Abstract
Alzheimer's disease (AD) is the most common cause of late-life dementia characterized by progressive neurodegeneration and brain deposition of amyloid-β (Aβ) and phosphorylated tau. The APOE ε2 encoding apolipoprotein E (APOE2) is a protective allele against AD among the three genotypes (APOE ε2, ε3, ε4), while APOE4 is the strongest genetic factor substantially increasing AD risk. APOE regulates brain lipid homeostasis and maintaining synaptic plasticity and neuronal function, where APOE2 has a superior function compared to APOE3 and APOE4. Gene therapy that increases APOE2 levels in the brain is, therefore, a promising therapeutic strategy for AD treatment. We previously reported that PEGylated liposomes conjugated with transferrin and a cell-penetrating peptide Penetratin sufficiently deliver chitosan-APOE2 cDNA plasmid complex into the brain of wild-type mice. Here, we investigated how brain-targeting liposome-based APOE2 gene delivery influences Aβ-related pathologies in amyloid model AppNL-G-F knockin mice at 12-month-old. We found a trend of reductions of insoluble Aβ levels in the mouse cortices 1 month after APOE2 gene therapy. Furthermore, in the AppNL−G-F knockin mice that received the APOE2 gene therapy, brain transcriptome analysis through RNA-sequencing identified the upregulation of genes/pathways related to neuronal development. This was supported by increases of Dlg4 and Syp mRNAs coding synaptic proteins in the experimental group. On the other hand, we found that APOE2 gene delivery increased soluble Aβ levels, including oligomers, as well as exacerbated neurite dystrophy and decreased synaptophysin. Together, our results suggest that brain-targeting liposome-based APOE2 gene therapy is potentially beneficial for synaptic formation at the transcriptional level. Forced APOE2 expressions, however, may exacerbate Aβ toxicity by increasing the dissociation of Aβ oligomers from aggregates in the presence of considerable amyloid burden.
Keywords: Alzheimer's disease, Dystrophic neurites, APOE2, Gene therapy, Nanoparticles
Highlights
-
•
Brain-targeting liposome-based APOE2 gene therapy increases soluble amyloid-β oligomers in AppNL−G-F mice.
-
•
Brain-targeting liposome-based APOE2 gene therapy exaggerates dystrophic neurite formation in AppNL−G-F mice.
-
•
Brain-targeting liposome-based APOE2 gene therapy upregulates genes related to neuronal formation in AppNL−G-F mice.
-
•
Abstract (275 words).
1. Introduction
Among the three human APOE genotypes (ε2, ε3, ε4) coding responsible each apolipoprotein E (APOE) isoform, carrying the APOE ε2 allele (APOE2) is protective for Alzheimer's disease (AD), while APOE4 substantially increases the risk of AD [[1], [2], [3]]. Though brain accumulation and deposition of amyloid-β (Aβ) peptides cleaved from amyloid precursor protein (APP) is a central defining factor of AD [4,5], APOE has been shown to contribute to AD pathogenesis through multiple pathogenic mechanisms [6]. Of note, APOE2 is associated with mitigated amyloid pathology compared with APOE3 and APOE4 [7]. Furthermore, APOE2 has been shown to prevent age-related cognitive decline among non-demented individuals regardless of Aβ deposition [8]. Thus, enhancing APOE2 expression and function in the brain might be an effective approach to treat AD and AD-related conditions. Indeed, APOE2 gene delivery through intracerebral adeno-associated virus (AAV) injection ameliorates brain amyloid pathology in PDAPP mice with murine Apoe background [9] and APP/PS1 mice with APOE4 [10]. Although gene therapy provides an excellent tool for treating various neurodegenerative diseases including AD, the clinical application of AAV as a delivery agent is impeded by the associated immune response and cytotoxicity as well as relatively less delivery efficiency into the central nervous system [11]. There is a need to establish non-viral approaches which allow more safe and efficient gene delivery into the brain. Thus, we utilized functionalized liposomes conjugated with polyethylene glycol (PEG), transferrin, and a cell-penetrating peptide Penetratin to establish gene delivery across the blood-brain barrier (BBB). Liposome surface modification with PEG enhances liposome concentration in circulation by reducing its non-specific interactions with the vascular endothelium and serum proteins [12,13]. Conjugation of transferrin on the liposomal surface facilitates liposome delivery into the brain, as transferrin receptor is abundant in cerebral capillary endothelial cells [14]. Penetratin-conjugated liposome enhances the translocation of liposome across the cellular membrane [15,16]. By encapsulating chitosan-APOE2 cDNA plasmid complexes into the functionalized liposomes, we have reported successful gene delivery and protein expression of APOE2 in mouse brains through intravenous injection of liposomes [17]. In this study, by using amyloid model AppNL−G-F knockin mice, we investigated the potential therapeutic effect of liposome-based APOE2 gene delivery into the brain on AD-related phenotypes [18].
2. Materials and methods
2.1. Materials
DOTAP and DOPE were purchased from Cayman Chemical (Michigan, USA, 15110 and 15091, respectively). Holo-Tf was purchased from Bio Vision incorporated (CA, USA, 7542-100) and Penetratin was purchased from Life Tein (New Jersey, USA, LT8103). Generation of Pen-Tf liposomes containing cDNA plasmids were described in “Preparation of lipid nanoparticles”. Antibodies are used in this manuscript are described in each section of “Methods”. Cholesterol assay and Prussian blue staining were purchase from Abcam (ab65390 and ab150674, respectively).
2.2. Preparation of lipid nanoparticles
GFP and APOE2 cDNA plasmids were incorporated into DSPE-PEG-liposomes conjugated with Penetratin (Pen) (RQIKIWFQNRRMKWKK) and holo-transferrin (Tf) according to our previous report [16]. Briefly, 45 mol % of DOTAP, 45 mol % of DOPE, 4 mol % of DSPE-PEG-Pen, and 2 mol % of cholesterol were solubilized in chloroform:methanol in 2:1 (v/v) ratio and subsequently dried using a rotavapor. Next, the thin-film was hydrated with HEPES buffer (pH 7.4) containing cDNA plasmids with chitosan (N:P = 5:1) and sonicated for 15 min to procure Pen-liposomes. Following this, DSPE-PEG-Tf (4 mol %) micelles were incorporated into Pen-liposomes by stirring overnight. The resulting Pen-Tf liposomes containing cDNA plasmids was then purified through a Sephadex G-100 column. Lyophilized liposomes were resuspended in HEPES buffer (pH 7.4) and sonicated for 30 min at room temperature prior to their use.
2.3. Animals
Both male and female homozygous AppNL−G-F knockin (AppNL−G-F) mice provided by Drs. Saito and Saido at RIKEN [18] were utilized in the experiments. The liposomes containing GFP or APOE2 cDNA-chitosan complex (15.2 μM liposome and 1 mg plasmid/kg body weight) were injected into AppNL−G-F mice through tail vein at 12-month-old [16]. Six males and seven females were injected with the APOE2 cDNA liposomes, and 10 mice (4 males and 6 females) were injected with the GFP cDNA liposomes as controls. The mice were subjected to behavioural tests one month after the injection, followed by sample collections for further biochemical experiments after being euthanized. The mice were housed in a 12-h light-dark cycle at 25 °C according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All animal experiments were approved by the Mayo Clinic Institutional Animal Care and Use Committee (A00001911-16-R22).
2.4. Neurobehavior tests
Fear conditioning test: The test was performed in a sound-attenuated chamber containing a grid floor, which delivers electric shocks. Freezing caused by the electric shocks was captured by an overhead camera. Mouse behavior was measured by the FreezeFrame software (Actimetrics). Mice stayed in the chamber without disturbance for 2 min, which and the baseline of freezing behavior was recorded. After recording the baseline, an 80-dB white noise, serving as the conditioned stimulus (CS), was given for 30 s. At the final 2 s of the noise, the experimental mouse received an electric shock (0.5 mA) on its feet, serving as the unconditioned stimulus (US). Another CS–US pair was given to the mouse after 1 min resting. The mouse was removed from the chamber 30 s later after the second CS-US pair. For contextual learning, the experimental mice were placed back into the training context 24 h after training and freezing was scored for 5 min. For cued learning, the mice were placed in different contexts, including novel odor, cage floor and visual cues, for 3 min. After that the auditory CS was provided and freezing of mice was recorded for 3 min. To avoid animal variability, the final scores of the context and cued tests were subtracted baseline of freezing [8,19].
Elevated plus maze test: Mice were placed in an apparatus composed of two opposing open arms and two opposing closed arms (50 × 10 cm). The closed arms equipped with 40 cm roofless gray walls that was connected by a central square platform. This apparatus was placed 50 cm above the ground. Mice were placed in the open arm, facing towards another open arm. The moving tracks were captured by an overhead camera for 5 min and analyzed by AnyMaze software [8,19].
Open field test: To test the exploratory behavior and anxiety, the mice were put in an open field chamber (40 × 40 × 30 cm) and monitored for 15 min. A 20 × 20 cm region was digitally designated as the large center. The track movement in this open field was captured by an overhead camera, and the distance in the center and total distance were analyzed by AnyMaze software [8,19].
2.5. ELISA
Mouse cortical samples were sequentially extracted in Tris-buffered saline (TBS), 1 % Triton X in TBS (TBS-X), and Guanidine hydrochloride (GDN-HCl) [20]. All the extraction buffers contained protease and phosphatase inhibitors. Human Aβ40 or Aβ42 levels in each fraction were measured using in-house ELISAs, in which mAb 2.1.3 or mAb 13.1.1 antibody was used as the capturing antibody for human Aβx-42 or Aβx-40, respectively, and mAb Ab5 (human Aβ1-16–specific) conjugated with HRP (ab274112, Abcam) was used as detection antibody [20]. For human APOE measurement, anti-human APOE monoclonal WUE-4 and anti-human APOE (K74180B; Meridian Life Sciences) conjugated biotin antibodies were used as capturing and detection antibody, respectively [21]. The ELISA plates were developed with 3,3′,5,5′-tetramethylbenzidine substrate (T5569, Sigma-Aldrich) and 85 % O-phosphoric acid was used to stop the reaction. Colorimetric quantification was recorded at 450 nm by a Synergy HT plate reader (BioTek Instruments Inc). The values in each sample were normalized to their protein concentrations determined by a BCA kit (23225, Thermo scientific).
2.6. Cholesterol measurement
Total cholesterol, HDL, and LDL/VLDL amounts were measured in the mouse liver following the manufacturer's protocol (ab65390, Abcam). Liver was homogenized in cholesterol assay buffer and centrifuged for 10 min at 4 °C, 13,000 g. Supernatant was the total cholesterol and transferred to a new tube. Pellet was resuspended in precipitation buffer and incubated for 10 min at room temperature, followed by centrifugation (2000 g for 10 min) at 4 °C. The supernatant and the pellet resuspended in PBS represent the HDL fraction and LDL/VLDL fraction, respectively. Duplicated standard and samples were loaded onto a 96-well plate. Each well was incubated with cholesterol reaction mix for 60 min at 37 °C. Colorimetric quantification was determined at 570 nm by a Synergy HT plate reader (BioTek Instruments Inc).
2.7. Immunohistochemistry
Mouse hemispheres were embedded in paraffin after the fixation with 10% formalin. The brain slices (5-μm thickness) were incubated with the following primary antibodies overnight at 4 °C: anti-GFAP (Pu020-UP, BioGenex), anti-Iba1 (019–19741, FUJIFILM Wako), and anti–pan-Aβ 33.1.1 (human Aβ1-16–specific) antibody. The sections were incubated with secondary antibodies (ABC kits, PK-6101 or PK-6102, Vector Laboratories) for 30 min at room temperature, and developed using 3, 3 -diaminobenzidine (DAB, SK-4100, Vector Laboratories). The images were scanned by an Aperio Slide Scanner (Leica Biosystems). The positivity of staining was quantified using Aperio ImageScope software (Leica Biosystems).
For immunofluorescent staining, samples were incubated with the following primary antibodies overnight at 4 °C: anti-fibrinogen antibody (ab34269, Abcam), anti-pan Aβ (MOAB2) antibody (MABN254, Sigma-Alrich), and/or anti-human APOE antibody (#13366, Cell Signaling Technology). The sections were incubated with secondary antibodies (A-11011 or A-31553, Thermo Fisher) for 1 hour at room temperature. To assess neurite dystrophy, the paraffin sections were sectioned in 15-μm thickness. The samples were stained with X34 (SML1954, Sigma-Aldrich) in 40 % EtOH/PBS at room temperature for 20 min followed by incubation with anti-LAMP1 (ab25245, Abcam) at 4 °C overnight. The sections were incubated with secondary antibodies (A11077, Invitrogen) for 1 h at room temperature. Images were captured by the Zeiss LSM 880 confocal microscope (ZEISS) and quantified by ImageJ software.
2.8. Prussian blue staining
By following the manufacturer's protocol (ab150674, Abcam), paraffin sections were deparaffined and rehydrated, followed by incubation in the iron stain buffer for 3 min and risen in water. Sections were counterstained in nuclear fast red for 2 min, dehydrated in alcohol, mounted using resin, and scanned by the Aperio Slide Scanner (Leica Biosystems). For positive staining control, mouse spleen was used in this study.
2.9. Western blot
Mouse cortical samples in TBS and TBSX fractions were used for Western blot analysis. Proteins were applied onto a 4–20 % precast polyacrylamide gel (5671095, Bio-Rad) and then transferred onto the PVDF membranes (1620177, Bio-Rad). The membranes were incubated with the following primary antibodies after blocking in 5 % milk: mouse APOE (#68587, Cell Signaling Technology), PSD95 (ab18258, Abcam), SNAP25 (PA5-85396, Thermo Fisher), Synaptophysin (ab16659, Abcam), α-tubulin (62204, Thermo Fisher), and 4G8 (800702, Biolegend) overnight at 4 °C. The membranes were incubated with secondary HRP-conjugated anti-mouse or rabbit antibodies (ab 6728 or ab6721, Abcam). The immunoreactivities were detected by SuperSignal Substrate (34095, Thermo Fisher) through a ChemiDoc imaging system (Bio-Rad) and quantified by ImageJ software.
2.10. Reverse transcription-quantitative PCR (RT-qPCR)
Mouse right cortical samples were subjected to total RNA extraction using a RNeasy Mini Kit (74106, QIAGEN). The cDNA was synthesized using SuperScript III reverse transcriptase (18080051, Invitrogen). Primer sequences in this study were as follows: 1) Actb-forward: 5′-GTGACGTTGACATCCGTAAAGA, Actb-reverse: 5′-GCCGGACTCATCGTACTCC; 2) Dlg4-forward: TCCGGGAGGTGACCCATTC, Dlg4-reverse: 5′-TTTCCGGCGCATGACGTAG; 3) Syp-forward: 5′-AGACATGGACGTGGTGAATCA, Syp-reverse: 5′-ACTCTCCGTCTTGTTGGCAC; 4) Snap25-forward: 5′-GCAAGGCGAACAACTGGAAC, Snap25-reverse: 5′-GGCCACTACTCCATCCTGATT; 5) Mgst1-forward: 5′-CTCAGGCAGCTCATGGACAAT, Mgst1-reverse: 5′- GTTATCCTCTGGAATGCGGTC; 6) Acadl-forward: 5′- TCTTTTCCTCGGAGCATGACA, Acadl-reverse: 5′- GACCTCTCTACTCACTTCTCCAG. Real-time PCR was performed with SYBR Green Supermix (1725271, Bio-Rad). Relative gene expression levels were quantified by the 2−ΔΔCT method normalized to Actb expression.
2.11. Brain transcriptome profiling
RNA-sequencing (RNAseq): Total RNA was extracted from mouse cortex by using a RNeasy Mini Kit (74106, QIAGEN). The RNA quantity and quality were measured by an Agilent 2100 bioanalyzer. A total of 23 samples were used for RNAseq using Illumina HiSeq 4000 at the Mayo Clinic. To generate raw gene counts, the MAP-RSeq (Mayo Clinic RNA-sequencing analytic pipeline) was used by mapping to the mouse genome mm10 [22]. To address gene length differences and achieve similar quantile-by-quantile distributions of gene expression across samples, the conditional quantile normalization (CQN) on the raw gene counts was applied [23]. Based on the bimodal distribution of CQN-normalized and log2-RPKM gene expression values, the detection threshold was defined by genes with an average log2 RPKM more than1 in at least one group.
Weighted correlation network analysis (WGCNA): WGCNA was performed using CQN-normalized and log2-transformed RPKM values adjusted for sex and RIN [24]. To establish a scale-free topology using signed network, the power of 7 was selected based on the relationship between power and scale independence. Hybrid dynamic tree cutting was used with a minimum module size of 60 genes and a minimum height of 0.2 for merging modules. Module was summarized by ME and assigned a unique color identifier. Modules were annotated using WGCNA R function anRichment. Gene-gene connections among top-ranked genes with high connectivity were regarded as hub genes that are visualized using VisANT. Gene-gene interactions among genes involved in gene ontologies and hubgenes in the tan module were hypothesized using GeneMANIA.
Gene set enrichment analysis (GSEA): Gene functional enrichment analysis was conducted using the camera function in limma package [25] to screen for ‘enriched’ MSigDB GO annotation gene set. The top 10 downregulated or upregulated GO terms were visualized using R (version 3.6.3).
2.12. Statistical analyses
GraphPad Prism software or R (version 3.6.3) was used for all statistical analyses. Quantitative data distributions were examined, and outliers were excluded in statistical analyses. Mean values for each group were compared by two-way Analysis of variance (ANOVA) after adjusting for sex or the two-tailed unpaired Student t-test. A p < 0.05 was considered statistically significant.
3. Results
3.1. Altered anxiety-like behavior after liposome-based APOE2 gene delivery in AppNL−G-F mice
To determine how brain APOE2 expression influences cognitive function, we intravenously injected the functionalized liposomes containing APOE2 cDNA plasmids into 12-month-old AppNL−G-F mice, then performed neurobehavior tests one month after the injection. The liposomes containing GFP cDNA plasmids were used as a control. We confirmed human APOE protein expression in the GDN fraction of mouse brains treated with the APOE2 cDNA-conjugated liposomes while human APOE was faintly detected in the brain by immunostaining (Supplementary Figs. 1A–B). There was no evident effect on endogenous levels of mouse APOE protein in the brain (Supplementary Fig. 1C). The contextual fear conditioning test found that freezing time increased in male mice from the APOE2 group, but this was not observed in the female mice (Fig. 1A). No evident changes between the APOE2 and control GFP groups were observed in the cued fear conditioning test (Fig. 1B) or open field test (Fig. 1C). The mice in the APOE2 group stayed for a longer time on open arms compared to the controls in the elevated plus maze test (Fig. 1D). These findings indicate that brain APOE2 gene delivery ameliorates fear learning and memory performance, and anxiety-related phenotype, although further studies are necessary due to the relatively small cohort size.
Fig. 1.
APOE2 gene delivery alters anxiety-like behavior in AppNL-G-Fmice. 12-month-old AppNL−G-F mice were treated with lipid nanoparticles incorporating GFP (circle) or APOE2 (square) cDNA plasmids and subjected to neurobehavior analyses one month after treatment. The percentage of time with freezing behavior in the contextual (A) and cued (B) fear conditioning (FC) tests, the large center to total distance ratio (C) in the open filed assay (OFA), and the ratio of the time spent in open arms to closed arms in the elevated plus maze (EPM) test (D) are shown. Closed and open symbols indicate male and female mice, respectively. Outliers based on quantitative data distributions were excluded in statistical analyses. Data are shown as mean ± SEM (N = 9–15/group). ∗p-value <0.05; ∗∗ p-value <0.01 by two-way ANOVA or two-tailed student's t-test. (A) t = 2.353, df = 9, p = 0.043; (D) F = 9.573, p = 0.006; t = 2.769, df = 10, p = 0.020.
3.2. Increased soluble Aβ in AppNL−G-F mice after the liposome-based APOE2 gene delivery
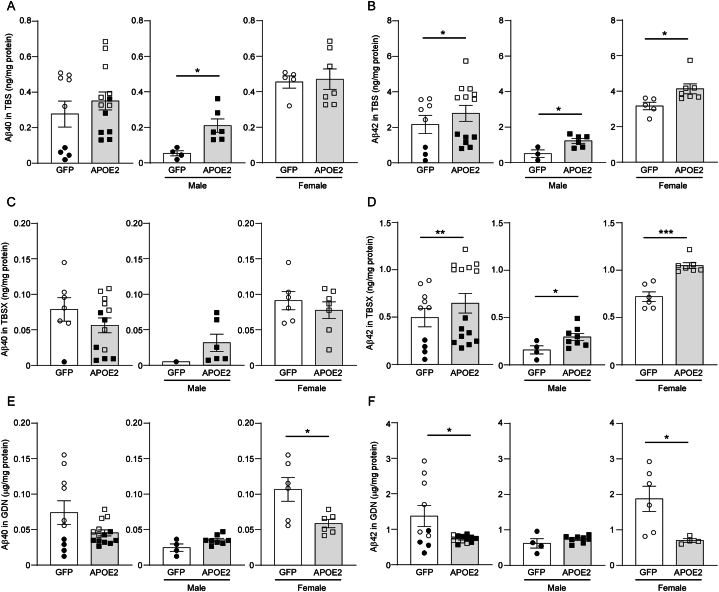
We next examined how APOE2 expression influences Aβ accumulation in the mouse brains (Fig. 2A–F). ELISA showed increased soluble Aβ40 levels in the TBS fraction of cortical samples from male AppNL−G-F mice that received the APOE2 cDNA liposomes (Fig. 2A). However, higher soluble Aβ42 levels in TBS (Fig. 2B) and TBSX fractions (Fig. 2D) were detected in both male and female mice in the APOE2 group than those in the controls. On the other hand, insoluble Aβ40 (Fig. 2E) and Aβ42 levels (Fig. 2F) in GDN fraction decreased in female mice in the APOE2 group. These findings imply that brain APOE2 gene delivery disaggregates insoluble Aβ into soluble Aβ. Consistently, we also found an increase in TBS-soluble Aβ oligomers in the AppNL−G-F mouse brains treated with the APOE2 cDNA liposomes by Western blotting (Supplementary Fig. 2). Immunohistochemical analysis using pan-Aβ antibody did not detect evident differences in the brain parenchymal Aβ depositions between the two groups. Similarly, no significant difference of cerebral amyloid angiopathy (CAA) in the leptomeningeal arteries was observed between two groups (Supplementary Fig. 3). When BBB leakage and brain hemorrhage were assessed by immunostaining for fibrinogen and Prussian blue staining, respectively, we did not detect evident deleterious effects of the administrations on those cerebrovascular phenotypes (Supplementary Fig. 4), although APOE2 is associated with the risk of CAA and CAA related hemorrhage [2]. Immunoreactivities for neurons (NeuN), astrocytes (GFAP), and microglia (Iba1) were also not affected by brain APOE2 gene delivery in AppNL−G-F mice (Supplementary Fig. 5). While AAV-based APOE2 gene delivery has been shown to reduce microgliosis [10], our liposome-based approach may have modest effects on glial activation.
Fig. 2.
APOE2 gene delivery increases soluble Aβ in AppNL-G-Fmice. 12-month-old AppNL−G-F mice were treated with lipid nanoparticles incorporating GFP (circle) or APOE2 (square) cDNA plasmids. Aβ40 and Aβ42 were sequentially extracted in TBS (A, B), TBS-X (C, D), and GDN-HCl (E, F) from mouse cortices one month after treatment, then measured by ELISA and normalized to the protein concentrations in each fraction. Closed and open symbols indicate male and female mice, respectively. Quantitative data distributions were examined, and outliers were excluded in statistical analyses. Data are shown as mean ± SEM (N = 7–15/group). ∗p-value <0.05; ∗∗ p-value <0.01; ∗∗∗p-value <0.001 by two-way ANOVA or two-tailed student's t-test. (A) t = 3.223, df = 8, p = 0.012; (B) F = 6.682, p = 0.019∗, t = 2.957 (male) and 2.524 (female), df = 7 (male) and 10 (female), p = 0.012 (male) and 0.030 (female). (D) F = 11.38, p = 0.003, t = 2.260 (male) and 5.778 (female), df = 10 (male) and 11 (female), p = 0.047 (male) and 0.0001 (female); (E) t = 2.728, df = 10, p = 0.021; (F) t = 2.610, df = 8, p = 0.031.
3.3. Exaggerated dystrophic neurites after liposome-based APOE2 gene delivery in AppNL−G-F mice
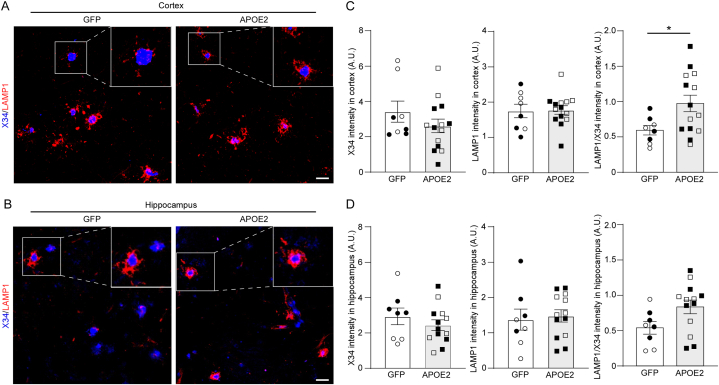
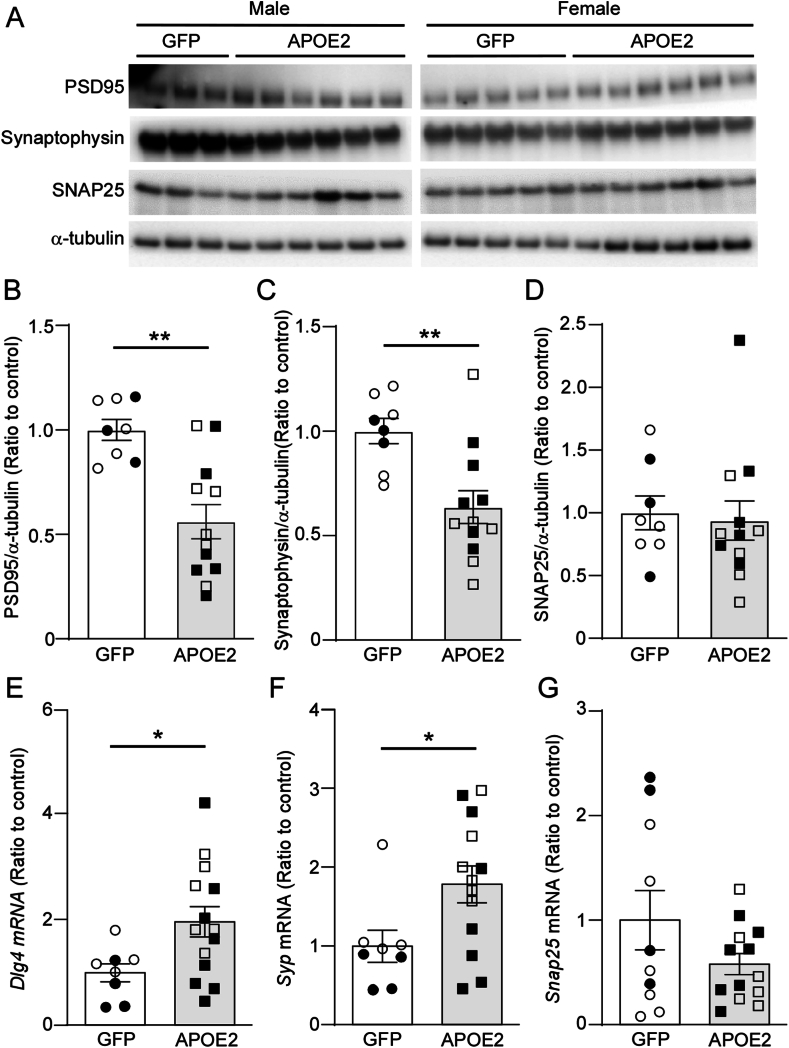
To determine the effect of brain APOE2 expression on neurons, neurite dystrophies were assessed by co-staining with LAMP1 antibody and amyloid specific X34 dye in brain samples from the AppNL−G-F mice. We found that LAMP1 fluorescence intensity normalized by X34 intensity was increased in the cortices from the APOE2 group by comparing to the controls (Fig. 3A–D). This suggests that neurite dystrophy was exacerbated by APOE2 expression in the brain. In addition, Western blotting also detected decreased synaptophysin and PSD95 protein levels in the cortical samples from mice in APOE2 group, whereas there were no differences in SNAP25 protein level between the groups (Fig. 4A–D). Conflictingly, mRNA levels of synaptophysin (Syp) and PSD95 (Dlg4), but not SNAP25 (Snap25), were upregulated by APOE2 cDNA liposome administration in the mouse brains (Fig. 4E–G).
Fig. 3.
APOE2 gene delivery exaggerates neuritic dystrophy in AppNL-G-Fmice. 12-month-old AppNL−G-F mice were treated with lipid nanoparticles incorporating GFP (circle) or APOE2 (square) cDNA plasmids. One month after treatment, the brain sections were stained with LAMP1 antibody for dystrophic neurites and with X34 for amyloid plaque in the cortex (A) and hippocampus (B). Scale bar: 20 μm. The immunoreactivities for X34 and LAMP1, and LAMP1/X34 ratio in the cortex (C) and hippocampus (D) were quantified. Closed and open symbols indicate male and female mice, respectively. Quantitative data distributions were examined, and outliers were excluded in statistical analyses. Data are shown as mean ± SEM (N = 8–14/group). ∗p-value <0.05 by two-way ANOVA, F = 5.326, p = 0.032.
Fig. 4.
APOE2 gene delivery reduces synaptic synaptophysin in AppNL-G-Fmice. 12-month-old AppNL−G-F mice were treated with lipid nanoparticles incorporating GFP (circle) or APOE2 (square) cDNA plasmids. (A–D) One month after treatment, protein amounts of PSD95 (B), synaptophysin (C), and SNAP25 (D) in mouse cortices were measured by Western blotting. The mRNA expression levels of corresponding genes, Dlg4 for PSD95 (E), Syp for synaptophysin (F), and Snap25 (G), were measured by RT-qPCR. Closed and open symbols indicate male and female mice, respectively. Quantitative data distributions were examined, and outliers were excluded in statistical analyses. Data are shown as mean ± SEM (N = 7–13/group). ∗p-value <0.05 and ∗∗p-value <0.01 by two-way ANOVA. (B) F = 15.648, p = 0.001; (C) F = 10.658, p = 0.005; (E) F = 6.180, p = 0.022; (F) F = 6.165, p = 0.023.
3.4. Altered brain transcriptomes after liposome-based APOE2 gene delivery in AppNL−G-F mice
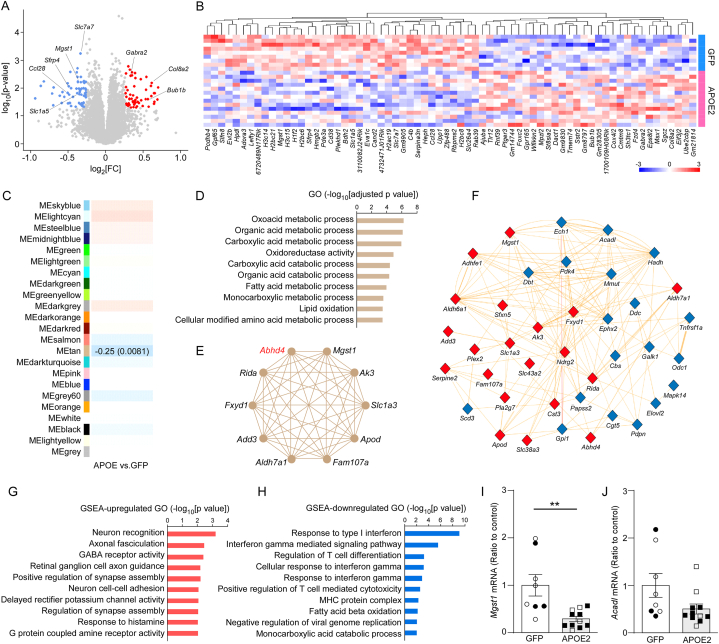
RNA-seq in the cortical samples identified 39 downregulated and 30 upregulated protein-coding differentially expressed genes (DEGs) by APOE2 cDNA liposome administration (adjusted p value < 0.05, |fold change| ≥ 1.2), such as Mgst1, Gabra2, and Sfrp4 (Fig. 5A–B). In addition, the weighted gene correlation network analysis (WGCNA) identified one significantly downregulated module (MEtan). Gene ontology (GO) revealed that MEtan was enriched for genes related to metabolic process pathways such as “oxoacid metabolic process,” “organic acid metabolic process,” and “catabolic acid metabolic process.” Top 20 hub genes in MEtan included Mgst1 and Acadl (Fig. 5C–F). To further investigate coordinated changes in gene ontology terms, gene set enrichment analysis (GSEA) was also performed. The upregulated GO terms were enriched for neuronal activities such as “neuron recognition,” “axonal fasciculation,” and “GABA receptor activities.” On the other hand, interferon gamma-related pathways were downregulated in the GSEA analysis. Consistently, “fatty acid beta oxidation” was also identified as one of the downregulated GO by GSEA analysis (Fig. 5G–H). We validated the downregulations of Mgst1 and Acadl mRNA levels by RT-qPCR (Fig. 5I–J).
Fig. 5.
APOE2 gene delivery modulates brain transcriptome in AppNL-G-Fmice. 12-month-old AppNL−G-F mice were treated with the lipid nanoparticles incorporating GFP or APOE2 cDNA plasmids, and the cortical brain samples were subjected to bulk RNA-seq one month after treatment. Volcano plot (A) and heat map (B) for the DEGs are shown. (C–F) WGCNA was performed in the RNA-seq. Correlations of module eigengenes (MEs) with the liposome administration are shown. Values in the heat map are the Pearson correlation coefficient and P value (C). Gene ontology (GO) pathways (D) and network plot of the hub genes with high intramodular connectivity (E). Interaction of genes involved in the gene ontologies (blue nodes) and hub genes (red nodes) (F) in MEtan are shown. (G, H) Gene set enrichment analysis (GSEA) identified the upregulated (G) and downregulated (H) GO pathways. (I, J) The mRNA expression levels of Mgst1 (I) and Acadl (J) were measured by RT-qPCR. Closed and open symbols indicate male and female mice, respectively. Quantitative data distributions were examined. Data are shown as mean ± SEM (N = 9–14/group). ∗∗p-value <0.01 by two-way ANOVA, F = 11.719, p = 0.003.
4. Discussion
The development and progression of AD is substantially affected by APOE genotypes [[1], [2], [3]], where carrying APOE2/2 and APOE4/4 is the lowest and highest risk for Alzheimer's dementia respectively (APOE2/2 < 2/3 < 3/3 < 2/4 < 3/4 < 4/4) [26]. Neuropathological phenotypes represented by CERAD score and Braak stage were also eased in the presence of APOE2 [26]. Gene therapy that increases brain APOE2 levels is an attractive therapeutic approach to treat AD. Several mouse experiments have shown that forced expression of APOE2 by AAV or lentivirus-based gene delivery systems reduce both soluble and insoluble Aβ levels in the brains of human APP transgenic amyloid mouse models [9,27,28]. Indeed, the clinical trial (NCT03634007) to assess safety and toxicity of intrathecal administration of AAV expressing APOE2 cDNA (LX1001) is ongoing in AD cases with APOE4 homozygotes without serious adverse events. However, inconsistent with these reports, our study found that liposome-based APOE2 cDNA plasmid gene delivery upregulates soluble Aβ42 levels in AppNL−G-F mouse brains accompanied with exacerbated neurite dystrophy and reduced synaptic proteins. While soluble Aβ oligomers were increased, there was a trend of decreases in insoluble Aβ levels after APOE2 cDNA plasmid delivery, particularly in female AppNL−G-F mice. APOE has been shown to interact with Aβ oligomers [29], APOE2 expression may stabilize Aβ oligomers and prevent their conformational change into insoluble aggregates. In addition, apoE peptide-conjugated liposomes can likely disaggregate Aβ fibrils [30]. Thus, it is possible that the combination of lipid nanoparticles and APOE2 expression accelerates Aβ disaggregation in our approach, though further studies are needed. Since soluble Aβ oligomers are more toxic than fibrils, which directly disturb the cellular membrane integrity [31,32] and trigger cellular stresses, such as oxidative stress and endoplasmic reticulum stress (ERS) [33], neuronal damages observed in mice from the APOE2 group may be due to the increase in soluble Aβ oligomers.
Indeed, WGCNA in the bulk RNA-seq identified several hub genes related to metabolic process including Mgst1. Microsomal Glutathione S-Transferase 1 (MGST1) localizes in the endoplasmic reticulum and outer mitochondrial membrane, protecting from oxidative stress [34]. Acadl, which encodes Acyl-CoA dehydrogenase long chain, was identified as a major downregulated gene in the GO analysis. ACADL is a mitochondrial matrix enzyme contributing to fatty acid beta oxidation [35]. Indeed, the downregulation of ACADL has been observed in AD brains [36]. Thus, the reduction of Mgst1 and Acadl may be involved in the Aβ oligomer-mediated cellular stress responses. Conflictingly, our results found that APOE2 cDNA-conjugated liposomes administration leads to the upregulation of Dlg4 and Syp mRNAs that code synaptic proteins, despite the signs of aggravated neurodegeneration. Gabra2, which encodes GABA type A receptor subunit alpha 2, was also one of the upregulated DEGs in the mouse cortices from the APOE2 group. GABRA2 has been reported to be significantly lower in AD brains than age-matched controls [37]. Thus, brain APOE2 expression may facilitate synaptic formation at transcription levels. Indeed, GSEA found that several pathways related to neuronal functions are upregulated by the APOE2 cDNA-conjugated liposome administration. In addition, among the DEGs, downregulation of Sfrp4 and upregulation of Fzd4 were detected in the APOE2 group mice. While secreted frizzled related protein 4 (SFRP4) serves as a Wnt antagonist, Frizzled receptor 4 (FZD4) functions as a Wnt co-receptor [38]. SFRP4 and FZD4 mediate synaptic maintenance through Wnt signaling [39], which is downregulated in AD brain [39]. Thus, our results suggest that increasing brain APOE2 expression may cause beneficial effects on synaptic integrity by activating Wnt signaling.
In summary, we developed a novel APOE2 cDNA plasmid delivery system using functionalized liposomes conjugated with PEG, transferrin, and Penetratin. We demonstrated that liposome-based APOE2 gene therapy reduces insoluble Aβ, but rather increase soluble Aβ oligomers and exacerbates neuronal damage in AppNL−G- mice. However, there are several limitations in our study. First, brain APOE2 expression through our liposome-based gene delivery system is relatively low as we cannot detect APOE in soluble fractions of mouse brains. Since virus-based approaches can induce more APOE2 expressions in the brain, the conflicting results may be explained by potentially different APOE2 expression levels. Further optimizations of our liposome formulations and administration route/dosage are necessary to improve brain APOE2 gene delivery. Second, we utilized only AppNL−G-F knockin mice to overcome possible artificial effects in APP transgenic mice. As AppNL−G-F mice express Aβ with Arctic mutation (E22G), the APOE-related Aβ clearance or degradation may differ from normal Aβ species. Third, we treated AppNL−G-F mice with the APOE2 cDNA liposomes at the age of 12 months. It is possible that excess amounts of soluble Aβ may be dissociated from Aβ plaques beyond the clearance capacity when APOE2 expression is induced at the late stage of Aβ deposition. Fourth, we used the mice with murine Apoe background. In APP/PS1 mice with APOE4, AAV-based APOE2 gene delivery can ameliorate brain Aβ pathology and suppress microglia activation [10]. Beneficial effects of APOE2 gene therapy possibly become more apparent in the presence of APOE4 by mitigating APOE4-related microglial dysregulation. Therefore, our future studies should investigate how early intervention with brain APOE2 expression by using lipid nanoparticle-based gene delivery system influence AD-related phenotypes in additional amyloid AD model mice with APOE4.
CRediT authorship contribution statement
Ni Wang: Writing – review & editing, Writing – original draft, Visualization, Investigation, Formal analysis, Conceptualization. Tammee M. Parsons: Investigation, Formal analysis. Yingxue Ren: Formal analysis, Data curation. Yining Pan: Formal analysis, Data curation. Aishe Kurti: Investigation. Skylar C. Starling: Investigation, Formal analysis. Chinenye Muolokwu: Resources, Methodology, Investigation. Jagdish Singh: Writing – review & editing, Supervision, Methodology, Investigation, Funding acquisition, Conceptualization. Takahisa Kanekiyo: Writing – review & editing, Writing – original draft, Visualization, Supervision, Investigation, Funding acquisition, Conceptualization.
Data availability
The bulk RNA-seq data are deposited in the Gene Expression Omnibus repository under accession number GSE279476. All other data are available upon reasonable request.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This work was supported by NIH grants R01AG068034 and R01AG083981 (to J.S. and T.K.), RF1AG081203, U19AG069701, R01AG071226, and RF1AG057181 (to T.K.), and a Cure Alzheimer’s Fund grant (to T.K.). We thank Drs. Takashi Saito and Takaomi C Saido from RIKEN for providing the AppNL-G-F mice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e39607.
Abbreviations
| AD | Alzheimer's disease |
| APOE | Apolipoprotein E |
| AAV | Adeno-associated virus |
| PEG | Polyethylene glycol |
| BBB | Blood-brain barrier |
| Pen | Penetratin |
| Tf | Transferrin |
| DSPE | 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine |
| DOTAP | 1,2-dioleoyl-3-trimethylammonium-propane |
| DOPE | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine |
| CS | Conditioned stimulus |
| US | Unconditioned stimulus |
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Yamazaki Y., et al. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 2019;15(9):501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., et al. APOE2: protective mechanism and therapeutic implications for Alzheimer's disease. Mol. Neurodegener. 2020;15(1):63. doi: 10.1186/s13024-020-00413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raulin A.C., et al. ApoE in Alzheimer's disease: pathophysiology and therapeutic strategies. Mol. Neurodegener. 2022;17(1):72. doi: 10.1186/s13024-022-00574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepp K.P., et al. The amyloid cascade hypothesis: an updated critical review. Brain. 2023;146(10):3969–3990. doi: 10.1093/brain/awad159. [DOI] [PubMed] [Google Scholar]
- 6.Martens Y.A., et al. ApoE Cascade Hypothesis in the pathogenesis of Alzheimer's disease and related dementias. Neuron. 2022;110(8):1304–1317. doi: 10.1016/j.neuron.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen W.J., et al. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA. 2015;313(19):1924–1938. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinohara M., et al. APOE2 eases cognitive decline during Aging: clinical and preclinical evaluations. Ann. Neurol. 2016;79(5):758–774. doi: 10.1002/ana.24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L., et al. Intracerebral adeno-associated virus gene delivery of apolipoprotein E2 markedly reduces brain amyloid pathology in Alzheimer's disease mouse models. Neurobiol. Aging. 2016;44:159–172. doi: 10.1016/j.neurobiolaging.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Jackson R.J., et al. APOE2 gene therapy reduces amyloid deposition and improves markers of neuroinflammation and neurodegeneration in a mouse model of Alzheimer disease. Mol. Ther. 2024;32(5):1373–1386. doi: 10.1016/j.ymthe.2024.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Au H.K.E., Isalan M., Mielcarek M. Gene therapy advances: a meta-analysis of AAV usage in clinical settings. Front. Med. 2021;8 doi: 10.3389/fmed.2021.809118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taher M., et al. PEGylated liposomes enhance the effect of cytotoxic drug: a review. Heliyon. 2023;9(3) doi: 10.1016/j.heliyon.2023.e13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenchov R., Sasso J.M., Zhou Q.A. PEGylated lipid nanoparticle formulations: immunological safety and efficiency perspective. Bioconjugate Chem. 2023;34(6):941–960. doi: 10.1021/acs.bioconjchem.3c00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomsen M.S., et al. Blood-brain barrier transport of transferrin receptor-targeted nanoparticles. Pharmaceutics. 2022;14(10) doi: 10.3390/pharmaceutics14102237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koren E., Torchilin V.P. Cell-penetrating peptides: breaking through to the other side. Trends Mol. Med. 2012;18(7):385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos Rodrigues B., Kanekiyo T., Singh J. ApoE-2 brain-targeted gene therapy through transferrin and Penetratin tagged liposomal nanoparticles. Pharm. Res. (N. Y.) 2019;36(11):161. doi: 10.1007/s11095-019-2691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues B.D., Kanekiyo T., Singh J. ApoE-2 brain-targeted gene therapy through transferrin and Penetratin tagged liposomal nanoparticles. Pharmaceut. Res. 2019;36(11) doi: 10.1007/s11095-019-2691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito T., et al. Single App knock-in mouse models of Alzheimer's disease. Nat. Neurosci. 2014;17(5):661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 19.Oue H., et al. LRP1 in vascular mural cells modulates cerebrovascular integrity and function in the presence of APOE4. JCI Insight. 2023;8(7) doi: 10.1172/jci.insight.163822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachibana M., et al. APOE4-mediated amyloid-beta pathology depends on its neuronal receptor LRP1. J. Clin. Invest. 2019;129(3):1272–1277. doi: 10.1172/JCI124853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y., et al. Apolipoprotein E lipoprotein particles inhibit amyloid-beta uptake through cell surface heparan sulphate proteoglycan. Mol. Neurodegener. 2016;11(1):37. doi: 10.1186/s13024-016-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalari K.R., et al. MAP-RSeq: Mayo analysis pipeline for RNA sequencing. BMC Bioinf. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen K.D., Irizarry R.A., Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13(2):204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiman E.M., et al. Exceptionally low likelihood of Alzheimer's dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 2020;11(1):667. doi: 10.1038/s41467-019-14279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodart J.C., et al. Gene delivery of human apolipoprotein E alters brain Abeta burden in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2005;102(4):1211–1216. doi: 10.1073/pnas.0409072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudry E., et al. Gene transfer of human Apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci. Transl. Med. 2013;5(212):212ra161. doi: 10.1126/scitranslmed.3007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garai K., et al. The binding of apolipoprotein E to oligomers and fibrils of amyloid-beta alters the kinetics of amyloid aggregation. Biochemistry. 2014;53(40):6323–6331. doi: 10.1021/bi5008172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bana L., et al. Liposomes bi-functionalized with phosphatidic acid and an ApoE-derived peptide affect Abeta aggregation features and cross the blood-brain-barrier: implications for therapy of Alzheimer disease. Nanomedicine. 2014;10(7):1583–1590. doi: 10.1016/j.nano.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Kayed R., Lasagna-Reeves C.A. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis. 2013;33(Suppl 1):S67–S78. doi: 10.3233/JAD-2012-129001. [DOI] [PubMed] [Google Scholar]
- 32.Sengupta U., Nilson A.N., Kayed R. The role of amyloid-beta oligomers in toxicity, propagation, and immunotherapy. EBioMedicine. 2016;6:42–49. doi: 10.1016/j.ebiom.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontana I.C., et al. Amyloid-beta oligomers in cellular models of Alzheimer's disease. J. Neurochem. 2020;155(4):348–369. doi: 10.1111/jnc.15030. [DOI] [PubMed] [Google Scholar]
- 34.Aniya Y., Imaizumi N. Mitochondrial glutathione transferases involving a new function for membrane permeability transition pore regulation. Drug Metab. Rev. 2011;43(2):292–299. doi: 10.3109/03602532.2011.552913. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X., et al. ACADL plays a tumor-suppressor role by targeting Hippo/YAP signaling in hepatocellular carcinoma. npj Precis. Oncol. 2020;4:7. doi: 10.1038/s41698-020-0111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuner S.M., et al. Hippocampal proteomics defines pathways associated with memory decline and resilience in normal aging and Alzheimer's disease mouse models. Behav. Brain Res. 2017;322(Pt B):288–298. doi: 10.1016/j.bbr.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govindpani K., et al. Impaired expression of GABA signaling components in the alzheimer's disease middle temporal gyrus. Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3. doi: 10.1038/s41392-021-00762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomer E., Buechler J., Salinas P.C. Wnt signaling deregulation in the aging and alzheimer's brain. Front. Cell. Neurosci. 2019;13:227. doi: 10.3389/fncel.2019.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data are deposited in the Gene Expression Omnibus repository under accession number GSE279476. All other data are available upon reasonable request.