Abstract
The increase in demand for natural rubber has led to the search for alternative sources. Lactuca serriola is emerging as a promising candidate, as the quality of the natural rubber it produces is comparable to that of the Pará Rubber Plant, Hevea brasiliensis. This study examines the effect of methyl jasmonate (MeJA), a known elicitor, on the expression of key rubber biosynthesis pathway genes (HMGR1, HMGS1, CPT2, and SRPP1) in the latex of L. serriola plants. The expression levels of these genes increased significantly after the foliar application of 200 and 400 µM MeJA. The highest relative expression level for HMGR1, HMGS1, CPT2 and SRPP1 was 3.74, 18.56, 11.91and 16.59 fold respectively. Furthermore, the rubber content in L. serriola showed a significant rise post-treatment compared to the control with increasing the level of MeJA (6.19%, 7.24% and 7.85% which correspond to 0, 200 and 400 µM). Gel permeation chromatography revealed an augmentation in the molecular weight of extracted natural rubber from treated plants. Samples treated with 400 µM of MeJA had the highest molecular weight (1570 kg mol−1) compared to control (1186 kg mol−1). This study has demonstrated that MeJA, through the regulation of rubber biosynthesis genes, is capable of enhancing the quality and quantity of natural rubber extracted from alternative sources, such as L. serriola.
Keywords: Cis-polyisoprene, Lactuca serriola, Natural rubber biosynthesis, Gene expression, Methyl Jasmonate, Rubber molecular weight
Subject terms: Biotechnology, Plant sciences
Introduction
Natural rubber (NR) is sourced from latex, a unique cytoplasmic substance found in rubber-producing plants. Latex is synthesized within either the laticiferous cell network or individual parenchymal cells across an extensive range of approximately 12,500 plant species1. Scientifically referred to as cis-1,4-polyisoprene, NR is used in a diverse array of products including a variety of industrial and medical products2. It possesses exceptional qualities such as resilience, impact resistance, elasticity and efficient heat dispersion3. Because of these characteristics, NR is irreplaceable in various applications including medical devices and heavy-duty tires4–7. There are over 2500 known rubber-producing plants, however, the majority exhibit low rubber yields and inadequate molecular weight8,9. Currently, Hevea brasiliensis, is the primary source of commercially feasible NR2,10, but its particular cultivation conditions and sensitivity to fungal infections make cultivation challenging.
With recent rapid economic growth, the demand for NR has surged, prompting search for alternative sources. In light of potential NR shortages and economic predictions, there is an immediate need for improvements in production11. This involves exploring possible rubber-producing species2 and employing genetic engineering techniques8. One such source is Lactuca serriola L., commonly known as Prickly lettuce—a plant species identified as a potential source of NR11. This species belongs to the Asteraceae family and is widely distributed across Europe, Asia, and North America. Cornish et al.12 demonstrated that L. serriola yields high molecular weight (Mw) rubber that is comparable to NR from Pará rubber trees. The plant is an annual that yields substantial biomass with concentrated rubber content and has a short growth cycle, which makes it easy to work with11,13.
The NR molecule is a polymer composed of units of isoprene derived from isopentenyl diphosphate (IPP) in the cis- configuration. The surface of rubber particles within laticifer cells serves as the site for NR metabolism14. Laticifers are specialized tissues for biosynthesis and storage in most rubber producing plants12.
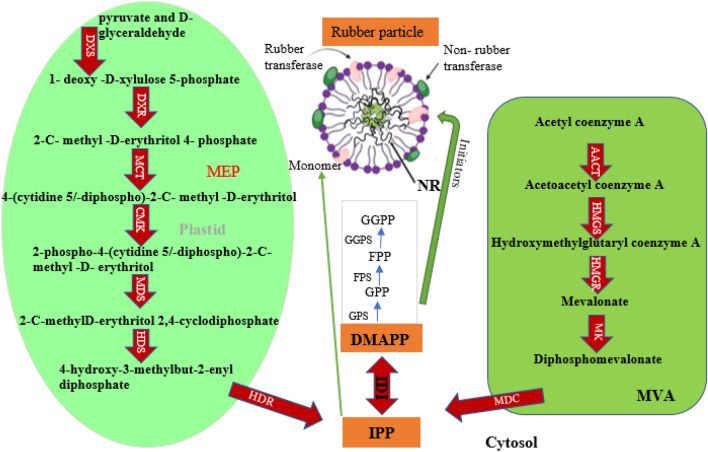
The process of rubber biosynthesis involves both the mevalonate (MVA) pathway, which converts pyruvic acid into isopentenyl diphosphate (IPP) and predominantly takes place within the cytosol, as well as the 2C-methyl-d-erythritol 4-phosphate (MEP) pathway, which transforms pyruvic acid and glycerate 3-phosphate into IPP and occurs in plastids15,16. IPP is a critical substrate for the production of various terpenes, including NR17. The MVA pathway is especially important, with key enzymes such as hydroxymethylglutaryl-CoA reductase (HMGR) and hydroxymethylglutaryl-CoA synthase (HMGS)5 (Fig. 1).
Fig. 1.
The biosynthesis pathway of natural rubber (cis-1,4-polyisoprene). AACT: acetyl coenzyme A acetyltransferase; HMGS: hydroxymethylglutaryl coenzyme A synthase; HMGR: hydroxymethylglutaryl coenzyme A reductase; MK: mevalonate kinase; MDC: diphosphomevalonate decarboxylase; DXS: 1- deoxy -D-xylulose 5-phosphate synthase; DXR: 1- deoxy -D-xylulose 5-phosphate reductoisomerase; MCT: 2-C- methyl -D-erythritol 4-phosphate cytidylyltransferase; CMK: 4-(cytidine 5/-diphospho)-2-C- methyl -D-erythritol kinase; MDS: 2- C- methyl -D-erythritol 2,4-cyclodiphosphate synthase; HDS: 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase.
The incorporation of IPP units into a prenyl chain is an essential step in the process, which requires initiators such as farnesyl pyrophosphate (FPP)12. The synthesis of NR involves the use of IPP precursors through the collaboration of various enzymes and proteins. These include farnesyl pyrophosphate synthase (FPS), cis-prenyltransferase (CPT), small rubber particle protein (SRPP), rubber elongation factor (REF), and other unidentified proteins working in synergy18.
One approach to cultivating alternative rubber crops involves the identification of essential regulatory genes in the processes of rubber biosynthesis. The regulation of gene expression is achieved by utilizing elicitors such as jasmonates within the laticiferous tissues, where NR synthesis occurs19. Jasmonates and its variants such as methyl jasmonate (MeJA) are cellular regulators that originate from lipids and play a vital role in developmental processes and help plants respond to biotic and abiotic stressors19. They can trigger secondary metabolism by activating transcription20.
Jasmonic acid (JA) plays a crucial role in encouraging the differentiation of laticiferous cells12. Transcriptional responses to JA, ethylene, and MeJA, were indicated in a cell line expressing laticifer-specific genes. Moreover, the induction of secondary laticifer differentiation, responsible for latex production in H. brasiliensis, is attributed to linolenic acid and JA21,22. In small-scale field trials this resulted in increased latex production23. The present study investigates the application of exogenous MeJA to L. serriola plants to induce secondary metabolism and understand its effects on the expression of rubber biosynthesis genes (HMGR, HMGS, CPT and SRPP). The objective is to assess changes in the expression of key rubber biosynthesis genes, quantifying and characterizing the rubber extracted. It is the first time that transcriptomics is being investigated in L. serriola with the plant specific designed primers.
Materials and methods
Plant materials and MeJA application
L. serriola seeds were gathered from Karaj, Iran (35°46′ N, 51°00′ E). To create working stock and eliminate environmental effects, a single seed from the collection was grown in the research greenhouse of the Iranian Biological Research Center. Plants were initiated by seeding the working stock collections in plastic pots filled with potting media (peat moss, coco peat and perlite) and maintained at temperatures of 26 °C during the day and 22 °C at night. Additional sodium vapor lighting was used to supplement natural light, providing a 14-h daytime photoperiod that matched the temperature profile. Plants were given sub-irrigation as required.
MeJA (Sigma-Aldrich) was prepared in a 0.1% (v/v) ethanol solution at three different concentrations (0, 200, and 400 µM) in 1000 mL volumetric flasks. The solution was applied to the plants approximately two weeks before the bolting stage using a sprayer until the runoff stage was reached (30 ml per plant). To ensure uniform application, the same number of sprayer cycles was used for each plant. The spraying process was repeated three times with three replicates, with one-week intervals, leading up to the latex harvest. Control plants were sprayed with distilled water containing 0.1% (v/v) ethanol24. The plants had an average height of approximately 150 cm, with stem diameters up to 12 mm. Latex was collected from the bolting stem of each individual in three replicates. For molecular analysis, latex was obtained at three separate time intervals following treatment: 6, 12, and 24 h in three biological replicates. In the assessment of rubber quality and quantity, latex sampling took place one week after the final MeJA application to examine the impact of MeJA concentration on rubber characteristics and the extracted latex was stored at − 20 °C for further analysis.
Latex sampling and rubber extraction
The proportions of water, resin, rubber, and insoluble material in the latex of each plant were assessed through gravimetric analysis13. Latex collection involved making 8 mm diagonal incisions on the stem using a razor, and the dripping latex was gathered in a pre-weighed microcentrifuge tube. The tube was then capped and re-weighed to determine the initial weight of the collected latex. The latex was dried for 48 h at 35 °C under vacuum to remove water. The rubber extraction was conducted exactly according to the method of Bell et al.13.
Gel permeation/size exclusion chromatography
The rubber component of the latex was analyzed using gel permeation size exclusion chromatography (GPC/SEC) with refractive index detection to determine the length of polymer chains. The dried rubber fractions were dissolved in tetrahydrofuran at a concentration of approximately 5 mg mL-1 at room temperature and left overnight. The procedure was done according to the method of Ramirez-Cadavid et al.25.
Fourier transform infrared analysis
For FTIR analysis, 15 to 20 mg of extracted L. serriola rubber was used. The functional groups of the samples were determined using KBr cells on an ALPHA II Bruker device. FTIR spectra were acquired in 16 scans over a range of 4000 to 400 cm−1 and a resolution of 4 cm−1.
Bioinformatics and plant specific primer design
No sequences for the target genes in L. serriola were available, therefore, existing RNA sequences for designing our target plant-specific primers in databases were used. Paired-end short reads from total RNA-seq data of L. serriola were obtained from the NCBI-SRA (Sequence Read Archive). To ensure the accuracy of the transcriptome assembly, initial quality assessment of raw reads in all samples was performed using FastQC software (Version 0.11.9;26). Initially, substandard bases, reads, and potential adapter impurities were removed using trimmomatic software. This involved setting the minimum phred score (TRAILING) to 20 and the minimum length (MINLEN) to 5027. Following trimming, the resulting high-quality reads, denoted as "clean reads," served as the primary input sequences for transcript assembly.
The transcript assembly was carried out using Trinity software (version 2.8.5), employing the de Bruijns graph-based strategy, following the methodology described by26. Functional annotation of the transcriptome was conducted to unveil functionality using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and UniProt (https://www.uniprot.org/). After selecting the best transcripts, primer designing was performed using Primer 3 software (version0.4.0). The designed primers were specific for the HMGR1, HMGS1, CPT2, and SRPP1 genes. The sequences of primers utilized for qRT-PCR are listed in Table 1.
Table 1.
Primer nucleotide sequences used in RT-PCR.
| Target genes | Primer sequencing (sequence in 5’-3’ direction) | Fragment size (bp) |
|---|---|---|
| CPT2 | F: CCAAAACCGCCTGAACAAGA | 153 |
| R: GAGTGACACCTGACCCTGAT | ||
| HMGR1 | F: TCCCCTCTATCCAGTTCACG | 129 |
| R: TGGTGTCAAAGGGTGTTCAA | ||
| HMGS1 | F: ATGGCTCCTCAAAACGTCGG | 248 |
| R: CTTCCTACTTCCAAGCGACCA | ||
| SRPP1 | F: AGTTGTCCTTCGCATACCCA | 145 |
| R: GCCGAAAACGATGTTCCTGT | ||
| Tubulin | F: CCATAAGTTTGATCTCATGTATGC | 101 |
| R: CAAGGTCCTCACGAGCCT |
Real-time RT-PCR
For RNA isolation and transcription analysis approximately 100 mg of latex was mixed with 1 ml of TRIzol reagent (Invitrogen). The RNA extraction procedure was according to manufacturer guidelines. To eliminate genomic DNA contamination, all RNA samples underwent treatment with the RNase-Free DNase Set (Fermentas, Waltham, MA). The quality and quantity of RNA were assessed through agarose gel electrophoresis and nanodrop ND-1000, respectively.
For cDNA synthesis, 3–5 μg of purified total RNA was employed in accordance with the manufacturer’s instructions using the Easy cDNA Synthesis Kit (Pars Tous, Iran). Plant-specific primer pairs were used for quantitative real-time polymerase chain reaction (RT-PCR). The RT-PCR analysis of NR biosynthesis genes’ relative expression was conducted using SYBR green Master Mix 2X (Pars -Tous, Iran) on a QIAGEN real-time PCR system.
The RT-PCR cycling conditions were adjusted as follows: an initial denaturation at 95 °C for 10 min, followed by 40 three-step cycles of 95 °C for 20 s, 20 s at an annealing temperature ranging from 54 to 58 °C (based on the individual annealing temperature required for each primer pair), and an extension at 72 °C for 20 s. The tubulin gene served as the internal control. All reactions were conducted in three biological replicates and three technical replicates. The results are expressed as mean ± SE and the gene expression relative changes were quantified by the comparative CT method28.
Statistical analysis
Statistical analysis was carried out using SAS 9.4. The research followed a factorial design based on a completely randomized setup with three replications. Mean comparisons were conducted using Fisher’s least significant differences at either the 0.05 or 0.01 probability thresholds. Furthermore, the standard error was calculated to provide a measure of precision. Data correlation and cluster analysis (heatmap) was performed using R package to assess relationships between variables.
Results
Gene expression analysis
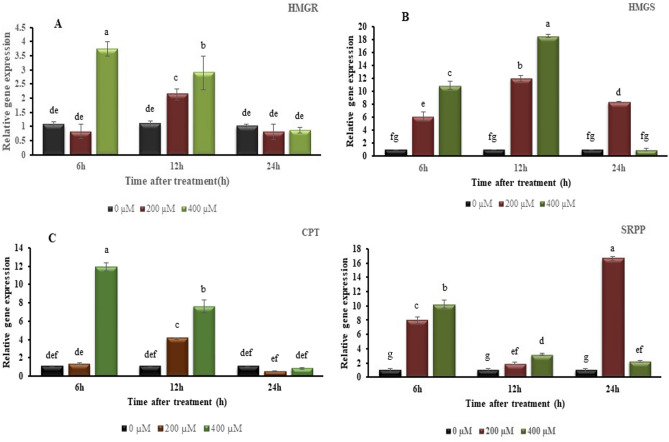
To assess the impact of MeJA treatment on gene expression within the rubber biosynthesis pathway, quantitative real-time (qRT-PCR) analysis was conducted. The relative expression levels of HMGR1, HMGS1, CPT 2 and SRPP1 genes were analyzed at 6, 12 and 24 h after treatment with MeJA at three different concentration levels (0, 200 and 400 µM). Statistically significant differences were observed in the expression levels of all genes in response to the elicitor treatments (Fig. 2).
Fig. 2.
Effect of different concentrations of methyl jasmonate at different exposure times on expression levels of (A) HMGR1, (B) HMGS1, (C) CPT 2 and (D) SRPP1 genes in rubber biosynthesis pathway in L. serriola. Error bars are shown as ± SE (n = 3). Means followed by the same letter are not significantly different according to the Duncan’s multiple range test at p = 0.01.
In HMGR1, after treatment with 400 µM MeJA the transcription value increased about 3.5-fold at 6 h, before dropping to around threefold at 12 h and 0.8-fold at 24 h. With 200 µM MeJA, the transcription value doubled at 12 h, but at 6 h and 24 h, no statistical increase was observed (Fig. 2A).
The HMGS1 results demonstrated a substantial increase at all time points, with a particularly notable 12-fold rise at 12 h after exposure to 200 µM MeJA, compared to the control. At the highest MeJA concentration of 400 µM, the expression of HMGS1 displayed a contrasting response, with a substantial increase at 6 h (10.9-fold) reaching its highest peak (18.6-fold) at 12 h post treatment (Fig. 2B).
For CPT2 transcription results at 200 µM MeJA, a significant increase occurred, peaking at 4.1-fold at 12 h (Fig. 2C). For 400 µM MeJA, a significant raise was observed at 6 h post-treatment (11.9-fold), then dropping to 7.59-fold at 12 h but still significantly higher compared to the controls, and further declined at 24 h (0.84-fold).
At 200 µM MeJA, a striking increase in expression level of SRPP1 was observed at 24 h (16.2-fold) and 6 h (7.8-fold), while it only moderately increased at 12 h (1.9-fold). At the 400 µM MeJA concentration, the expression pattern commenced with a significant increase of tenfold at 6 h, followed by another notable rise of threefold at 12 h, relative to the control. Additionally, a more moderate increase of twofold was observed at 24 h (Fig. 2D).
Effect of methyl jasmonate on rubber content and molecular weight
The analysis of rubber content and molecular weight provides insights into the influence of MeJA on rubber biosynthesis in L. serriola. Our findings reveal significant alterations in both rubber content and molecular weight, demonstrating the regulatory role of MeJA in rubber production and quality.
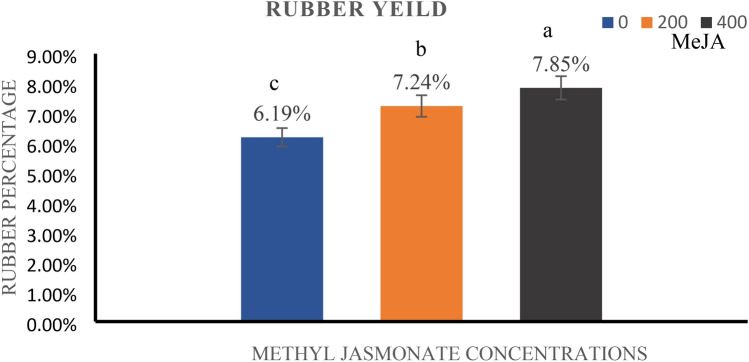
Impact of methyl jasmonate on rubber content
Three concentrations of MeJA (0 µM, 200 µM, and 400 µM) were administered to L. serriola plants to examine the impact of MeJA on the extracted rubber and other latex components. A substantial increase in rubber content was observed in treated plants compared to the control plants (Fig. 3). The rubber content in the control group was 6.2%, whereas the post-treatment rubber content for 200 µM and 400 µM was 7.2% and 7.8%, respectively. Treatment with MeJA resulted in a 14–16% increase in rubber yield, which was statistically significant (p < 0.01). Additionally, the acetone-soluble resin material of latex was also quantified in the treatment groups (Table 2). This change demonstrates the potential of MeJA to modulate the rubber biosynthesis pathway.
Fig. 3.
Effect of different concentrations of methyl jasmonate on the extracted rubber content in L. serriola. Error bars are shown as ± SE (n = 3). Means followed by the same letter are not significantly different according to the Duncan’s multiple range test at 0.01 probability level.
Table 2.
Effect of different concentrations of methyl jasmonate on some attributes of latex and its extracted rubber in L. serriola.
| Samples | Mw (kg/mol) | Mw/Mn (PDI) | Latex Water (%wt/wt) | Latex Resin (%wt/wt) | Latex Insolubles (%wt/wt) |
|---|---|---|---|---|---|
| Control | 1186 | 1.34 | 41** ± 0.15 | 17.25** ± 0.20 | 35.5ns ± 0.18 |
| MeJA 200 µM | 1510 | 1.34 | 38.84** ± 0.16 | 18.13** ± 0.15 | 35.79ns ± 0.39 |
| MeJA 400 µM | 1570 | 1.28 | 37.6** ± 0.24 | 18.78* ± 0.10 | 35.77ns ± 0.18 |
** and * are significant at P < 0.01 and P < 0.05 respectively. ns is not significant. n = 3.
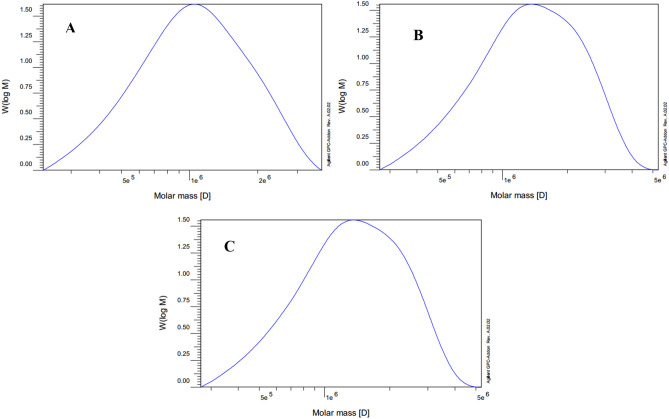
Influence of methyl jasmonate on molecular weight
The molecular weight (Mw) of treated and non-treated rubber from a mixture of three biological replicates were measured at the bolting stage. MeJA treatment had a distinct impact on the molecular weight of the synthesized rubber. Notably the Mw of rubber produced in the MeJA-treated plants exhibited a marked increase compared to the control group. The Mw of control group was 1186 kg/mol, while these figures for the treated plants with 200 µM and 400 µM were 1510 kg/mol and 1570 kg/mol, respectively (Table 2). The rubber samples investigated in this study had Mws above 1000 kg/mol, a characteristic of a high-quality rubber. The polydispersity values were 1.34, 1.34, and 1.28 for the control, 200 µM treatment, and 400 µM treatment, respectively (Table 2; Fig. 4).
Fig. 4.
GPC/SEC chromatograms of extracted rubber from latex of methyl jasmonate treated L. serriola. (A) control plants, (B) 200 µM of MeJA, (C) 400 µM of MeJA.
FTIR analysis
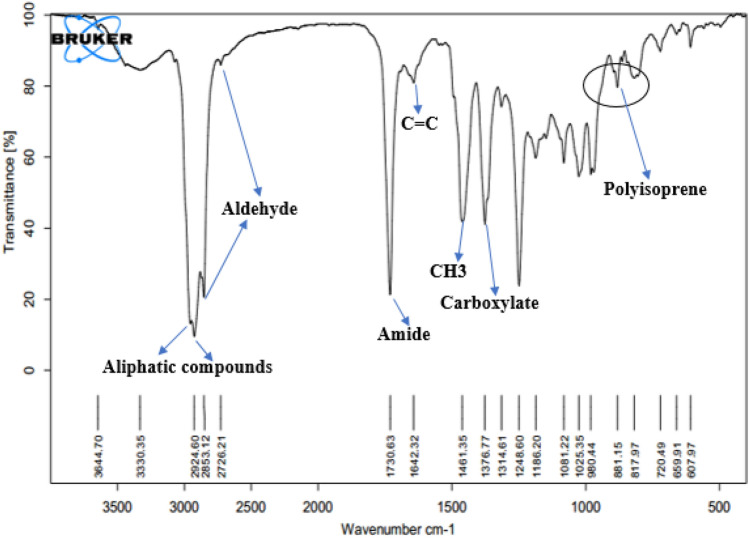
The FTIR analysis of NR extracted from L. serriola showed distinctive chemical structures spanning the range of 500–4000 cm-1. Notably, distinct peaks were observed at the wavelengths 2963, 2924, and 2853 cm−1 (Fig. 5). Additionally, a prominent sharp peak at 1730 cm−1 and a characteristic polyisoprene peak at 860 cm−1 were evident in the infrared spectrum of NR. These apparent peaks are associated with specific compounds as shown in Fig. 5.
Fig. 5.
FTIR spectrum of cis-1,4-polyisporene derived from of L. serriola.
Mutual correlations among rubber biosynthesis gene expression levels and rubber yield in L. serriola
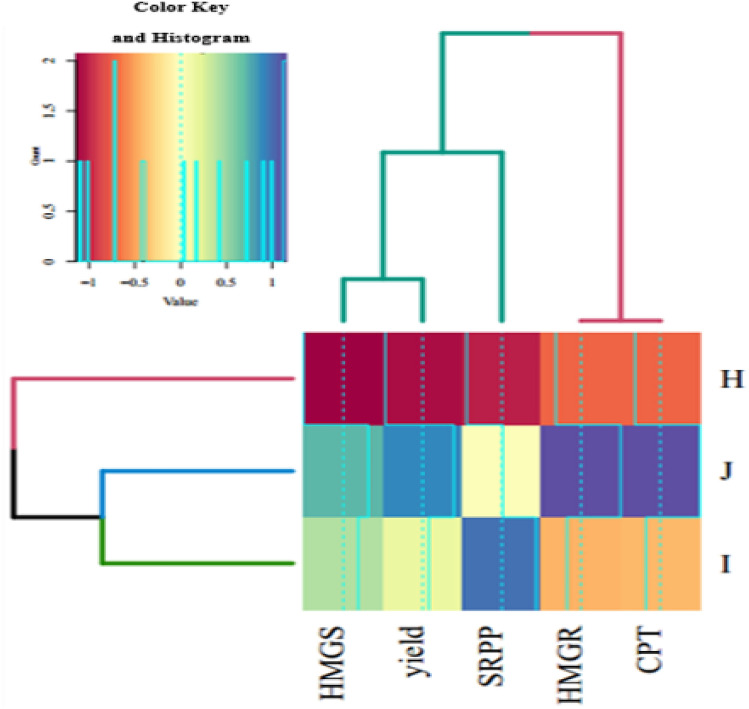
To study potential correlations and cluster analysis between the expression profile of studied genes (HMGR1, HMGS1, CPT2, and SRPP1) and rubber yield, heatmap and correlation analysis were conducted. The findings suggest that treatment with MeJA led to a rise in the expression levels of the examined genes and an increase in rubber yield. These characteristics exhibited positive correlations with each other, with the exception of yield and SRPP1 at specific MeJA concentrations (Fig. 6). Cluster analysis identified that HMGS1 and yield were grouped together in one cluster, followed by SRPP1 in a subsequent level of classification. Furthermore, HMGR1 showed correlation with CPT2 in the subsequent tier of classification. Moreover, MeJA treatments at different levels (200 µM and 400 µM) were clustered together.
Fig. 6.
Shows the heatmap and cluster analysis of the expression levels of rubber biosynthesis genes (HMGR1, HMGS1, CPT2 and SRPP1) and rubber yield under 3 different levels of MeJA (H:0 µM, I: 200 µM and J:400 µM). The visualization combines a heatmap and cluster analysis to show how MeJA affects NR yield and rubber biosynthesis genes in plants. Darker colors in the heatmap indicate higher values for those parameters, while lighter colors represent lower values. The clustering helps identify groups of parameters that respond similarly to MeJA. The distance between dendrograms on the top and left sides of the heatmap reflects the degree of similarity between parameters or MeJA treatments.
The correlation analysis (Fig. 7) aimed to determine whether these genes collaborate to co-regulate rubber synthesis and accumulation. The temporal gene expression levels of HMGR1, HMGS1 and CPT2 exhibited highly significant positive associations. SRPP1 did not show any correlation with HMGR1 and CPT2.
Fig. 7.

Correlation among the expression levels of rubber biosynthesis pathway genes and rubber yield in L. serriola. Darker colors indicate higher values, while lighter colors represent weaker correlations.
The HMGR1, HMGS1, and CPT2 expression levels all showed a strong correlation with rubber yield. However, a weaker correlation was observed between SRPP1 and rubber yield.
Discussion
The rubber biosynthesis pathway, a complex network of biochemical processes crucial for NR production in rubber-producing plant species, was the focal point of this study. The impact of MeJA on the expression of four rubber biosynthesis genes—HMGR1, HMGS1, CPT2, and SRPP1—in L. serriola and effect on rubber yield and quality were investigated.
When plants are treated with exogenous JA or MeJA, they stimulate their own synthesis by metabolizing α-linolenic acid (α-LeA). The internal JA serves as the substrate, undergoing a transformation through a process which activates the pathway of JA signaling, consequently, the regulation in expression of JA-responsive genes and also downstream genes linked to the biosynthesis of NR, occurs due to the transcription factors’ release18, ultimately leading to an increase in rubber content29.
The investigated genes in this study, exhibited clear upregulation in response to MeJA treatment and implies a positive regulatory role in reinforcing rubber production in L. serriola (Fig. 2). HMGR1, HMGS1 and CPT2 had higher expression levels after application of 400 µM MeJA, but SRPP1 had much higher expression at 200 µM MeJA. The genes responsible for vital enzymes and proteins in natural latex metabolism showed inducible expression in latex derived from L. serriola plants at least at one-time point. These genes expression levels in plants exposed to exogenous MeJA, showed variability dependent on the stimuli concentration (200 µM, 400 µM), duration of induction (6, 12 and 24 h), and gene type29. The variation implies a potential association between increased expression of the genes and production of latex, potentially contributing to a systematic defense mechanism within laticifers against plant stress30.
HMGS1 and HMGR1, displayed significant changes in expression levels after MeJA treatment. Sirinupong et al.31 demonstrated that there is a positive correlation between content of latex rubber with HMGS activity. HMGS, which catalyzes the irreversible conversion of acetoacetyl-CoA to HMG-CoA, appears to play a crucial role and possibly acts as a key regulatory point in the MVA pathway in plants32. Rate-limiting HMGR in the MVA pathway, results in diminished IPP availability when its activity decreases, resulting in a reduced rate of rubber synthesis. Therefore, a strong correlation between the rubber production rate and HMGR expression level is possible33. Our results are consistent with those of Du et al.34, who investigated the identification of multiple HMGR genes, revealing their evolutionary relationships and expression patterns in response to 1 mmol/L MeJA and ethylene treatments over four time points (0, 3, 6, 12, and 24 h). They found that the expression of HMGR genes was significantly influenced by MeJA, which is known to regulate plant defense responses and secondary metabolite production. Notably, HMGR1 expression in roots of Taraxacum- koksaghyz peaked 12 h after MeJA treatment.
CPT, responsible for the production of cis-polyisoprene, exhibited a marked increase in expression at both MeJA concentrations, suggesting its involvement in rubber biosynthesis regulation through catalyzing the condensation of an allylic prenyl diphosphate to IPP. According to Cherian et al.8, CPT genes were reported to be essential for rubber biosynthesis in T. kok-saghyz35, L. sativa36, and H. brasiliensis37. The application of RNAi to inhibit CPT in T. brevicorniculatum resulted in a decrease in levels of cis-1,4-polyisoprene leading to the virtual elimination of extended chain molecules33, indicating the involvement of CPT in regulating molecular weight.
SRPP, a protein associated with rubber particle formation, demonstrated the most significant changes in expression, particularly at 200 µM MeJA and 24 h post-treatment. This suggests a potential link between SRPP and rubber particle dynamics influenced by MeJA. There is substantial evidence confirming SRPP's role in latex coagulation, contributing to the sealing of wound sites and preventing potential infections by fungal or bacterial pathogens38–40. The involvement of SRPP in producing rubber with high molecular weight, rather than in the rubber biosynthesis process itself, is supported by lack of SRPP homologs observed in Ficus carica and Ficus benghalensis8,41,42. This suggests that SRPP influences both rubber quantity and the stability of rubber molecular weight in plants43. However, Chakrabarty et al.44 established that the homologs of SRPP in lettuce did not play rate-limiting catalytic roles in the biosynthesis of natural rubber.
The role of SRPP is also crucial in MVA, MEP, and FPP synthesis, as well as in integrating IPP units into the prenyl chain. In particular, SRPP has been suggested to enhance the stability of rubber particles without compromising the integrity of cellular membrane.
To determine the presence of a synergistic effect among the rubber biosynthesis genes and rubber yield, a correlation analysis was conducted. The significant positive associations among HMGR1, HMGS1 and CPT2 indicate a coordinated regulation of these genes implicated in rubber accumulation throughout the latex metabolic process (Figs. 6 and 7). The high and significant correlation observed between HMGR1, HMGS1, and CPT2 with rubber yield suggests that these genes likely play important roles in rubber synthesis in L. serriola. However, SRPP may not act as a limiting factor29. Despite the increase in the number of SRPP1 transcripts, weaker correlation with rubber yield was evident. Additionally, there was no observed synergistic effect between SRPP1 and two other studied genes (HMGR1 and CPT2). This contrast might be explained by the concept that SRPP may play a role in the regulation of rubber molecular weight rather than direct rubber biosynthesis8.
Besides its potential involvement in regulating the molecular weight of rubber, SRPPs might be triggered by stress, thereby enhancing stress tolerance in plants45. The higher expression levels could be a response to elicitor stress, and it is plausible that SRPP transcript levels are post-transcriptionally regulated. Our findings align with those of Wu et al.29, who reported similar results in H. brasiliensis. They noted a strong correlation between HMGR, HMGS, and CPT with dry rubber yield, but that SRPP showed no significant correlation.
The potential of MeJA as a signaling molecule to stimulate the production of diverse secondary metabolites by triggering the expression of relevant genes is well established (e.g., Refs.46–48). According to Wei et al.49, MeJA significantly upregulates genes involved in terpene biosynthesis, underscoring its role as a potent elicitor of secondary metabolites. This upregulation is driven by the activation of transcription factors that regulate biosynthetic gene expression, a mechanism also observed in other plants like Medicago truncatula and Taxus. The analysis of rubber content and molecular weight provides insights into the influence of MeJA on rubber biosynthesis in L. serriola. Our findings reveal significant alterations in both rubber content and molecular weight, indicating the regulatory role of MeJA in rubber production.
Regarding rubber content, our results indicate that treatment with MeJA (400 µM) significantly increased rubber content by approximately 16% compared to the control samples (Fig. 3). Analysis of the expression profiles of rubber biosynthesis genes revealed elevated relative expression levels of CPT2, HMGR1, and HMGS1, demonstrating a correlation with the induced rubber accumulation.
Application of exogenous MeJA not only stimulates the density of laticifer cells but also triggers the JA signaling pathway responsible for regulating rubber biosynthesis in H. brasiliensis18,50. In addition, inducing the differentiation of secondary laticifers is a promising strategy for increasing rubber yield, as the number of laticifers directly affects latex production. While genetically controlled, laticifer formation can be induced by external signals, including exogenous JA and MeJA. Studies have shown that JA and MeJA promote secondary laticifer differentiation, with mechanical damage during tapping and reduced turgor pressure also rapidly increasing JA levels51.
Our findings align with the discoveries of Saeedi et al.52, who examined the impact of MeJA concentrations (300 µM and 600 µM) on rubber content in Taraxacum kok-saghyz plants. They demonstrated the substantial and positive effects of MeJA treatment, which resulted in heightened rubber yields. In a separate study, Wu et al.29 also reported an increase in rubber yield as a result of the effect of MeJA on rubber trees.
According to Bell et al.13, the rubber content among the investigated biotypes ranged from 2 to 12%, with the most frequent percentages being between 4 and 5%, aligning with our findings. However, our research outcomes showed a significant difference with the results reported by11, who reported a rubber content of 54% in L. serriola plants. This disparity in rubber content can be attributed to our rubber extraction method, which involved extraction from the whole latex, leading to a lower rubber percentage.
Molecular weight (Mw) stands out as a crucial parameter in determining the quality of rubber, exhibiting a positive correlation between quality and molecular weight. The capacity to transform rubber into a high-quality product characterized by exceptional resistance to abrasion and tensile strength is linked to the Mw of the initial rubber polymer chain53.
The rubber samples investigated in this study had all Mws above 1000 kg/mol, which is one of the characteristics of a high-quality rubber. A Mw of 1000 kg/mol or higher stands as the primary and most significant determinant of usable rubber1. L. serriola naturally synthesizes a high molecular weight NR, but it could be influenced by substrate concentration and type and the ratio of the initiator FPP to IPP5. These can be attributed to the modulation of enzyme activity and modifications in the levels of gene expression related to chain elongation and termination of rubber, which were enhanced after application of MeJA in this study. Our findings reveal an increase in the molecular weight of rubber following treatment with MeJA (Table 2 and Fig. 4). This heightened molecular weight in response to MeJA is likely attributed to the influence of MVA pathway enzymes, such as HMGR1 and HMGS1, which play a direct role in IPP production.
Genes such as SRPP1, CPT2, and HMGR1 could be involved in the regulation of Mw, as their expression levels responded to MeJA application. Previous studies have demonstrated the pivotal role of CPT, SRPP, and HMGR in regulating Mw across various rubber-producing plants33,43.
Mw uniformity is assessed by the polydispersity index54. Values of polydispersity were calculated through the division of the weight-averaged molecular weight by the number-averaged molecular mass (Mn). Elevated rate indicates either an uneven distribution of Mw or a blend of molecules of rubber varying in size55. In this investigation, all analyzed rubber samples exhibited a narrow Mw range (Table 2). These are consistent with the polydispersity value of 1.1 for rubber extracted from L. serriola determined by Bushman et al. (2015). It is commonly assumed that, while keeping other molecular features unchanged, reduced polydispersity indices lead to advantageous characteristics, including heightened tensile strength, softening point and impact strength56.
The FTIR analysis of NR from L. serriola in this study (Fig. 5) showed distinct chemical structures in the range of 500–4000 cm-1 associated with saturated aliphatic compounds, aligning with prior research methodologies57. Notably, in the spectral range of 1738 to 2850 cm-1, a weak band at 2726 cm-1 was observed, assigned to C-H stretching mode linked to aldehyde compounds58. The sharp peak at 1730 cm-1, indicating the symmetric stretching mode of C=O (amide), corroborated our observations and aligned with prior studies59. Peaks at 1642, 1461, and 1376 cm-1 corresponded to C=C stretching, CH3 symmetric deformation, and carboxylate functionalities, respectively consistent with earlier research52,60. The distinctive polyisoprene peak observed at 881 cm-1, associated with the C-H out-of-plane bending vibration of cis-1,4-polyisoprene units, aligns with the results reported by Aielo et al.61 which reported nearly the same results. Our FTIR results contribute to understanding NR composition, aligning with prior research60. However, due to lack of references for FTIR of L. serriola natural rubber, the polyisoprene peak wave number is little higher (881 cm−1) which could be the plant characteristic.
This research highlights the influence of MeJA in modulating gene expression in rubber biosynthesis pathway, consequently impacting both the quantity and quality of the produced rubber. Subsequent research is warranted to clarify the precise molecular processes behind these observed responses and their relevance in enhancing rubber production in rubber-producing plant species.
Conclusions
This study provides valuable insights into the potential application of MeJA as a strategic tool to enhance rubber yield in non-traditional rubber-producing plants, exemplified by L. serriola. The research effectively illustrates the modulatory effect of MeJA in influencing the expression of rubber biosynthesis genes and its consequential impact on both the quantity and quality of rubber in L. serriola. MeJA demonstrated a notable increase in the expression of critical rubber biosynthesis genes (HMGR1, HMGS1, CPT2, and SRPP1), resulting in a concurrent elevation of rubber yield and molecular weight. To deepen our understanding, future research should delve into the intricate molecular mechanisms underlying the MeJA-mediated upregulation of these genes.
Furthermore, it is essential to conduct thorough investigations into the long-term effects of MeJA treatment on both the rubber yield and the quality of the produced rubber. MeJA appears to be a promising tool with the potential to significantly enhance rubber yield in L. serriola by skillfully regulating the expression of key rubber biosynthesis genes. This research not only sheds light on the molecular intricacies involved but also unveils new pathways for improving rubber production in non-traditional rubber-producing plants. The implications of such advancements could potentially influence practices in the broader rubber industry and contributing to more sustainable rubber sourcing.
Acknowledgements
The authors would like to thank Urmia University and the University of Tehran for providing the research facilities. Moreover, this study was supported by the RUDN University Strategic Academic Leadership Program.
Author contributions
All authors contributed to the study conception and design. The initial idea was presented by MR.N. and M.A. The design of the experiments and the execution of the experiments were carried out by M.A and A.F., P.A and R.G. Data analysis was done by A.F and M. Z. M.A., MR.N. and A.F. wrote and edited the manuscript. All authors discussed the results and contributed to the final manuscript.
Data availability
The data that support the findings of this study are available within the paper. Any other supporting data are available from the corresponding author upon request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Salehi, M., Cornish, K., Bahmankar, M. & Naghavi, M. R. Natural rubber-producing sources, systems, and perspectives for breeding and biotechnology studies of Taraxacum kok-saghyz. Ind. Crops Prod.170, 113667. 10.1016/J.indcrop.2021.113667 (2021). [Google Scholar]
- 2.Van Beilen, J. B. & Poirier, Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol.25(11), 522–529. 10.1016/j.tibtech.2007.08.009 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Mooibroek, H. & Cornish, K. Alternative sources of natural rubber. Appl. Microbiol. Biotechnol.53, 355–365. 10.1007/s002530051627 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Cornish, K. Similarities and differences in rubber biochemistry among plant species. Phytochemistry57, 1123–1134. 10.1016/S0031-9422(01)00097-8 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Cornish, K. Biochemistry of natural rubber, a vital raw material, emphasizing biosynthetic rate, molecular weight and compartmentalization, in evolutionarily divergent plant species. Nat. Prod. Rep.18, 182–189. 10.1039/A902191D (2001). [DOI] [PubMed] [Google Scholar]
- 6.Mofidi, S. S. H. et al. Effect of drought stress on natural rubber biosynthesis and quality in Taraxacum kok-saghyz roots. PLoS ONE19(1), e0295694. 10.1371/journal.pone.0295694 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau, N. S. et al. The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci. Rep.6(1), 28594. 10.1038/srep28594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherian, S., Ryu, S. B. & Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J.17, 2041–2061. 10.1111/pbi.13181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita, S. & Takahashi, S. Molecular mechanisms of natural rubber biosynthesis. Annu. Rev. Biochem.89, 821–851. 10.1146/annurev-biochem-013118-111107 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Men, X., Wang, F., Chen, G. Q., Zhang, H. B. & Xian, M. Biosynthesis of natural rubber: Current state and perspectives. Int. J. Mol. Sci.20(1), 50. 10.3390/ijms20010050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman, B. S. et al. Identification and comparison of natural rubber from two Lactuca species. Phytochemistry67, 2590–2596. 10.1016/j.phytochem.2006.09.012 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Cornish, K., Castillón, J. & Scott, D. J. Rubber molecular weight regulation, in vitro, in plant species that produce high and low molecular weights in vivo. Biomacromolecules1, 632–641. 10.1021/bm000034z (2000). [DOI] [PubMed] [Google Scholar]
- 13.Bell, J. L., Burke, I. C. & Neff, M. M. Genetic and biochemical evaluation of natural rubber from Eastern Washington prickly lettuce (Lactucaserriola L.). J. Agric. Food. Chem.63, 593–602. 10.1021/jf503934v (2015). [DOI] [PubMed] [Google Scholar]
- 14.Brown, D. et al. Subcellular localization and interactions among rubber particle proteins from Hevea brasiliensis. J. Exp. Bot.68, 5045–5055. 10.1093/jxb/erx331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow, K. S. et al. Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J. Exp. Bot.63, 1863–1871. 10.1093/jxb/err363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi, S., Lee, H. J., Yamashita, S. & Koyama, T. Characterization of cis-prenyltransferases from the rubber producing plant Hevea brasiliensis heterologously expressed in yeast and plant cells. J. Plant Biotechnol.29(4), 411–417. 10.5511/plantbiotechnology.12.0625a (2012). [Google Scholar]
- 17.Tholl, D. Biosynthesis and biological functions of terpenoids in plants. In Biotechnology of Isoprenoids. Advances in Biochemical Engineering/Biotechnology Vol. 148 (eds Schrader, J. & Bohlmann, J.) 63–106 (Springer, 2015). [DOI] [PubMed] [Google Scholar]
- 18.Dong, G. et al. Transcriptome analysis of Taraxacum kok-saghyz reveals the role of exogenous methyl jasmonate in regulating rubber biosynthesis and drought tolerance. Gene867, 147346. 10.1016/j.gene.2023.147346 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Wasternack, C. & Hause, B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot.111(6), 1021–1058. 10.1093/aob/mct067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, J. P. et al. Transcriptome analysis of Hevea brasiliensis in response to exogenous methyl jasmonate provides novel insights into regulation of jasmonate-elicited rubber biosynthesis. Physiol. Mol. Biol. Plants24(3), 349–358. 10.1007/s12298-018-0529-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh, S. C., Othman, A. S. & Veera Singham, G. Identification and characterization of jasmonic acid-and linolenic acid-mediated transcriptional regulation of secondary laticifer differentiation in Hevea brasiliensis. Sci. Rep.9(1), 14296. 10.1038/s41598-019-50800-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian, W. M., Yang, S. G., Shi, M. J., Zhang, S. X. & Wu, J. L. Mechanical wounding-induced laticifer differentiation in rubber tree: An indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol.182, 95–103. 10.1016/j.jplph.2015.04.010 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Duan, C., Zeng, R. & Li, Y. Regulation of plant hormones on biosynthesis of natural rubber in Hevea brasiliensis. Chin. J. Trop. Agric.24(5), 61–68 (2004). [Google Scholar]
- 24.Kiani, H. S., Noudehi, M. S., Shokrpour, M., Zargar, M. & Naghavi, M. R. Investigation of genes involved in scent and color production in Rosa damascena Mill. Sci. Rep.14(1), 20576 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Cadavid, D. A. et al. Development of novel processes for the aqueous extraction of natural rubber from Taraxacum kok-saghyz (TK). J. Chem. Technol. Biotechnol.94(8), 2452–2464. 10.1002/jctb.6027 (2019). [Google Scholar]
- 26.Mehta, A. et al. In silico microRNA identification from Stevia rebaudiana transcriptome assembly. Eur. J. Med. Plants.15(2), 1–14. 10.9734/EJMP/2016/25221 (2016). [Google Scholar]
- 27.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120. 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods25(4), 402–408. 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Wu, C., Lan, L., Li, Y., Nie, Z. & Zeng, R. The relationship between latex metabolism gene expression with rubber yield and related traits in Hevea brasiliensis. BMC Genom.19, 1–18. 10.1186/s12864-018-5242-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan, C. et al. Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis. Tree Physiol.30(10), 1349–1359. 10.1093/treephys/tpq066 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Sirinupong, N., Suwanmanee, P., Doolittle, R. F. & Suvachitanont, W. Molecular cloning of a new cDNA and expression of 3-hydroxy-3- methylglutaryl-CoA synthase gene from Hevea brasiliensis. Planta221, 502–512. 10.1007/s00425-004-1463-7 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Tang, C. et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants2(6), 1–10. 10.1038/nplants.2016.73 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Post, J. et al. Laticifer-specific cis-prenyltransferase silencing affects the rubber, triterpene, and inulin content of Taraxacum brevicorniculatum. Plant Physiol.158(3), 1406–1417. 10.1104/pp.111.187880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du, P. et al. Genome-wide identification and characterization of the HMGR gene family in Taraxacumkok-saghyz provide insights into its regulation in response to ethylene and methyl Jsamonate treatments. Plants13(18), 2646. 10.3390/plants13182646 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epping, J. et al. A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants1(5), 1–9. 10.1038/nplants.2015.48 (2015). [Google Scholar]
- 36.Qu, Y. et al. A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem.290(4), 1898–1914. 10.1074/jbc.M114.616920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita, S. et al. Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. elife5, e19022. 10.7554/eLife.19022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran, X., Liu, Y. & Zhao, D. The relationship between EuSRPP1 gene expression and rubber biosynthesis in Eucommia ulmoides Oliver (Du-zhong). Ind. Crops Prod.175, 114246 (2022). [Google Scholar]
- 39.Schmidt, T. et al. Molecular cloning and characterization of rubber biosynthetic genes from Taraxacum koksaghyz. Plant Mol. Biol. Rep.28, 277–284. 10.1007/s11105-009-0145-9 (2010). [Google Scholar]
- 40.Wititsuwannakul, R., Rukseree, K., Kanokwiroon, K. & Wititsuwannakul, D. A rubber particle protein specific for Hevea latex lectin binding involved in latex coagulation. Phytochemistry69(5), 1111–1118. 10.1016/j.phytochem.2007.12.007 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Dai, L., Yang, H., Zhao, X. & Wang, L. Identification of cis conformation natural rubber and proteins in Ficus altissima Blume latex. Plant Physiol. Biochem.167, 376–384 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Singh, A. P., Wi, S. G., Chung, G. C., Kim, Y. S. & Kang, H. The micromorphology and protein characterization of rubber particles in Ficus carica, Ficus benghalensis and Hevea brasiliensis. J. Exp. Bot.54(384), 985–992. 10.1093/jxb/erg107 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Collins-Silva, J. et al. Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry79, 46–56. 10.1016/j.phytochem.2012.04.015 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Chakrabarty, R., Qu, Y. & Ro, D. K. Silencing the lettuce homologs of small rubber particle protein does not influence natural rubber biosynthesis in lettuce (Lactuca sativa). Phytochemistry113, 121–129. 10.1016/j.phytochem.2014.12.003 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Balbuena, T. S., Salas, J. J., Martinez-Force, E., Garces, R. & Thelen, J. J. Proteome analysis of cold acclimation in sunflower. J. Proteome Res.10, 2330–2346. 10.1021/pr101137q (2011). [DOI] [PubMed] [Google Scholar]
- 46.Ruan, J. et al. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci.20(10), 2479. 10.3390/ijms20102479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing, B. et al. Phenolic acid production is more effectively enhanced than tanshinone production by methyl jasmonate in Salvia miltiorrhiza hairy roots. Plant Cell Tissue Organ Cult.134, 119–129. 10.1007/s11240-018-1405-x (2018). [Google Scholar]
- 48.Yousefian, S., Lohrasebi, T., Farhadpour, M. & Haghbeen, K. Effect of methyl jasmonate on phenolic acids accumulation and the expression profile of their biosynthesis-related genes in Mentha spicata hairy root cultures. Plant Cell Tissue Organ Cult.142(2), 285–297. 10.1007/s11240-020-01856-9 (2020). [Google Scholar]
- 49.Wei, Q. et al. Transcriptome analysis reveals regulation mechanism of methyl jasmonate-induced terpenes biosynthesis in Curcuma wenyujin. PLoS ONE17(6), e0270309. 10.1371/journal.pone.0270309 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng, X. et al. Jasmonate signaling in the regulation of rubber biosynthesis in laticifer cells of rubber tree, Hevea brasiliensis. J. Exp. Bot.69, 3559–3571. 10.1093/JXB/ERY169 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Florez-Velasco, N., Ramos, V. F., Magnitskiy, S., & Balaguera-López, H. Ethylene and jasmonate as stimulants of latex yield in rubber trees (Hevea brasiliensis): molecular and physiological mechanisms. A systematic approximation review. Advanced Agrochem (2024).
- 52.Saeedi, F., Naghavi, M. R., Sabokdast, M. & Jariani, P. Taraxacum kok-saghyz LE Rodin, as a novel potential source of natural rubber in Iran: A good candidate for commercial use. Iran. Polym. J.32(10), 1257–1269. 10.1007/s13726-023-01204-6 (2023). [Google Scholar]
- 53.Swanson, C. L., Buchanan, R. A. & Otey, F. H. Molecular weights of natural rubbers from selected temperate zone plants. J. Appl. Polym. Sci.23(3), 743–748. 10.1002/app.1979.070230309 (1979). [Google Scholar]
- 54.Salehi, M., Bahmankar, M., Naghavi, M. R. & Cornish, K. Rubber and latex extraction processes for Taraxacum kok-saghyz. Ind. Crops Prod.178, 114562. 10.1016/j.indcrop.2022.114562 (2022). [Google Scholar]
- 55.Wood, D. F. & Cornish, K. Microstructure of purified rubber particles. Int. J. Plant Sci.161(3), 435–445. 10.1086/314269 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Agrawal, S. L. et al. Mathematical correlation of polydispersity using gel permeation chromatography and rubber process analyzer for raw rubbers. J. Elastomers Plast.38, 31–41. 10.1177/0095244306054684 (2006). [Google Scholar]
- 57.Fernández-Berridi, M. J., González, N., Mugica, A. & Bernicot, C. Pyrolysis-FTIR and TGA techniques as tools in the characterization of blends of natural rubber and SBR. Thermochim. Acta444(1), 65–70. 10.1016/J.TCA.2006.02.027 (2006). [Google Scholar]
- 58.Rosli, N. A., Ahmad, I., Anuar, F. H. & Abdullah, I. Mechanical and thermal properties of natural rubber-modified poly (lactic acid) compatibilized with telechelic liquid natural rubber. Polym. Test54, 196–202. 10.1016/J.POLYMERTESTING.2016.07.021 (2016). [Google Scholar]
- 59.Kumutha, K., Alias, Y. FTIR spectra of plasticized grafted natural rubber–LiCF3SO3 electrolytes. Spectrochim. Acta, Part A: Molecular and Biomolecular Spectroscopy, 64(2), 442–447 (2006). 10.1016/j.saa.2005.07.044 [DOI] [PubMed]
- 60.Ali, A. M. M. et al. Grafted natural rubber-based polymer electrolytes: ATR-FTIR and conductivity studies. Ionics14, 491–500. 10.1007/s11581-007-0199-3 (2008). [Google Scholar]
- 61.Aielo, P. B. et al. Evaluation of sodium diclofenac release using natural rubber latex as carrier. Mater. Res.17, 146–152 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within the paper. Any other supporting data are available from the corresponding author upon request.