NB cells adopt a panel of transcriptome states aligned on the SNPCs-to-neuroblast differentiation branch and that the primary tumor site determines their dissemination path. By crossing this complete view of gene expression dynamics across the metastatic process with transcriptomic data from paired primary tumor / bone marrow NB patient samples, we identify a set of therapeutic candidate genes whose expression is consistent with an active contribution to the growth of bone marrow metastasis.
Results
NB transcriptomic plasticity across disease progression in an embryonic microenvironment
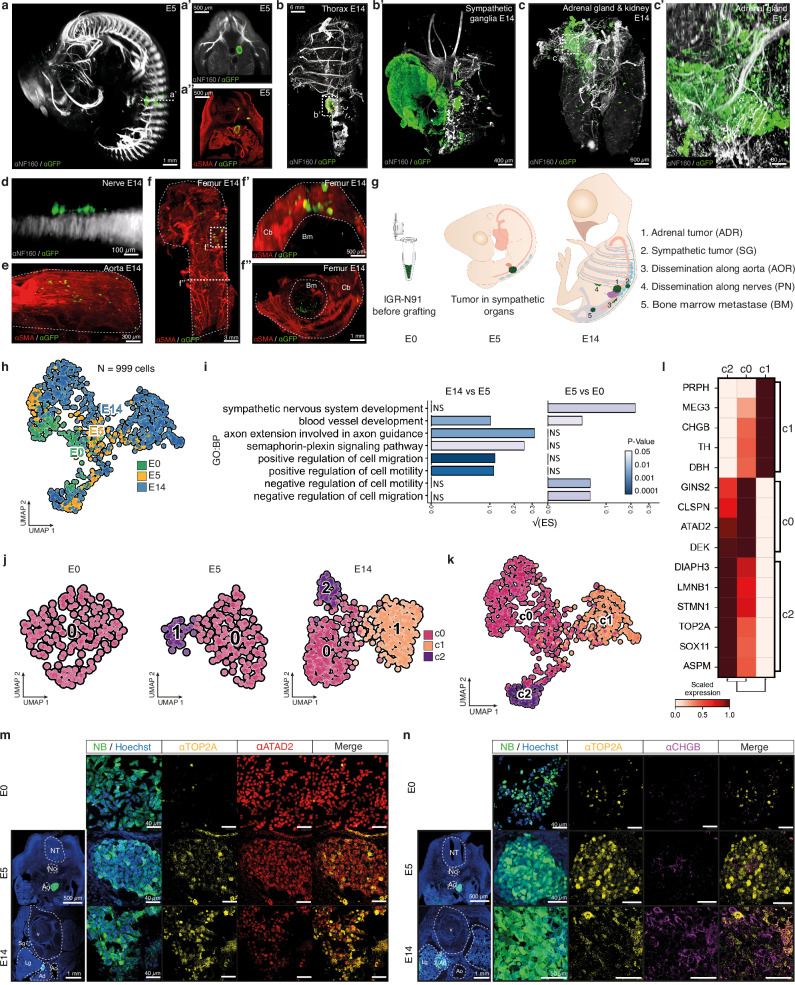
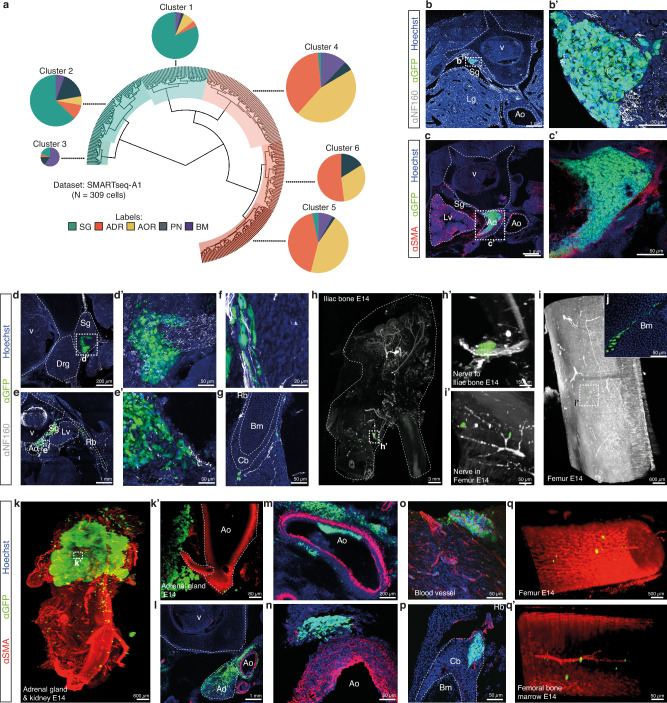
We set up 3D imaging approaches for E14 chick embryos with the objective to investigate whether NB secondary dissemination could drive the formation of metastatic foci, that, in patients, mostly consist in bones and bone marrow involvement. We engrafted IGR-N91::GFP stage 4 NB cell line in series of E2 avian embryos and collected embryos at E5, a time-point preceding the onset of metastasis for this NB cell line22–24, and then at E14 to analyze the localization and features of NB cells by 3D imaging. At E5, IGR-N91 cells exclusively formed tumors within the developing SG and ADR, that lie in close proximity, consistent with our previous work (Fig. 1a, Supplementary movie 1)22–24. Notably at E14, primary tumors in SG or ADR tissues were even more clearly distinct (Fig. 1b, c, Supplementary movies 2-6). Moreover, while clusters of disseminating cells could still be detected along aortic and peripheral nerves paths at E14 (Fig. 1d, e, Supplementary movies 7-9), we also observed the systematic presence of NB foci into the bone parenchyma -ribs, long bones- and into the bone marrow hence mimicking typical features of metastatic NB in patients (Fig. 1f, Supplementary movies 10-12).
Fig. 1. l NB transcriptomic plasticity across disease progression in an avian embryonic microenvironment.
a–f Representative lightsheet imaging of chick embryos grafted with IGR-N91::GFP cells at E2 and harvested at E5 (N = 11) (a) or E14 (N = 7) from 3 independent experiments (b–f) and labelled with α-NF160, or α-SMA, and α-GFP antibodies. E: Embryonic day. Optical sections at E5 in a’ and a”. b, c illustrate E14 rib cage with sympathetic ganglia tumor (in N = 5/7 embryos, enlargement in b’) and/or adrenal tumor (in N = 7/7, optical section c’). d, e show NB cells on nerves (in N = 4/7 embryos) (d), and on the aorta (in N = 7/7 embryos) (e). f illustrates bone invasion (in N = 7/7 embryos) with optical sections highlighting cortical (f’) and marrow (f”) metastases. Scale bars are indicated. g Overview of IGR-N91::GFP cell location harvested for scRNAseq. h Integration of 998 IGR-N91::GFP cells (E0 cells in culture, and from N = 22 E5 and N = 15 E14 embryos from 5 independent experiments) in a UMAP colored by timepoint. i Bar plot showing pathway enrichment score (ES) (Gene Ontology biological processes, p-value < 0.05) based on gene differential expression in E5 vs E0 and in E14 vs E5 (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.1, min.pct > 0.1). Square root of the ES is shown. j UMAP of NB cells colored by transcriptomic state (c0, c1, c2) with Louvain clustering annotation, at the same resolution (res = 0.3) for E0 (162 cells), E5 (178 cells) and E14 (658 cells) stages. k UMAP of IGR-N91::GFP cells integrated overtime colored by transcriptomic clusters (c0, c1, c2). l Heatmap of scaled expression values of genes markers for clusters shown in k (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.25, min.pct > 0.1) grouped by phenotype. m, n Representative images of IGR-N91::GFP cells in culture and in sections of E5 (N = 10 embryos from 4 independent experiments) and E14 (N = 8 embryos from 5 independent experiments) embryos labeled with α-ATAD2 (c0), α-TOP2A (c2) and α-CHGB (c1) antibodies. Source data are provided as a source data file. Adr Adrenal gland, Ao Aorta, Bm Bone marrow, Cb Cortical bone, Lg Lung, No Notochord, NT Neural Tube, Sg Sympathetic ganglia, V Vertebrae.
To explore the spatio-temporal sequence of transcriptomic adaptations that occur in NB cells across disease progression, we next micro-dissected and sorted GFP+-NB cells from series of E5 and E14 chick embryos engrafted at E2 with IGR-N91::GFP cells. In E5 embryos, prior to metastatic onset, NB cells were harvested from sympatho-adrenal primary tumors while at E14, we microdissected out NB cells from five physical sites representing different steps of the disease progression: SG tumors, ADR tumors, cells migrating on peripheral nerves (PN), in/on the dorsal aorta (AOR), and metastatic foci in the bone marrow (BM) (Fig. 1g). We performed three series of high depth single cell RNA-seq experiments (two Smart-Seq2 and one seqWell datasets) with the aim of capturing subtle adaptations over time (SMARTseq-A2 and seqWell datasets, with NB cells prior to the graft, from sympatho-adrenal tumors at E5 and from ADR and SG tumors at E14) and to the different embryonic microenvironments (SMARTseq-A1 dataset corresponding to NB cells harvested from E14 embryos and located in the ADR, SG, PN, AOR and BM foci) (Fig. 1h and Supplementary Fig. 1a–d). Biological processes significantly regulated by the primary tumor microenvironment between E0 (IGR-N91::GFP cells in culture) and E5 were mostly related to developmental processes (nervous system, vessels, regulation of migration) and metabolic adaptations. At E14, the major changes reflected a progressive adaptation of NB migratory and metabolic features, and cell-cell dialogs within the different embryonic microenvironments (Fig. 1iand Supplementary Fig. 1e). When embedding IGR-N91::GFP cells in UMAPs at each time point separately, while cells in culture -E0 step- showed a single transcriptomic profile, using the same clustering parameters, we observed a progressive complexification of NB transcriptomic states upon exposure to the embryonic microenvironment with two distinct clusters at E5 and three clusters at E14 (Fig. 1j). This suggests a progressive emergence of three co-existing states, starting from a homogeneous cell population in culture. Interestingly, the combined integration of E0, E5 and E14 NB cell data resulted in three transcriptomic clusters (c0, c1, c2) that did not correspond to E0/E5/E14 time points (Fig. 1k). To further characterize these three states, we extracted key gene markers of each cluster. Strikingly, most are reported markers of cycling progenitor populations -such as ASPM25 and TOP2A26 for c2 cluster or ATAD227 and CLSPN28 for c0- or of the development of the sympatho-adrenal lineage -such as CHGB, TH, DBH, SOX11 for c18- thus suggesting transcriptome specificities linked to their original progenitor lineage (Fig. 1l). We could validate the expression profile and temporal dynamics of some markers of each cluster -c0: ATAD2, c1/CHGB, c2: LMNB1/TOP2A- by immunofluorescence on IGR-N91 cells in culture (E0) and on sympatho-adrenal IGR-N91 tumors (both at E5 and E14) (Fig. 1m, n and Supplementary Fig. 1f). Moreover, their expression was also observed in tumors formed by two additional NB cell lines, SHEP::GFP and SH-SY5Y::GFP, engrafted in the avian embryo, suggesting they reflect gene expression programs, thereafter called “transcriptome states”, shared between NB cells (Supplementary Fig. 1g–k).
NB transcriptomic diversity mimics early states of the SA developmental trajectory
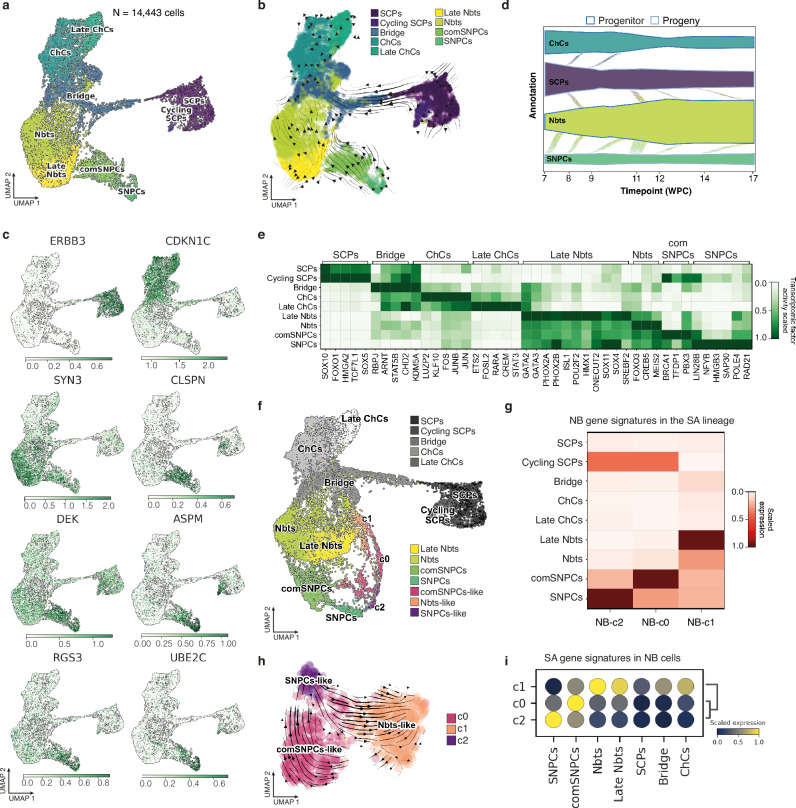
Our embryonic model of NB allows to reproduce NB transcriptome adaptations over a spatio-temporal window that covers the main steps of the physiological development of the sympatho-adrenal (SA) lineage, including the two neural crest-related differentiation axes that give rise to SA tissues, coming from the early ventromedial SA precursors -Sympathetic Neural Progenitor Cells (SNPCs)- and late ventrolateral precursors -Schwann Cell Precursors (SCPs)-8. As we assumed that the transcriptome dynamics of NB cells might relate to subtle transitions linked to their original lineage, we generated a single cell transcriptomic atlas of the human SA lineage by integrating key published datasets15,16, with the underlying objective to cover as exhaustively as possible transient and early states of the lineage. From this upgraded atlas, we could rebuild transcriptome states and transitions previously described (Fig. 2a and Supplementary Fig. 2a-c). The method used to integrate these two datasets (described in the Methods section) not only strengthened the statistical weight of each cluster, but also allowed to correct batch bias from each dataset processing and consequently to highlight yet undescribed and subtly different cell states common to both studies. In particular, we could re-assign part of the previously identified neuroblasts and cycling neuroblasts to two distinct clusters separated from the neuroblast population. Analysis of transcriptomic cell fate vectors highlighted these two clusters as early states preceding the neuroblastic commitment, thus pointing at neuroblastic precursor states (Fig. 2b). Extraction of markers of both clusters pointed at two distinct cycling progenitor states, fully separated from SCPs, bringing them closer to early ventromedial migrating progenitors depicted as SNPCs and transitional committed-SNPCs (comSNPCs)8,29 (Fig. 2c). In silico analysis of transcriptomic transitions via probability mass flows from one cell state to another predicted the progenitor status of SNPCs and their potential to mostly fuel the neuroblast compartment. The second SA root, SCPs, was predicted to contribute over time to both chromaffin and neuroblast populations (Fig. 2d). In addition to cell cycle specificities, SNPCs and comSNPCs states were highlighted by specific sets of transcription factors, distinguishing them from any other SA progenitor or from the neuroblastic state (Fig. 2e and Supplementary Fig. 2d).
Fig. 2. NB cell plasticity in avian embryonic tissues relies on the adoption of features of the human SNPCs-to-neuroblasts differentiation program.
a UMAP embedding of 14,443 single cells of human physiological sympatho-adrenal cells colored by phenotypic state. The UMAP results from the integration of Jansky et al., 2021 and Kameneva et al., 2021 datasets. b UMAP embedding of human SA atlas with arrows highlighting the directions of state-to-state transitions. c Expression of key marker genes in UMAP embedding of the human SA atlas; color scales indicate log-normalized gene expression. d Probability mass flow as a function of time post-conception from either SCP or SNPC progenitor state using cellular growth and death score, combined with cell transcriptomic similarity to compute differentiation directions. e Heatmap of the scaled transcription factor activity for key genes in each physiological SA phenotypical state. Gene lists were extracted using differential expression with two-sided Wilcoxon rank sum test (p-value < 0.05, log fold change > 0.25, min.pct > 0.1). f Unimodal projection UMAP of NB cells -colored by transcriptomic states- mapped onto the human SA lineage atlas. g Heatmap of scaled expression values for NB-c0, NB-c1 and NB-c2 gene signature scores into cell populations of the SA lineage. NB-c0, NB-c1 and NB-c2 gene signatures were built from the intersection of NB gene markers grouped by phenotype (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.25, min.pct > 0.1) with anchor genes used for the unimodal projection UMAP shown in (f). h RNA velocity estimates for NB cells harvested at E14 on a UMAP embedding using dynamical modeling. i Dot plot illustrating SCPs-related or SNPCs-related differentiation dynamic gene signature scores into NB cell transcriptomic states. Cell cycle-related genes (listed in gene ontology GO:0007049 biological process) were removed from all signatures. Source data are provided as a source data file. ChCs Chromaffin Cells, Nbts Neuroblasts, SCPs Schwann Cell Precursors, SNPCs, Sympathetic Neuron Progenitor Cells, comSNPCs committed SNPCs.
Next, we projected our NB dataset onto the upgraded SA atlas to characterize potential transcriptomic similarities. Notably, NB cells exclusively projected onto the SNPCs to neuroblasts differentiation axis (Fig. 2f). From this projection dataset, we extracted gene signatures highlighting the proximities of NB clusters with the SA lineage. The resulting signatures (NB-c0, NB-c1, NB-c2) were scored in each cell population of the SA lineage, showing the highest expression of NB-c2 in SNPCs; NB-c0 in committed-SNPCs and NB-c1 in late Neuroblasts (Fig. 2g).
These relationships were further confirmed by looking at the expression profile of key markers of SA identities into NB c0/c1/c2 clusters. Notably, SCPs emblematic markers -ERBB3, SOX10, PLP1- were all negative (Supplementary Fig. 2e). Moreover, while the regression of cell cycle genes modified both SA lineage and NB UMAP embeddings, transcriptomic similarities between NB cell clusters and SNPCs/Nbts-related states remained stable, suggesting state proximities that went beyond cycling properties (Supplementary Fig. 2f-i). Therefore, we annotated the c0, c1 and c2 clusters as comSNPCs-like, Nbts-like and SNPCs-like states respectively.
We further questioned the transcriptome dynamics in between SNPCs/Nbts-related NB clusters by performing a computational inference of temporal dynamics using scRNA-Seq (RNA velocity). As suggested by the sequence of clusters emergence from E0 to E14 stage -comSNPCs-like, SNPCs-like and finally Nbts-like NB cells- RNA velocity vectors predicted that NB cells did not solely follow the physiological direction of SNPCs-to-Nbts differentiation but rather went back and forth in between the three phenotypic states (Fig. 2h). Thus, in the embryonic SA microenvironment, NB tumor plasticity is manifested in their ability to re-play and back up a portion of SNPCs differentiation path. We next assessed whether the nature of transcriptomic transitions occurring in NB cells, could also be related to physiological transitions occurring in the human SA lineage, beyond cell cycle transitions. From our refined atlas of the SA lineage, we subset the two branches that end in a neuroblast state, starting from each SA progenitor populations -the SCPs and the SNPCs- (Supplementary Fig. 3a, b). We extracted gene signatures that mark up the differentiation trajectories starting from each SA progenitor, to capture dynamic gene expression variations highlighting the transition from one transitory state to another (Supplementary Fig. 3c–j). We removed cell cycle-related genes in this set of gene signatures and scored them in SNPCs/Nbts-like NB cell states. This unambiguously showed that the transcriptomic dynamics in between NB SNPCs/Nbts-like clusters is reminiscent of SNPCs, comSNPCs and Nbts physiological transitions (Fig. 2i).
Patient intratumor heterogeneity is based on co-existing SA-related transcriptome states
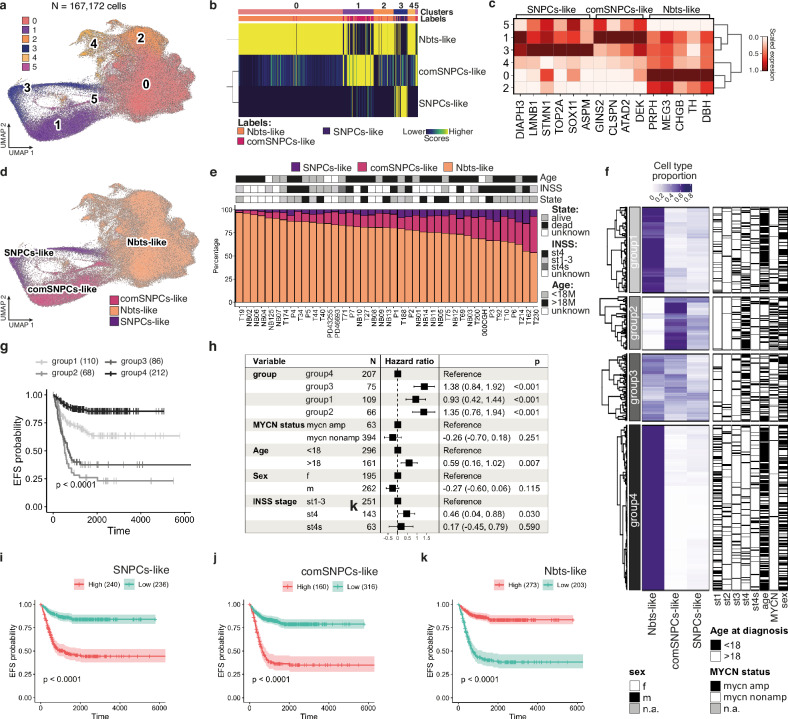
We further wondered whether NB plastic behavior and associated heterogeneity of transcriptome states could relate to clinical heterogeneity of patient tumors. We re-analyzed published single cell RNA-seq datasets of NB patient samples14,15,20, and completed the study with 7 patients for which both the primary tumor and matched bone marrow metastasis could be sequenced (CEL-Seq2 protocol) (Supplementary Tables 1 and 2). scRNA-seq data from these 41 patient samples were embedded to build a neuroblastoma atlas restricted to tumor cells (167,172 cells) (Fig. 3a and Supplementary Fig. 4a, b). Unsupervised clustering highlighted 6 clusters for which similarity scores with SNPCs/comSNPCs/Nbts-like clusters were computed. Notably, each cluster unambiguously matched with one of the three SA-related transcriptome states without any overlap, hence suggesting that NB intratumor transcriptomic heterogeneity could relate to the adoption of features of SNPCs-related interconnected states (Fig. 3b). This was further confirmed by analyzing the expression of top markers of SNPCs/comSNPCs/Nbts-like states in each NB patient cluster, with the exception of a single cluster (c5) that matched with both SNPCs-like and comSNPCs-like states (Fig. 3c, d). Similar results were obtained whatever the initial anatomical location of the tumor sample (Supplementary Fig. 4c, d). Accordingly, the expression of markers of the physiological SA lineage confirmed the proximity between NB cells and SNPCs/Nbts-related states while markers of the SCPs branch were absent (Supplementary Fig. 4e, f). Gene signatures highlighting the dynamic mesenchymal (MES) and noradrenergic (NOR) states of neuroblastoma cells11 were not found differentially expressed in between NB clusters (Supplementary Fig. 4g, h). Interestingly, the mapping of patient cells, patient by patient, on the refined SA physiological atlas, led to a perfect matching of NB cells with the SNPCs-to-neuroblasts differentiation branch for 39 patients, with two samples that displayed a minor overlap with cycling SCPs, which could be due to the weight of cell cycle genes for these samples with low amounts of cells (Supplementary Fig. 5). Although the relative fractions of NB cells in the 3 states followed the same tendency for the 41 patient samples, we noticed that these proportions were variable in between samples (Fig. 3e). We thus assessed whether the relative proportion of the three transcriptomic states could have a clinical impact. We deconvolved bulk RNA-seq data from two large NB cohorts (SEQC: 498 samples30, Kocak 649 samples31) to study the behavior of SNPCs/comSNPCs/Nbts-like gene signatures together with established clinical criteria. Interestingly in both cohorts, the hierarchical clustering of NB transcriptomes by their relative abundance of SNPCs/comSNPCs/Nbts-like states, highlighted four subgroups of tumors (Fig. 3f and Supplementary Fig. 6a). In accordance with published results, a high fraction of Nbts-like cells (Groups 1 and 4) was associated with a better prognosis while a high content of SNPCs-like and comSNPCs-like cells (Groups 2 and 3) was related to a poor prognosis. Notably, the group having the highest content in comSNPCs-like cells (Group 2) also showed the worse prognosis, suggesting that this highly plastic and transitory cell state crucially impacts on clinical outcome (Fig. 3g and Supplementary Fig. 6b). Interestingly, multivariate analyses confirmed that a high proportion of Nbts-like cells together with low proportions of comSNPCs/SNPCs-like cells independently predicted the most favorable outcome while the hazard ratio significantly increased together with the fraction of comSNPCs-like cells. Hazard ratios were also significantly and independently impacted by disease stage and age although to a lesser extent (Fig. 3h and Supplementary Fig. 6c). Accordingly, in both cohorts, stage 4 metastatic NB displayed significantly lower fractions of Nbts-like cells and higher proportions of comSNPCs-like cells as compared to localized stages, while such differences were less clear regarding the fraction of SNPCs-like cells (Supplementary Fig. 6d, e). This observation again argued for the idea that the transitory and highly plastic nature of comSNPCs-like cells impacts on disease outcome, as also highlighted by the significant difference in event free survival (EFS), in both cohorts, when considering high versus low comSNPCs-like cell fraction (Fig. 3i–k and Supplementary Fig. 6f–h).
Fig. 3. Patient intra-tumor heterogeneity reflects SA lineage-related transcriptome plasticity.
a UMAP plot of 167,172 NB cells from patient samples colored by unsupervised clustering. Only NB cells were kept for the analysis. b Comparison of transcriptomic states in NB patient single cell data with annotated clusters of NB cells exposed to embryonic microenvironments in the avian model. Similarity score was computed based on Spearman correlation across reference markers. c Heatmap of the scaled expression values for marker genes of human SNPCs/Nbt-like clusters of NB cells exposed to the embryonic microenvironment in NB patient sample transcriptomic clusters. d UMAP plot of NB cells from patient samples colored and labeled by SA lineage-related transcriptome states. e Bar plot of fractions of NB cells assigned to SA-related transcriptomic states per patient sample with associated clinical information. f Heatmap with hierarchical clustering of 649 RNA-seq patient samples from Kocak’s cohort with associated clinical information based on deconvolution, using patient NB scRNA-seq atlas as a reference to assess proportion of NB transcriptomic states. g Kaplan–Meier analysis of EFS in NB patients according to SA lineage-related hierarchical clustering (group1 to 4). Log-rank test with Bonferroni adjustment was performed to compare survival curves. h Forest plot of hazard ratio depending on SA lineage-related NB profiles (group 1 to 4) from Cox regression EFS analyses in multivariable analyses adjusted for age, sex, MYCN status and INSS classification. Error bars represent the 95% confidence interval of the likelihood test performed on the calculated hazard ratio (two-sided test). i–k Kaplan–Meier analysis of EFS in NB patients according to the relative proportion of SNPCs-like (i), comSNPCs-like (j) or Nbts-like states (k). Log-rank test was performed. Error bands show the 95% confidence interval. Source data are provided as a source data file.
Metastasizing NBs maintain SA-related states but adapt to their microenvironment
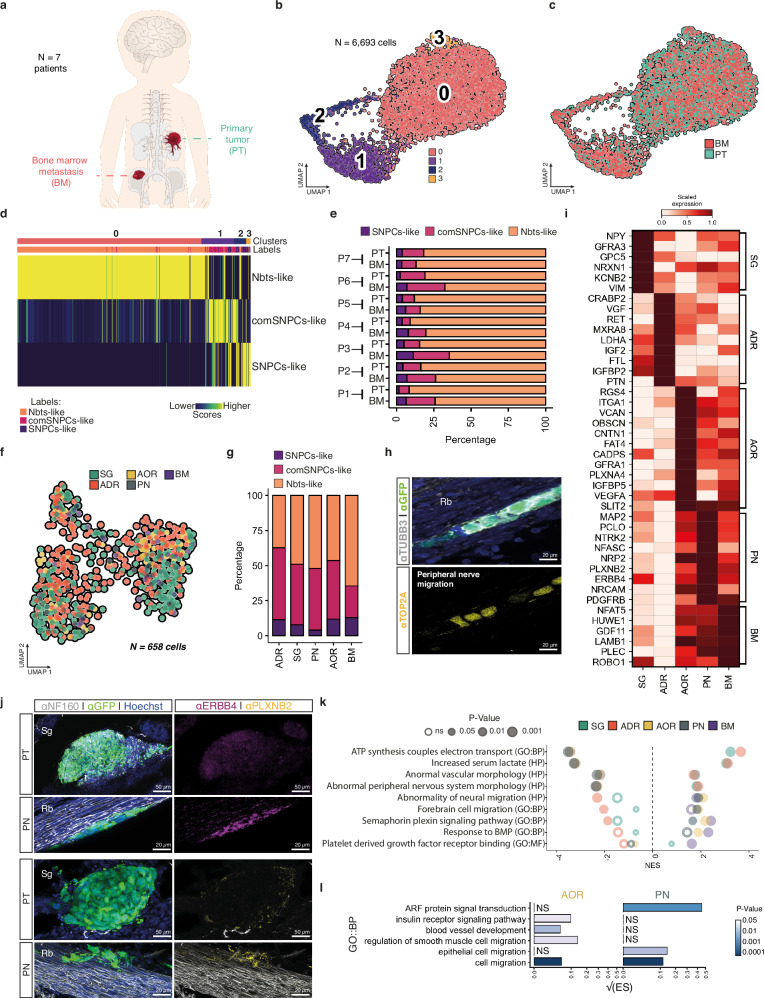
We next assessed whether variations in lineage-related NB states could underlie NB metastatic progression. We further examined the scRNA-seq data obtained from the seven pairs of NB primary tumor (PT) and matched bone marrow aspirates (BM) obtained at diagnosis and generated in this study (Supplementary Table 2). Tumor cells were selected based on the detection of copy number variations (CNVs) and the lack of expression of emblematic markers of non-tumoral cell populations of the microenvironment -i.e.: mostly fibroblasts, endothelial and immune cells in PTs-, and mesenchymal cells and immune cells in BMs (Fig. 4a and Supplementary Fig. 7a–h). The fraction of tumor cells was between 38% and 78% in PTs. For BM samples, an enrichment of tumor cells was performed based on a panel of cell surface markers (see in the Methods section) leading to a final fraction of tumor cells comprised between 40% and 74%. A total of 6693 tumor cells from both PTs and BMs was further sub-sampled for deeper analysis. Tumor cells embedded in UMAP followed the same type of organization as the complete NB tumor cell atlas with three main transcriptome clusters that matched with SNPCs-like, comSNPCs-like and Nbts-like states, irrespective of the patient and of the PT/BM origin of the sample (Fig. 4b–e and Supplementary Fig. 7i, j).
Fig. 4. SA-lineage-related NB states are maintained across the metastatic dissemination.
a Illustration of patient samples from paired primary tumor (PT) and bone marrow (BM). b, c UMAP of 6,693 NB cells sub-sampled from 7 integrated paired PT/BM colored by transcriptomic clusters (b) and by tissue location (c). d Transcriptomic comparison of PT/BM clusters (in b) with NB SA-related clusters exposed to avian embryonic microenvironments. Similarity scores were computed with Spearman correlation across reference markers. e Bar plot of fractions of NB cells assigned to SA-related states per patient and per sample type. f UMAP of IGR-N91::GFP cells colored by cell location in E14 embryos -257 cells from SG tumors, 261 cells from ADR tumors, respectively 25 and 84 cells on PN and AOR, and 31 cells in the BM. g Bar plot with fractions of cells shown in (f) and assigned to SA-related states for each NB cell location. h Representative image of IGR-N91::GFP cells on PN showing heterogeneous expression of the c2 marker TOP2A in E14 embryos (observed in N = 3/3 embryos from 2 independent experiments). Sections were labeled with α-GFP, α-TUBB3, and α-TOP2A antibodies. i Heatmap of scaled expression values of marker genes extracted from genes differentially expressed between locations (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.1 min.pct > 0.1). j Representative images of IGR-N91::GFP cells in E14 embryos, in primary tumors or in contact with PN labeled with α-GFP, α-NF160 and α-PLXNB2 or α-ERBB4 antibodies. k, Gene set enrichment analysis from differentially expressed genes for each location. Pathways were extracted from ontology gene sets of molecular signature database (nperm = 1000, ranked by log fold change, abs(NES) > 1.3, Fischer’s exact test). l Bar plot showing pathway enrichment score (ES) (GO biological processes, p-value < 0.05) in NB cells migrating on PN or AOR based on differential gene expression (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.1, min.pct > 0.1). Square root of the ES is shown. Scale bars are indicated. Source data are provided as a source data file. Rb Rib, Sg Sympathetic ganglia.
Similarly, the three lineage-related NB transcriptome states were detected in NB cells harvested at E14 from each physical position of the metastatic cascade -ADR, SG, AOR, PN, BM- in the avian graft model, including in SG and ADR primary tumor sites, despite established differences in the origin of neuroblasts that populate these two developing tissues3–5 (Fig. 4f, g). This result was further supported by the immunodetection of key markers of SNPCs-like and comSNPCs-like clusters such as TOP2A and LMNB1 in NB cells that were actively disseminating (Fig. 4h and Supplementary Fig. 7k). Thus, NB plasticity that manifests in the primary tumor microenvironment is maintained across metastatic progression in embryonic tissues. We next wondered which type(s) of phenotypic adaptations could underlie and/or sustain NB progression at each step of the process. The extraction of top markers expressed by NB cells at each step of disease progression -ADR and SG primary tumors; PN and AOR routes of dissemination; BM secondary site- defined specific adaptations related to a given microenvironment, namely: neuronal-related features at the SG level (NRXN1, GFRA3, NPY, VIM); response to nutrients and extracellular signals at the ADR level (VFG, LDHA, PTN, IGFBP2); cell adhesion at AOR level (OBSCN, GFRA1, ITGA1, CNTN1); neuronal migration at PN level (NRCAM, ERBB4, NTRK2, PLXNB2) and gliogenesis at the BM level (NFAT5, LAMB1, PLEC) (Fig. 4i). In support, ERBB4 and PLXNB2 proteins were highly detected in NB cells migrating on nerves (Fig. 4j). Interestingly, pathway analysis revealed common types of NB adaptations in primary tumors, that concerned changes in cell metabolism, but also at the active dissemination steps (nerves and aorta) where embryonic and neuronal-related migration signaling were enriched (Fig. 4k). Specific adaptations concerned NB cells in the BM with signaling pathways that might highlight dialogs with the nascent medullary niche -response to BMPs and PDGFR binding pathways- (Fig. 4k). In particular, NB cells migrating along nerves or via the aorta expressed specific gene programs highlighting distinct migration mechanisms in each of this tissue microenvironment (Fig. 4l). Therefore, NB cells rapidly and specifically adapt to the successive embryonic microenvironment they interplay with.
Combined single cell genetic and transcriptomic analyses to trace NB dissemination dynamics
To obtain a dynamic and exhaustive picture of NB transcriptome evolution across the metastatic progression, we further integrated the genetic information of transcriptomic regions (single nucleotide polymorphisms and indel alterations). As such information is only accessible and informative from full lenght RNA sequencing, we focused on SMARTseq2 datasets, with the aim of relating the clonal evolution of NB cells into the embryo to their physical path, starting from each primary tumor site. By extracting genetic variants from the two SMARTseq2 datasets (A1 and A2), performed on two different batches of IGR-N91 cells, we could verify that the genetic evolution was sufficiently rapid to distinguish cells from each experiment although harvested in the same tissues (SG, ADR, Supplementary Fig. 8a) and from each time point (E0, E5, E14) (Supplementary Fig. 8b) based on this genetic information. This illustrated the possibility to use genetic information to trace the trajectory of NB cells over a short period of time, in a given scRNA-seq dataset. Thus, we further used the SMARTseq-A1 dataset to trace the spatial trajectory of NB cells. Informative genetic variants consisted in a majority of single nucleotide polymorphisms (SNPs), leading to missense mutations that for the top 10 mutated genes concerned >60% of cells (Supplementary Fig. 8c-e). While these genes were all found expressed in the SA lineage, we did not observe any marked tendency for SNPCs-related specific genes, except for LBR and NEFM (Supplementary Fig. 8f). With this set of variants, NB cells harvested from SG and ADR primary tumors could almost perfectly be distinguished by different clusters of genetic variants (Supplementary Fig. 8b, g). Following the same logic, we built a single cell-based parsimony tree of cells harvested from each physical position to assess their clonal relationship with cells located in ADR- and SG-primary tumors. This approach confirmed the segregation of NB cells into two subclonal populations having either a SG or an ADR origin (Fig. 5a). Interestingly, AOR cells were mostly embedded in ADR-derived clusters while PN and BM cells were evenly distributed in both SG and ADR-derived clusters. These parsimony tree characteristics suggested that primary tumors located in the sympathetic chain or in the adrenal medulla both disseminated via the peripheral nerves while the aortic path preferentially concerned tumors of the adrenal medulla. Moreover, both types of primary tumor and dissemination roads were predicted to feed the BM niche with metastatic cells, with similar predicted efficiency (Fig. 5a and Supplementary Fig. 8h). Confocal analysis of sections or 3D imaging of E14 chick embryos engrafted at E2 with IGR-N91::GFP cells allowed to complement this prediction as key physical transitions from one tissue to another could be detected in imaging data (Fig. 5b–q and Supplementary Fig. 9, Supplementary Movies 10–12).
Fig. 5. Genetic tracing of NB cells across the dissemination process allows mapping their physical trajectories in avian tissues.
a Parsimony tree showing the genetic relationships between NB cells located in adrenal tumors (orange arm) or sympathetic tumors (blue arm) and subclones located in the dissemination paths (AOR, PN) and in the bone marrow (BM). Genetic information was extracted from SMART-Seq2 single cell data obtained from E14 avian embryos grafted at E2 with IGR-N91::GFP cells (SMARTseq-A1 dataset, N = 9 embryos). Genetic proximity between cells was computed using single nucleotide polymorphisms and indel alterations of transcriptomic regions to build a cell-to-cell genetic divergence matrix based on variant allele frequency. b–q Representative images of the physical paths taken by IGR-N91::GFP cells in E14 embryos (8 embryos from 5 independent experiments analyzed). Tissue sections or whole embryos were labeled with α-GFP and α-NF160 or α-SMA antibodies. E: Embryonic day. b, c show IGR-N91 primary tumors located in the SG (observed in N = 6/8 embryos) (b, enlargement in b’) and in the ADR (observed in N = 6/8 embryos) (c, enlargement in c’). d, e show NB cells transiting to the PN (observed in N = 8/8 embryos) from a sympathetic tumor (d, enlargement in d’) and from an adrenal tumor (e, enlargement in e’). f–i show NB cell migration along nerves at distance from primary tumor sites (f) and reaching innervated bones (g–i) and the BM (observed in N = 4/8 embryos) (j). k–n show adrenal tumors transiting via the AOR (observed in N = 6/8 embryos). k (enlarged optical section in k’) illustrates the proximity of adrenal tumors with the AOR and l-n are transverse sections of the adrenal/aorta region. o–q show NB cells migration along blood vessels (o) at distance from adrenal tumors and reaching the bones (p) and the BM (q, optical section in q’). b–j, l–p are sections of embryos imaged with confocal microscopy. h–k, q were imaged with lightsheet microscopy. Scale bars are indicated. Source data are provided as a source data file. Adr Adrenal gland, Ao Aorta, Bm Bone marrow, Drg Dorsal root ganglia, Lg Lung, Lv Liver, Sg Sympathetic ganglia, V Vertebrae.
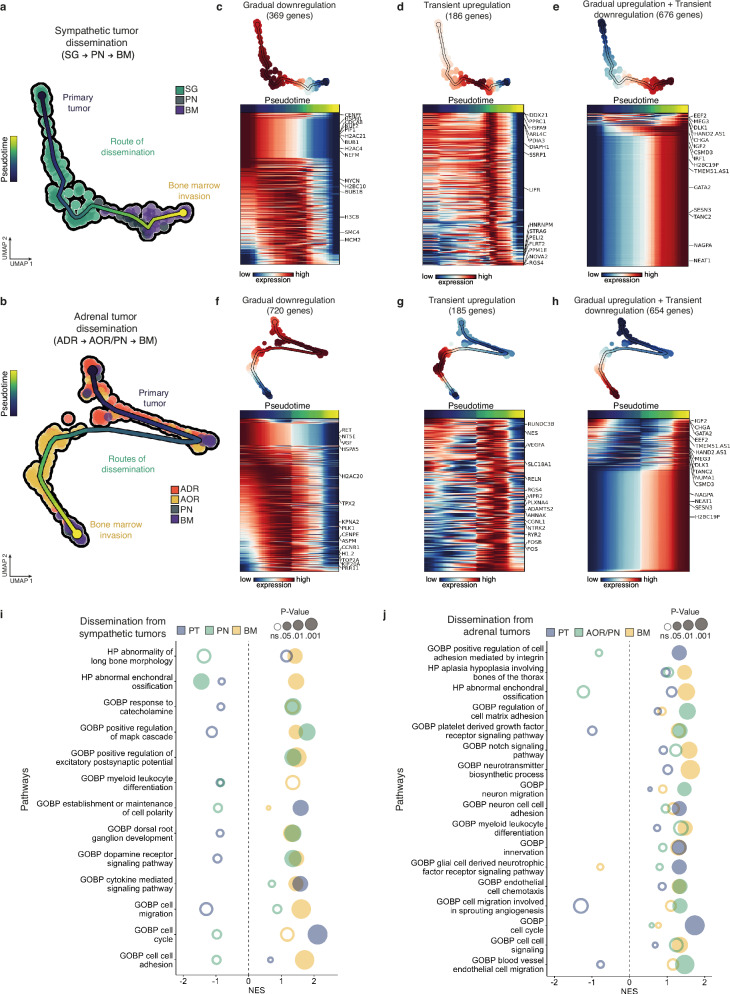
We next combined single cell genomic (variant allele frequency data) and transcriptomic data in a single matrix to build the trajectories of NB transcriptomic profiles across dissemination, starting from each primary tumor site (Fig. 6a, b). For both trajectories -i.e.: SG-to-PN-to-BM and ADR-to-PN/AOR-to-BM- we performed an exhaustive extraction of genes that signed each NB transition from an embryonic tissue to another (Fig. 6c–h). Gene enrichment analysis performed on these signatures for both routes of dissemination highlighted key biological processes specific to each transition and to embryonic tissue hosting NB cells, suggesting that NB cell dissemination was sustained by active dialogs with the successive environments they crossed (Fig. 6i, j).
Fig. 6. Dynamic transcriptomic adaptations of NB cells across their dissemination in avian embryonic tissues.
a, b UMAP plots based on the integration of both variant allele frequency and transcriptomic single cell data to map the trajectory of NB cells disseminating from a sympathetic ganglia (a,122 cells) or an adrenal (b, 200 cells) primary tumor site to the bone marrow (BM) via the peripheral nerves (PN) and/or the aorta (AOR). Each trajectory was determined using parsimony tree extracted from cell-to-cell genetic divergence clustering. c–h Sets of genes -and corresponding heatmaps- which expression significantly changes along linear pseudotime of NB cell trajectories using Moran’s I test. Genes following the same type of dynamics (gradual downregulation, transient upregulation, gradual upregulation and transient downregulation) were grouped in independent heatmaps. i, j Gene set enrichment analysis (GSEA) from genes listed to be dynamically regulated in NB cell trajectories from the primary tumor sites to the bone marrow. Pathways were generated with ontology gene sets of molecular signature database (nperm = 1000, ranked by dispersions, abs(NES) > 1.3). Fischer’s exact test was performed. Source data are provided as a source data file.
BM invasion is featured by a functional and dynamic gene set of potential clinical interest
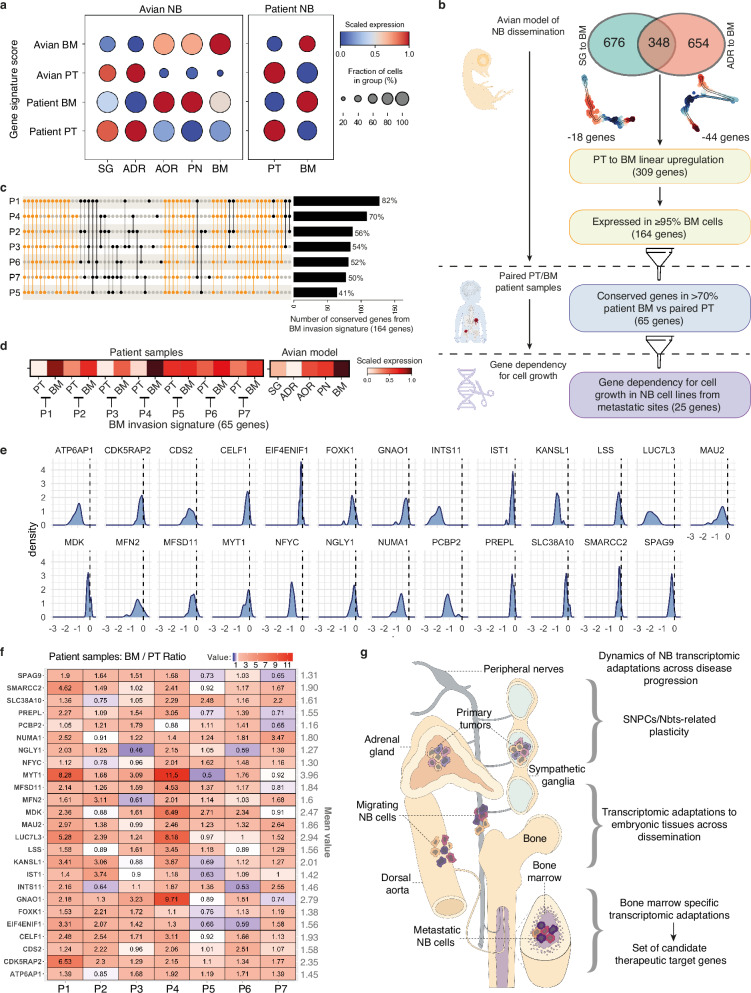
To document the proximity between gene programs in the avian model and in patients, we extracted NB gene signatures from differential gene expression analysis in both avian and patient contexts, in PT and in BM samples. We scored these signatures in NB cells extracted from avian and patient samples. Interestingly, BM and PT signatures from each context had the highest expression score in the corresponding anatomical site (Fig. 7a). Moreover, while only partially overlapping, avian- and patient-derived NB signatures expressions were significantly correlated in NB cells extracted from patients or from the avian model (Supplementary Fig. 10a), suggesting proximities regarding biological mechanisms involved.
Fig. 7. NB metastasis to the BM is featured by a functional and dynamic gene set having potential clinical interest.
a Dot plot showing the level of expression of NB gene signatures built from the avian model (avian-PT, avian-BM) and from patient data (patient-PT and patient-BM) in NB cells located in primary tumors or in the bone marrow in each context (avian model and patients) (two-sided Wilcoxon rank sum test; p-value < 0.05, log fold change > 0.1, min.pct > 0.1). b Decision tree to extract a list of candidate genes functionally involved in BM invasion by NB cells. c Upset plot of the 164 genes enriched during BM invasion in the embryonic model of NB dissemination (and expressed in patient samples) in the 7 patient BM compared to paired PT. Yellow bars indicate pools of genes which expression profile is shared by at least 5 out of the 7 patients. Bar plot of the fraction of genes from signature conserved in each patient is shown on the right. d Heatmap of the expression of BM invasion signature refined from patient samples data (65 genes) in paired PT/BM patient samples and in NB cells at each step of dissemination. e Distribution of effect scores of the 65 BM-genes calculated using Chronos algorithms on CRISPR/Cas9 data from DepMap portal on 18 NB cell lines obtained from metastatic sites. Graphs are shown for the 25 genes associated with significant NB dependency for cell growth as detailed in the methods section. Data are normalized using annotated sets of common-essential and non-essential genes. No effect on cell growth has a score of 0, and a negative score is indicative of gene dependency for cell growth. f Heatmap of gene expression ratios (BM over PT) of the 25 genes extracted in (e). Values of BM/PT ratio for each gene and for each patient are indicated on the heatmap. g Scheme summarizing the types of dynamic transcriptomic adaptations occurring in NB cells across disease progression in an entire growing organism. Source data are provided as a source data file.
We could identify 348 genes that were commonly enriched in both SG-to-BM and ADR-to-BM trajectories in NB cells transitioning to a BM localization in the avian embryo (Fig. 7b). This list of genes, selected on their dynamic regulation across NB progression, and not accessible from clinical data only, could have a functional impact on the dissemination and establishment of NB cells in the nascent bone marrow niche. We next thought to intersect our analysis of dynamic trajectories across NB metastatic progression in the avian model with PT/BM patient data, with the objective of extracting a functional gene set. We performed serial cross analyses driven by the aim to extract a set of genes specifically expressed in metastatic cells and showing transcriptome dynamics compatible with the targeting -i.e., that are not predicted to undergo transient downregulation during NB dissemination as targeting them could instead boost the active dissemination phase. Starting from the 348-gene signature dynamically enriched at the BM level, we subtracted 62 genes predicted to display a transient downregulation at any point of the active dissemination step (Fig. 7b and Supplementary Fig. 10b,c). Then, we selected all genes showing a significant upregulation in >95% of cells reaching the BM in the avian model to focus on frequent events. 164 genes remained after this filtering step (Fig. 7b). The analysis of gene expression variations in PT/BM pairs highlighted 65 of these genes (40%) that were significantly upregulated in BM cells as compared to matched PT cells, in at least 5 patients (out of 7 patients) (Fig. 7c). Interestingly, the 65-gene signature was indeed upregulated at the BM level in all of the 7 NB patient pairs and its expression score at each step of metastatic progression in the avian model was compatible with a gradual upregulation, without any transient downregulation (Fig. 7d). As a final filtering step, we assessed NB dependency for cell growth to each of these 65 genes using published data of genome-wide CRISPR loss-of-function screens (DepMap, https://depmap.org/portal). We analyzed data from 18 human NB cell lines derived from metastatic sites, and could identify 25 out of the 65 genes whose expression was required for metastatic NB cell growth (Fig. 7e and Supplementary Fig. 10d). We also analyzed the 62 additional genes that we did not retain in our final screening as they were transiently down-regulated during metastatic dissemination, thus suggesting that their targeting could have deleterious pro-dissemination effect. Interestingly, among the 62 genes that we subtracted from our screening, 22 were found associated with NB dependency for cell growth following the same approach (Supplementary Fig. 10e), highlighting the added value of our model allowing the tracing of gene dynamics across the full metastatic process. Thus, we report here a refined set of 25 genes of potential clinical interest to target NB metastatic cells, identified via a multimodal analysis of NB cell behaviors, transcriptome dynamics and gene dependency across disease progression.
Discussion
The identification of the precise developmental stage(s) at which NB emerges remains partial, especially given the challenges to experimentally access embryonic processes. Nevertheless, recent studies of NB tumor genomic features combined with mathematical modeling have made it possible to trace the evolutive history of NBs. It provided evidence for very early and temporally-confined occurrence of initiating oncogenic event(s), within the first three gestational months5, thus fully concomitant to the morphogenesis of sympatho-adrenal organs16,18,32,33. Analysis of clonal evolution during NB metastatic spread confirmed and extended the view of early—likely prenatal- metastatic seeding from primary tumors to local and distant sites, prior to diagnosis6. In previous work, we provided evidence that human NB cells implanted in the trunk neural crest of the chicken embryo underwent ventrally-oriented migration, stopping in the nascent sympatho-adrenal territories to form primary tumors22–24. In a next step, some detached from the tumor mass, selecting the dorsal aorta and peripheral nerves as dissemination routes21,22. It was let open whether this embryonic model could also recapitulate the metastatic disease, with secondary foci in sites homologs to those of the patients. In this study, we set up imaging procedures to analyze avian embryos at late developmental stages (E14 rather than E9 in refs. 22,23). We could demonstrate that the dissemination routes convey NB cells to the bones and the bone marrow, which thus reproduces the typical metastatic pattern of NB in patients. Fine analyses with light sheet and confocal microscopy to characterize the embryonic substrates driving the cells to the metastatic site show NB cells migrating along peripheral nerves and internal and external surfaces of the dorsal aorta, thus coopting developing axon bundles and vessels to infiltrate the bone and the bone marrow. These observations also support that the NB metastatic disease has an embryonic occurrence that is concomitant to organogenesis processes, which is consistent with the reported timing of NB tumor evolution5,6. However, we took a non-exhaustive standard dissection approach to describe the routes that convey NB cells to the bone marrow. Additional secondary metastatic sites -such as the liver or the brain-, could rely on different dissemination patterns and underlying phenotypic adaptations.
Having an experimental access to NB evolution in the embryonic context of origin with our avian model, we found that the transcriptome heterogeneity manifested by NB cells over disease progression shares key features with a restricted portion of the developing sympatho-adrenal lineage. Interestingly this diversity progressively emerged from an initially homogeneous state of cultured cells, when NB cells settle in the embryonic sympatho-adrenal tissues, thus suggesting an involvement of embryonic cues. We characterized three connected states matching the physiological SNPCs-to-neuroblasts trajectory initiated from SA progenitors. The latter have the bi-potentiality of giving rise to sympathetic neurons and adrenal cells. This may explain our observations that grafted NB cells are capable of recapitulating migration towards both types of SA derivatives.
We found that the temporal sequence of state occurrence in NB was aberrant, with first the comSNPCs-like state, second, the earlier SNPCs-like state, and lastly the Nbts-like state. Moreover, interconversions between all three states were predicted from RNA velocity and dynamic transcriptome programs. Remarkably, tumor-specific plasticity remained framed within this restricted portion of the SA axis, whatever NB cells were engaged into metastatic dissemination or not. Interestingly, our findings are in line with a recent patient sample-based study demonstrating the conservation of cellular plasticity upon metastasis21. Analysis of key markers of these states in several other NB cell lines engrafted in the avian model showed similar dynamic expression over disease progression. Thus, in the embryonic context of origin, NB cells may follow a common program of transcriptomic adaptations, whatever the nature of their genomic alterations. Whether this transcriptomic landscape covers the spectrum of states that NB cells can take remains open. Indeed, transitions through different states may occur that we did not capture at the embryonic stages of experimental manipulations and investigated time points. Strikingly however, the transcriptome states revealed by our avian model of NB, in addition to be depicted in patient tumors, also recapitulated the spectrum of intratumor and interpatient heterogeneity. Moreover, we detected no obvious features shared between NB and the physiological SCPs differentiation branch, neither in our model of NB disease nor in patient tumors. As well, acquisition of mesenchymal or SCPs-like transcriptomic traits by NB cells engaging into the metastatic process was not observed, which is in line with other scRNA-seq-based studies14,21. This confirms the idea that NB cell plasticity manifests itself primarily through the expression of SNPCs/Nbts-like features, but not SCPs. Whether this reflects the identity of NB cell of origin or rather the expression of malignant plasticity remains to be demonstrated experimentally. Whether and how these transcriptome states are differentially adapted depending on genomic features of NB cells, whether these features impact on the plasticity between and within states and what would be their functional outcomes remain enigmatic. Interestingly for 6 out of 7 patient samples, the fraction of comSNPCs-like and SNPCs-like cells was increased in the BM as compared to the matched PT, suggesting enhanced plasticity and pathological regulations of transcriptomic dynamics in favor of progenitor-like states in the bone marrow niche.
By combining scRNA-seq and tracing of NB subclones with genetic variants, we could reconstitute the physical routes conveying NB cells from the two primary tumor sites to their final bone marrow destination. Interestingly, this analysis revealed that both nerve and aorta substrates deliver NB cells from adrenal tumors to the bone marrow, while the nerves were by far the predominant path taken from the sympathetic ganglia site. This may reflect organ-specific architecture controlling the access to the two routes, or different tissue maturation states. Although our analyses did not highlight a precise, conserved genetic landscape that would drive NB cells to specific physical sites during the metastatic process, we cannot exclude that some genetic variants functionally impact NB dissemination properties. Alternatively, specific local microenvironment provided by the two primary tumor sites could induce distinct adaptations of NB cells, driving disseminating NB cells preferentially to one or the other route. In support, we found differential gene expressions in NB cells located in adrenal and sympathetic ganglia tumors, some coding for distinct receptors of extracellular signals. Similarly, we found transcriptomic adaptations to the nerve or aorta substrate that highlight specific dialogs between NB cells and these embryonic environments. Likewise, RHO-GEF-containing obscurin1 (OBSCN) transcript, enriched in NB cells migrating on the dorsal aorta is thought to regulate “microtentacle” processes formed by metastatic cancer cells to attach to the endothelium34. In NB cells migrating along the nerves, a set of emblematic genes involved in neuronal migration -i.e., NRCAM, ERBB4, NTRK2- and/or axon guidance mechanisms -i.e., NRP2, PLXNB2 NFASC-, was found specifically enriched35–37. Interestingly, we observed that rather than acquiring mesenchymal or SCPs-like transcriptomic traits, metastatic NB cells undergo a series of phenotypic adaptations to each of the physical substrates successively met. Future deeper investigations of these microenvironment-specific adaptations should reveal unexpected pro-metastatic molecular dialogs. Moreover, inter-specie differences also have to be taken into consideration, and human specificities may exist that could be translated into additional molecular signaling mediating cancer cell-environment communications lacking in the avian embryo model. Additional levels of heterogeneity among IGR-N91 cells exist that we did not study. For example, following NB cell grafting, we observed a continuum of migrating cells spanning the sympatho-adrenal path. Cells at the rear did not reach the target at the first time (E5) of our tumor microdissections. These cells may manifest some transcriptional differences that we did not capture by focusing on primary tumor sites. Moreover, at E5, sympathetic and adrenal derivatives already emerge as physically distinct territories. However, for technical feasibility, NB cells were collected indifferently of their sympathetic or adrenal location. Thus, we may have missed some early location-specific transcriptional features.
Nevertheless, by crossing experimental data of NB gene expression dynamics across the complete metastatic process with patient tumor scRNA-seq data, we could extract a set of candidate target genes predicted to support the growth of bone marrow metastasis. We could refine this gene set by subtracting genes predicted to be upregulated in NB cells reaching the bone marrow, but showing transient downregulation during dissemination, thus which targeting could drive adverse effect in steps preceding bone marrow invasion. Of note, among the 25 genes that were finally selected as candidate therapeutic targets, we identified Midkine (MDK), a pleiotropic growth factor highly expressed during embryogenesis, and aberrantly activated in a range of pathological contexts, including in cancer38–40. Specifically, Midkine was found to be overexpressed in aggressive forms of NB41 and was recently proposed to be functionally involved in pro-tumoral cell-cell dialogs between NB and myeloid cells residing in the bone marrow21. Thus overall, our study provides insights into yet inaccessible steps of NB progression in patients. In addition, our characterization of gene expression dynamics across the full metastatic process using the avian NB model lays the foundation for innovative therapeutic approaches driven by a comprehensive and functional view of NB disease.
Methods
Our research complies with all relevant ethical regulations.
Chick embryos
Naked Neck strain embryonated eggs were obtained from a local supplier (Elevage avicole du Grand Buisson, Saint Maurice sur Dargoire, France). Laying hen’s sanitary status was regularly checked by the supplier according to French laws. Eggs were housed in an 18 °C incubator until use. They were next incubated at 38.5 °C in a humidified incubator until the desired developmental stage, calculated according to Hamburger Hamilton Stages (HH), – i.e., HH14 (E2) for the graft step, HH28 (E5) for 3 days and HH40 (E14) for 12 days post-graft analyses (E: Embryonic day). Experiments on avian embryos were carried out in a development window ranging from day 2 to day 14, in compliance with the 2013 ethical guidelines 86/609/EEC and the French application of the law, authorizing unregulated manipulations during the first two-thirds of gestation in avian species. Given this directive, maximal tumor size/burden was not considered in this study.
Cell lines
Human stage 4 neuroblastoma IGR-N91 and SHEP cells were kindly gifted by Dr. J. Bernard (Gustave Roussy Institute, Paris, France) and Dr. M Schwab (Institute for Experimental Pathology, Heidelberg, Germany) respectively. SH-SY5Y were obtained from ATCC (ATCC® CRL-2266). IGR-N91, SH-SY5Y and SHEP cell lines stably expressing GFP (IGR-N91::GFP, SH-SY5Y::GFP and SHEP::GFP) were previously generated and described in Delloye-Bourgeois22. IGR-N91::GFP and SH-SY5Y::GFP NB cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) GlutaMAXTM (Life Technologies). SHEP::GFP cells were cultured in RPMI 1640 GlutaMAXTM (Life Technologies). Media were each supplemented with 10% Fetal Bovine Serum (FBS), 25 U/mL Penicillin Streptomycin (Sigma), 2.5 mg/mL Amphotericin B (Sigma). Cell lines were maintained in sterile conditions in a 37 °C-, 5% CO2-incubator.
Human NB patient samples
Ethics statement
Primary tumor biopsies and bone marrow aspirates were taken at the timepoint of diagnosis prior to chemotherapeutic treatment. After routine diagnostic procedures, remaining material was used if written informed consent from patients and/or parents was obtained. The use of these samples was reviewed and approved by the biobank committee at the Princess Máxima Center. Primary tumor and bone marrow samples of 7 high-risk patients between the age of 11 and 151 months were included in this cohort. Clinical characteristics of the selected patients are shown in Supplementary Table 2.
Primary tumor patient samples
Primary tumor samples were processed fresh within 4 h after the surgery. The tissue was cut into pieces of <1 mm3 with a scalpel and subsequently digested with Collagenase type I, II and IV (each 2.5 mg/mL, Life Technologies) for up to 1 h at 37 °C. The sample was filtered through a 70 μm cell strainer, washed with DMEM (300 g for 10 min at 4 °C) and further dissociated into single cells with NeuroCult cell dissociation kit according to manufacturer’s protocol (Stemcell Technologies #05707). Cells were stained with DAPI (Merck #D9542) and DRAQ5 (Biolegend #424101). Live cells (DAPI-negative, DRAQ5-positive) were sorted into 384-well plates containing 10 μL of mineral oil, 50 nL of RT primers, deoxynucleotide triphosphates (dNTPs) and synthetic mRNA Spike-Ins on a SONY cell sorter SH800S. Plates were spun down, snap‐frozen on dry ice, and stored in ‐80⁰C until further processing.
Bone marrow patient samples
Mononuclear cells were isolated from diagnostic bone marrow samples by Ficoll-Paque (Cytiva) and viably cryopreserved in IMDM + 20%, FCS + 10% DMSO. For analysis they were thawed in IMDM + 20% FCS +DNAse (100 µg/mL)+ MgCl2 (5mM). Cells were mechanically dissociated by resuspending in FACS buffer (PBS + 0.5% FCS +2 mM EDTA) and filtering through a 70 µM cell strainer. Consecutive stainings with nearIR (15 min), surface markers (25 min) and Draq5 (15 min) were performed at 4 °C and at a cell concentration of 10.106 cells/ml. Details of the surface marker antibodies are listed in the table below. They were validated in previous studies42. Draq + /NearIR- cells were selected, then CD45-/CD34- cells. Finally, to enrich the fraction of NB tumor cells, two parallel selections were performed to sort CD90 + /GD2 + , CD90 + /GD2- and CD90-/GD2+ cells. Complementary to these markers used for the sorting, other markers were assessed to validate the gating strategy: CD3 (T Lymphocytes), CD56/CD81 (tumor markers), CD13/CD73/CD105/CD146/CD271 (mesenchymal markers).
Using BD AriaIII, cells were analyzed and sorted into 384-well plates containing 10 μL of mineral oil, 50 nL of RT primers, deoxynucleotide triphosphates (dNTPs) and synthetic mRNA Spike-Ins. Plates were spun down, snap‐frozen on dry ice, and stored in ‐80 °C until further processing.
| Marker | Fluorophore | Dilution factor | Manufacturer | Catalogue number |
|---|---|---|---|---|
| CD13 | BV421 | 100 | BD | 562596 |
| CD45 | BV510 | 800 | BD | 563204 |
| CD34 | BV605 | 200 | BD | 745247 |
| CD81 | BV650 | 100 | BD | 740590 |
| CD90 | BV711 | 200 | BD | 740786 |
| GD2 | BV786 | 400 | BD | 744073 |
| CD105 | FITC | 25 | Biorad | MCA 1557F |
| CD146 | PerCP Cy5.5 | 200 | BD | 746081 |
| CD56 | PE | 200 | Biolegend | 355504 |
| CD271 | PE-CF594 | 200 | BD | 563452 |
| CD73 | PE-Cy7 | 400 | BD | 561258 |
| CD3 | APC | 50 | BD | 561804 |
| Live/dead nearIR | 20,00 | Invitrogen | L10119 | |
| Draq5 DNA dye | 25,000 | Abcam | ab108410 |
Neuroblastoma patient samples processing—PMC pipeline
Cells were further processed by scDiscoveries (Utrecht, Netherlands), following the SORT-seq protocol, a partially robotized version of the CEL-seq2 protocol43 that consists in combining FACS sorting and reverse transcription reaction with a robotic liquid handler44. Paired-end 2 × 75 bp read length was used for sequencing on the Illumina NovaSeq6000 sequencer at the single-cell facility of Princess Maxima Center (Utrecht, Netherlands) and on the Illumina NextSeq500 at the Hartwig Medical Foundation (Utrecht, Netherlands), respectively for the bone marrow and primary patient samples. Raw data was processed with the Sharq pipeline45. Read mapping was performed using STAR (v2.6.1), on the hg38 Patch 10 human genome. Function featureCounts (v1.5.2) was used to assign reads based on GENCODE (v26) to generate count tables.
NB cell lines graft in ovo
E2 (stage HH14) chick embryos were grafted with ~2500 fluorescent NB cells at the neural crest level, -i.e., between somite 18 and presumptive somite 24. NB cells were implanted with a glass capillary connected to a pneumatic PicoPump (PV820, World Precision Instruments) under a fluorescence stereomicroscope. Briefly, for each glass capillary used, the number of cells ejected with a single pump pulse was determined and the concentration/number of pulses adjusted to graft ~2500 cells per embryo. Eggs were closed with solvant-free tape and placed back in the humidified incubator until E5 (HH28) or E14 (HH40). The survival rate of grafted embryos was comprised between 72 and 96% at E5, and between 35 and 51% at E14. The grafting efficiency (corresponding the fraction of living embryos with sympathetic and/or adrenal tumors) was above 94% both at E5 and at E14.
A single, independent graft experiment, referred to a series of grafts performed with the same batch of NB cells (same culture dish) and the same batch of synchronized embryonated eggs.
Immunofluorescence on Slices
Chick embryos were harvested and fixed in 4% Paraformaldehyde (PFA). Embryos were embedded in 7.5% gelatin - 15% sucrose in PBS to perform 50 µm transverse cryosections. Antigen retrieval was done in citrate buffer (pH6, 1 0mM, Millipore – Sigma-Aldrich, #C8532). Permeabilization and saturation of sections were performed in PBS-BSA 4%-Triton 0.5%. The following primary antibodies were applied to sections: anti-NF160 mouse IgG (1/200, Thermo Fisher Scientific, Life Technologies, #130700), anti-SMA mouse IgG (1/200, Millipore—Sigma-Aldrich, #A2547), anti-TOP2A mouse IgG (1/200, Novus Biologicals, #53281), anti-LMNB1 mouse IgG (1/200, Abcam, #ab8982), anti-ATAD2 rabbit IgG (1/200, Thermo Fisher Scientific - Invitrogen, #720138), anti-CHGB rabbit IgG (1/200, Millipore – Sigma-Aldrich, #HPA008759), anti-GFP goat IgG (1/500, Abcam, #ab5450), Alexa 555 anti-mouse IgG (1/500, Thermo Fisher Scientific, Life Technologies), FP647 anti-rabbit IgG (1/500, Interchim) and Alexa 488 anti-goat IgG (1/500, Thermo Fisher Scientific, Life Technologies) were used as secondary antibodies. Nuclei were stained with Hoechst (H21486, Thermo Fisher Scientific, Invitrogen). Slices were imaged with a confocal microscope (Olympus, FV1000, X81) using either a 4X objective for whole slice imaging or a 40X objective.
Tissue clearing, whole-mount immunofluorescence and Selective Plane Illumination Microscopy (SPIM) Imaging
PFA-fixed E5 (HH28) or E14 (HH40) embryos were dehydrated in successive increasing methanol concentrations, incubated overnight with 2% H2O2 in methanol and rehydrated following the reverse protocol. Whole-mount immunofluorescence was performed using a blocking solution composed of 20% DMSO, 0.5% Triton 100X, 4% Bovine Serum Albumine (BSA) and 100 mM glycine, in PBS. Primary and secondary antibodies were applied in the blocking solution. The following primary antibodies were used: anti-GFP rabbit IgG (1/200, Thermo Fisher Scientific, #A11222), anti-Neurofilament 160kDa, RMO-270 mouse monoclonal IgG (1/200, Thermo Fisher Scientific, #13-0700), anti-SMA mouse IgG (1/200, Millipore—Sigma-Aldrich, #A2547), anti-L1CAM goat IgG (1/50, R&D, #AF277), anti-ERBB4 rabbit IgG (1/50, Millipore – Sigma-Aldrich, #HPA012016), anti-PLXNB2 (1/100, Proteintech, #10602-1-AP), anti-TUBB3 rabbit IgG (1/200, Biolegend, #802001), and the following secondary antibodies: Alexa 555 anti-mouse IgG (1/500, Lige Technologies #A31570) and FP-647H anti-rabbit IgG (1/500, Interchim, #FP-SC5110). Embryos were then cleared using a modified iDISCO protocol46. Embryos were dehydrated in successive methanol baths before clearing with dichloromethane followed by dibenzyl ether baths. Cleared samples were imaged using a light sheet UltraMicroscope Blaze (Miltenyi Biotec), using either a 4X objective for whole organ imaging (laser power set at 5 – 10%, sheet NA 0.035, sheet width 60%) or a 12X objective (laser power set at 1–5%, sheet NA 0.0135, sheet width 20%).
Analysis of 3D light sheet images
3D images were built and quantified with Arivis Vision4D software.
NB cell harvesting for single cell RNA-seq (scRNA-seq)
IGR-N91::GFP cells were harvested before engraftment by trypsination and from chick embryos using a fluorescence stereomicroscope. The “E0” step corresponded to NB cells prior to the graft. Tumor cells were dissected out from E5 chick embryos in sympatho-adrenal tissues and from E14 chick embryos at different localizations: primary tumor sites (adrenal medulla, ADR, and sympathetic ganglia, SG), secondary metastatic site (bone marrow, BM), and NB cells on spreading routes (dorsal aorta, AOR, and embryonic peripheral nerves, PN). Specifically, as the size of NB foci located in the BM, PN and AOR was variable, we performed a systematic dissection of the femoral, iliac and costal bones to collect the bone marrow (BM), intercostal nerves (PN), and the infrarenal, suprarenal and descending portions of the aorta (AOR). For each independent graft experiment, the dissected tissues from living embryos were pooled per anatomical localization. Harvested tissue were kept on ice in F12 medium throughout the manipulation, then dissociated with type IV Collagenase (1.25 mg/ml, Sigma-Aldrich) and DNAse I (0.1 mg/mL, Sigma-Aldrich) for 10 min at 37 °C followed by trypsin (2.9 mg/mL, Sigma-Aldrich) dissociation for 5 min at 37 °C. The reaction was stopped with DMEM GlutaMAXTM (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS). Dissociated samples were frozen in DMEM containing 20% FBS and 10% DMSO.
Three scRNA-seq datasets were generated:
SMARTseq-A1: NB cells retrieved from E14 embryos, covering the different tissue localizations: ADR, SG, AOR, PN, BM
SMARTseq-A2 and seqWell: NB cells collected prior to the graft (E0) and from primary tumors formed at E5 (sympatho-adrenal tumors) and at E14 (ADR and SG tumors separately).
Generation of scRNA-seq data from the avian embryonic model of NB
Pre-processing and library preparation
Samples were sorted with the cellenONE technology (Cellenion Bico) to discard non-fluorescent chick cells and to dispatch human single cells in 96-well plates. Full-length RNA-seq was performed with SMART-Seq2 kit according to the standard protocol47 or with the 3’ scRNA-seq seqWell kit (seqWell). Single cell libraries were generated with the Nextrera XT DNA Library Preparation Kit (NextSeq500, Illumina) exploiting single end mode and paired end (75 nucleotides), respectively for SMART-seq2 and seqWell kits. Cells were multiplexed via dual index-strategy. Bcl2fastq (v2.17.1.14) was used to generate one FASTQ file per cell containing the genetic sequence of each read (sequenced cDNA) associated with a quality score for each nucleotide. Trimming process to remove the step of low-quality nucleotides and adapter sequences was carried out with the CutAdapt software (v1.9.1). Detection of cDNA contaminants was done with the FastQ Screen (v0.5.2). Sequenced libraries were mapped on pre-indexed GRCh38 human genome with ENSEMBL annotation (v93) through STAR software (v2.7) and 2-pass option allowing to increase the spliced reads mapping detection to novel junctions. Afterwards, using HTSeq-count software (v0.12.4), count matrices were produced and used as input for single cell transcriptomic analyses.
Quality control of NB cells in the avian embryonic microenvironment
Seurat package (v4.0.1) was used to compute the quality control metrics based on the number of both distinct and total count genes, and mitochondrial genes to have an overview of cell viability. Dispersion of each parameter was visualized to remove cells exhibiting extensive variability compared to the overall dispersion for each sequenced condition. Data were normalized for sequencing depth, scaled to 10,000 RNA counts per cell and log-transformed. The 2,000 most variable genes were defined to compute principal component analysis (PCA) using 50 dimensions. Relative proximities between cells were assessed with 20 nearest neighbors. Stable groups were identified as clusters insensitive to small changes in the resolution parameter displayed through Clustree package (v0.4.3). At last, dimensional reduction was performed via the uniform manifold approximation and projection method (UMAP).
Removing doublet cells from datasets
The first 20 principal components were used for UMAP dimension reduction. In the step for estimating doublet rate using the doubletFinder48 (v2.0), the doublet rate was assumed to be 0.075 and adjusted according to the estimated proportion of homotypic doublets from Poisson statistics. pN was set at 0.25, pK was set at 0.09. The doublet label predicted by DoubletFinder was used to eliminate doublet cells from the downstream analysis.
Multi-datasets integration
Data generated from NB cells in the avian model (NB at E0, E5, E14), and published data from the human SA lineage and patient tumor samples were integrated thanks to Seurat methods49 to match shared cell populations across datasets. Data were prepared for the integration using SCTransform normalization with 3000 variable features. Set of anchors between data were extracted (k = 5, dims = 50) before final integration using weighting anchors of 50 for published data and 100 for NB cells from the avian model. Afterwards, a standard protocol of cell clustering and plotting was used (dim = 20, res = 0.1, 0.1, 0.5 respectively NB cells from avian embryo, patient samples, and SA lineage).
Unimodal UMAP projection
A set of tools developed in Seurat was applied to find anchors and project query labels onto a UMAP structure of the reference. Tools consist in determining a set of shared anchors between a reference and a query object with FindTransferAnchors, in order to classify the query cells based on reference data using TransfertData function. Next, the number of neighbors to use for finding anchors was adjusted according to each dataset since the choice of K is dependent on the quality of input data (noise / number of cells / sequencing depth). Then, query cells embedding were corrected onto the reference UMAP with 20 dimensions using MapQuery function.
Cell type annotation
Set of strategies was deployed to best annotate cells. By performing unbiased cell type recognition with SingleR package (v1.0.5)50 based on Spearman correlation method between an annotated reference dataset to a label free query. Marker genes that defined clusters by differential gene expression (DEG) were extracted using the Seurat FindAllMarkers function (P-value < 0.05, log-fold change > 0.25, method = Wilcoxon). Cell signature scores were generated with AddModuleScore Seurat function.
To annotate NB tumor cells from patient samples, we combined copy number variations (CNVs), with validated markers of other cells of the tumor microenvironment. Markers used to annotate patient cells are detailed in Supplementary Fig. 7b and f. CNVs at the single cell level were called with R package InferCNV (v1.10.0) (https://github.com/broadinstitute/inferCNV) using default parameters. Non-tumoral cells from the microenvironment were used as reference cells for patient tumor samples.
Cell cycle analysis
Cell cycle scores were based on the expression of G /M and S phase markers obtained from a published study based on murine hematopoietic progenitors51 using the Seurat CellCycleScoring tool.
Transcription factors analysis
SCENIC (v1.1.2)52 pipeline was applied to infer Gene Regulatory Networks and their associated cell states from scRNA-seq data. SCENIC was used with a normalized matrix extracted from Seurat pipeline combined with RcisTarget databases (hgnc19, 500pb upstream, transcription start site +/- 10 kbp). GENIE3 package (v3.16) with a Random Forests approach was employed to build a co-expression network. Finally, co-expression modules were obtained with TF-motif enrichment analysis, to score the regulons activity on individual cells.
RNA velocity
scVelo module (v0.2.5)53 was used to estimate the time derivative of the gene expression for each cell in a phenotypic state. Estimation was computed by distinguishing unspliced and spliced RNAs from expression matrices obtained from velocyto software (v0.17)54. Spliced and unspliced matrices were normalized and log-transformed with a minimum of share features to 20, keeping the 2000 most variable genes. Moments for velocity estimation were computed for each cell across its 30 nearest neighbors. Full splicing kinetics of specified genes through dynamical model were recovered considering both transcription rates and cell-specific latent time. Finally, cell velocity was mapped on cell embedding extracted from UMAP clustering.
Directed cell fate beyond RNA velocity
Cellrank (v1.5.1) was used to draw directed cell fate without RNA velocity information55. The human SA atlas dataset was processed using Scanpy module (v1.9.1) to keep the 15,000 most variable genes, normalize total counts per cell, log transform and compute the nearest neighbors (dims = 10, k = 10). CytoTRACE pseudotime was then employed to detect initial, intermediate, and terminal populations56. Next, the estimation of cell-cell transition probabilities based on transcriptomic similarity was computed to obtain transition matrix based on Key Nearest Neighbor (KNN) graph and pseudo-temporal ordering (method = soft, nu = 0.8, b = 20). Projection of the transition matrix was mapped on cell embedding extracted from UMAP clustering.
Temporal cell trajectories
Temporal cell trajectories of the SA lineage were inferred with CellRank combined to Waddington OT (WOT) pipeline57. By representing a mass flow of cell fate over time, temporal cell trajectories give information about a probability that a progenitor cell at a time t gives rise to a progeny at a time t + 1. Probability mass flow in time (min-flow = 0.15) was obtained by estimating the initial growth rates based on human pre-compiled sets of proliferation- and apoptosis-related genes and by simulating random walks on the Markov chain by iteratively choosing the next cell based on the current cell’s transition probabilities (n-sims = 100, max-iter = 500).
Pseudotime tree inference
Raw data from the SA lineage atlas were processed with Scanpy module (v1.9.1), by filtering a minimum of 10 cells expressing the gene, normalizing total counts per cell, log transformation and computing the nearest neighbors (dims = 20, k = 20). Each branch of differentiation from either Schwann Cell Progenitors (SCPs) or Sympathetic-Neural Progenitor Cells (SNPCs) was subset and recomputed using PAGA-initialization on UMAP clustering for downstream analysis58. Tree inference and advanced pseudotime analysis were managed by scFates module (v1.0)59. For SNPCs differentiation trajectory, the learning curve was drawn with 20 nodes using ElpiGraph algorithm and converted from hard to soft assignment using 10 iterations of SimplePPT. For SCPs differentiation trajectory, the learning tree was drawn with 10 nodes using ElpiGraph algorithm. Genes significantly associated with each trajectory were determined (Benjamini-Hochberg adjustment (FDR) < 0.0001 and amplitude > 0.3) and fitted for the trajectories by employing mgcv package (v1.8).
Cell cycle-related genes (listed in gene ontology GO:0007049 biological process) were removed from all these dynamic signatures to calculate their expression score in NB cell clusters.
Pathway analysis
Functional enrichment analysis was led by Gprofiler2 package (v0.2.1) using biological processes from gene ontology60. Gene set enrichment analysis (GSEA) was conducted on ontology gene sets from the molecular signatures database with fgsea package (v1.16.0) comparing gene lists obtained from DEG (P-value < 0.05, log-fold change > 0.25, method = Wilcoxon) or from pseudotime analysis, respectively sorted according to the log-fold change or FDR.
Deconvolution of bulk RNA-seq datasets
NB patient scRNA-seq atlas was used to deconvolute bulk RNA-seq data (SEQC Shi and Fisher’s cohort, N = 498 samples, GEO ID: GSE62564; Kocak’s cohort, N = 649, GEO ID: GSE45547). Bulk RNA-seq data were processed by performing background correction, normalization with the ‘quantile’ method, and filtering out control probes by exploiting limma package (v3.28.14). Estimations of cell type proportions from bulk sequencing data were extracted with MuSiC package (v1.0)61. Deconvolution was done on key markers of SA lineage-related transcriptome states from NB patient scRNA-seq atlas (P-value < 0.05, log-fold change > 0.25, method = Wilcoxon).
Survival analysis
Survival package (v3.5.0) was employed to perform Kaplan-Meier and multivariate survival analysis. Event-Free Survival (EFS) was binarized according to the criteria of event and no event over time for each patient with available clinical information -i.e., respectively 498 and 476 for SEQC and Kocak cohorts. Univariable analyses were managed by comparing groups with log-rank test. Hazard ratio was computed from cox regression in multivariable analyses adjusted for age, sex and MYCN status, depending on SA lineage-related NB profiles.
Variant calling on scRNA-seq data
Starting from aligned sequences to human genome, GATK software (v4.1.4) was employed to extract variant from RNA sequences. Optical and PCR duplicates were highlighted with the MarkDuplicatesSpark tool of picard-tools software (v2.1.0). Post-processing RNA reads aligned against the full reference were optimized with the GATK SplitNCigarReads tool. Then, score recalibration was performed with GATK BaseRecalibrator - ApplyBQSR tool, applying a machine learning tool to detect and correct patterns of systematic errors in the quality scores of bases. Calling Single Nucleotide Polymorphisms (SNPs) and indels as variants detection was performed with GATK HaplotypeCaller that produced one variant calling format (VCF) file per cell. VCF contains information for each variant of a sample, such as the genomic position or the nature and type of the mutation. Per-sample VCF files produced by HaplotypeCaller was next combined with GATK GenomicsDBImport tool. The minimum phred-scaled confidence threshold at which variants should be called was set to 30. The last step comprised a joint genotyping with the tool GenotypeGVCFs.
Evolutionary tree construction
Starting from SNPs and indel information extracted with GATK protocol, variants affecting genes expressed in >99% of sequenced cells were taken into account. Maftool package (v2.17) was used to summarize variants information. DENDRO (v0.1.1) was applied to filter out variants with variants allele frequency <0.05 (too rare) or greater 0.95 (too common). A genetic divergence matrix was built to compute hierarchical clustering for the parsimony tree, depicting the evolutive relationships between subclones62.
scRNA-seq dissemination trajectory
To combine variant information with RNA data, matrices weight was adjusted to be comparable by multiplying by 100 the values of SNPs and indel matrix. Scanpy module (v1.9.1) was used for downstream analysis of cell clustering, combined to scFates module (v1.0) to compute pseudotime inference and extract a set of genes significantly associated with each trajectory of dissemination.
Analysis of genes dependency for cell growth
Gene dependency for cell growth was computed using Chronos algorithms on CRISPR-Cas9 screening database from DepMap portal (https://depmap.org/portal/) and normalized using annotated sets of common-essential and non-essential genes on 18 NB cell lines obtained from metastatic sites.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Julien Falk and Muriel Bozon for light sheet microscopy advices, the React4kids French national network in fundamental research in pediatric oncology for stimulating discussions and the r2 platform support team (r2.amc.nl) for their help in accessing RNAseq data. We thank Fabrice Lavial and Cédric Maurange for their advice on the project in the frame of B.V. PhD work. This work has been supported by the Fondation Bettencourt-Schueller (V.C.), and by grants from the INCa (PLBIO18-161) (V.C.), Fondation ARC pour la recherche sur le cancer Programmes Labellisés N° ARCPGA12021020003088_3559 (V.C.) et ARC PJA 20181207900 (C.D.-B.), Association Hubert Gouin – Enfance & Cancer (C.D.-B.) and AVIESAN PhD Fellow (B.V.). This work was conducted within the framework of the LABEX CORTEX and LABEX DevWeCAN of Université de Lyon, within the program Investissements d’Avenir’ (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR) (V.C.).
Author contributions
V.C. and C.D.B. designed and oversaw the study. C.D.B., B.V., F.R., K.T. and E.T. made experiments on the avian embryo model. B.V. performed the bioinformatic analyses of scRNA-seq datasets from avian the embryo model with support from OI, under the supervision of CDB. JJM and GT oversaw the obtention of matched primary tumor-bone marrow patient samples. A.B., J.J.M., G.T. and I.T. generated the scRNA-seq data from patient samples. B.V. and A.B. conducted the bioinformatic analyses of the matched patient samples and the analyses combining avian and patient datasets. S.C., J.L., I.T. and G.A. contributed to sample preparation and scRNA-seq. V.C., C.D.B., G.T., J.J.M. reviewed analysis results and interpretation of findings. B.V., C.D.B., V.C. wrote the manuscript with input from J.J.M. and A.B.
Peer review
Peer review information
Nature Communications thanks Domenico Ribatti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. Information and data provided in the present manuscript, figures, supplementary information and source data is sufficient to assess whether the study claims are supported by the data. Raw lightsheet and confocal microscopy files generated in the study are available upon simple request. NB raw sequencing data from pairs of primary tumor and invaded bone marrow have been deposited in GEO database under the accession number GSE245175 (patients #1 to #7 data were analyzed in the present study). Raw sequencing data from NB implanted in the avian chick embryo have been deposited in GEO database under the accession number (GSE237881). ScRNA-seq data of NB patient samples from Dong et al.14 and from Kildisiute et al.20 used in this study were respectively obtained from the GEO (GSE137804) and from the European Genome-phenome Archive (EGAD00001008345). scRNA-seq data of human embryonic adrenal glands and NB patient samples from Jansky et al.15 were obtained from the European Genome-phenome Archive (EGAS00001004388; and EGAD00001006624). scRNA-seq data of human embryonic adrenal glands from Kameneva et al.16 were obtained from the GEO (GSE147821). Bulk RNA-seq data of NB from SEQC cohort were obtained from the GEO (GSE49711)30. Bulk RNA-seq data of NB from Kocak cohort were obtained from the GEO (GSE45547)31. Source data are provided with this paper.
Competing interests
V.C. and C.D.B. declare the following competing interest: both are scientific advisors for the European Research Biological Center (ERBC) group. G.A. is employed by Cellenion, a BICO company. B.V., F.R., A.B., O.I., K.T., E.T., I.T., S.C., J.L., J.J.M., and G.T. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arjan Boltjes, Florie Reynaud.
These authors jointly supervised this work: Céline Delloye-Bourgeois, Valérie Castellani.
Contributor Information
Céline Delloye-Bourgeois, Email: celine.delloye@lyon.unicancer.fr.
Valérie Castellani, Email: valerie.castellani@univ-lyon1.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53776-3.
References
- 1.Filbin, M. & Monje, M. Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat. Med.25, 367–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kattner, P. et al. Compare and contrast: pediatric cancer versus adult malignancies. Cancer Metastasis Rev.38, 673–682 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Jiang, M., Stanke, J. & Lahti, J. M. The connections between neural crest development and neuroblastoma. Curr. Top. Dev. Biol.94, 77–127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maris, J. M. Recent advances in neuroblastoma. N. Engl. J. Med.362, 2202–2211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Körber, V. et al. Neuroblastoma arises in early fetal development and its evolutionary duration predicts outcome. Nat. Genet.55, 619–630 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundem, G. et al. Clonal evolution during metastatic spread in high-risk neuroblastoma. Nat. Genet.55, 1022–1033 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeineldin, M., Patel, A. G. & Dyer, M. A. Neuroblastoma: When differentiation goes awry. Neuron110, 2916–2928 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponzoni, M. et al. Recent advances in the developmental origin of neuroblastoma: an overview. J. Exp. Clin. Cancer Res.41, 92 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delloye-Bourgeois, C. & Castellani, V. Hijacking of embryonic programs by neural crest-derived neuroblastoma: From physiological migration to metastatic dissemination. Front. Mol. Neurosci.12, 52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeva, V. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet.49, 1408–1413 (2017). [DOI] [PubMed] [Google Scholar]
- 11.van Groningen, T. et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet.49, 1261–1266 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Thirant, C. et al. Reversible transitions between noradrenergic and mesenchymal tumor identities define cell plasticity in neuroblastoma. Nat. Commun.14, 2575 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan, X. et al. Single-cell profiling of peripheral neuroblastic tumors identifies an aggressive transitional state that bridges an adrenergic-mesenchymal trajectory. Cell Rep.41, 111455 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Dong, R. et al. Single-cell characterization of malignant phenotypes and developmental trajectories of adrenal neuroblastoma. Cancer Cell38, 716–733.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Jansky, S. et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 53, 683–693 (2021). [DOI] [PubMed]
- 16.Kameneva, P. et al. Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin. Nat. Genet.53, 694–706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedoya-Reina, O. C. et al. Single-nuclei transcriptomes from human adrenal gland reveal distinct cellular identities of low and high-risk neuroblastoma tumors. Nat. Commun.12, 5309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kastriti, M. E. et al. Schwann cell precursors represent a neural crest-like state with biased multipotency. EMBO J.41, e108780 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanemaaijer, E. S. et al. Single-cell atlas of developing murine adrenal gland reveals relation of Schwann cell precursor signature to neuroblastoma phenotype. Proc. Natl Acad. Sci. USA118, e2022350118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kildisiute, G. et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci. Adv.7, eabd3311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fetahu, I. S. et al. Single-cell transcriptomics and epigenomics unravel the role of monocytes in neuroblastoma bone marrow metastasis. Nat. Commun.14, 3620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delloye-Bourgeois, C. et al. Microenvironment-deriven shift of cohesion/detachment balance within tumors induces a switch toward metastasis in neuroblastoma. Cancer Cell32, 427–443.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Ben Amar, D. et al. Environmental cues from neural crest derivatives act as metastatic triggers in an embryonic neuroblastoma model. Nat. Commun.13, 2549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akkermans, O. et al. GPC3-Unc5 receptor complex structure and role in cell migration. Cell185, 3931–3949.e26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, X.-L. et al. The microtubule-associated protein ASPM regulates spindle assembly and meiotic progression in mouse oocytes. PLoS One7, e49303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assou, S. et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genom.10, 10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morozumi, Y. et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J. Mol. Cell Biol.8, 349–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madgwick, S. et al. Claspin haploinsufficiency leads to defects in fertility, hyperplasia and an increased oncogenic potential. Biochem. J.479, 2115–2130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirino, K., Nakahata, T., Taguchi, T. & Saito, M. K. Efficient derivation of sympathetic neurons from human pluripotent stem cells with a defined condition. Sci. Rep.8, 12865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su, Z. et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol.15, 523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kocak, H. et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis.4, e586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito, D., Takase, Y., Murai, H. & Takahashi, Y. The dorsal aorta initiates a molecular cascade that instructs sympatho-adrenal specification. Science336, 1578–1581 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Furlan, A. et al. Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science357, eaal3753 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry, N. A., Vitolo, M. I., Martin, S. S. & Kontrogianni-Konstantopoulos, A. Loss of the obscurin-RhoGEF downregulates RhoA signaling and increases microtentacle formation and attachment of breast epithelial cells. Oncotarget5, 8558–8568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birchmeier, C. ErbB receptors and the development of the nervous system. Exp. Cell Res.315, 611–618 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Sakurai, T. The role of NrCAM in neural development and disorders–beyond a simple glue in the brain. Mol. Cell. Neurosci.49, 351–363 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Zang, Y., Chaudhari, K. & Bashaw, G. J. New insights into the molecular mechanisms of axon guidance receptor regulation and signaling. Curr. Top. Dev. Biol.142, 147–196 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai, Y.-Q. et al. Multiple pathophysiological roles of midkine in human disease. Cytokine135, 155242 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Winkler, C. & Yao, S. The midkine family of growth factors: diverse roles in nervous system formation and maintenance. Br. J. Pharmacol.171, 905–912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filippou, P. S., Karagiannis, G. S. & Constantinidou, A. Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene39, 2040–2054 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Kishida, S. & Kadomatsu, K. Involvement of midkine in neuroblastoma tumourigenesis. Br. J. Pharmacol.171, 896–904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hochheuser, C. et al. The metastatic bone marrow niche in neuroblastoma: altered phenotype and function of mesenchymal stromal cells. Cancers (Basel)12, 3231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol.17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muraro, M. J. et al. A single-cell transcriptome Atlas of the human pancreas. Cell Syst.3, 385–394.e3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Candelli, T. et al. Sharq, a versatile preprocessing and QC pipeline for single cell RNA-seq. bioRxiv10.1101/250811 (2018).
- 46.Renier, N. et al. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Picelli, S. et al. Full-length RNA-seq from single cells using smart-seq2. Nat. Protoc.9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 48.McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst.8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol.20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood128, e20–e31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol.38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 54.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods19, 159–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science367, 405–411 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiebinger, G. et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell176, 928–943.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol.20, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faure, L. et al. Single cell RNA sequencing identifies early diversity of sensory neurons forming via bi-potential intermediates. Nat. Commun.11, 4175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J. & Peterson, H. gprofiler2 – an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Research9, ELIXIR–709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun.10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, Z., Xu, B., Minn, A. & Zhang, N. R. DENDRO: genetic heterogeneity profiling and subclone detection by single-cell RNA sequencing. Genome Biol.21, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Source data are provided with this paper. Information and data provided in the present manuscript, figures, supplementary information and source data is sufficient to assess whether the study claims are supported by the data. Raw lightsheet and confocal microscopy files generated in the study are available upon simple request. NB raw sequencing data from pairs of primary tumor and invaded bone marrow have been deposited in GEO database under the accession number GSE245175 (patients #1 to #7 data were analyzed in the present study). Raw sequencing data from NB implanted in the avian chick embryo have been deposited in GEO database under the accession number (GSE237881). ScRNA-seq data of NB patient samples from Dong et al.14 and from Kildisiute et al.20 used in this study were respectively obtained from the GEO (GSE137804) and from the European Genome-phenome Archive (EGAD00001008345). scRNA-seq data of human embryonic adrenal glands and NB patient samples from Jansky et al.15 were obtained from the European Genome-phenome Archive (EGAS00001004388; and EGAD00001006624). scRNA-seq data of human embryonic adrenal glands from Kameneva et al.16 were obtained from the GEO (GSE147821). Bulk RNA-seq data of NB from SEQC cohort were obtained from the GEO (GSE49711)30. Bulk RNA-seq data of NB from Kocak cohort were obtained from the GEO (GSE45547)31. Source data are provided with this paper.