Abstract
Mutations in the RNA helicase DDX3X, implicated in various cancers and neurodevelopmental disorders, often impair RNA unwinding and translation. However, the mechanisms underlying the impairment and the differential interactions of DDX3X mutants with wild-type (WT) X-linked DDX3X and Y-linked homolog DDX3Y remain elusive. This study reveals that specific DDX3X mutants more frequently found in disease form distinct hollow condensates in cells. Using a combined structural, biochemical, and single-molecule microscopy study, we show that reduced ATPase and RNA release activities contribute to condensate formation and these catalytic deficits result from inhibiting the catalytic cycle at multiple steps. Proteomic investigations further demonstrate that these hollow condensates sequester WT DDX3X/DDX3Y and other proteins crucial for diverse signaling pathways. WT DDX3X enhances the dynamics of heterogeneous mutant/WT hollow condensates more effectively than DDX3Y. These findings offer valuable insights into the catalytic defects of specific DDX3X mutants and their differential interactions with wild-type DDX3X and DDX3Y, potentially explaining sex biases in disease.
Subject terms: RNA, Enzyme mechanisms
Here the authors find that DDX3X mutants with catalytic impairments form hollow condensates in cells that have slower dynamics when co-condensed with DDX3Y than with WT DDX3X, revealing potential mechanisms of sex biases in DDX3X-related diseases.
Introduction
The DEAD-box RNA helicase DDX3X is a crucial component of numerous cellular and developmental processes1 that is conserved from yeast to humans2. Whole-body knockout of Ddx3x leads to embryonic lethality in mice3, and knockdown of Ddx3x in the developing murine nervous system disrupts cortex development4, highlighting the essential nature of DDX3X. DDX3X contains a helicase core common to all DEAD-box proteins that is flanked by two intrinsically disordered regions (IDRs) (Fig. 1a)5. The non-processive RNA unwinding activity of the helicase core helps DDX3X facilitate translation initiation for specific transcripts via unwinding their highly structured 5’ untranslated regions (5’UTRs) under unstressed conditions6. Upon cellular stress, the IDRs of DDX3X (particularly its N-terminal IDR) drive it to undergo liquid-liquid phase separation (LLPS) and participate in stress granule formation, which leads to the translation repression of sequestered transcripts7.
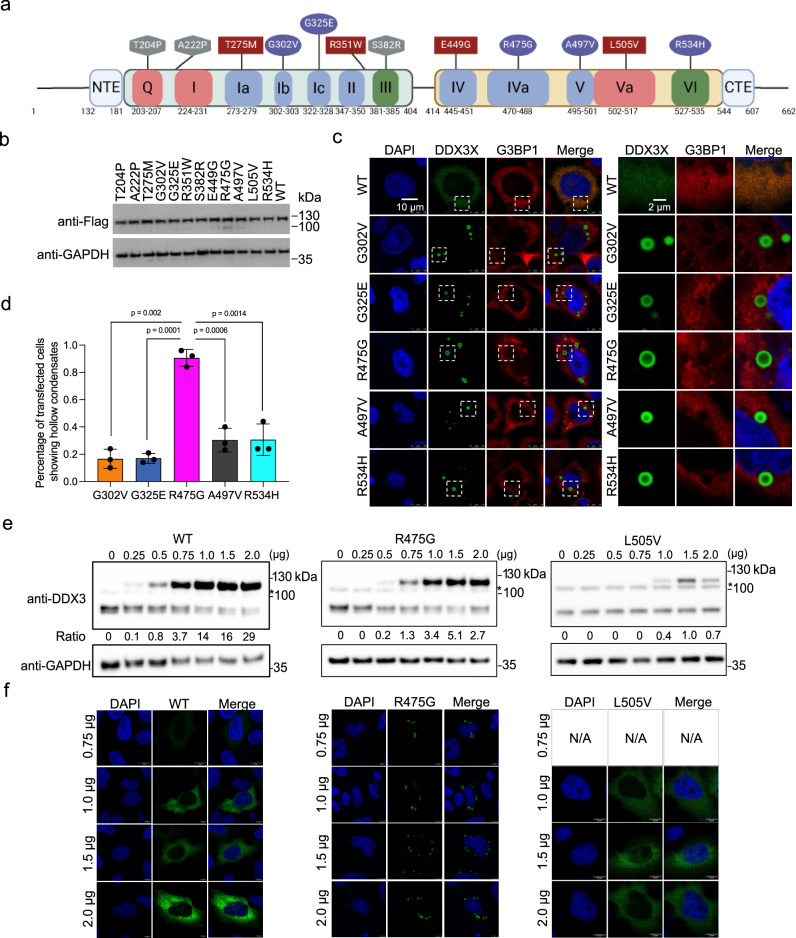
Fig. 1. A subset of DDX3X disease mutants form unique hollow condensates in cells.
a Schematic of the conserved domains and motifs of DDX3X and the locations of each disease variant used in this study. Motifs responsible for ATP binding and hydrolysis (Q, I, and Va) are in red, RNA binding (Ia, Ib, Ic, II, IV, IVa, V) in blue, and coordination of ATPase and RNA binding (III and VI) in green. Diffuse mutants are shown in squares, speckled mutants in hexagons, and hollow mutants in ovals. N-terminal IDR = a.a. 1–131, N-terminal extension (NTE) = a.a. 132–181, N-terminal RecA-like domain = a.a. 182–403, linker = a.a. 405–413, C-terminal RecA-like domain = a.a.414–543, C-terminal extension (CTE) = a.a. 545–607, C-terminal IDR = 608–662. Created in BioRender. Owens (2023) BioRender.com/k65s989. b Western blots showing that the mClover3-tagged DDX3X variants were expressed at similar levels in HeLa cells. c Representative images showing the localization of mClover3-tagged WT DDX3X or the indicated variants and mCherry-tagged G3BP1 in HeLa cells. The white boxes indicate regions zoomed on the right. Scale bars, 10 µm and 2 μm. d Quantified percentage of HeLa cells containing DDX3X variant hollow condensates. Values represent means ± s.d. from three independent measurements of at least 20 cells each. Significance was calculated using a two-tailed t-test e Representative Western blots showing expression of exogenous mClover3-tagged WT, R475G, and L505V DDX3X and endogenous WT DDX3X. * indicates a non-specific band produced by this antibody. Ratios of exogenous/endogenous band intensity included below each lane. Titrations were performed in triplicate, additional blots can be found in the Supplementary Fig. 1. f Immunofluorescence imaging showing cellular distribution patterns of WT, R475G, and L505V DDX3X at the indicated plasmid concentrations. Source data are provided as a Source Data file.
Recent studies have reported over 100 recurrent mutations in DDX3X across numerous cancers, including medulloblastoma8–10, Burkitt’s lymphoma11, chronic lymphocytic leukemia, and natural killer-T cell lymphoma12–14. Mutations in DDX3X are also linked to DDX3X syndrome, which accounts for 1-3% of otherwise unexplained intellectual disability cases4,15–17. These mutations often cause defects in the enzymatic activities (RNA binding, ATPase activity, and RNA unwinding), although the degree varies among pathogenic mutants4,10,18. As a consequence of disrupting the catalytic activity, several mutants of DDX3X negatively impact translation10, particularly the translation of DDX3X targets4. Although most of these mutations are in the catalytic core of DDX3X and not in its IDRs, disease mutants of DDX3X frequently aberrantly phase separate in cells without adding an external stressor4,10. DDX3Y, the sexually dimorphic homolog of DDX3X, is both a weaker ATPase and a stronger phase separator than DDX3X19. In concurrent work, we found that RNA plus WT DDX3X or DDX3Y form nano-sized RNA protein clusters (RPCs) with numerous copies of the proteins that optimally enhance helicase activity and may be nuclei for phase separation20. These results suggest that the enzymatic and phase separation properties of DDX3X and DDX3Y are intertwined. However, much work remains to decipher the mechanism of catalytic defects and how such defects contribute to the development of the corresponding disorders.
The diseases associated with DDX3X mutants, as well as a wide range of other diseases, often manifest in a sex-biased manner. For example, DDX3X syndrome is significantly more common in XX individuals than in XY individuals4, while XX individuals with pediatric medulloblastoma have a higher survival probability than XY individuals21. Recently, a growing body of evidence suggests that some of these sex biases may stem from the different biochemical and biophysical properties of sexually dimorphic protein homologs19,22–24. DDX3X and its Y chromosome-encoded homolog, DDX3Y, are two of these sexually dimorphic homologs. Because mutants of DDX3X are often heterozygous25,26, the mutant allele will almost always be co-expressed with either wild-type (WT) DDX3X (in XX individuals) or with DDX3Y (in XY individuals). However, it is not known whether the enzymatic and condensation defects of DDX3X disease mutants are differently altered by co-expression with WT DDX3X or DDX3Y.
Here, we focused on understanding how a range of DDX3X disease mutants impact enzyme catalysis, condensation properties, and interactions with WT DDX3X or DDX3Y to interfere with cellular functions. The results show that a specific subset of these mutants that occur more frequently across several cancers and DDX3X-related neurodevelopmental disorders than other mutants promote the formation of prominent cell condensate structures. These mutants sequester WT DDX3X, DDX3Y, and additional proteins essential for various cellular functions, possibly explaining their more severe pathogenic outcomes. Our study also reveals the mechanism underlying the formation of condensate structures under non-stressed conditions. These observations provide insights into the molecular basis of DDX3X-related pathogenesis in humans and reveal DDX3X/3Y-based differences in human disease.
Results
DDX3X disease mutants present with distinct cellular distribution patterns
Like other members of the DEAD-box family, the catalytic core of DDX3X consists of two RecA-like domains [amino acids (a.a.) 182–403 and 414–544] flanked by N- and C-terminal intrinsically disordered regions (IDRs) (a.a. 1–131 and 608–662). Within the two RecA-like domains are twelve conserved motifs that have been implicated in either binding and hydrolyzing ATP (Q, I, and Va motifs, in red), binding RNA (Ia, Ib, Ic, II, IV, IVa, and V motifs, in blue) or coordinating these functions (III and VI motifs, in green)5 (Fig. 1a). In this work, we studied one disease mutation in (or near) each of these twelve conserved motifs to investigate the effects of mutants across the DDX3 catalytic core.
To begin, we expressed either WT DDX3X or one of twelve mutants (each tagged with N-terminal Flag and C-terminal mClover3) (Fig. 1a) to similar levels in Henrietta Lacks’ (HeLa) cells (Fig. 1b). WT DDX3X showed diffuse localization throughout the cytoplasm in unstressed cells, which is consistent with previous studies4,10,19 (Fig. 1c). Strikingly, five of the mutants (G302V, G325E, R475G, A497V, and R534H) formed spherical cytoplasmic puncta in which the mutant was enriched in the outer shell but depleted in the center (Fig. 1c). Hereafter, we refer to these mutants as “hollow” mutants. This morphology was also observed in HEK293T cells for all hollow mutants (Supplementary Fig. 1a). The percentages of transfected cells with hollow puncta varied among these five hollow mutants; while four formed hollow puncta in ~20% of cells, R475G formed hollow puncta in more than 90% of cells (Fig. 1d). R475G has previously been linked to severe neurodevelopmental phenotypes4, raising the possibility that disease severity and this high propensity for forming hollow puncta are correlated. Orthogonal projections of R475G further support the hollow structure of the puncta with the mutant protein enriched in the shells (Supplementary Fig. 1b). To ensure that hollow morphology was not only present when the mutant was overexpressed, we titrated R475G-mClover3 and assessed protein expression using Western blotting. R475G hollow puncta were observed at the same concentrations as endogenous WT. The number of hollow condensates increased along with increasing protein expression levels (Fig. 1e, f). Across this concentration range, both WT and the non-hollow mutant L505V remained diffuse throughout the cytoplasm (Fig. 1e, f). Additionally, Flag-tagged G325E (lacking mClover3) also formed hollow puncta in cells, indicating that the fluorescent tag did not influence the morphology (Supplementary Fig. 1c). Like WT and L505V, a few other mutants used in this study (T275M, R351W, E449G) were diffuse in the cytoplasm; these mutants are hereafter referred to as “diffuse” mutants (Supplementary Fig. 1d). Additionally, we observed that the remaining three mutants (T204P, A222P, and S382R) presented with small foci with irregular shape throughout the cytoplasm; these mutants are hereafter referred to as “speckled” mutants (Supplementary Fig. 1d).
WT DDX3X undergoes LLPS and enters stress granules (SGs)7 upon cellular stress. This led us to consider whether the hollow puncta we observed were, in fact, SGs. Under unstressed conditions, none of the mutants’ puncta colocalized with the canonical SG marker G3BP127 (Fig. 1c and Supplementary Fig. 1d). Upon arsenite treatment, all but the speckled DDX3X mutants colocalized with G3BP1, showing that they entered SGs (Supplementary Fig. 1e). Changes in the cellular environment upon stress, including but not limited to phosphorylation of eIF2α28 and accumulation of untranslated mRNA and free 40S ribosomal subunits29 possibly disrupt the hollow condensates and drive the mutant DDX3X to SGs. These results suggest that the differences in the cellular environments under unstressed and stressed conditions impact the morphology and condensation properties of DDX3X puncta. Collectively, these results suggest that DDX3X mutants have distinct cellular condensation propensities, expression patterns, and possibly different protein interactomes compared to WT DDX3X.
DDX3X hollow mutants display decreased ATPase and RNA release activities
To test how the DDX3X mutants disrupt enzyme catalysis, we purified WT DDX3X and each mutant (Supplementary Fig. 2a). While we were able to purify all the diffuse and hollow mutants, the speckle mutants always aggregated during purification attempts. The three speckled mutations (T204P, A222P, and S382R) sit in the N-terminal RecA-like domain. In the apo state structure (PDB 5E7I) (Fig. 2a, top), Thr204 is at the N-terminus of an alpha helix, flanked by two prolines. T204P thus results in a stretch of three proline residues in a row. Ala222 and Ser382 are both in the central beta-sheet that runs through the middle of the N-terminal RecA-like domain, and the backbone carbonyl of Ala222 hydrogen bonds with the backbone amine of Ser382. The A222P and S382R mutations may disrupt the formation of this beta-sheet, causing protein instability. When expressed in HeLa cells, the speckled mutants precipitated to the cell pellet after lysis using radioimmunoprecipitation assay (RIPA) buffer (Fig. 2a, bottom). These cellular precipitations and the resistance to purification of these three mutants imply that these mutations likely result in misfolding of the N-terminal RecA-like domain and protein instability.
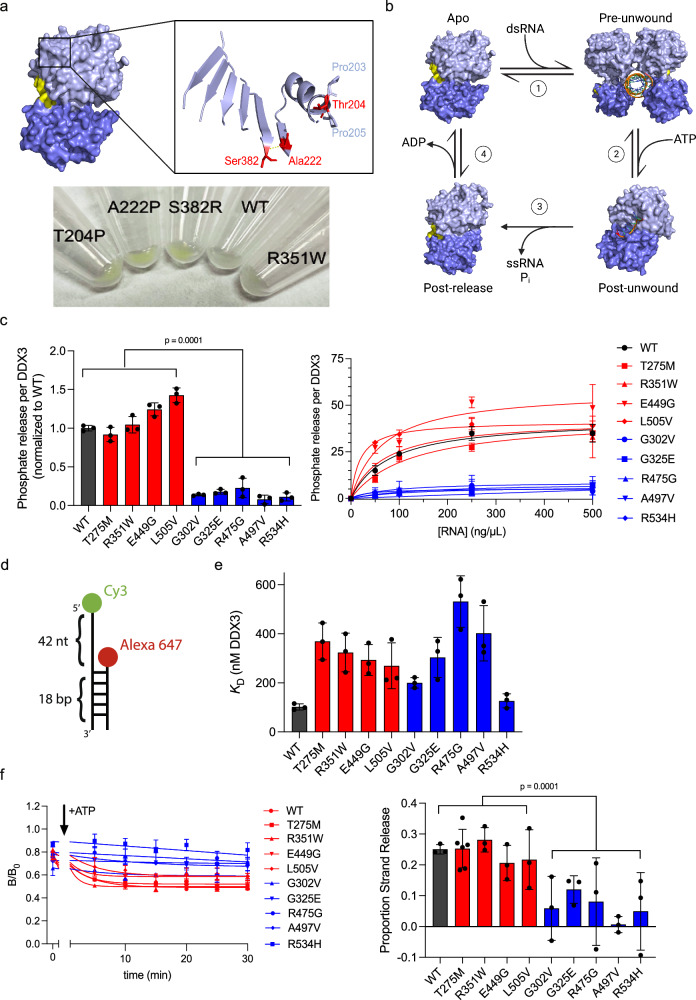
Fig. 2. Hollow mutants are characterized by their decreased ATPase and RNA strand release activities.
a Top, DDX3X apo structure zoomed into the three speckle mutants (red). Pro203 and Pro205 flanking Thr204 are indicated in light blue. The yellow line indicates the hydrogen bond between Ala222 and Ser382. Bottom, insoluble pellets of HeLa cells expressing mClover3-tagged WT DDX3X or the indicated mutants, after RIPA lysis and centrifugation. The green color of the speckle mutant pellets indicates their insolubility relative to WT DDX3X and R351W (a diffuse mutant). b Current model of the DDX3X helicase reaction. Light blue = N-terminal RecA-like domain, dark blue = the C-terminal RecA-like domain, orange = RNA. In the apo state (PDB 5E7I), a DDX3X dimer binds dsRNA (1) to form the pre-unwound state (PDB 6O5F). Upon addition of ATP, each monomer performs strand separation (2), converting to the post-unwound state (PDB 2DB3). Following ATP hydrolysis, DDX3X releases both the inorganic phosphate and the product ssRNA strand (3), forming the post-release state (PDB 4PXA). ADP release (4) returns DDX3X to apo state. c Left, ATPase activity of WT DDX3X or indicated mutants, measured as described in Methods, normalized to WT. Values represent means ± s.d., n = 3 replicates. Significance calculated using two-way ANOVA comparing diffuse mutants (and WT) to hollow mutants. Right, ATPase activities at the indicated concentration of total RNA. Values fit to Michaelis–Menten. Values = means ± s.d., n = 3 replicates. d Schematic of the dsRNA probe. e Summarized KD values for the indicated WT and mutant DDX3X. Values represent mean ± s.d., n = 3 independent reactions for WT and diffuse mutants, n = 6 for hollow mutants (n = 5 for G325E). f Left, time courses tracing the B/B0 (proportion strand release) for short strand RNA over, fit as described in Methods. Right, change in B/B0 signal from 0 to 30 min for each protein. Significance calculated using two-way ANOVA comparing diffuse mutants (and WT) to hollow mutants. Values represent means ± s.d., n = 3 independent replicates (n = 6 for T275M). Source data are provided as a Source Data file.
The current model of the DDX3X catalytic cycle is as follows (Fig. 2b): apo DDX3X (PDB 5E7I)30 binds to dsRNA as a multimer (without IDRs, the crystallization construct forms a dimer, but evidence for higher order assemblies exist for full-length protein20,31) to adopt the pre-unwound state (PDB 6O5F)32. ATP binding produces the post-unwound state (represented in Fig. 2b by the related DEAD-box protein Vasa with ssRNA and AMPPNP, PDB 2DB3)33. DDX3X releases its single-stranded product upon ATP hydrolysis, adopting the post-release state (DDX3X with ADP, PDB 4PXA)18. ADP release completes the cycle (Fig. 2b and Supplementary Movie 1). Given the crucial role of ATP hydrolysis and that the ATPase activity of DEAD-box enzymes often influences their condensation properties34, we assayed the ATP hydrolysis activity (Fig. 2b, step 3) of all the mutants we were able to purify using malachite green to detect phosphate release as previously described19 (Fig. 2c, left). When tested at equal protein concentration, all hollow mutants exhibited significantly lower ATPase activities than the diffuse mutants and WT DDX3X, suggesting that ATPase activity may correlate with their propensity to form hollow condensates in cells. Because the ATPase activity of DEAD-box proteins is RNA-stimulated, we questioned whether these differences in ATPase activity could be due to decreased RNA binding affinity (Fig. 2b, step 1). A mutant whose sole problem is a deficiency in RNA binding relative to WT is expected to reach the same maximum phosphate release rate as the WT at saturating RNA concentration. To test this, we performed electrophoretic mobility shift assays (EMSAs) with each mutant using a dsRNA probe (Fig. 2d). We found that all the mutants, except for R534H, had RNA binding affinities weaker than WT and that, unlike ATPase activity, RNA binding did not correlate with cellular morphology (Fig. 2e and Supplementary Fig. 2b). We then repeated the malachite green ATPase assay at several total RNA concentrations (Fig. 2c, right), finding that the ATPase activity of WT DDX3X and the diffuse mutants saturated at a much higher level than the hollow mutants (~30 μM phosphate after 30 min for WT/diffuse vs. ~5 μM for hollow, or ~1 μM/min for WT/diffuse vs. ~0.2 μM/min for hollow).
Given that ATP hydrolysis triggers RNA strand release (Fig. 2b, step 3), we next studied whether the ATPase deficiency impairs strand release using a single-molecule fluorescence time-resolved anisotropy approach. Briefly, the same dsRNA used from EMSA experiments (Fig. 2d) was incubated with either WT DDX3X or the indicated mutant (Supplementary Fig. 2c). Since DDX3X is a non-processive helicase, it must unbind and rebind RNA as it progresses through the enzymatic cycle. This activity can be monitored with fluorescence anisotropy as the short-strand RNA is released (and tumbles more quickly) by adding ATP. This activity is interpreted as strand release activity from either the DDX3X enzyme and/or escape from RNA-protein clusters (catalytically active nano-sized RNA-protein assemblies) of DDX3X20. Specifically, the fraction of anisotropic (slow tumbling) RNA (B/B0) was monitored for the different species of RNA (long single-stranded, short single-stranded, and duplex RNA) (Fig. 2f, left). The fraction of anisotropic RNA decreased for the short strand RNA over time with the WT DDX3X and diffuse mutants; however, the fraction of anisotropic RNA for the short strand decreased significantly less with the hollow mutants (Fig. 2f, right). On the other hand, the fraction of anisotropic RNA for the long strand and duplex species remained high with and without the addition of ATP for WT, diffuse, and hollow DDX3X, indicating stably bound protein (Supplementary Fig. 2d, e). These results suggest that hollow mutants of DDX3X have severe defects in turnover involving ATP hydrolysis and RNA strand release, while diffuse mutants are comparable to WT in these activities.
Specific defects attributable to individual hollow mutants
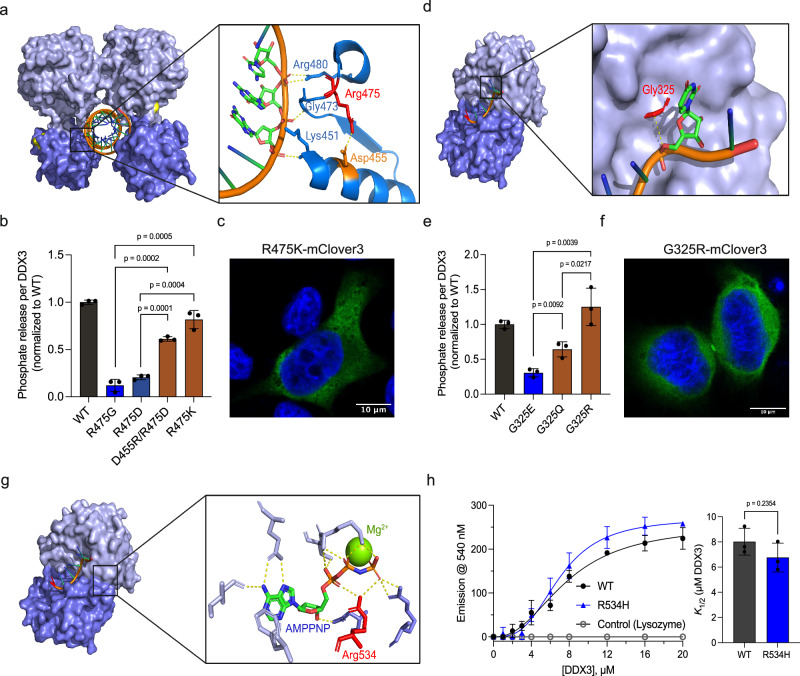
The diminished catalysis we observed in Fig. 2 could result from difficulties progressing through any (or multiple) step(s) of the DDX3 catalytic cycle. To obtain a thorough mechanistic understanding of how individual mutations disrupt catalysis, we traced the interactions of the mutated residues throughout the DDX3X catalytic cycle. For example, Arg475 forms a salt bridge with Asp455 in the pre-unwound state, which seems to position several residues for RNA binding (Fig. 3a). This salt bridge is not present in subsequent states. Flipping the residues of this salt bridge (D455R/R475D, Supplementary Fig. 3a) resulted in partial rescue of both ATPase activity (Fig. 3b) and RNA-binding activity (Supplementary Fig. 3b, c). However, restoration of the positive charge (R475K, Supplementary Fig. 3a) showed an equivalent rescue of RNA binding to the salt bridge swap (Supplementary Fig. 3b, c) but a more robust rescue of ATPase activity (Fig. 3b). This suggests that, while the Asp455/Arg475 salt bridge is crucial for RNA binding in the pre-unwound state, other steps of the catalytic cycle likely require a positive charge at position 475. Thus, R475G may disrupt the formation of the pre-unwound state and the formation of one or more subsequent states.
Fig. 3. Mechanistic studies reveal how DDX3X mutants interfere with catalysis.
a Pre-unwound structure of DDX3X zoomed on Arg475 (red) and the surrounding RNA-binding residues. Asp455 is noted in orange, and the dashed yellow line indicates a putative salt bridge between these residues. Their respective yellow dashed lines indicate interactions between Lys451, Gly473, and Arg480 and the RNA backbone. b ATPase activity of mCherry-tagged WT DDX3X, R475G, R475D, D455R/R475D, and R475K (normalized to WT). Significance was calculated using a two-tailed t-test. Values represent mean ± s.d., n = 3 independent reactions. c A representative image of R475K-mClover3 in HeLa cells, scale bar = 10 μm. d Post-unwound structure of Vasa zoomed in on Gly325 (red, numbered according to DDX3X homology). The dashed line indicates the 3.6 Å distance between the alpha carbon of Gly325 and the RNA backbone. e ATPase activity of mCherry-tagged WT DDX3X, G325E, G325Q, and G325R normalized to WT. Significance was calculated using a two-tailed t-test. Values represent mean ± s.d., n = 3 independent reactions. f A representative image of G325R-mClover3 in HeLa cells, scale bar = 10 μm. g Post-unwound structure of DDX3X zoomed in on the ATP-binding pocket, showing residues involved in ATP binding and Mg2+ cation (yellow sphere). Arg534 (numbered with DDX3X homology) is indicated in red. Yellow dashed lines indicated interactions between residues and ATP. h TNP-ATP binding curve for MBP-tagged WT DDX3X, R534H, and negative control (lysozyme), fit as described in methods. Right, K1/2 values for WT and R534H. Significance was calculated using a two-tailed t-test. Values represent mean ± s.e.m., n = 3 independent measurements. Source data are provided as a Source Data file.
Gly325 sits in a positive cleft on the N-terminal RecA-like domain. In the pre-unwound state, this cleft is solvent accessible, but during the transition to the post-unwound state (Fig. 2b, step 2), this cleft accepts the newly unwound RNA (Fig. 3d), which we hypothesize is blocked by the G325E mutation. To evaluate whether G325E hinders catalysis due to steric or electrostatic effects, we generated DDX3X mutants G325Q and G325R and conducted further ATPase assays (Supplementary Fig. 3a). While G325Q (uncharged but as large as glutamate) partially rescued ATPase activity, G325R (positively charged and larger than glutamate) showed a full rescue (Fig. 3e). These results suggest that the negative charge introduced by the disease mutant G325E is a major disruptor of DDX3X catalysis. This is supported by time-resolved fluorescence anisotropy studies, as RNA mixed with G325R showed a higher change in the fraction of anisotropic molecules than RNA mixed with G325E, indicating a higher degree of strand release comparable to WT (Supplementary Fig. 3d).
The Arg534 residue is located in the C-terminal RecA-like domain. In the post-unwound state, Arg534 coordinates the ATP’s α and γ phosphates where it likely participates in hydrolysis (Fig. 3g). The R534H mutation most likely disrupts only the ATP hydrolysis step (Fig. 2b, step 3), as Arg534 is distal from the ATP-binding site during all of the other states. Supporting this, R534H can bind both dsRNA (Fig. 2e) and the ATP analog TNP-ATP35 (Fig. 3h) as efficiently as WT DDX3X, indicating that R534H can form both the pre-unwound (dsRNA-bound) and post-unwound (ssRNA and ATP-bound) complexes as readily as WT DDX3X.
In contrast to the hollow mutants illustrated above, the four diffuse mutations do not disrupt ATP hydrolysis or strand release (Fig. 2). In the pre-unwound state, Thr275 and Arg351 are located on the solvent-accessible face of the N-terminal RecA-like domain, where their residues do not form direct interactions with the RNA (Supplementary Fig. 3e, left). Glu449 in the pre-unwound state is near the RNA-binding surface of the C-terminal RecA-like domain but does not associate with the dsRNA (Supplementary Fig. 3e, right). Leu505 is not resolved in the pre-unwound structure. In the post-unwound state, Thr275, Arg351, and Glu449 all form hydrogen bonds with the ssRNA backbone (Supplementary Fig. 3f, left). Mutation of these residues (T275M, R351W, and E449G) would disrupt these hydrogen bonds and possibly weaken the binding of the ssRNA in the post-unwound state; however, these residues would not electrostatically clash with the RNA in the same manner as G325E; thus these mutations do not disrupt catalysis in the same manner as G325E. Leu505 sits between the N- and C-terminal RecA-like domains in the post-unwound state (Supplementary Fig. 3f, right) in a region where either a leucine or a valine should be sterically tolerated.
Next, we expressed R475K and G325R in cells to assess whether rescued ATP hydrolysis was concurrent with a loss of hollow morphology (Fig. 3c, f). Indeed, both rescue mutations restored the diffuse cytoplasmic localization present in WT DDX3X (hollow puncta were not observed). Given that ATP hydrolysis is necessary for the release of bound RNAs18,30,33,36,37, it is possible that the RNA trapped in the protein functions as a multivalent platform that recruits additional DDX3X molecules and other proteins, contributing to hollow puncta formation. These data support a model in which mutations that disrupt strand release after unwinding form hollow condensates (G302V, G325E, R475G, A497V, and R534H), but mutations that do not disrupt strand release (T275M, R351W, E449G, and L505V) do not form hollow condensates.
RNA-binding affinity influences inter- and intramolecular dynamics of DDX3X mutant hollow condensates
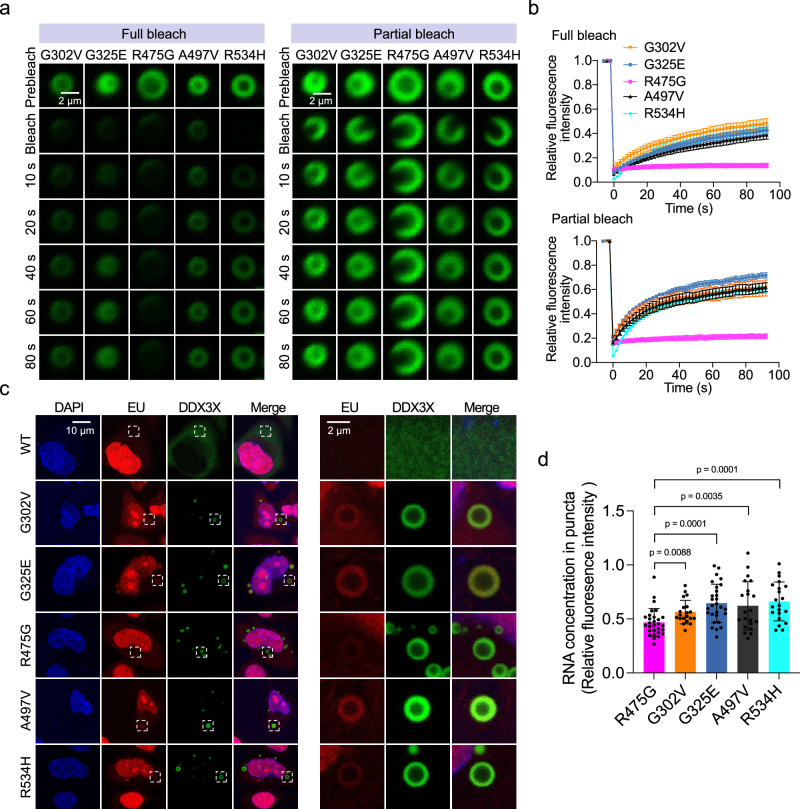
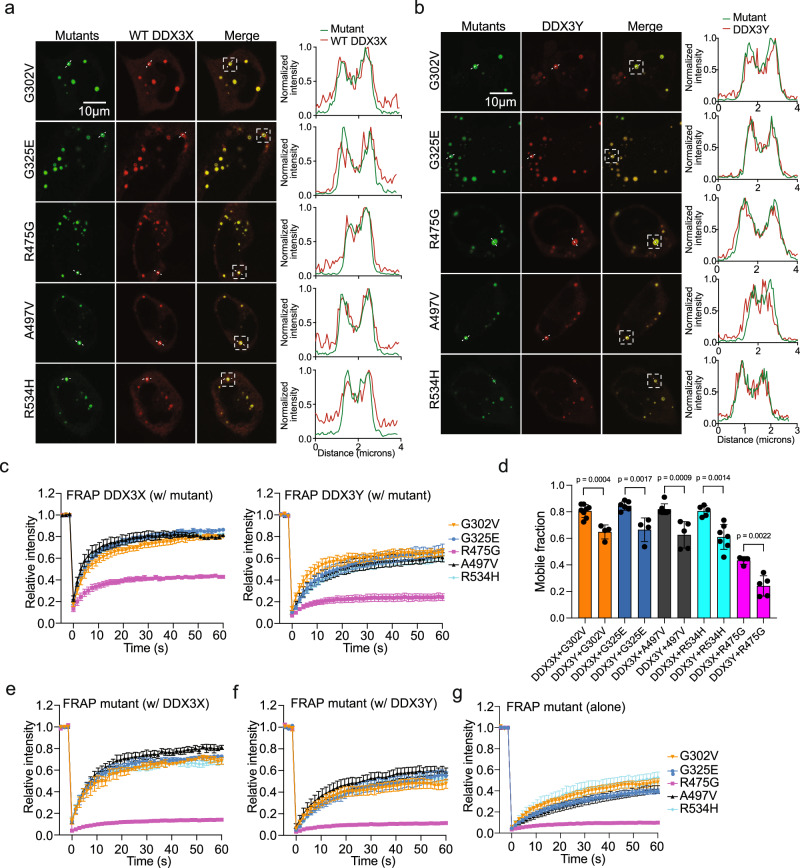
Unlike WT DDX3X, which only undergoes LLPS in cells under stressed conditions7, select DDX3X mutants have increased phase separation propensities leading to puncta formation under unstressed conditions4,10. Hollow puncta are likely formed via LLPS, as we were able to observe the fusion of two hollow condensates while performing live-cell imaging (Supplementary Fig. 4a). In addition to fusion, a key trait of liquid-liquid phase separated condensates is that they are in a dynamic equilibrium with their surroundings38. To directly study the molecular exchange between the cytoplasm and the hollow condensates formed by DDX3X mutants, we performed “full bleach” fluorescence recovery after photobleaching (FRAP) experiments, in which we bleached an entire hollow condensate and measured the dynamics of fluorescence signal recovery (Supplementary Fig. 4b, left). We found that the fluorescence of G302V, G325E, A497V, and R534H hollow condensates recovered gradually, while the fluorescence of R475G hollow condensates recovered very little in the 100 s experimental timeframe (Fig. 4a, left and 4b, top). Additionally, “partial bleach” experiments were carried out to reveal the mobility of molecules within the hollow condensates (Supplementary Fig. 4b, right)39. Similarly, the fluorescence of R475G hollow condensates showed almost no recovery after partial bleaching, unlike the other mutants, which is consistent with the full bleach results (Fig. 4a, right and 4b, bottom). The lower inter- and intramolecular dynamics of the R475G hollow condensates sets them apart from the other mutant hollow condensates. The uniquely low dynamics of R475G are in line with our finding that the percentage of cells expressing hollow condensates was much higher for R475G than for the other mutants (Fig. 1d). Binding deficiencies in RNA-binding proteins such as FUS (fused in sarcoma) decrease the dynamics of their protein-RNA condensates40. While all the hollow mutants (save for R534H) displayed weaker RNA-binding affinity than WT DDX3X, R475G had the weakest RNA affinity (Fig. 2e). This suggests that the decreased RNA binding of R475G may decrease the dynamics of its hollow condensates relative to the other hollow mutants.
Fig. 4. RNA-binding affinity differentiates the inter- and intramolecular dynamics of DDX3X hollow puncta.
a Time-lapsed images of representative FRAP experiments on hollow condensates of the indicated mClover3-tagged DDX3X variants expressed in HeLa cells. Full bleach (left) and partial bleach (right) experiments were performed to measure the inter- and intramolecular dynamics (respectively) of each condensate. Scale bar, 2 μm. b Recovery curves for the indicated mClover3-tagged DDX3X variants in HeLa cells. Top: recovery plot for full bleach experiments. Bottom: recovery plot for partial bleach experiments. The traces of the FRAP data represent means ± s.e.m. For full bleach, n = 6 for G325E and R534H, n = 5 for G302V, n = 4 for R475G, and n = 3 for A497V. For partial bleach, n = 6 for R475G, G02V, A497V, and R543H and n = 7 for G325E. Data were fit as described in methods. c Left, imaging of EU-labeled RNAs (red) in HeLa cells expressing mClover3-tagged WT or DDX3X hollow variants. Scale bar, 10 μm. Right, zoomed-in view of the boxed area from the left figures. Scale bar, 2 μm. d Quantification of EU fluorescence intensity for each of the hollow condensates represented in c. Values were normalized using the fluorescence intensity in the nucleoplasm of each cell. Values represent means ± s.d. from 20 cells. Significance was calculated between R475G and the other variants using a two-tailed t-test. Source data are provided as a Source Data file.
Furthermore, to assess whether the decreased RNA-binding affinities measured in vitro correspond to decreased RNA binding in cells, we utilized click chemistry to fluorescently label nascent RNA in HeLa cells after incorporation of 5-ethynyluridine (5-EU) (Supplementary Fig. 4c). The results showed that the cytoplasmic RNAs were enriched in the outer shells of hollow condensates relative to the cytoplasm and the center of the condensates (Fig. 4c). After quantifying the RNA signal intensity in the condensates’ shells of each of the hollow mutants (normalizing to the RNA signal in the nucleoplasm), we found that R475G hollow condensates had the weakest 5-EU signals of all the tested mutants (Fig. 4d), consistent with the in vitro EMSA data (Fig. 2e).
R475G hollow condensates are enriched with proteins in various signaling pathways and lack translation machinery
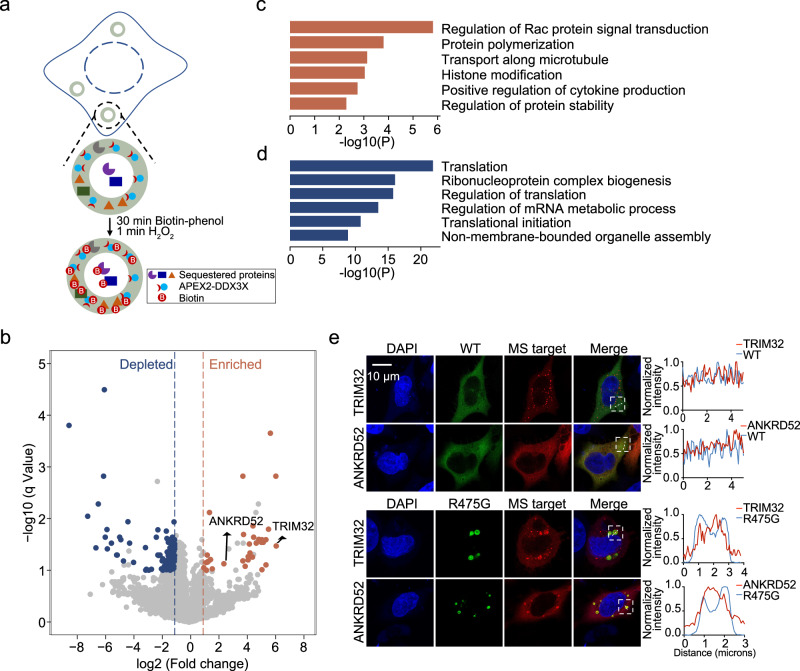
Under unstressed conditions, DDX3X mutant hollow condensates lack the SG marker protein G3BP1, indicating that these condensates are not SGs (Fig. 1c). This raised the possibility that these condensates may sequester unique proteins and have functions divergent from those of SGs. To understand the potential biological functions of these hollow condensates, we sought to identify their protein components using an adapted ascorbate peroxidase (APEX2)-based proximity labeling method41–43 (Fig. 5a). The labeling efficiency and specificity of APEX2-tagged DDX3X were examined as previously shown19. In this study, we prioritized comparing the protein networks of R475G and WT DDX3X, as R475G displayed the highest propensity to form hollow condensates among all the mutants tested.
Fig. 5. R475G hollow condensates enrich proteins in various signaling pathways and deplete translation machinery.
a Schematic of APEX2-mediated proximity labeling of proteins in R475G hollow condensates. b Volcano plots showing differential protein enrichment via streptavidin pull-downs from APEX2-R475G compared to APEX2-WT expressing cells. Differentially enriched proteins are shown in orange (adjusted p-value < 0.1, log2 fold change >1), and differentially depleted proteins are shown in blue (adjusted p-value < 0.1, log2 fold change < −1). The rest of the proteins are shown in light gray. c Gene Ontology (GO) analysis of the differentially enriched proteins in R475G condensates, illustrated as orange dots in b. d Gene Ontology (GO) analysis of the differentially depleted proteins in R475G condensates, illustrated as blue dots in b. p value was calculated using Student’s two-tailed t test with Benjamini–Hochberg FDR correction. e Representative images of localization of mCherry-tagged TRIM32 and ANKRD52 in mClover3-tagged WT- or R475G-expressing HeLa cells. Imaging was repeated in biological triplicate. Scale bar, 10 μm.
APEX2-WT DDX3X and APEX2-R475G were expressed in cells at a similar level (Supplementary Fig. 5a). After incubating with biotin-phenol for 30 min, cells were exposed to H2O2 for 1 min to trigger biotin labeling by the APEX2 moiety. Cells were then lysed, and the labeled proteins were enriched via streptavidin pulldown. Western blots analyzed protein labeling and enrichment before being subjected to proteomics analysis (Supplementary Fig. 5b). The Western blots and Coomassie stain results showed that the biotinylated proteins were enriched in pulldown fractions compared to the flowthrough and the input samples (Supplementary Fig. 5b).
Unsupervised hierarchical clustering of the APEX2-MS protein abundances resulted in two separate clusters for the biological triplicates of APEX2-WT and APEX2-R475G, indicating that the R475G mutation was the main determinant of differences between the interactomes (Supplementary Fig. 5c). Proteins with significantly altered abundance were selected based on the following criteria: (1) minimum fold-change of 2 in either direction, adjusted p-value < 0.1, (2) identified by a minimum of 2 peptides, and (3) detected in at least two replicates (Supplementary Data 1).
Analysis of these significant proteins revealed that, compared to the APEX2-WT DDX3X pulldown, 33 proteins were enriched in APEX2-R475G hollow condensates while 128 proteins were enriched in the APEX2-WT pulldown (Fig. 5b). Gene ontology (GO) analysis suggests that the proteins enriched in R475G condensates participate in the regulation of Rac protein signal transduction and protein polymerization (formation of cytoskeletal polymers and ubiquitin chains), while proteins depleted in R475G condensates mainly play roles in translation regulation and ribonucleoprotein complex biogenesis (Fig. 5c, d).
Because these hollow condensates appear biphasic, with the potential for proteins to either accumulate in the hollow core or the spherical shell of the puncta, we next sought to determine where in the condensates the proteins identified in our proteomics screen might reside. A previous study of TDP-43 hollow condensates determined that conventional antibody-based immunofluorescence cannot be used to detect the components in the hollow core, as fixation produces a barrier that prevents antibody penetration44. Given the low dynamics of R475G hollow condensates, we felt this would also likely be the case for these condensates. Thus, TRIM32 (an E3 ubiquitin ligase) and ANKRD52 (a regulatory protein of protein phosphatase 6), two top targets enriched in APEX2-R475G mass spec pulldown (Fig. 5b), were constructed with a mCherry tag and then subjected to fluorescence imaging. In WT DDX3X-expressing cells, TRIM32 and ANKRD52 were mostly diffuse in the cytoplasm, with TRIM32 showing small foci. In contrast, in cells expressing R475G, TRIM32 and ANKRD52 were sequestered inside of R475G hollow condensates (Fig. 5e). The trapping of these proteins in the relatively undynamic R475G condensates may disrupt the cellular pathways in which they normally participate.
DDX3X mutant hollow condensates recruit WT DDX3X and DDX3Y
Most DDX3X disease mutants are expressed heterozygously4,43, so we wondered whether DDX3X disease-related mutants co-condensed with WT DDX3X or DDX3Y in cells. To assess this, we first co-expressed mCherry-tagged WT DDX3X with each of the mClover3-tagged hollow mutants at a similar level in HeLa cells to look for co-condensates. When co-expressed, WT DDX3X was recruited to the shells of all the hollow condensates formed by each mutant (Fig. 6a). The presence of these co-condensates without the addition of a stressor suggests that these DDX3X mutants could contribute to disease (at least partially) by trapping WT DDX3X within their hollow condensates. To assess the ability of WT DDX3X molecules to move between the hollow condensates and the cytoplasm, we measured the dynamics of WT DDX3X sequestered in the hollow condensates using full bleach FRAP experiments. WT DDX3X recovered to ~80% of its original intensity in co-condensates with G302V, G325E, A497V, and R534H. This is comparable to the level of FRAP recovery of DDX3X in SGs19. However, WT DDX3X only recovered to ~40% of its original intensity when in co-condensates with R475G (Supplementary Fig. 6a and Fig. 6c, left). While all the hollow mutant condensates interfere with the cellular localization of WT DDX3X, condensates of R475G appeared to trap WT more strongly than other mutants.
Fig. 6. Mutant hollow condensates trap WT DDX3X and DDX3Y.
a Representative images showing co-localization of mCherry-tagged WT DDX3X and mClover3-tagged DDX3X hollow variants. White boxes indicate the condensates used to generate the corresponding intensity plots along the dotted white line. Scale bar, 10 μm. b Representative images showing co-localization of mCherry-tagged DDX3Y and mClover3-tagged DDX3X hollow variants. White boxes indicate the condensates used to generate the corresponding intensity plots along the dotted white line. Scale bar, 10 μm. c Recovery curves for WT DDX3X (left) and DDX3Y (right) sequestered in the hollow condensates formed by mClover3-tagged DDX3X hollow mutants in HeLa. FRAP traces data represent means ± s.e.m. Left: n = 8 for G302V, 5 for R534H, 6 for G325E, 4 for R475G, and 7 for A497V. Right, n = 4 for G302V, 7 for R534H, 4 for G325E, 5 for R475G, and 5 for A497V. d Comparison of the mobile fractions of mCherry-tagged WT DDX3X or DDX3Y in condensates with mClover3-tagged DDX3X variants, taken from the FRAP analysis in c. Mobile fraction calculated as described in Methods. Values represent means ± s.d., n values as described in c. Significance was calculated using a two-tailed t-test. e Recovery curves for each mClover3-tagged DDX3X variant in co-condensates with mCherry-tagged WT DDX3X in HeLa from the cellular FRAP. FRAP traces represent means ± s.e.m. n = 3 for G302V, 6 for R534H, 7 for G325E, 4 for R475G, and 3 for A497V. f Recovery curves for each mClover3-tagged DDX3X variant in hollow co-condensates with mCherry-tagged DDX3Y in HeLa cells from the cellular FRAP experiments. Traces represent means ± s.e.m., n = 5 for G302V, 8 for R534H, 3 for G325E, 5 for R475G, and 5 for A497V. g Recovery curves for each mClover3-tagged DDX3X variant in condensates when expressed alone in HeLa from the cellular FRAP. Traces represent means ± s.e.m. n = 5 for G325E, 4 for R475G, 6 for A497V, 6 for G302V, and 4 for R534H. All FRAP curves were fit as described in Methods. Source data are provided as a Source Data file.
We next performed these experiments with DDX3Y. Co-expression of mCherry-tagged DDX3Y and each of the mClover3-tagged DDX3X mutants showed that, as with WT DDX3X, DDX3Y was recruited to the shells of each mutant’s hollow condensate (Fig. 6b). In our previous work, we found that condensates of DDX3Y (both in vitro and in cells) were less dynamic than condensates of DDX3X19. We wondered whether the same would be true for DDX3Y in co-condensates with DDX3X mutants. Thus, we repeated the full bleach FRAP in HeLa cells co-expressing DDX3Y and each DDX3X mutants (Supplementary Fig. 6b and Fig. 6c, right). The results with DDX3Y largely mirrored those with WT DDX3X: the sequestered DDX3Y in the G302V, G325E, A497V, and R534H condensates was more dynamic than in R475G condensates (Fig. 6c, right). However, DDX3Y only recovered to ~60% of its original intensity when sequestered in G302V, G325E, A497V, and R534H hollow condensates, and ~20% when sequestered into R475G condensates. The mobile fraction of DDX3Y was significantly lower than the mobile fraction of WT DDX3X in each type of co-condensate measured (Fig. 6d), although there was no significant difference in recovery halftime for G302V, A497V, and R475G mutants when they co-condense with either DDX3X or DDX3Y (Supplementary Fig. 6c). The reduced dynamics of these co-condensates suggests that sequestration of WT DDX3X or DDX3Y within hollow condensates may inhibit their ability to facilitate translation, as both WT DDX3X and DDX3Y participate in translation initiation45.
Reciprocally, we performed full bleach FRAP on DDX3X hollow mutants in co-condensates with WT DDX3X or DDX3Y. When co-expressed with mCherry-tagged WT DDX3X (Supplementary Fig. 6d and Fig. 6e), all the hollow mutants showed improved mobile fractions compared to when they were expressed alone (Supplementary Figs. 6f and 6g, Fig. 6g). The magnitude of this effect was much smaller for R475G than it was for the other hollow mutants. However, when co-expressed with mCherry-tagged DDX3Y (Supplementary Fig. 6e and Fig. 6f), only G325E showed significant improvement in its mobile fraction (Supplementary Fig. 6g). Additionally, both DDX3X and DDX3Y were able to reduce the recovery halftime for all the hollow mutants except for R475G (Supplementary Fig. 6h). These data indicate that co-condensates of WT DDX3X and mutant DDX3X (which would be present in XX individuals with a mutated DDX3X allele) are more dynamic than co-condensates of DDX3Y and mutant DDX3X (which would be present in XY individuals with a mutated DDX3X allele).
DDX3X hollow mutants are over-represented in cases of DDX3X-related cancers and neurodevelopmental disorders
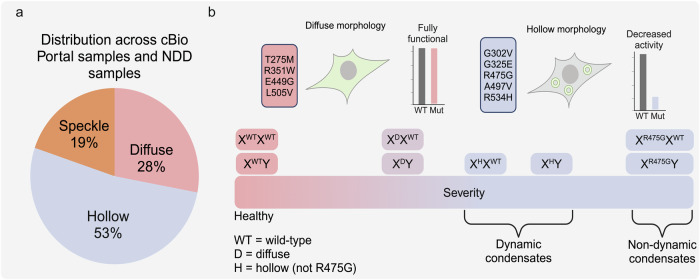
Given the distinct morphological and catalytic phenotypes observed in our hollow mutants, we questioned whether these mutants may be correlated more strongly with DDX3X-related diseases than the diffuse mutants. We examined the cBio Portal database46 and ten studies of DDX3X-related neurodevelopmental disorder (NDD)4,25,26,47–53. We counted the number of patient samples in which the mutations appeared (and mutations within three amino acids). Across this data set, mutations at the positions of the hollow mutants accounted for 53% of total appearances. In comparison, mutations at the positions of the diffuse and speckled positions represented only 28% and 19% of appearances, respectively (Fig. 7a). If the mutants of the three morphologies were all equally likely to appear in disease, one would expect hollow mutants to appear ~42% of appearances (as there are five hollow mutants out of the twelve investigated in this study), indicating that hollow mutants are over-represented in disease. This suggests that mutations that both disrupt catalytic activity and cause aberrant cellular phase separation may be more strongly associated with the development and severity of DDX3X-related diseases.
Fig. 7. Distribution of mutants across DDX3X-related diseases and a proposed working model.
a Distribution of DDX3X mutants in this study across cBioPortal samples and ten studies of DDX3X-related neurodevelopmental disorders. The number of patient samples in which the mutations at the position of each mutant appeared, in addition to mutations at positions within three amino acids of our selected mutants, were counted. b Proposed working model. DDX3X mutants that disrupt ATPase and strand release activities form hollow condensates in cells without external stress, while mutants that do not disrupt those catalytic steps remain diffuse. The severity of disease related to these mutations is correlated with the dynamics of these condensates. When coexpressed with WT DDX3X, hollow condensates are more dynamic than when coexpressed with DDX3Y, which may explain sex bias in DDX3X-related diseases. Created in BioRender. Owens (2023) BioRender.com/j70v272.
Discussion
Our work introduces an integrative approach across structural, biochemical, biophysical, and cellular methods to categorize DDX3X disease mutants. In doing so, we found that a specific group of DDX3X mutants over-represented in human diseases form cellular hollow condensates. We further revealed the molecular mechanism underlying the formation of these condensates: deficient ATPase and RNA strand release activities relative to wild-type protein. Hollow condensates sequestered WT DDX3X, DDX3Y, and proteins in diverse cell signaling pathways, possibly explaining why these mutants are over-represented among diseases associated with DDX3X mutations. Furthermore, we found that the presence of WT DDX3X improved the dynamics of heterogenous hollow condensates ~20% more than the presence of DDX3Y, underscoring that differential interactions with WT DDX3X and DDX3Y possibly underlie sex biases in DDX3X-related disease (Fig. 7b).
Specific DDX3X mutants form unique cytoplasmic hollow condensates
Forming membraneless compartments within cells via LLPS is crucial for properly regulating several cellular processes, ranging from gene expression to signal transduction54,55. Some condensates, such as SGs, have a uniform spherical morphology in which each component is fairly well-distributed56, while others have distinct sub-compartments, such as the nucleolus57. In addition, a different class of multilayered condensates has been reported where the condensate forms a vesicle-like structure with a seemingly lower content (“hollow”) internal space39,44,58. Until this point, these hollow cellular condensates have only been observed in the nucleus when seen in cells44. In our current study, we found that a specific subset of disease mutants of DDX3X can form hollow condensates in the cytoplasm, and these condensates may disturb the function of WT DDX3X, DDX3Y, and protein signal transduction (Figs. 5 and 6).
Formation of these hollow condensates can be either a passive process regulated by the protein’s biochemical and/or biophysical properties or an active process driven by “chaperone” proteins44. In the case of in vitro condensates of protamine, hollow condensates are formed via the LLPS of protein-RNA complexes when the RNA:protein ratio is either >1.87 or <0.07558. On the other hand, RNA binding-deficient mutants of TDP-43 require the ATPase activity of HSP70 to form hollow condensates44. For the DDX3X mutants tested in this study, we found that the propensity to form hollow condensates was correlated with the proteins’ decreased ATPase and strand release activities (Fig. 2) and that the dynamics of these hollow condensates may be linked to their RNA-binding affinities (Fig. 4). We did not find any chaperone-like proteins inside of the hollow condensate in our APEX2-MS analysis (Fig. 5). However, DDX3X is an ATPase, unlike TDP-43. It is possible that mutants with diminished strand release activity form hollow condensates because they are unable to release bound RNA.
Although it has been previously shown that increased RNA concentrations in condensates of the DEAD-box helicase Dhh1 resulted in decreased dynamics59, this is not the case for R475G, which has the lowest dynamics of all mutants tested. The relationship between internal RNA concentration and condensate dynamics may differ for human DDX3X and its mutants than for yeast Dhh1. Their intrinsically disordered regions may drive this difference, as we have previously shown that differences in IDR sequences are responsible for the different dynamics of DDX3X and DDX3Y condensates19. Because the hollow mutants are over-represented in disease (Fig. 7), revealing the cause of their condensation may provide important insights for future therapeutic development.
Hollow mutations of DDX3X cause disruptions at specific steps in the catalytic cycle
Based on the reaction scheme (Fig. 2b and Supplementary Movie 1), we could directly assess how different DDX3X mutations impair the catalytic cycle. In particular, we found that R475G disrupts the transition from the apo state to the pre-unwound state (Fig. 2b, step 1) by disrupting RNA binding in addition to other potential downstream disruptions (Fig. 3b), G325E disrupts the transition from the pre-unwound state to the post-unwound state (Fig. 2b, step 2) by blocking the deposition of ssRNA in the acceptor cleft (Fig. 3e), and R534H explicitly blocks ATP hydrolysis (Fig. 2e, step 3). These defects stall the catalytic cycle and limit ATP hydrolysis and strand release (Fig. 2c, f). R475G has been noted to cause severe neurodevelopmental defects, as has also been noted with mutations near G325E (T323I and R326H) and R534H (T532M)4. These defects all cause decrease in both ATP hydrolysis and RNA product strand release (Fig. 2). Taken together, we present the following model for aberrant condensate formation for these mutants: Under unstressed conditions, WT protein or mutations that do not disrupt catalysis (the diffuse mutants) form transient, multivalent protein-RNA interactions (because of the catalytic turnover) and do not form condensates. For mutants that disrupt catalysis (the hollow mutants), the protein-RNA interactions are longer-lived (because of the impaired ATP-driven RNA release) and drive condensate formation without the addition of cellular stress, as catalysis is necessary for the release of RNAs from DEAD-box protein condensates34. Once formed, the dynamics of these aberrant condensates are determined by their RNA contents, as R475G (the mutant with the weakest affinity for RNA) forms nearly static condensates. Under stressed conditions (such as arsenite treatment) and the subsequent changes in the cellular environment, DDX3X, RNA, and other resident proteins entered stress granules. Because of the altered cellular environment, including different RNA and protein components upon stress, DDX3X mutants no longer maintain the hollow morphology.
These disease-related mutants provided us with natural experiments to test the role of particular DEAD-box residues and the current model of DDX3-mediated catalysis (Fig. 2b). Taken together with our concurrent in-depth investigation of how the intrinsically disordered regions of DDX3X and DDX3Y both contribute to enzyme catalysis and differentiate the activities of these sexually dimorphic homologs20, our work provides the most complete picture to date of how the structured and intrinsically disordered regions of DDX3X and DDX3Y contribute to enzyme activity.
Hollow condensates of DDX3X mutants may contribute to disease via sequestration of other proteins
The five hollow mutants formed their cellular condensates without adding an external stressor, and these hollow condensates do not contain G3BP1, meaning they are not “canonical” SGs56. The APEX2 proteomics results revealed that R475G hollow condensates sequester a subset of proteins that do not normally interact with WT DDX3X. R475G condensates are also depleted of proteins relating to protein translation, further separating them from SGs56. A previous study suggested that R475G localizes to FMRP-positive granules in N2a cells without stress4, suggesting that the condensation properties of these mutants might vary between cell types.
One of the most enriched proteins in R475G condensates was the E3 ubiquitin ligase TRIM32. TRIM32 drives neural progenitor differentiation during neuronal development by preferentially accumulating in one daughter cell during division. The TRIM32-positive daughter cell will differentiate into a mature neuron, while the TRIM32-negative daughter cell will remain a progenitor60. Expression of R475G in neural progenitor cells might disrupt the proper segregation of TRIM32, as its hollow condensates are significantly less dynamic than condensates of other mutants (Figs. 4 and 6). This possibly contributes to R475G’s more severe neurodevelopmental phenotypes than other DDX3X mutants. In addition to TRIM32, many other proteins enriched in R475G condensates regulate Rac signaling. DDX3X mutants may thus also dysregulate Rac signaling, which has been implicated in several cancers61. The disease implications of these sequestered proteins warrant further investigation.
The distinct properties of DDX3X mutant hollow condensates may correlate with sex biases in disease incidence and severity
The disruption of cellular condensate dynamics can often lead to tumorigenesis, as with mutant forms of UTX23 and AKAP9562. Additionally, a mutant of DDX3X not included in this study (L566S) was recently found to form cellular condensates with dynamics comparable to R475G and to form amyloid-like fibrils in vitro63, further supporting a link between DDX3X condensation and disease.
DDX3X mutant hollow condensates sequester WT DDX3X or DDX3Y when co-expressed in cells (Fig. 6). The heterogeneous condensates containing mutant DDX3X and WT DDX3X are more dynamic than those of mutant DDX3X and DDX3Y (Fig. 6). This difference in dynamics mirrors the difference in dynamics for condensates of WT DDX3X or DDX3Y alone, which is determined by sequence differences between both proteins’ N-terminal IDRs19. The propensity for phase separation is correlated with ATPase activity for DEAD-box proteins34, and our previous work suggests this relationship also extends to dynamics of the condensates formed by LLPS (lower activity = less dynamic)19. R475G, which is impaired at multiple points along its catalytic cycle (Figs. 2 and 3), is the most catalytically impaired mutant and, as such, produces the least dynamic condensates (Fig. 4). Thus, WT DDX3 (and other co-condensed proteins) are more strongly trapped by this mutant than by the other hollow mutants. These differing condensation properties could possibly contribute to the sex biases in the incidence and severity of DDX3X-related disorders, which are frequently biased against XY individuals4,21. Based on our results, we hypothesize that DDX3X mutant hollow condensates formed in XY individuals would be less dynamic and more able to sequester the remaining DDX3Y (and other constituent proteins) than condensates formed in XX individuals. As such, we propose that mutants of DDX3X that form less dynamic condensates likely contribute to more severe disease and that this effect is exacerbated by co-expression with DDX3Y, as in any person with a Y chromosome (Fig. 7b). Future work should prioritize the study of these mutants in both XX and XY chromosomal sex-defined genetic backgrounds.
Methods
Cell culture, transfection, and Escherichia coli strains
HeLa (ATCC CCL-2) and HEK293T (ATCC CRL-3216) cells were cultured in DMEM + GlutaMAX (GIBCO) with 10% FBS (GIBCO) and 1% Pen/Strep (Corning) in a humidified incubator with 5% CO2 at 37 °C. Lipofectamine 2000 (Invitrogen) was used for plasmid transfection. Experiments were conducted on transfected cells after 24 h transfection. For bacterial cell culture, TOP10 and BL21(DE3)-RIL chemically competent bacterial strains were grown in lysogeny broth containing the corresponding antibiotics at 200 rpm, 37 °C.
Constructs
The DDX3X-mCherry plasmid was generated in our previous study19 and the DDX3X-mClover3 was constructed by substituting the fluorescent tag using fusion PCR. The different mClover3-tagged DDX3X mutants were generated using fusion PCR with the wild-type DDX3X-mClover3 plasmid as template. For the APEX2-MS experiments, the DNA fragment encoding APEX2 was PCR amplified from pcDNA5/FRT/TO APEX2-GFP (Addgene) and fused to the N-terminus of DDX3X using fusion PCR. To verify the specificity of APEX2 labeling, TRIM32 and ANKRD52 genes were cloned from HeLa cDNA, fused with a mCherry tag at the C-terminus, and inserted into the pcDNA3 vector for mammalian cell expression. G3BP1 was also cloned from HeLa cDNA, fused with an mCherry tag at the C-terminus, and inserted into the pPB vector. All plasmids were validated by Sanger sequencing. All oligonucleotides sequences are listed in Supplementary Data 2.
Protein purification
Wild-type and mutant DDX3X tagged with mCherry or MBP were purified according to our previous report19. Briefly, the plasmids were transformed into Escherichia coli strain BL21-RIL to express the recombinant proteins. The bacteria were cultured in lysogeny broth at 37 °C until OD600 nm = 0.8 before administration of 1 mM IPTG and shaken at 200 rpm at 16 °C for 16 h after induction. The pellets from 2 L bacterial culture were resuspended with 80 mL binding buffer (25 mM Tris-HCl, pH 7.5, 10% glycerol, 500 mM NaCl for mCherry-tagged and 25 mM Tris-HCl pH and 500 mM NaCl for MBP-tagged) and subjected to sonication. After centrifuging at 31,000 × g for 30 min to remove the cell debris, the supernatant was loaded onto a Ni-NTA column. White boxes indicate the condensates used to generate the corresponding intensity plots along the dotted white line. Scale bar, 10 μm. Next, 10 column volumes of the binding buffer supplemented with 50 mM imidazole was used to wash away the non-specifically bound proteins. Another 10 column volumes of a high salt buffer (25 mM Tris-HCl, pH 7.5, 10% glycerol, and 2 M NaCl for mCherry-tagged and 25 mM Tris-HCl pH 7.5 and 2 M NaCl for MBP-tagged) was used to remove the associated RNAs from wild-type DDX3X and DDX3X mutants. Finally, four column volumes of the binding buffer supplemented with 500 mM imidazole was used to elute the bound proteins. mCherry-tagged proteins were dialyzed into the storage buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, and 2 mM DTT) with TEV enzyme cleavage of the His-tag simultaneously for 1 hr at room temperature. Then, the proteins were concentrated using Amicon Ultra-15 (Millipore) tubes before loading to a Superdex 75 column, with storage buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, and 2 mM DTT for mCherry-tagged and 25 mM Tris-HCl pH 8.0, 500 mM NaCl for MBP-tagged). The purity of the proteins was analyzed by SDS-PAGE. Purified proteins were aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C. Once thawed, aliquots were never refrozen. All purification steps after sonication were performed at room temperature.
Electrophoretic mobility shift assay (EMSA)
To measure the RNA binding affinities of different DDX3X mutants, EMSAs were carried out. A double-stranded RNA probe with 5’ overhang, to which DDX3X is known to bind19,64, with following sequences: 5’-biotin/ACCGCUGCCGUCGCUCCG/AlexF647N/−3’ and 5’-/Cy3/UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUCGGAGCGACGGCAGCGGU-3’ was used. Both strands were ordered from IDT and annealed by heating the equimolar mixtures of both strands at 65 °C for 5 min, then slowly cooling the sample to room temperature over 3 h. Various concentrations of each mCherry-tagged DDX3X mutant proteins were incubated with 100 nM double-stranded RNA probe at room temperature for 30 min in a buffer consisting of 50 mM Tris-HCl pH 7.5, 150 mM KCl, 2 mM MgCl2, 10 mM beta-mercaptoethanol and 1 mg/mL BSA before loading to a homemade 6% polyacrylamide gel (37.5:1) in 0.5× TBE buffer. The gel was run at 120 V for 2 h at 4 °C. The gel was then scanned using an Amersham Typhoon using the Cy5 channel. Fiji was used to quantify intensity of the bottom free RNA band. Fraction bound was calculated using the following equation:
| 1 |
Where, “In” is the intensity of the free probe band at a given concentration and “Io” is the intensity of the free probe band at 0 nM protein. Fraction bound was plotted versus concentration in GraphPad Prism and fit with the Hill equation to give KD.
Fluorescence imaging
HeLa cells were seeded to a 6-well plate with a coverslip in each well and cultured overnight. The various DDX3X plasmids were transfected to cells with Lipofectamine 2000 the next morning. After 24 h in culture, the cells were washed once with PBS and then fixed using 4% paraformaldehyde in PBST (PBS with 0.05% Tween-20) at room temperature for 15 min. Then, the cells were washed twice by PBST and permeabilized by 0.5% Triton in PBST at room temperature for 20 min. After being washed once with PBST, the cells were blocked with 1% BSA in PBST at room temperature for 30 min. For anti-Flag imaging, mouse anti-Flag (Invitrogen MA1-91878) was diluted 1:1000 in PBST and added to the cells overnight at 4 °C. Following this, cells were washed four times with PBST and then treated with anti-mouse 594 secondary antibody (Invitrogen A32742) diluted 1:1000 in PBST for 1 hour at room temperature After being washed three times with PBST, the cells were incubated with 0.5 μg/mL DAPI for 1 min. After 4 PBST washes, an antifade reagent (Invitrogen) was used to mount the slides. The images were taken using a Leica TCS SP8 confocal microscope.
Quantification of hollow puncta
To quantify the percentage of cells showing hollow puncta, a slide with HeLa cells transiently transfected with each DDX3X mutant was scanned left to right and top to bottom. The images for the first 20–26 transfected cells with puncta were taken. We then calculated the number of cells with hollow puncta divided by the total 20–26 cells. For each DDX3X mutant, three biological replicates were carried out. The results were plotted using GraphPad Prism.
EU labeling of RNA in cells
To quantify the RNAs in DDX3X mutant condensates, the cellular RNAs were first labeled with 5-ethynyl uridine (EU). An Alexa Fluor 594 fluorophore was then added through a click chemistry reaction using Click-iT RNA image kits (ThermoFisher). Briefly, HeLa cells were seeded to a 6-well plate. The plasmids of DDX3X mutants were transfected into the cells using Lipofectamine 2000. After 24 h in culture, the medium was removed and fresh medium containing 1 mM EU was added. After a 1 hr incubation, the cells were fixed and permeabilized as stated above. Alexa 594 was conjugated to EU using the Click-iT reaction. After three PBST washes, the cells were incubated with 1 mL Hoechst 33342 to stain the DNA. After 4 PBST washes, an antifade reagent (Invitrogen) was used to mount the slides. The images were taken using a Leica TCS SP8 confocal microscope. The average Alexa594 intensity in hollow puncta of each DDX3X mutant was quantified using Fiji65, with the average Alexa594 intensity in the corresponding nucleoplasm used for normalization.
Fluorescence recovery after photobleaching (FRAP)
The FRAP assays were conducted using the bleaching module of a Zeiss LSM 880 confocal microscope. The 488 nm laser was used to bleach the mClover3 signal, and the 561 nm laser was used to bleach the mCherry signal. HeLa cells were seeded on 35 mm poly-D-lysine coated glass-bottomed dishes (Mattek), and the plasmids of DDX3X mutants were transfected into the HeLa cells. After 24 h culture, the normal DMEM medium was replaced with FluoroBrite DMEM (GIBCO) medium with 10% FBS. For full bleaching assay to dissect the molecule exchange between puncta and surroundings, an entire punctum was selected to bleach with 100% laser power and time-lapse images were collected afterward. To probe the molecule exchange inside of the puncta, partial bleaching was performed by focusing on a circular region of interest (ROI) of a punctum using 100% laser power, and time-lapse images were collected afterward as well. The fluorescence intensity was directly measured in the Zen software. The values were reported relative to pre-bleaching time points. GraphPad Prism was used to plot the data. The halftime for each replicate was calculated using the following formula:
| 2 |
Where, “a” is the slow recovery fraction, “c” is the rapid diffusion fraction, and “b” is the recovery rate. The halftime is ln(2)/b, and the mobile fraction is a + c. Significance was calculated using an unpaired Student’s two-tailed t test.
Malachite green ATPase assay
ATPase measurements were taken using the Malachite Green Phosphate Assay Kit (Sigma) according to the manufacturer’s instructions. Briefly, 1 μM of each mCherry tagged DDX3X mutant protein was incubated with 100 ng/μL total RNA extracted from HeLa cells (or the indicated concentration) for 15 min in the reaction buffer (25 mM Tris-HCl, pH 8, 200 mM NaCl, 1 mM DTT, and 2 mM MgCl2) before the addition of 2 mM ATP. The reaction was incubated at 16 °C for 30 min. Then the reaction was quenched by the addition of malachite green mixture and left for an additional 30 min to develop the color. The samples were then loaded into a clear-bottom 384-well plate and the absorbance at OD620 nm was measured. Values were converted from absorbance units to μM free phosphate using a standard curve generated with the kit’s phosphate standard. Background values (free phosphate detected from reactions lacking RNA) were subtracted from the values from reactions with RNA, and interference from increased RNA concentrations (apparent absorbance due to scattering from RNA-only reactions) was subtracted from each point. Data were plotted in GraphPad Prism. Significance was calculated using an unpaired Student’s two-tailed t test.
Multi-parameter confocal fluorescence time-resolved microscopy and spectroscopy
All single molecule measurements were performed on multi-parameter confocal time-correlated single photon counting (TCSPC) microscopy and spectroscopy (MicroTime-200; PicoQuant, GmbH). Double stranded RNA: an 18-mer (5’-biotin/ACCGCUGCCGUCGCUCCG/Alexa647N/−3’) annealed to a 42-mer (5’-/Cy3/UUUUUUUUUUUUUUUUUUUUUUUUCGGAGCGACGGCAGCGGU-3’) (IDT), labeled with the donor (Cy3) and acceptor (Alexa647) dyes were excited with 532 nm and 637 nm pulsed diode lasers (LDH-D-TA-532, LDH-D-TA-637, PicoQuant) in pulsed-interleaved excitation (PIE) mode at a repetition rate of 20 MHz. The beam passed through an excitation dichroic filter ZT532/637 (Chroma Technology) and a water immersion objective lens Olympus UPLanSApo 60×/1.2 w with collar correction for coverslip thickness. All the measurements were carried out with laser focused 20 μm above the coverslip, while maintaining the power of both the lasers below 20 μW. Fluorescence signals from the sample after passing through 50 μm pinhole were split into vertical and horizontal channels by a polarized beam splitter (U-MBF3-Olympus). Cy3 and Alexa647 emission signals were separated by identical dichroic filters (T635 lpxr, Chroma Technology) in each pathway. Polarized and wavelength-selected photons through bandpass filters ET582/64 (for donor, Cy3) and ET690/70 (for acceptor, Alexa647) were projected onto four single-photon avalanche photodiode (SPAD) detectors and cataloged by a HydraHarp TCSPC time-interval analyzer in (PicoQuant). Data was acquired for the labeled dsRNA alone followed by several 5-min recording periods (over the course of 1–2 h) after adding the protein to the RNA. Subsequently, data was collected for 5-min periods over the course of another thirty minutes following the addition of 1 mM MgATP. Nunc Lab-Tek chambers (ThermoFisher-155411) with borosilicate coverslip bottoms were used. These chambers were treated with 50% (w/v) PEG-8000 solution, incubated at room temperature for 3–4 h, followed by several washes with FRET buffer (50 mM Tris, pH 7.5, 125 mM NaCl and 2 mM MgCl2). Confocal detection volume for 532 and 637 nm lasers were calibrated and determined using rhodamine6-G and ATTO637N dyes.
Time-resolved fluorescence anisotropy
The files acquired from the Microtime-200 fluorometer were converted into universal photon HDF5 file format using the “phconvert” module of the FRETBursts software (v0.7)66. HDF5 files were further analyzed using custom python scripts67. The parallel and perpendicular ns fluorescence intensity decays for DO particles, DL particles and AO particles within the respective selected regions of E vs. S plots and burst selection filters were used to calculate the time-resolved anisotropy decay (r(t)) using the equation below. DO particles used donor excitation (FDexDem). DL and AO particles used acceptor excitation (FAexAem).
| 3 |
Where, r is the anisotropy, III(t) is the parallel fluorescence intensity, I⊥(t) is the perpendicular fluorescence intensity, G is the correction factor for difference in sensitivity of the two detectors. Intensities were corrected for background counts. The time resolved anisotropy curves (r(t)) were fitted with the equation below to obtain rotational correlation time (Φ) and the proportion of molecules with hindered rotation (B/B0):
| 4 |
Where, B0 is the anisotropy value ~ 1/2 ns after the laser pulse and completion of internal (segmental) motions within the RNA construct.
Net Proportion of Strand Release (ΔB/B0) was calculated by subtracting the end point B/B0 (30 min after the addition of 1 mM ATP) from the initial B/B0 right before 1 mM ATP was added.
TNP-ATP binding assay
WT DDX3X or R534H (MBP-tagged) was serially diluted to twice the indicated concentrations in 1X reaction buffer (25 mM Tris-HCl, pH 8, 200 mM NaCl, 1 mM DTT, and 2 mM MgCl2). TNP-ATP was then diluted to 4 μM in reaction buffer. 50 μL of protein was then mixed with 50 μL TNP-ATP to produce the indicated protein concentration with 2 μM TNP-ATP in 1X buffer. This mixture was then incubated on ice for 10 min. The reactions were the loaded into a black bottom 384-well plate and fluorescent emission from 500 to 600 nm was measured with excitation at 410 nm. To analyze data, the emission value at 500 nm was first subtracted from each trace. Then, the value of the no protein sample at 540 nm was subtracted from the other concentration’s values at 540. These values were then plotted vs concentration using GraphPad Prism 9 and fit with a Hill binding curve. Lysozyme, which does not bind ATP, was used as a negative control.
Cellular APEX labeling
To capture the proteins in the hollow puncta formed by R475G DDX3X, a recently established proximity labeling method was carried out followed by mass spectrometry19,41. Briefly, one 10 cm plate of HEK 293T cells was transfected with 3.5 μg APEX2 fused wild-type DDX3X- or R475G-expressing plasmid. After 24 h culture, the normal DMEM medium was replaced with DMEM medium supplemented with 500 mM biotin-phenol for 30 min. Then, H2O2 (Sigma-Aldrich) was added to each cell culture dish at 1 mM final concentration for exactly 1 min with gentle agitation. To stop the labeling, the culture medium was removed, and the quenching solution (10 mM sodium ascorbate, 10 mM sodium azide, and 5 mM Trolox in PBS) was immediately used to wash the cells three times. Finally, 1 mL quenching solution was applied to cover the cells. Cells were then collected with a cell scraper. The unlabeled control samples were prepared in parallel under the same procedure as aforementioned, without the addition of the H2O2.
Mass spectrometry sample preparation
The cell pellets collected after APEX labeling were lysed by gentle pipetting in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% (wt/vol) SDS, 0.5% (wt/vol) sodium deoxycholate and 1% (vol/vol) Triton X-100) supplemented with 1x protease inhibitor cocktail, 1 mM PMSF and quenchers (10 mM sodium azide, 10 mM sodium ascorbate and 5 mM Trolox). After resuspension and on ice for ~2 min, the lysates were clarified by centrifuging at 15,000 × g for 10 min at 4 °C. The concentration of the cell lysate was quantified by using the Pierce 660-nm assay. The same amount of cell lysate was incubated with RIPA buffer washed Pierce Streptavidin magnetic beads for 1 hr at room temperature on a rotator. The beads were pelleted using a magnetic rack and washed sequentially with 1 mL RIPA buffer twice, once with 1 M KCl, once with 0.1 M Na2CO3, once with 2 M urea in 10 mM Tris-HCl, pH 8.0, and twice with RIPA lysis buffer. The wash buffers were kept on ice throughout the procedure. The biotinylated proteins were eluted from the beads by boiling each sample in 30 μL of 3× protein loading buffer supplemented with 2 mM biotin and 20 mM DTT for 10 min. Then, the samples were placed on a magnetic rack to pellet the beads and to collect the eluate. All the eluate was loaded to 10% SDS-PAGE gel and ran 1 cm into the gel. Then, the entire stained gel regions were excised, reduced with tris(2-carboxyethyl)phosphine (TCEP), alkylated with iodoacetamide, and digested with trypsin. Tryptic digests were analyzed using a 95 min LC gradient on the Thermo Q Exactive HF mass spectrometer as described previously68.
Mass spectrometry data analysis
MS/MS data were searched with full tryptic specificity against the Swiss Prot human proteome database (06/04/2021), the wild-type DDX3X and R475G protein sequences, and a common contaminant database using MaxQuant 1.6.17.069. Search parameters used include two missed cleavages, static carbamidomethylation of Cys, and variable modifications of protein N-terminal acetylation, Asn deamidation, and Met oxidation. Consensus identification lists were generated with false discovery rates set at 1% for protein, and peptide identifications. High confidence identification of proteins with significant change refers to proteins satisfying the following criteria: minimum fold-change of 2 in either direction, adjusted p-value < 0.1, identified by a minimum of 2 razor + unique peptides, and detected in at least two of the three replicates. High confidence proteins were subjected to gene ontology (GO) analysis using Metascape v3.570.
Protein quantification and Western blot
Protein concentrations of the samples were calculated using the Bradford Assay (Thermo Fisher, 23246). Protein samples were boiled at 95 °C in Laemmli sample buffer for 10 min. After brief centrifugation, the samples were loaded onto SDS-PAGE gels. After running at 180 V for 1 h, the gels were transferred to membrane by semi-dry transfer apparatus at 20 V for 50 min. Then, the membrane was blocked with 5% milk or BSA in 1× PBST for 30 min at room temperature or 4 °C overnight. The membrane was then incubated in 3% milk or BSA in 1× PBST containing the corresponding primary antibody overnight at 4 °C [rabbit anti-FLAG HRP-conjugated (Invitrogen MA1-91878-HRP, 1:1,000), rabbit anti-DDX3Y (Invitrogen PA5-90055, 1:3,000), rabbit anti-GAPDH (Invitrogen PA1-16777, 1:1,000)]. After washing three times with 1× PBST, the horseradish peroxidase (HRP)-conjugated secondary antibody (Abcam ab6721, 1:20,000) in 1% of milk was applied and incubated at room temperature for 1 h. After washing three times with PBST, the membranes were visualized using ECL Western Blotting Detection Kit (Thermo Fisher).
Quantification and statistical analysis
Images were analyzed using Fiji v2.14.0/1.54f. All data are presented as the mean ± standard error of mean (s.e.m.) or standard deviation (s.d.) from the independent measurements. The statistical analyses were performed using GraphPad Prism 9 (GraphPad Software). p values below 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by the National Institutes of Health (R35GM133721 and R01HL160726 to K.F.L., R35GM133721-03S1 to A.Y., R50CA221838 to H.Y.T, T32GM132039 to A.Y. and M.C.O., and R35GM118139 to Y.E.G.) and the National Science Foundation (DGE-2236662 to E.L.). K.F.L. is supported by the American Cancer Society (RSG-22-064-01-RMC), the Damon Runyon Innovator Award, and the Linda Pechenik Montague Investigator Award. We thank Dr. Matthew Kayser for sharing the Leica SP8 confocal microscope. We would also like to thank Drs. Roger Greenberg, Ben Black, and Nancy Bonini for their helpful comments while drafting this manuscript. Figures 1a and 7 were created with Biorender.com. We thank the pilot grant from Penn Center for Genome Integrity for supporting this work.
Author contributions
M.C.O., H. Shen., and K.F.L. designed the experiments. H. Shen, A.Y., M.C.O., and E.L. generated the constructs and the mammalian cell lines used in this study. H. Shen., A.Y., M.C.O., E.L., and X.W. purified the recombinant DDX3X and DDX3X mutants used in this study. H.Shen. and A.Y. performed the immunofluorescence imaging and FRAP experiments and analyzed the data. M.C.O. performed the ATP binding assay, the ATPase assays, and the EMSA assays together with E.L. M.C.O. performed the structural analysis and generated Supplementary Movie 1. M.S.M.F. and M.C.O. performed the plasmid titration Western blots and imaging in Fig. 1. A.Y. performed the fluorescence anisotropy experiments with guidance from Y.E.G. and H. Shweta. H. Shen performed the APEX2 proteomics and H-Y.T. performed the proteomic data analysis. All authors participated in writing, discussing, and editing the manuscript.
Peer review
Peer review information
Nature Communications thanks Rick Russell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The APEX2-MS proteomics data generated in this study have been deposited in the MassIVE database under accession code MSV000090789. All other data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Code availability
All code used to analyze the multiparameter confocal spectroscopy data has been deposited into the Zenodo database under accession code 13334020.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Michael C. Owens, Hui Shen.
Contributor Information
Yale E. Goldman, Email: yegoldman@ucdavis.edu
Kathy Fange Liu, Email: liufg@pennmedicine.upenn.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53636-0.
References
- 1.Mo, J. et al. DDX3X: structure, physiologic functions and cancer. Mol. Cancer20, 38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma, D. & Jankowsky, E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol. 49, 343–360 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Chen, C.-Y. et al. Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet.25, 2905–2922 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Lennox, A. L. et al. Pathogenic DDX3X Mutations Impair RNA Metabolism and Neurogenesis during Fetal Cortical Development. Neuron106, 404–420.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singleton, M. R., Dillingham, M. S. & Wigley, D. B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem.76, 23–50 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Calviello, L. et al. DDX3 depletion represses translation of mRNAs with complex 5’ UTRs. Nucleic Acids Res49, 5336–5350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih, J.-W. et al. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J.441, 119–129 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Jones, D. T. W. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature488, 100–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patmore, D. M. et al. DDX3X suppresses the susceptibility of hindbrain lineages to medulloblastoma. Dev. Cell54, 455–470.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentin-Vega, Y. A. et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep.6, 25996 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, C. et al. Sequential inverse dysregulation of the RNA helicases DDX3X and DDX3Y facilitates MYC-driven lymphomagenesis. Mol. Cell81, 4059–4075.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Jiang, L. et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet.47, 1061–1066 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Ojha, J. et al. Identification of recurrent truncated DDX3X mutations in chronic lymphocytic leukaemia. Br. J. Haematol.169, 445–448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, K. et al. Clinical implications of cancer gene mutations in patients with chronic lymphocytic leukemia treated with lenalidomide. Blood131, 1820–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadek, M., Sherr, E. H. & Floor, S. N. The variant landscape and function of DDX3X in cancer and neurodevelopmental disorders. Trends Mol. Med. 10.1016/j.molmed.2023.06.003 (2023). [DOI] [PubMed]
- 16.Boitnott, A. et al. Developmental and behavioral phenotypes in a mouse model of DDX3X syndrome. Biol. Psychiatry90, 742–755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy, T. et al. DDX3X syndrome: summary of findings and recommendations for evaluation and care. Pediatr. Neurol.138, 87–94 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Epling, L. B., Grace, C. R., Lowe, B. R., Partridge, J. F. & Enemark, E. J. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J. Mol. Biol.427, 1779–1796 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen, H. et al. Sexually dimorphic RNA helicases DDX3X and DDX3Y differentially regulate RNA metabolism through phase separation. Mol. Cell82, 2588–2603.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanas, A., Shweta, H., Owens, M. C., Liu, K. F. & Goldman, Y. E. DDX3X and DDX3Y constitutively form nano-sized RNA-protein clusters that foster enzymatic activity. BioRxivhttps://www.biorxiv.org/content/10.1101/2023.11.29.569239v1.full (2023).
- 21.Weil, M. D., Lamborn, K., Edwards, M. S. & Wara, W. M. Influence of a child’s sex on medulloblastoma outcome. JAMA279, 1474–1476 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, T. A. et al. A Cluster of Autism-Associated Variants on X-Linked NLGN4X Functionally Resemble NLGN4Y. Neuron106, 759–768.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi, B. et al. UTX condensation underlies its tumour-suppressive activity. Nature597, 726–731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godfrey, A. K. et al. Quantitative analysis of Y-Chromosome gene expression across 36 human tissues. Genome Res.30, 860–873 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, H. et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry21, 133–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snijders Blok, L. et al. Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am. J. Hum. Genet.97, 343–352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markmiller, S. et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedersha, N. et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell13, 195–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R. & Parker, R. Distinct stages in stress granule assembly and disassembly. Elife5, e18413 (2016). [DOI] [PMC free article] [PubMed]
- 30.Floor, S. N., Condon, K. J., Sharma, D., Jankowsky, E. & Doudna, J. A. Autoinhibitory Interdomain Interactions and Subfamily-specific Extensions Redefine the Catalytic Core of the Human DEAD-box Protein DDX3. J. Biol. Chem.291, 2412–2421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, D., Putnam, A. A. & Jankowsky, E. Biochemical Differences and Similarities between the DEAD-Box Helicase Orthologs DDX3X and Ded1p. J. Mol. Biol.429, 3730–3742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, H. & Ji, X. The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nat. Commun.10, 3085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell125, 287–300 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature573, 144–148 (2019). [DOI] [PubMed] [Google Scholar]
- 35.LaConte, L. E. W., Srivastava, S. & Mukherjee, K. Probing Protein Kinase-ATP Interactions Using a Fluorescent ATP Analog. Methods Mol. Biol.1647, 171–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, F., Putnam, A. & Jankowsky, E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl. Acad. Sci. USA105, 20209–20214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, Y. et al. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc. Natl. Acad. Sci. USA105, 20203–20208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alberti, S., Gladfelter, A. & Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt, H. B. & Rohatgi, R. In Vivo Formation of Vacuolated Multi-phase Compartments Lacking Membranes. Cell Rep.16, 1228–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niaki, A. G. et al. Loss of Dynamic RNA Interaction and Aberrant Phase Separation Induced by Two Distinct Types of ALS/FTD-Linked FUS Mutations. Mol. Cell77, 82–94.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fazal, F. M. et al. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell178, 473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marmor-Kollet, H. et al. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by sumoylation and links to ALS pathogenesis. Mol. Cell80, 876–891.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padrón, A., Iwasaki, S. & Ingolia, N. T. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol. Cell75, 875–887.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, H. et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science371, eabb4309 (2021). [DOI] [PMC free article] [PubMed]
- 45.Venkataramanan, S., Gadek, M., Calviello, L., Wilkins, K. & Floor, S. N. DDX3X and DDX3Y are redundant in protein synthesis. RNA27, 1577–1588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, X. et al. Phenotypic expansion in DDX3X - a common cause of intellectual disability in females. Ann Clin Transl Neurol5, 1277–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai, Y. et al. Expansion of clinical and genetic spectrum of DDX3X neurodevelopmental disorder in 23 chinese patients. Front. Mol. Neurosci.15, 793001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dikow, N. et al. DDX3X mutations in two girls with a phenotype overlapping Toriello-Carey syndrome. Am. J. Med. Genet. A173, 1369–1373 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Nicola, P. et al. De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am. J. Med. Genet. A179, 570–578 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Tang, L. et al. Prospective and detailed behavioral phenotyping in DDX3X syndrome. Mol. Autism12, 36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scala, M. et al. Three de novo DDX3X variants associated with distinctive brain developmental abnormalities and brain tumor in intellectually disabled females. Eur. J. Hum. Genet.27, 1254–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boehning, M. et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol.25, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, P. et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alshareedah, I., Moosa, M. M., Raju, M., Potoyan, D. A. & Banerjee, P. R. Phase transition of RNA-protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. USA117, 15650–15658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linsenmeier, M. et al. Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat. Commun.13, 3030 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicklas, S. et al. A complex of the ubiquitin ligase TRIM32 and the deubiquitinase USP7 balances the level of c-Myc ubiquitination and thereby determines neural stem cell fate specification. Cell Death Differ26, 728–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mack, N. A., Whalley, H. J., Castillo-Lluva, S. & Malliri, A. The diverse roles of Rac signaling in tumorigenesis. Cell Cycle10, 1571–1581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, W. et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol.22, 960–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Castro Fonseca, M. et al. Molecular and cellular basis of hyperassembly and protein aggregation driven by a rare pathogenic mutation in DDX3X. iScience24, 102841 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim, Y. & Myong, S. RNA remodeling activity of DEAD box proteins tuned by protein concentration, RNA length, and ATP. Mol. Cell63, 865–876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ingargiola, A., Lerner, E., Chung, S., Weiss, S. & Michalet, X. FRETBursts: An Open Source Toolkit for Analysis of Freely-Diffusing Single-Molecule FRET. PLoS One11, e0160716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shweta, H. & Goldman, Y. E. Software for the analysis of FRETBursts anisotropy data. Zenodo, 10.5281/zenodo.13334020 (2024).
- 68.Yan, Q. et al. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat. Commun.13, 53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol.26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun.10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The APEX2-MS proteomics data generated in this study have been deposited in the MassIVE database under accession code MSV000090789. All other data generated in this study are provided in the Source Data file. Source data are provided with this paper.
All code used to analyze the multiparameter confocal spectroscopy data has been deposited into the Zenodo database under accession code 13334020.