Abstract
Evolutionary innovations in chemical secretion—such as the production of secondary metabolites, pheromones, and toxins—profoundly impact ecological interactions across a broad diversity of life. These secretory innovations may involve a “legacy-plus-innovation” mode of evolution, whereby new biochemical pathways are integrated with conserved secretory processes to create novel products. Among secretory innovations, bioluminescence is important because it evolved convergently many times to influence predator–prey interactions, while often producing courtship signals linked to increased rates of speciation. However, whether or not deeply conserved secretory genes are used in secretory bioluminescence remains unexplored. Here, we show that in the ostracod Vargula tsujii, the evolutionary novel c-luciferase gene is co-expressed with many conserved genes, including those related to toxin production and high-output protein secretion. Our results demonstrate that the legacy-plus-innovation mode of secretory evolution, previously applied to sensory modalities of olfaction, gustation, and nociception, also encompasses light-producing signals generated by bioluminescent secretions. This extension broadens the paradigm of secretory diversification to include not only chemical signals but also bioluminescent light as an important medium of ecological interaction and evolutionary innovation.
Keywords: novelty, evolution, bioluminescence, WGCNA, complex traits, evo-devo
Introduction
Exocrine glands are specialized secretory structures, often representing lineage-specific innovations that produce diverse chemical secretions. These secretions—including secondary metabolites, pheromones, and toxins—have been instrumental in evolving new ecological interactions using behaviors like intraspecific communication (e.g. courtship signaling and caste social systems) and predator–prey interactions (Jackson and Morgan 1993; Trhlin and Rajchard 2011; Ellis and Oakley 2016). Examples of secretory innovations include courtship glands of salamanders, venom glands of snakes and remipedes, defensive tergal glands of rove beetles, and ink and opaline glands of mollusks (Johnson et al. 2006; Rollins and Staub 2017; von Reumont et al. 2017; Barua and Mikheyev 2021; Brückner et al. 2021). Secretory innovations may often use a shared “genetic toolkit” (Oakley 2024) for secretion that was modified in different lineages by adding new genes to produce diverse products (Brückner and Parker 2020; Zancolli et al. 2022), a mode of evolution we call “legacy-plus-innovation.” Although exocrine glands clearly exhibit widespread morphological convergence, only a few studies have characterized gene expression to support the use of a shared genetic toolkit and the legacy-plus-innovation model (Brückner and Parker 2020; Barua and Mikheyev 2021; Brückner et al. 2021; Zancolli et al. 2022), limiting our understanding of the historical constraints that shape the broad range of secretory novelties.
Thus far, the legacy-plus-innovation mode of secretory evolution is proposed for secretory outputs perceived by olfactory, gustatory, and nociceptive mechanisms (Brückner and Parker 2020; Zancolli et al. 2022), leaving the genetic underpinnings of light-producing secretions largely unexplored, despite the ecological importance of visual interactions (reviewed in Schaefer 2010). Chemical secretions that generate light are produced by bioluminescent systems, which have evolved convergently many times, using an impressive variety of structural and functional forms (Haddock et al. 2010; Lau and Oakley 2021). Bioluminescent glands facilitate many ecological interactions, including courtship signals, antipredation “burglar alarm” displays, and various predation strategies (Ellis and Oakley 2016; McCloskey et al. 2017; Morin 2019; Claes et al. 2020; Goodheart et al. 2020). For example, bioluminescent glands of some marine crustaceans and mollusks and syllid worms discharge glowing mucus into the water as antipredation and courtship displays, and the bioluminescent glands of pocket sharks secrete illuminated lures to attract prey (Verdes and Gruber 2017; Morin 2019; Claes et al. 2020). The widespread evolution of secretory bioluminescence and the ecological consequences associated with visual interactions of light displays motivate a quest to understand the genetic underpinnings of these adaptive and often beautiful secretions.
Secretory bioluminescence highlights vision as an important sensory modality within the framework of secretory evolution, but the extent to which a shared secretory toolkit also underlies secreted bioluminescence remains obscure. To test whether “legacy-plus-innovation” underlies secretory bioluminescence, we analyze gene expression in the bioluminescent upper lip (BUL) of cypridinid ostracods (crustaceans), a secretory innovation with important ecological, cellular, and biochemical functions (Morin 1986; Huvard 1993; Abe et al. 2000). Since its origin with the clade Luminini at least 237 Ma (Ellis et al. 2022), luminine bioluminescence has had important ecological functions, including deterring predation and signaling for courtship. Nearly all Luminini release large amounts of bioluminescent mucus during predation attempts, often escaping unscathed, due either to the light itself and/or to the unpalatability of the mucus (Morin 2019). Within Luminini, a subclade called Luxorina also produces complex bioluminescent signals for courtship, which only evolved once (Cohen and Morin 2003; Ellis et al. 2022). The bioluminescence of Luminini is produced by secretory cells and nozzle-like appendages embedded in the upper lip (labrum; Fig. 1; Huvard 1993; Abe et al. 2000). While the upper lip as a whole is homologous between bioluminescent and nonbioluminescent ostracods, the luminous upper lip is a light organ unique to Luminini, and therefore may be considered a radical transformation of an existing body part, which Wagner (2015) terms a Type II novelty. Compared with non-BUL, BUL have more cell types, including those that synthesize and secrete mucus that contains light-emitting compounds (Huvard 1993; Abe et al. 2000). The two compounds include evolutionarily novel (Oakley 2005), biochemically well-characterized enzymes called c-luciferases (Thompson et al. 1990; Nakajima et al. 2004; Hensley et al. 2021), and a novel small molecule that is oxidized to produce light, a luciferin often called vargulin (Kishi et al. 1966).
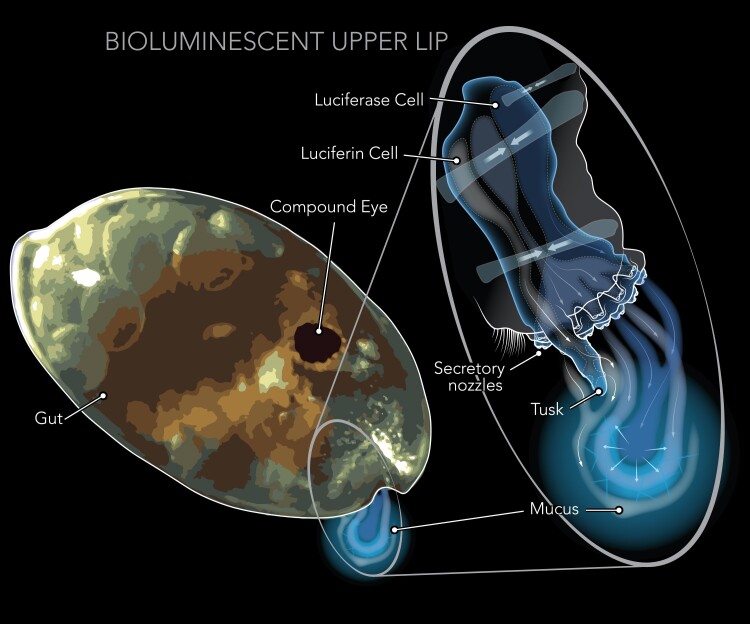
Fig. 1.
An illustration of a bioluminescent ostracod, highlighting the specialized light-secreting gland, the BUL, along with the compound eye and gut. The BUL of ostracods is positioned in front of the mouth, where it releases light-producing compounds into the surrounding water. The light reaction involves a substrate, luciferin, and an enzyme, luciferase, each synthesized in separate secretory cells of the upper lip, and discharged from the tusks and nozzles into a mucous-like substance, as illustrated in the diagram. The diagram of the BUL is adapted from the Science article (DOI: 10.1126/science.ade5292) and used with permission.
In addition to the established roles of novel c-luciferases and vargulin in evolving secretory bioluminescence, we herein report that the upper lip of a bioluminescent ostracod expresses deeply conserved genes of secretory pathways, despite vast evolutionary distances separating this bioluminescence system from other secretory innovations. Using the bioluminescent ostracod Vargula tsujii, we analyzed genes co-expressed with c-luciferase, which we call a “Bioluminescent Co-Expression Network” (BCN). We find this BCN to contain many conserved and putatively secreted genes, some of which resemble toxin-like products, as well as nonsecreted housekeeping genes involved in pathways related to protein secretion and associated stress responses. We also compared differential gene expression of entire upper lips of luminous and nonluminous ostracods, finding distinct patterns of DE in the luminous upper lip. The DE analyses further emphasize the use of conserved toxin-like genes and other conserved secretory pathways. At the same time, we report that many novel genes besides c-luciferase are expressed in the BCN and luminous upper lip. Together, these results reveal legacy-plus-innovation evolution in the BUL, whereby elements of a deeply conserved secretory toolkit are deployed along with new genes. Our results extend this secretory innovation paradigm (Brückner and Parker 2020; Zancolli et al. 2022) to also encompass secreted light as a crucial medium of ecological interaction and evolutionary innovation.
Results
To understand the contributions of new and existing genes and their co-expression patterns to an evolutionarily novel BUL, we conducted two major classes of analyses, with the sample sets summarized here. First, we conducted DE in 2 species, 1 luminous and 1 nonluminous, by sampling for each species 5 biological replicates of 3 tissue types: dissected upper lip, compound eye, and gut of adults; for a total of 30 RNA expression profiles. For DE, we used a previously published transcriptome (Lau et al. 2024) for the luminous species, and generated a de novo transcriptome from a single individual for the nonluminous species, which was lacking. Second, we quantified gene co-expression networks in the luminous species using data from 57 RNA-seq samples, including all tissue types from the DE dataset (upper lip, compound eye, and gut), previously published upper lip samples, whole bodies of juvenile instar stages, and whole adult bodies, some of which were subjected to physical stimulation to produce bioluminescence. All sample information, sequencing and mapping statistics for both luminous and nonluminous ostracods are summarized in supplementary table S1, Supplementary Material online.
Co-expression of Conserved Secretory Pathways and Toxin-Like Genes with c-Luciferase
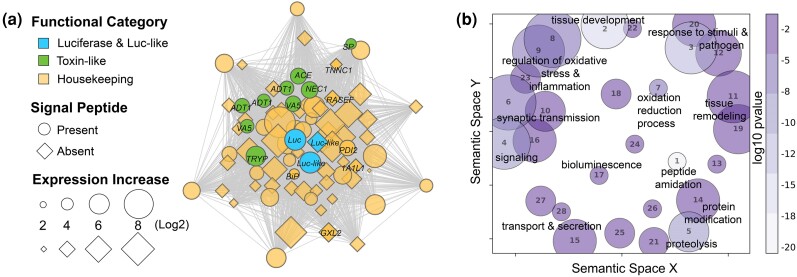
The genes co-expressed with c-luciferase across 57 tissues/stages of V. tsujii form a co-expression network (module) with 958 genes that we call the BCN, of which 35% exhibit high module membership (MM > 0.8; Fig. 2a, supplementary table S3, Supplementary Material online). The network revealed connections between c-luciferase, luciferase-like genes, toxin-like genes, other putatively secreted products (based on presence of a signal peptide), and nonsecreted housekeeping genes, many of which are strongly connected (MM > 0.8; Fig. 2a, supplementary table S3, Supplementary Material online). In addition to bioluminescence, significantly enriched Gene Ontology (GO) terms of the BCN include proteolysis, protein processing and modification, secretion, lipid regulation and transport, peptide modification, tissue development and remodeling, cell signaling, response to stimuli and cellular stress, and oxidative stress and inflammation (Fig. 2b, supplementary table S3, Supplementary Material online). Annotations of nonsecreted housekeeping genes include maintenance of tissue and cellular functions, involvement in protein secretory pathways and downstream targets, that respond to inflammation, cellular and oxidative stress, and regulate cell fate decisions (Fig. 2b). The concerted activity of these processes could be related to demands of protein synthesis and secretion consistent with the role of the BUL in high-volume secretion of c-luciferase and other proteins integral to the bioluminescent mucus. Other biological processes enriched in the BCN include synaptic transmission and nervous system regeneration (Fig. 2b), perhaps from labral nerves that innervate the upper lip (Budd 2021).
Fig. 2.
a) The BCN represents a group of co-expressed genes that are associated with the expression of luciferase, luciferase-like genes, toxin-like genes, and housekeeping genes some of which are constituents of stress and secretory protein pathways. The BCN comprises over 900 co-expressed genes and about one-third of the BCN genes are significantly upregulated uniquely in the upper lip of V. tsujii, of which 26% are considered integral to the network (MM > 0.8). Out of 277 co-expressed genes upregulated in the BUL, only genes with annotations (30%, 83 annotated genes) are plotted here to visualize connections and overall network topology. Bioluminescent genes, including c-luciferase (Luc) and similar but not functionally tested luciferase-like (Luc-like) genes are blue. Putative toxin-like genes are green: Venom Allergen 5 (VA5), A disintegrin and metalloproteinase with thrombospondin (ADT1), SP inhibitor (SP), trypsin-1 (TRYP), neuroendocrine convertase 1 (NEC1), and angiotensin-converting enzyme (ACE). Protein secretory pathway genes are in yellow with black labels: PDI2, BiP, RASEF, glucoside xylosyltransferase 2 (GXLT2), troponin C (TNNC1), and 1-aminocyclopropane-1-carboxylate synthase-like protein 1 (1A1L1). Genes that have a signal peptide and are presumably secreted outside the cell are indicated by a circle and genes that do not have a signal peptide are indicated by a diamond. Distance between nodes indicates strength connectivity. b) GO terms enriched in the BCN are visualized using GO figure. Each bubble represents a GO term or a cluster of GO terms summarized by a representative term reported in the supplement. The size of the bubble indicates the number of GO terms in each cluster and the color is the P-value or the average P-value of the representative GO term in that cluster. The GO terms for each bubble are found in supplementary table S3, Supplementary Material online.
Our analyses reveal for the first time the co-expression of c-luciferase with deeply conserved toxin-like domains and genes, which is of particular interest because ostracods use bioluminescence in antipredator responses. In V. tsujii, we find the BCN to have transcripts with signal peptides similar to genes expressed in venom glands and salivary glands of other animals. Out of the 121 BCN transcripts (supplementary table S3, Supplementary Material online) with a signal peptide and no transmembrane domain (and therefore likely to be secreted products), 29 have domains of known toxin gene families (von Reumont et al. 2017; supplementary table S6, Supplementary Material online). At least 16 (of known toxin gene families) of these toxin-like genes have high connectivity (MM > 0.8) in the BCN (i.e. hub genes), with several also significantly upregulated in the BUL (supplementary tables S3 and S6, Supplementary Material online). Similar to the global transcriptomic profiles of convergent venom glands (Zancolli et al. 2022) and the “metavenom network” expressed in the venom glands of snakes (Barua and Mikheyev 2021), we find many genes in the BCN with similarity to genes involved in the global endoplasmic reticulum (ER) stress (ERS) response, which also includes the protein secretory pathway. Genes that function as sensors of ERS in the unfolded protein response (UPR) pathway and genes in the ER-associated degradation (ERAD) pathway, which are responsible for high-output protein secretion, also have strong connections with c-luciferase in the BCN. Some of these genes include heat shock proteins (HSB), ER chaperone (BiP), receptor expression-enhancing protein 5 (REEP5), calpain-A (CANA), protein disulfide isomerases (PDIs), translation initiation factor 2 (IF2), and Ras and EF-hand domain-containing protein (RASEF; Fig. 2a, supplementary table S5, Supplementary Material online). We also identified deeply conserved genes associated with processes downstream of ERS response pathways, such as inflammatory responses, lipid regulation, detoxification, redox regulation of cell fate under ERS, and ER calcium signaling and transport (Fig. 2b, supplementary table S3, Supplementary Material online; Walter and Ron 2011; Senft and Ronai 2015; Carreras-Sureda et al. 2018; Zhang et al. 2019; Pick et al. 2021). In addition, we found several candidate sulfotransferases that are upregulated in the BUL, although not uniquely compared with the eye and gut, making them candidates for facilitating a storage form of vargulin in V. tsujii (supplementary table S2, Supplementary Material online; Lau et al. 2024).
DE of Conserved Secretory Pathway Genes and Toxin-like Gene Families
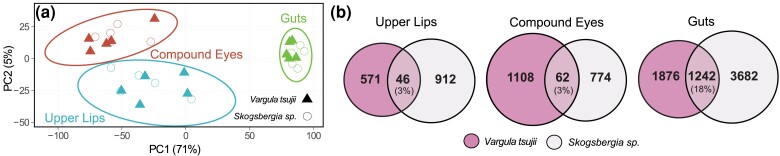
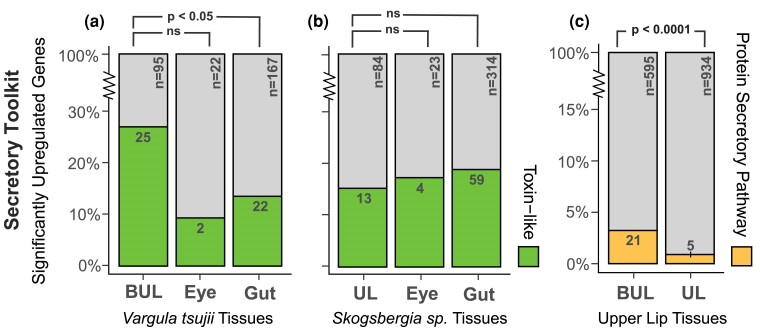
The results of expression analyses in upper lips of a luminous V. tsujii and nonluminous Skogsbergia sp. reveal an intriguing mix of legacy and innovation. The common evolutionary legacy of upper lips is reflected in the similar overall expression patterns of one-to-one orthologs between the upper lips of the two species, which cluster by tissues (Fig. 3a). In addition, both species show significantly upregulated genes that comprise an unexpectedly complex cocktail of transcripts coding for putative toxin-like proteins with signal peptides (Table 1). For the BUL, out of the total number of upregulated transcripts with signal peptides and without transmembrane domains, we found 25 transcripts (26%) representing at least 11 different toxin-like classes based on domain identity (Fig. 4a, Table 1, and supplementary table S6, Supplementary Material online). The 11 different toxin classes can be classified broadly into six functional categories: (ⅰ) neurotoxins: ShKT, (ⅱ) protease inhibitors: Kunitz, lipocalin, serpin, (ⅲ) serine proteases (SPs): peptidase S1, peptidase S8, (ⅳ) metalloproteinases: peptidase M2, metalloproteinase M12, (ⅴ) CAP domain proteins, and (ⅵ) other enzymes: CUB and C-type lectin. For the non-BUL, we found 13 transcripts (15%), including some from classes shared with the BUL: C-type lectin, ShKT, CUB, WAP, peptidase S1, peptidase S8, and metalloproteinase M12 (Fig. 4b, Table 1, and supplementary table S6, Supplementary Material online). For both species, we compared the ratio of putative toxin-like genes to secreted genes in both upper lips compared with two tissue types not involved in the secretory process of antipredator displays—the compound eye and gut. For the luminous V. tsujii, we found the ratio of putative toxin-like genes to secreted genes to be significantly higher in the BUL compared with the gut, but not significantly higher compared to the compound eye (BUL:Gut, Fisher's exact test, P < 0.05; BUL:Compound Eye, Fisher's exact test, P = 0.25; Fig. 4a, supplementary table S6, Supplementary Material online). In contrast, the nonluminous upper lip expressed a lower ratio of putative toxin-like genes to secreted genes compared with the compound eye and gut (Fig. 4b, supplementary table S6, Supplementary Material online). Additionally, the putative toxin-like genes identified through the ToxProt database revealed similar patterns in both luminous and nonluminous upper lips (supplementary table S6, Supplementary Material online).
Fig. 3.
Orthologous gene expression values from two species cluster by tissues, while differentially expressed genes mainly differ between tissue types for each species. a) Expression patterns of orthologous genes between upper lips of bioluminescent and nonbioluminescent ostracods are conserved. PCA clustered gene expression of 4,217 one-to-one orthologs by tissue, and differences among tissues explain >75% of the variation in the dataset. Ellipses represent 95% confidence intervals. b) Venn diagram illustrating the number of significantly upregulated genes shared across both upper lips versus compound eyes versus guts for each species, V. tsujii and Skogsbergia sp. The values in parentheses represent the percentage of shared genes found in the same orthogroup.
Table 1.
Secretory toolkit genes significantly upregulated across upper lips
| Species | Tissue | Putative toxin like | Secretory protein pathway | ||
|---|---|---|---|---|---|
| Protein family | No. of genes | Protein pathway Subsystem | No. of genes | ||
| Vargula tsujii | BUL | C-type lectin | 2 | Golgi glycosylation | 2 |
| Metalloproteinase M12 | 3 | Trafficking regulation | 3 | ||
| Peptidase S1 and S8 | 3 | Protein folding | 7 | ||
| Lipocalin | 2 | Post-Golgi trafficking | 3 | ||
| Kunitz | 3 | ERAD | 2 | ||
| CUB | 7 | UPR | 1 | ||
| CAP | 2 | Coat protein Complex II | 2 | ||
| ShKT | 1 | Translocation | 1 | ||
| Peptidase M2 | 1 | ||||
| Serpin | 1 | ||||
| Skogsbergia sp. | UL | C-type lectin | 2 | Golgi glycosylation | 1 |
| Metalloproteinase M12 | 2 | Post-Golgi trafficking | 1 | ||
| Peptidase S1 and S8 | 4 | ERAD | 1 | ||
| WAP | 2 | Coat protein Complexes I & II | 2 | ||
| CUB | 1 | ||||
| ShKT | 2 | ||||
The table summarizes the diversity of secretory toolkit genes, which encompasses both secretory protein pathway genes and putative secretory toxin-like genes, which are significantly upregulated in the BUL and non-BUL upper lip.
BUL, bioluminescent upper lip.
Fig. 4.
DE of toxin-like secretory and secretory protein pathway products that are expressed uniquely across luminous and nonluminous upper lips. a) Compared with the gut, the luminous upper lip has a significantly higher proportion of toxin-like genes to secreted genes, but not significantly higher compared with the compound eye. A statistically significant pairwise comparison between BUL:Gut is indicated by P < 0.05. b) Compared with the compound eye and gut, the nonluminous upper lip expressed a lower proportion of toxin-like genes to secreted genes. c) Compared with the nonluminous upper lip, the BUL expressed a significantly higher proportion of secretory protein pathway genes compared with all significantly upregulated genes uniquely expressed. A statistically significant pairwise comparison between BUL:UL is indicated by P < 0.0001. The bar plot represents the proportions as percentages, while the raw numbers are displayed inside the colored bars, with n indicating the total number. BUL, bioluminescent upper lip.
Although a similar overall expression of orthologs (Fig. 3a) reflects a shared evolutionary history between luminous and nonluminous upper lips, only a few homologous genes are significantly upregulated in both upper lips (Fig. 3b), likely due to functional differences like the innovation of light production. We found the upper lips of V. tsujii and Skogsbergia sp. share significantly fewer upregulated genes (3%, 46 homologs) in common than the guts of the same two species (18%, 1,242 homologs; Fisher's exact test, P < 0.0001; Fig. 3a, supplementary table S2, Supplementary Material online). In contrast, the number of significantly upregulated genes shared between both upper lips is comparable with the number shared (3%, 62 homologs) between the compound eyes of the same two species (Fisher's exact test, P = 0.8441; Fig. 3b, supplementary table S2, Supplementary Material online). Consistent with the few upregulated genes shared across upper lips, enrichment analysis revealed that these upregulated genes are associated with distinct GO terms (supplementary figs. S2 and S3, table S2, Supplementary Material online). Although most biological processes are unique to each upper lip, semantically similar GO terms include processes related to proteolysis, lipid metabolism, collagen synthesis, tissue development and signaling, and neurotransmission (supplementary fig. S4 and table S2, Supplementary Material online). Among the genes shared across upper lips, several are putatively secreted genes similar to toxin and anticoagulant proteins, as well as genes involved in collagen synthesis and neural signaling pathways (Fig. 3b, supplementary table S2, Supplementary Material online). One major difference in gene expression between the two upper lips is that the luminous upper lip contains a significantly higher number of upregulated genes of the deeply conserved protein secretory pathway (Fig. 4c and Table 1). The protein secretory pathway can be compartmentalized into subsystems responsible for protein folding, posttranslational modifications, and trafficking of proteins (Feizi et al. 2017). In the BUL, we found at least 21 genes that belong to 8 different subsystems including Golgi glycosylation, trafficking regulation, protein folding, ERAD, UPR, post-Golgi trafficking, coat protein Complex II and translocation (Fig. 4c and Table 1, supplementary table S5, Supplementary Material online). Similar to the BCN, the BUL upregulates many stress response genes related to processes downstream of the protein secretory pathway, including those involved in antioxidant stress responses (e.g. glutathione S-transferase and cytochrome P450), inflammation, apoptotic signaling, lipid biosynthesis, and other biosynthetic pathways (Walter and Ron 2011; Cao and Kaufman 2014; Senft and Ronai 2015). Although there are significantly more secretory protein pathway genes upregulated in V. tsujii upper lip compared with Skogsbergia upper lip (BUL:UL, Fisher's exact test, P < 0.0001), and the number of gene copies varied across a taxonomically wider set of bioluminescent and nonbioluminescent lineages, we did not observe a clear trend between gene copy number and bioluminescent lineages (Fig. 4c, supplementary fig. S5 and table S5, Supplementary Material online).
The Novel BUL Expresses a Higher Proportion of Novel Genes
In addition to ancient genes of a conserved secretory toolkit, the BUL and the BCN express many novel genes that originated similarly in time to bioluminescence itself (e.g. Luminini-specific genes, Fig. 5). To investigate the origin of V. tsujii genes restricted to the Luminini clade, we analyzed orthologous groups across four taxonomic groups: Arthropoda, Ostracoda, Luminini, and Luxorina. Out of the 14,690 proteins that clustered into orthogroups, our KinFin analysis identified V. tsujii proteins restricted to Arthropoda (n = 9,182, 62.5%), Ostracoda (n = 2,991, 20.4%), Luminini (n = 646, 4.4%), Luxorina (n = 747, 5.1%), and V. tsujii–specific proteins that were found in orthogroups of 2 or more sequences (n = 1,124, 7.7%). With the addition of singletons, the number of V. tsujii–specific proteins totaled a final count of 3,905 proteins (22.4% of V. tsujii proteome).
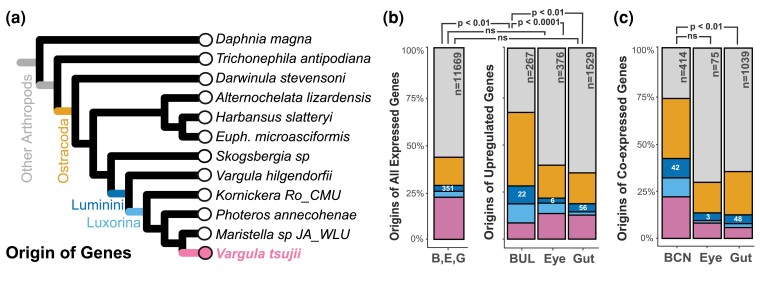
Fig. 5.
The contribution of novel and conserved genes co-expressed in the BCN and significantly upregulated genes uniquely expressed in the BUL, compound eye, and gut of V. tsujii restricted to V. tsujii, Luxorina, Luminini, Ostracoda, and Arthropoda. Conserved ostracoda/arthropoda genes dominate the set of genes in each dataset. a) Phylogeny of bioluminescent and nonbioluminescent ostracods and representatives of other arthropods are used to determine the origin of genes. b) Compared with the complete set of genes differentially expressed (minimum of two counts or more across biological replicates), only the upregulated genes of the BUL were characterized by a significantly higher proportion of Luminini-specific genes. Statistically significant pairwise comparison between BUL:B, E, G Dataset is indicated by P < 0.01. Statistical significance for pairwise comparisons between B, E, G (BUL, Eye, and Gut) Dataset, and tissue types was corrected for multiple comparisons using the Bonferroni method. Compared with the proportion of the total number of Luminini genes upregulated in the compound eye and gut, the BUL had a significantly higher proportion of Luminini genes compared with the gut and compound eye. Statistically significant pairwise comparisons between BUL:Eye and BUL:Gut are indicated by P < 0.0001 and P < 0.01, respectively. c) Compared with modules expressed in the compound eye and gut, the BCN contains a higher proportion of Luminini genes, though not statistically significant in the compound eye. Statistically significant pairwise comparison between BUL:Gut module is indicated by P < 0.01. The bar plot represents the proportions as percentages, while the raw numbers are displayed inside the colored bars, with n indicating the total number. B/BUL, bioluminescent upper lip; E, compound eye; G, gut.
First, a high proportion of all genes upregulated uniquely in the V. tsujii upper lip are Luminini specific. More precisely, the proportion of Luminini-specific genes upregulated in the BUL is significantly higher than the proportion of novel genes upregulated in the compound eye or gut, two tissues that are much older than the origin of bioluminescence (BUL:Compound Eye, Fisher's exact test, P < 0.0001; BUL:Gut, Fisher's exact test, P < 0.01; Fig. 5b, supplementary table S4, Supplementary Material online). Second, compared with the complete set of expressed genes (e.g. expressed in luminous upper lip, gut, and compound eye), only the upregulated genes of the BUL were characterized by a significantly higher proportion of Luminini-specific genes (BUL:B, E, G Dataset, χ2, P < 0.01; Compound Eye:B, E, G Dataset, χ2, P = 0.546; Gut:B, E, G Dataset, χ2, P = 0.476; Fig. 5b, supplementary table S4, Supplementary Material online). Most novel Luminini-specific genes upregulated in the BUL, including several genes with the highest log-fold change, are unannotated, but contain signal peptides and lack transmembrane domains (supplementary table S4, Supplementary Material online). Among the annotated, novel genes include those related to sulfotransferases, putative toxin-like genes, collagen synthesis, and neuronal processes related to neuropeptide synthesis and modulation of neuronal signaling (supplementary table S4, Supplementary Material online). Additionally, novel c-luciferase and luciferase-like genes are significantly upregulated in the BUL. Finally, both the upper lip and the BCN contain a high number of Luminini-specific genes. More precisely, the total number of Luminini-specific genes in the BCN is numerically higher than novel genes from co-expression modules of the compound eye and gut, although not significantly higher than the compound eye (BCN:Compound Eye module, Fisher's exact test, P = 0.1276; BCN:Gut module, Fisher's exact test, P < 0.01; Fig. 5c, supplementary table S4, Supplementary Material online).
Discussion
Through detailed analyses of gene expression, we show that an evolutionarily novel bioluminescence system (Ellis et al. 2022) uses deeply conserved genes of a secretory toolkit to impact ecological interactions by creating light. As such, the secreted bioluminescence of ostracods expands a growing case for the common re-deployment of a conserved secretory toolkit to a new sensory modality, like those found in secretory innovations across diverse and distantly related taxa. This secretory toolkit has integrated novel genes time and again during evolution to create innovative biological chemistries with dramatic effects on ecological interactions among species.
We found a suite of deeply conserved genes expressed in the upper lips and the BCN to include many toxin-like genes (Figs. 2a and 4). Bioluminescent mucus is conspicuously secreted during predation attempts on Luminini ostracods (Rivers and Morin 2012), presumably having impacted predator–prey interactions for hundreds of millions of years (Ellis et al. 2022). However, why fishes spit out ostracods is unknown. Bioluminescence may be aposematic (Underwood et al. 1997), sometimes startle would be predators (Harvey 1952; Morin 1983; Shimomura 2012), or makes them more vulnerable to larger predators (“burglar alarm effect”; Morin 1983; Haddock et al. 2010; Haskell and Bell 2021); but these remain untested hypotheses in ostracods. The mucus produced by ostracods also could contain unpalatable or toxic substances (Morin 2019), plausible because fish often expel an ostracod during predation attempts (Morin 2019). We find both a luminous and nonluminous species to express toxin-like genes in their upper lips, with a higher proportion of such genes expressed in the light-defended V. tsujii (Fig. 4a and b). Notably, toxin-like genes include some related to CAP, which play roles in defense, reproduction, and immune regulation, and include the allergenic Ag5 protein from Polybia paulista (Aparecido dos Santos-Pinto et al. 2015; Blank et al. 2020). Also expressed are CUB domains found in venomous animals (Krishnan et al. 2009; Walker et al. 2017, 2019), metalloproteinase M12 genes common in venom (Bond and Beynon 1995), and genes from the lipocalin and SP families, known for their roles in predation and defense (Fry et al. 2009). These findings suggest a potential defensive function of mucus in nonlight-defended ostracods, which should be further investigated through behavioral assays with fish or other predators and with detailed assessments of gene function. Currently, we only have gene expression data for two species, so future work characterizing secretomes across diverse ostracod species could test a hypothesis that the origin of secreted toxins preceded the origin of bioluminescence. Regardless of the outcome of future experiments on function, the expression of these toxin-like genes supports a role for long-conserved gene families in secreted bioluminescent systems.
Another class of deeply conserved genes that are highly expressed in the luminous upper lip and BCN are likely involved in mitigating cellular stress, perhaps to tolerate the high secretory load of bioluminescent secretion. Dedicated gland cells that secrete large amounts of proteins or small molecules have a high secretory load compared with other cells and may require a quality control system to accommodate mass production and secretion of products (Walter and Ron 2011; Senft and Ronai 2015; Brückner and Parker 2020; Zancolli et al. 2022). The high demand of light-emitting compounds and other secretory products probably requires activation of cellular and protein stress response mechanisms (ERS response pathways) to increase secretory capacity of cells (Brückner and Parker 2020). Having a secretory toolkit to control and regulate cellular processes could facilitate evolutionary transitions from low- to high-output secretory systems (Walter and Ron 2011; Senft and Ronai 2015; Brückner and Parker 2020; Barua and Mikheyev 2021). While we know large volumes of mucus are secreted during ostracod bioluminescence, we know of no similarly high-volume secretory function for nonluminous upper lips, which may function in digestion by smearing mucus on food (Abe and Vannier 1995; Vannier et al. 1998; Abe et al. 2000), a far cry from the large, rapid, and voluminous secretions of luminous ostracods during defense or courtship displays (Morin 1986; Morin and Cohen 1991). Consistent with facilitation of high secretory demand, the BUL upregulates more secretory toolkit genes than the nonluminous upper lip (Fig. 4c), with several as hub genes, strongly co-expressed with c-luciferase in the BCN (Fig. 2a). The GO enrichment analyses of BCN and luminous upper lip also reflect stress-related processes, such as protein processing and folding, modulation of ER stress, tissue remodeling, apoptosis, inflammation, redox regulation, and restoring cellular homeostasis; all of which are tightly associated with ER function (Fig. 1a and supplementary fig. S2, Supplementary Material online; Senft and Ronai 2015; Lee 2017). Intriguingly, we also found genes in the BCN and luminous upper lip implicated in lipid regulation and transport, which link to protein secretory pathways, mitigation of cellular stress, and have been functionally implicated in firefly bioluminescence by providing a source of energy (Tanguy et al. 2016; Jarc and Petan 2019). Although the secretory toolkit genes significantly upregulated in the luminous upper lip are expressed in the transcriptome of the nonluminous Skogsbergia sp., most of these genes are not significantly upregulated, further supporting the establishment during the evolution of a high-output bioluminescent system through the deployment of ancient, conserved machinery for secretory and quality control processes that mitigate cellular stress.
Coupled with legacy elements of a conserved secretory toolkit, we also document the expression of novel genes, which in secretory systems may often enable organisms innovative ways to chemically interact with other organisms and their environment (Walter and Ron 2011; Senft and Ronai 2015; Brückner and Parker 2020; Zancolli et al. 2022). As observed in other secretory and evolutionary novelties (Gould et al. 2008; Jasper et al. 2015; Babonis et al. 2016; Belcaid et al. 2019; Fig. 5), we found many Luminini-specific genes in the BCN and remarkably a high number in the novel BUL. Surprisingly, we found homologous and functionally similar compound eyes to exhibit a similarly small number of shared genes as the functionally distinct upper lips, in contrast to the guts of the same two species (Fig. 3). The low number of shared genes across the compound eyes could be related to differences in visual interactions of each species. Alternatively, similarities in gut expression could be more similar due to the shared scavenging niche between the two target species. In addition to c-luciferase, other novel genes could also be functionally critical. For example, about half of the putatively secreted genes upregulated in the BUL and co-expressed in the BCN are unannotated, with many of these Luminini specific, suggesting a possible role in bioluminescent phenotypes. Out of the novel genes annotated in the BUL, none were related to secretory processes, further supporting the use of conserved genes for secretion. Future functional studies aimed at establishing a direct role of novel genes in the evolution and development of cypridinid bioluminescence phenotypes would be valuable.
As cradles of evolutionary innovation, exocrine glands have catalyzed the emergence of diverse biological functions across many unrelated taxa (Brückner and Parker 2020). We report gene expression patterns in secreted bioluminescence of ostracods, despite being a lineage-specific innovation, use deeply conserved genes, including toxin-like genes and genes that are involved in secretory processes to protect against cellular stress. These genes may represent an ancient secretory toolkit for high-output secretory cells, similar to gene sets used in the few other secretory innovations studied in sufficient detail (Barua and Mikheyev 2021; Brückner et al. 2021; Zancolli et al. 2022). Secretory innovations may therefore routinely integrate a similar and conserved secretory toolkit with novel biosynthesis pathways. By leading to the production of myriad new small molecules and proteins, including pheromones, toxins, and bioluminescence, this process of secretory innovation has had a profound impact on ecological interactions throughout evolutionary history.
Materials and Methods
RNA Extraction, Sequencing, and Mapping
For both DE analyses and co-expression analyses, we isolated RNA from animals kept in lab cultures (Goodheart et al. 2020) at the University of California, Santa Barbara. Cultures of luminous V. tsujii originated by trapping with bait near Wrigley Marine Lab (Catalina Island, CA, USA; 33.444969, −118.484471). Cultures of nonluminous Skogsbergia sp. originated by trapping with bait in Thallasia beds near Southwater Caye (Belize; 16.812833, −88.083154), then kept using methods published for V. tsujii, except at 27 to 30 °C. For DE experiments, we extracted total RNA using Agencourt RNAClean magnetic beads (Beckman Coulter). We optimized a protocol using RNAClean magnetic beads to extract total RNA (see protocols.io for details) (Mesrop 2024). For co-expression analyses in V. tsujii, we began with an initial set of 63 RNA samples, which included 15 RNA-seq samples from the DE dataset and 9 upper lip RNA-seq samples downloaded from the NCBI database (PRJNA935772), and added 39 RNA samples, extracted using TRIzol (Invitrogen). The additional 39 RNA samples included 21 adult whole bodies and 18 juvenile V. tsujii samples comprising 3 to 6 biological replicates from each of the 5 juvenile instar stages (A-I male, A-I female, A-II, A-III, A-IV, A-V), but 6 samples were excluded from the co-expression analyses due to low counts. We used 3′ Tag RNA-seq to quantify RNA expression, with sequencing performed at the UT-Austin DNA Core Facility (Weng and Juenger 2022). We used published bioinformatics tools (github.com/Eli-Meyer/TagSeq_util) to trim 3′ Tag RNA-seq data using BBDUK (v38.18), remove low-quality reads (<20), and polymerase chain reaction duplicates. We used Bowtie2 (v2.4.2) to map cleaned reads to reference transcriptomes (Li and Dewey 2011; Langmead and Salzberg 2012). For all samples, weak and ambiguous reads were removed if alignments had <40 matching bp (Langmead and Salzberg 2012). All sample information and mapping statistics are summarized in supplementary table S1, Supplementary Material online.
Reference Transcriptomes, Annotation, and GO Analyses
For use in DE analyses, we generated and annotated a de novo transcriptome for the nonluminous species, Skogsbergia sp., with libraries prepared and sequenced by Novogene using NovaSeq 6000 PE150 platform. We trimmed reads with Trimmomatic (Grabherr et al. 2011), assembled with Trinity (v2.1.1), evaluated transcriptome completeness with BUSCO (Waterhouse et al. 2018; 89.8% using the Arthropoda_odb9 lineage database), then reduced redundancy by merging transcripts >90% similar using CD-HIT (v4.8.1; Chen et al. 2016). After CD-HIT, we extracted the longest isoform for each gene with Trinotate (v3.2.2; Grabherr et al. 2011) and identified candidate protein-coding regions with TransDecoder (v3.2.2; Bryant et al. 2017). All assembly statistics for the nonluminous de novo transcriptome are summarized in supplementary table S1, Supplementary Material online. For the luminous species V. tsujii, we used an existing de novo transcriptome assembly, following the methods in Lau et al. (2024) (SRR21201581 and SRR24065266). For both nonluminous and luminous reference transcriptomes, we generated GO terms with BLASTX (v2.10.1) with a default e-value of 1e−10 and the SwissProt database (downloaded April 2022) (Altschul et al. 1990; Grabherr et al. 2011). We used Trinotate to predict signal peptides, transmembrane domains, and protein domains with signalP (v4.1), TMHMM (v2.0), and Pfam (v35.0), respectively (Krogh et al. 2001; Nielsen 2017; Mistry et al. 2021). We examined GO enrichment with TopGO (v2.50.0; Alexa and Rahnenfuhrer 2020), using a reference transcriptome for the “universe” dataset and various differentially expressed or co-expressed gene sets as the “test” dataset. We then reduced the redundancy of GO terms, and clarified GO relationships (e.g. parent-to-child) to one another using the GO-Figure! Python package (Reijnders and Waterhouse 2021).
Tissue-level Analyses of Differential Gene Expression
For DE analyses, we determined in each study species the differentially upregulated genes of three tissue types—upper lips, compound eyes, and guts—using five biological replicates for each tissue/species combination (supplementary table S2, Supplementary Material online). We employed DEseq2 (v1.40.2) on each species separately, assuming a P < 0.05 and FC > 1.5 for the significance of differentially expressed genes using the Benjamini–Hochberg method to account for false discovery rate (Love et al. 2014). Within each species, pairwise comparisons were done across tissue types (i.e. upper lip to compound eye, upper lip to gut, gut to compound eye), but to determine tissue-specific differential upregulation, each tissue was compared with the other two (e.g. genes upregulated uniquely in the upper lip were determined by comparing upper lip expression to both compound eye and gut; supplementary fig. S1, Supplementary Material online). Genes upregulated uniquely in each tissue type were used for all downstream analyses. We performed Fisher's exact tests between species to determine whether there was a statistically significant number of shared genes between tissue types (supplementary table S2, Supplementary Material online).
Constructing Gene Co-Expression Networks
To quantify co-expression, we used WGCNA (v1.72.1) in R to determine weighted co-expression networks in V. tsujii, to identify a BCN, and to compare the BCN to gene interactions in a nonluminous relative (Langfelder and Horvath 2008). WGCNA facilitates the identification and characterization of co-expression networks by clustering genes based on shared co-expression patterns (Langfelder and Horvath 2008). Genes clustered together in the same module (network) are often co-regulated, meaning they are likely involved in similar biological processes (Langfelder and Horvath 2008). For each module, WGCNA identifies “hub genes”—genes with high network connectivity represented by a module membership value (MM > 0.8)—which are considered integral to regulating the expression of other genes in the module and associated with key processes represented by the module (Langfelder and Horvath 2008). The input expression matrix for V. tsujii co-expression included 57 genes as rows and 33,479 samples as columns with samples normalized with the variance-stabilizing transformation in DESeq2. Following the recommendation by WGCNA, we removed 6 samples from the original set of 63 RNA-seq samples due to low counts (Langfelder and Horvath 2008). Additionally, genes with zero counts and genes with less than five counts in more than three samples were excluded to reduce noise (Langfelder and Horvath 2008). We checked for batch effects between samples from different RNA-seq runs and extraction methods and found that results remained qualitatively consistent across conditions. Similarities in expression were calculated by Pearson correlation, creating a matrix that was transformed into an adjacency matrix by raising the correlation to a soft threshold power to preserve the strongest correlations and reduce noise. We chose a soft threshold of three using “pickSoftThreshold” and created an adjacency matrix as a “signed” network, where modules correspond to positively correlated genes. This threshold of three satisfied the scale-free topology criterion for our dataset, where the R2 index of scale-free topology is above 0.9 (Langfelder and Horvath 2008). We used a threshold of 0.05 and a minimum module size of 30 to merge similar expression profiles, leading to 20 co-expression modules in V. tsujii. Of these, we identified a BCN as the module containing a c-luciferase shown to function in bioluminescence (Hensley et al. 2021; supplementary table S3, Supplementary Material online).
Determining Orthologous Genes and Comparative Transcriptomics
To generate a cross-species expression matrix, we first determined one-to-one orthologs across the reference transcriptomes of V. tsujii and Skogsbergia sp. using OrthoFinder (Emms and Kelly 2019), which infers gene families that originated before each of the ancestral nodes of a species tree. For cross-species comparison, we used only the upper lip, gut, and eye expression data to allow comparable datasets between V. tsujii and Skogsbergia. To determine the similarity of expression between the upper lips across species, compared with the eye and gut tissue, we performed principal component analysis (PCA) using one-to-one orthologs expressed in both species. To compare across samples, expression counts of a cross-species expression matrix were filtered and normalized by adding a pseudo count of 1 × 10−5 to prevent log2(0) scores, followed by a log2 transformation using the log2 function in DESeq2 (Love et al. 2014). The batch effect caused by multiple species was removed using an empirical Bayes method performed by the ComBat function in sva (v3.19; Leek et al. 2012).
Assessment of Clade-Specific Genes
A recent phylogeny of cypridinid ostracods showed a single origin of bioluminescence and a single subsequent transition to using bioluminescence for courtship (Ellis et al. 2022). To identify genes in V. tsujii that originated just before cypridinid bioluminescence (Luminini), just before bioluminescent courtship signaling (Luxorina), or before the origin of ostracods (which we termed “Ostracoda” or “All Arthropods”), we used Orthofinder, adding previously published transcriptomes with taxon sampling summarized in supplementary table S4, Supplementary Material online (Emms and Kelly 2019). We analyzed orthologous groups using KinFin (v1.1) using four user-defined taxon sets: Arthropoda, Ostracoda, Luminini, and Luxorina (Laetsch and Blaxter 2017). For V. tsujii, we determined how many expressed genes are found in each clade and how many are clade-specific genes (without nested genes; Laetsch and Blaxter 2017). We performed Fisher's exact tests between tissue types and modules to determine whether there was a statistically significant number of Luminini specific (without nested genes) in the BCN and BUL compared with conserved genes shared with nonluminous ostracods and arthropods (supplementary table S4, Supplementary Material online).
Identifying Secretory Pathway Genes and Putative Secreted Transcripts
To identify protein secretory pathway genes in the BCN and significantly upregulated in the upper lips of V. tsujii and Skogsbergia sp., we created a reference database of protein secretory pathway genes found in Feizi et al. (2017). We performed a blast search with an e-value threshold of 1e–5 against this reference database and additionally searched for transcripts with relevant gene annotations and GO terms associated with the secretory protein pathway (supplementary table S5, Supplementary Material online). To identify putative secreted transcripts, we retrieved all transcripts with a signal peptide and without a transmembrane domain from the set of genes that are both co-expressed with c-luciferase in the BCN and also significantly upregulated in the bioluminescent and non-BUL. Transcripts were identified as putative toxin-like transcripts if they had a domain present in known toxin protein families related to toxins. We manually searched for InterProIDs for known protein toxin families or domains listed in von Reumont et al. (2017), as summarized in supplementary table S6, Supplementary Material online, and any GO terms related to toxin and venom (von Reumont et al. 2017). Next, to determine whether the number of putative toxin-like genes in the BUL is significant, we compared the number of toxin-like transcripts in the BUL to the number of toxin-like transcripts found in tissue types not expected to contain toxin-like transcripts (compound eye and gut). We used similar steps to identify putative toxin-like transcripts expressed in the non-BUL compared with the two tissue types not predicted to be involved in defense, the compound eye and gut of Skogsbergia sp. For each species, we performed Fisher's exact tests to determine if both upper lips express a significantly higher number of putative toxin-like transcripts compared with the compound eye and gut. We also expanded our search for putative toxin-like transcripts by performing a blast search against the ToxProt database which includes all genes expressed in venomous or poisonous tissues across different phyla, summarized in supplementary table S6, Supplementary Material online (Jungo and Bairoch 2005).
Supplementary Material
Acknowledgments
The authors thank T. Fallon, S. Yi, and L. Babonis for commenting on earlier drafts.
Contributor Information
Lisa Y Mesrop, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA.
Geetanjali Minsky, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA.
Michael S Drummond, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA.
Jessica A Goodheart, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA; Division of Invertebrate Zoology, American Museum of Natural History, New York, NY 10024, USA.
Stephen R Proulx, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA.
Todd H Oakley, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, CA 93106, USA.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Funding
This study was funded by the Society for the Study of Evolution (Graduate Research Exellence Award), the Wrigley Institute for Environmental Studies, University of Southern California (Wrigley Institute Graduate Fellowship), and the University of California, Santa Barbara (Worster Summer Research Fellowship). Use was made of computational facilities purchased with funds from the National Science Foundation (CNS-1725797) and administered by the Center for Scientific Computing (CSC). The CSC is supported by the California NanoSystems Institute and the Materials Research Science and Engineering Center (MRSEC; NSF DMR 2308708) at University of California, Santa Barbara. This work was supported by the US National Science Foundation grants DEB-2153773 and IOS-1754770 to T.H.O.
Data Availability
All code and additional figures and tables can be found at Github, https://github.com/lmesrop/BCN_publication and Zenodo, https://doi.org/10.5281/zenodo.13841021. All study data are included in the article and supplementary material. Raw sequencing reads for all samples used in the DE and WGCNA analyses, along with the de novo transcriptome assembly for the nonluminous Skogsbergia sp., have been deposited in the National Center for Biotechnology Information Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject: PRJNA1109557.
References
- Abe K, Ono T, Yamada K, Yamamura N, Ikuta K. Multifunctions of the upper lip and a ventral reflecting organ in a bioluminescent ostracod Vargula hilgendorfii (Müller, 1890). Hydrobiologia. 2000:419(1):73–82. 10.1023/A:1003998327116. [DOI] [Google Scholar]
- Abe K, Vannier J. Functional morphology and significance of the circulatory system of Ostracoda, exemplified by Vargula hilgendorfii (Myodocopida). Mar Biol. 1995:124(1):51–58. 10.1007/BF00349146. [DOI] [Google Scholar]
- Alexa A, Rahnenfuhrer J. TOPGO: enrichment analysis for gene ontology 2.40.0. R package version. 2020:2:2010.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aparecido dos Santos-Pinto JR, Delazari dos Santos L, Arcuri HA, Ribeiro da Silva Menegasso A, Pêgo PN, Santos KS, Castro FM, Kalil JE, De-Simone SG, Palma MS, et al. B-cell linear epitopes mapping of antigen-5 allergen from Polybia paulista wasp venom. J Allergy Clin Immunol. 2015:135(1):264–267.e8. 10.1016/j.jaci.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Babonis LS, Martindale MQ, Ryan JF. Do novel genes drive morphological novelty? An investigation of the nematosomes in the sea anemone Nematostella vectensis. BMC Evol Biol. 2016:16(1):1–22. 10.1186/s12862-016-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua A, Mikheyev AS. An ancient, conserved gene regulatory network led to the rise of oral venom systems. Proc Natl Acad Sci U S A.. 2021:118(14):e2021311118. 10.1073/pnas.2021311118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcaid M, Casaburi G, McAnulty SJ, Schmidbaur H, Suria AM, Moriano-Gutierrez S, Pankey MS, Oakley TH, Kremer N, Koch EJ, et al. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc Natl Acad Sci U S A. 2019:116(8):3030–3035. 10.1073/pnas.1817322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank S, Bazon ML, Grosch J, Schmidt-Weber CB, Brochetto-Braga MR, Bilò MB, Jakob T. Antigen 5 allergens of hymenoptera venoms and their role in diagnosis and therapy of venom allergy. Curr Allergy Asthma Rep. 2020:20(10):58. 10.1007/s11882-020-00954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Protein Sci. 1995:4(7):1247–1261. 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner A, Badroos JM, Learsch RW, Yousefelahiyeh M, Kitchen SA, Parker J. Evolutionary assembly of cooperating cell types in an animal chemical defense system. Cell. 2021:184(25):6138–6156.e28. 10.1016/j.cell.2021.11.014. [DOI] [PubMed] [Google Scholar]
- Brückner A, Parker J. Molecular evolution of gland cell types and chemical interactions in animals. J Exp Biol. 2020:223(Suppl_1). 10.1242/jeb.211938. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo T-H, Davis FG, et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017:18(3):762–776. 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd GE. The origin and evolution of the euarthropod labrum. Arthropod Struct Dev. 2021:62:101048. 10.1016/j.asd.2021.101048. [DOI] [PubMed] [Google Scholar]
- Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014:21(3):396–413. 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras-Sureda A, Pihán P, Hetz C. Calcium signaling at the endoplasmic reticulum: fine-tuning stress responses. Cell Calcium. 2018:70:24–31. 10.1016/j.ceca.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wan Y, Lei Y, Zobel J, Verspoor K. Evaluation of CD-HIT for constructing non-redundant databases. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2016. 2016:703–706. 10.1109/BIBM.2016.7822604. [DOI] [Google Scholar]
- Claes JM, Delroisse J, Grace MA, Doosey MH, Duchatelet L, Mallefet J. Histological evidence for secretory bioluminescence from pectoral pockets of the American Pocket Shark (Mollisquama mississippiensis). Sci Rep. 2020:10(1):18762. 10.1038/s41598-020-75656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AC, Morin JG. Sexual morphology, reproduction and the evolution of bioluminescence in ostracoda. Paleontol Soc Papers. 2003:9:37–70. 10.1017/S108933260000214X. [DOI] [Google Scholar]
- Ellis EA, Goodheart JA, Hensley NM, González VL, Reda NJ, Rivers TJ, Morin JG, Torres E, Gerrish GA, Oakley TH, et al. Sexual signals persist over deep time: ancient co-option of bioluminescence for courtship displays in cypridinid ostracods. Syst Biol. 2022:72(2):264–274. 10.1093/sysbio/syac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EA, Oakley TH. High rates of species accumulation in animals with bioluminescent courtship displays. Curr Biol. 2016:26(14):1916–1921. 10.1016/j.cub.2016.05.043. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019:20(1):1–14. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizi A, Gatto F, Uhlen M, Nielsen J. Human protein secretory pathway genes are expressed in a tissue-specific pattern to match processing demands of the secretome. NPJ Syst Biol Appl. 2017:3(1):22. 10.1038/s41540-017-0021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009:10(1):483–511. 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Goodheart JA, Minsky G, Brynjegard-Bialik MN, Drummond MS, Munoz JD, Fallon TR, Schultz DT, Weng J-K, Torres E, Oakley TH, et al. Laboratory culture of the California sea firefly Vargula tsujii (Ostracoda: Cypridinidae): developing a model system for the evolution of marine bioluminescence. Sci Rep. 2020:10(1):10443. 10.1038/s41598-020-67209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RM, Oakley T, Goldstone JV, Dugas JC, Brady ST, Gow A. Myelin sheaths are formed with proteins that originated in vertebrate lineages. Neuron Glia Biol. 2008:4(2):137–152. 10.1017/S1740925X09990238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011:29(7):644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock SHD, Moline MA, Case JF. Bioluminescence in the sea. Ann Rev Mar Sci. 2010:2(1):443–493. 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- Harvey EN. Bioluminescence. New York: Academic Press; 1952. [Google Scholar]
- Haskell EC, Bell J. A model of the burglar alarm hypothesis of prey alarm calls. Theor Popul Biol. 2021:141:1–13. 10.1016/j.tpb.2021.05.004. [DOI] [PubMed] [Google Scholar]
- Hensley NM, Ellis EA, Leung NY, Coupart J, Mikhailovsky A, Taketa DA, Tessler M, Gruber DF, De Tomaso AW, Mitani Y, et al. Selection, drift, and constraint in cypridinid luciferases and the diversification of bioluminescent signals in sea fireflies. Mol Ecol. 2021:30(8):1864–1879. 10.1111/mec.15673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvard AL. Ultrastructure of the light organ and immunocytochemical localization of luciferase in luminescent marine ostracods (Crustacea: Ostracoda: Cypridinidae). J Morphol. 1993:218(2):181–193. 10.1002/jmor.1052180207. [DOI] [PubMed] [Google Scholar]
- Jackson BD, Morgan ED. Insect chemical communication: pheromones and exocrine glands of ants. Chemoecology. 1993:4(3-4):125–144. 10.1007/BF01256548. [DOI] [Google Scholar]
- Jarc E, Petan T. Lipid droplets and the management of cellular stress. Yale J Biol Med. 2019:92:435–452. [PMC free article] [PubMed] [Google Scholar]
- Jasper WC, Linksvayer TA, Atallah J, Friedman D, Chiu JC, Johnson BR. Large-scale coding sequence change underlies the evolution of postdevelopmental novelty in honey bees. Mol Biol Evol. 2015:32(2):334–346. 10.1093/molbev/msu292. [DOI] [PubMed] [Google Scholar]
- Johnson PM, Kicklighter CE, Schmidt M, Kamio M, Yang H, Elkin D, Michel WC, Tai PC, Derby CD, et al. Packaging of chemicals in the defensive secretory glands of the sea hare Aplysia californica. J Exp Biol. 2006:209(1):78–88. 10.1242/jeb.01972. [DOI] [PubMed] [Google Scholar]
- Jungo F, Bairoch A. Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledgebase. Toxicon. 2005:45(3):293–301. 10.1016/j.toxicon.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Kishi T, Goto T, Hirata Y, Shimomura O, Johnson FH. Cypridina bioluminescence I sructure of cypridina luciferin. Tetrahedron Lett. 1966:7(29):3427–3436. 10.1016/S0040-4039(01)82806-9. [DOI] [Google Scholar]
- Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SV. The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb. Structure. 2009:17(4):611–619. 10.1016/j.str.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001:305(3):567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. KinFin: software for taxon-aware analysis of clustered protein sequences. G3. 2017:7(10):3349–3357. 10.1534/g3.117.300233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008:9(1):559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau ES, Goodheart JA, Anderson NT, Liu VL, Mukherjee A, Oakley TH. Similar enzymatic functions in distinct bioluminescence systems: evolutionary recruitment of sulfotransferases in ostracod light organs. Biol Lett. 2024:20(5):20230585. 10.1098/rsbl.2023.0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau ES, Oakley TH. Multi-level convergence of complex traits and the evolution of bioluminescence. Biol Rev Camb Philos Soc. 2021:96(2):673–691. 10.1111/brv.12672. [DOI] [PubMed] [Google Scholar]
- Lee JM. Nuclear receptors resolve endoplasmic reticulum stress to improve hepatic insulin resistance. Diabetes Metab J. 2017:41(1):10–19. 10.4093/dmj.2017.41.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012:28(6):882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011:12(1):323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey M, Schell J, Sandland GJ, Gerrish GA. Luminescent syllid (Odontosyllis spp.) courtship display densities vary across marine habitats around South Water Caye. Belize. 2017:28:40–45. 10.18785/gcr.2801.12. [DOI] [Google Scholar]
- Mesrop LY. Total RNA extraction protocol for marine ostracods (bioluminescent and non-bioluminescent ostracods). protocols.io. 2024. 10.17504/protocols.io.eq2lyw6dmvx9/v1. [DOI] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021:49(D1):D412–D419. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin JG. Coastal bioluminescence: patterns and functions. Bull Mar Sci. 1983:33:787–817. https://api.semanticscholar.org/CorpusID:89229837. [Google Scholar]
- Morin JG. Firefleas of the sea: luminescent signaling in marine ostracode crustaceans. Fla Entomol. 1986:69(1):105–121. 10.2307/3494749. [DOI] [Google Scholar]
- Morin JG. Luminaries of the reef: the history of luminescent ostracods and their courtship displays in the Caribbean. J Crustacean Biol. 2019:39(3):227–243. 10.1093/jcbiol/ruz009. [DOI] [Google Scholar]
- Morin JG, Cohen AC. Bioluminescent displays, courtship, and reproduction in ostracodes. In: Bauer Raymond T., Martin Joel W., editors. Crustacean sexual biology. New York: Columbia University Press; 1991. p. 1–16. [Google Scholar]
- Nakajima Y, Kobayashi K, Yamagishi K, Enomoto T, Ohmiya Y. cDNA cloning and characterization of a secreted luciferase from the luminous Japanese ostracod, Cypridina noctiluca. Biosci Biotechnol Biochem. 2004:68(3):565–570. 10.1271/bbb.68.565. [DOI] [PubMed] [Google Scholar]
- Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017:1611:59–73. 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- Oakley T. Building, maintaining, and (re-)deploying genetic toolkits during convergent evolution. Integr Comp Biol. 2024. doi: 10.1093/icb/icae114. [DOI] [PubMed] [Google Scholar]
- Oakley TH. Myodocopa (Crustacea: Ostracoda) as models for evolutionary studies of light and vision: multiple origins of bioluminescence and extreme sexual dimorphism. Hydrobiologia. 2005:538:179–192. 10.1007/s10750-004-4961-5. [DOI] [Google Scholar]
- Pick T, Beck A, Gamayun I, Schwarz Y, Schirra C, Jung M, Krause E, Niemeyer BA, Zimmermann R, Lang S, et al. Remodelling of Ca2+ homeostasis is linked to enlarged endoplasmic reticulum in secretory cells. Cell Calcium. 2021:99:102473. 10.1016/j.ceca.2021.102473. [DOI] [PubMed] [Google Scholar]
- Reijnders MJMF, Waterhouse RM. Summary visualizations of gene ontology terms with GO-figure! Front Bioinform. 2021:1:638255. 10.3389/fbinf.2021.638255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers TJ, Morin JG. The relative cost of using luminescence for sex and defense: light budgets in cypridinid ostracods. J Exp Biol. 2012:215(16):2860–2868. 10.1242/jeb.072017. [DOI] [PubMed] [Google Scholar]
- Rollins RE, Staub NL. The presence of caudal courtship-like glands in male and female ouachita dusky salamanders (Desmognathus brimleyorum). Herpetologica. 2017:73(4):277–282. 10.1655/Herpetologica-D-17-00003.1. [DOI] [Google Scholar]
- Schaefer HM. Visual communication: evolution, ecology, and functional mechanisms. In: Kappeler P, editor. Animal behaviour: evolution and mechanisms. Heidelberg: Springer Berlin; 2010. p. 3–28. [Google Scholar]
- Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015:40(3):141–148. 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura O. Bioluminescence: chemical principles and methods. Singapore: World Scientific; 2012. [Google Scholar]
- Tanguy E, Carmon O, Wang Q, Jeandel L, Chasserot-Golaz S, Montero-Hadjadje M, Vitale N, et al. Lipids implicated in the journey of a secretory granule: from biogenesis to fusion. J Neurochem. 2016:137(6):904–912. 10.1111/jnc.13577. [DOI] [PubMed] [Google Scholar]
- Thompson EM, Nagata S, Tsuji FI. Vargula hilgendorfii luciferase: a secreted reporter enzyme for monitoring gene expression in mammalian cells. Gene. 1990:96(2):257–262. 10.1016/0378-1119(90)90261-O. [DOI] [PubMed] [Google Scholar]
- Trhlin M, Rajchard J. Chemical communication in the honeybee (Apis mellifera L.): a review. Vet Med. 2011:56(6):265–273. 10.17221/1543-VETMED. [DOI] [Google Scholar]
- Underwood TJ, Tallamy DW, Pesek JD. Bioluminescence in firefly larvae: a test of the aposematic display hypothesis (Coleoptera: Lampyridae). J Insect Behav. 1997:10(3):365–370. 10.1007/BF02765604. [DOI] [Google Scholar]
- Vannier J, Abe K, Ikuta K. Feeding in myodocopid ostracods: functional morphology. Mar Biol. 1998:132(3):391–408. 10.1007/s002270050406. [DOI] [Google Scholar]
- Verdes A, Gruber DF. Glowing worms: biological, chemical, and functional diversity of bioluminescent annelids. Integr Comp Biol. 2017:57(1):18–32. 10.1093/icb/icx017. [DOI] [PubMed] [Google Scholar]
- von Reumont BM, Undheim EAB, Jauss R-T, Jenner RA. Venomics of remipede crustaceans reveals novel peptide diversity and illuminates the Venom's biological role. Toxins (Basel). 2017:9(8). 10.3390/toxins9080234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. Evolutionary innovations and novelties: let us get down to business!. Zool Anz J Comp Zool. 2015:256:75–81. doi: 10.1016/j.jcz.2015.04.006. [DOI] [Google Scholar]
- Walker AA, Madio B, Jin J, Undheim EAB, Fry BG, King GF. Melt with this kiss: paralyzing and liquefying venom of the assassin bug Pristhesancus plagipennis (Hemiptera: Reduviidae). Mol Cell Proteomics. 2017:16(4):552–566. 10.1074/mcp.M116.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AA, Robinson SD, Undheim EAB, Jin J, Han X, Fry BG, Vetter I, King GF. Missiles of mass disruption: composition and glandular origin of venom used as a projectile defensive weapon by the assassin bug Platymeris rhadamanthus. Toxins (Basel). 2019:11(11). 10.3390/toxins11110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011:334(6059):1081–1086. 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018:35(3):543–548. 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Juenger TE. A high-throughput 3′-tag RNA sequencing for large-scale time-series transcriptome studies. Methods Mol Biol. 2022:2398:151–172. 10.1007/978-1-0716-1912-4_13. [DOI] [PubMed] [Google Scholar]
- Zancolli G, Reijnders M, Waterhouse RM, Robinson-Rechavi M. Convergent evolution of venom gland transcriptomes across Metazoa. Proc Natl Acad Sci U S A. 2022:119(1). 10.1073/pnas.2111392119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang L, Zhou L, Lei Y, Zhang Y, Huang C. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol. 2019:25:101047. 10.1016/j.redox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code and additional figures and tables can be found at Github, https://github.com/lmesrop/BCN_publication and Zenodo, https://doi.org/10.5281/zenodo.13841021. All study data are included in the article and supplementary material. Raw sequencing reads for all samples used in the DE and WGCNA analyses, along with the de novo transcriptome assembly for the nonluminous Skogsbergia sp., have been deposited in the National Center for Biotechnology Information Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProject: PRJNA1109557.