Abstract
Herpes Simplex Virus 1 (HSV-1) is a member of the alpha-Herpesviridae family, with over 60 % of the global population being seropositive. HSV-1 can cause a range of symptoms, from mild discomfort to severe, life-threatening complications. The emergence of HSV-1-resistant strains and the diminishing effectiveness of current antiviral treatments highlight the urgent need for new antiviral drugs. Computational molecular docking studies can offer valuable insights for the development of novel antiviral compounds targeting HSV-1. To address this need, we have developed HerpDock, an open-source Graphical User Interface (GUI) program designed for molecular docking, high-throughput virtual screening, and absorption, distribution, metabolism, and elimination (ADME) studies. HerpDock stands out as a unique, user-friendly pipeline that extends beyond the capabilities of existing docking programs based on AutoDock Vina. It requires only the ligand name or structure from the user, as it already includes optimized structures of numerous HSV-1 viral proteins and relevant host proteins. With just a few clicks, HerpDock performs comprehensive docking studies, result analysis, and visualization, making it particularly valuable for Herpes researchers without a bioinformatics background. HerpDock is freely available at https://github.com/Sudhk24/Herpdock
Keywords: Docking, HSV-1, Antiviral, Computation Studies, Graphical User Interface
Graphical Abstract
1. Introduction
Herpesviridae is a family of enveloped double-stranded DNA viruses, comprising eight human herpesviruses (HHVs) categorized into three subfamilies: alpha-Herpesviridae (including herpes simplex virus (HSV) and varicella-zoster virus (VZV)), beta-Herpesviridae (including cytomegalovirus (CMV), HHV-6, and HHV-7), and gamma-Herpesviridae (represented by Epstein–Barr virus (EBV) and HHV-8) [1]. These viruses establish latent infections, with the potential for reactivation due to various endogenous or exogenous triggers, leading to a spectrum of clinical manifestations [2], [3], [4], [5], [6], [7]. Among them, Herpes Simplex Virus-1 (HSV-1) is particularly notable for its global prevalence, affecting approximately 66.6 % of the population aged 0–49 years worldwide [8], [9], [10], [11], [12], [13], [14]. Although many individuals infected with HSV-1 remain asymptomatic, the virus can cause a range of conditions, from mild orofacial blisters to severe complications such as infectious blindness and, in rare cases, encephalitis, particularly in immunocompromised individuals [15], [16], [17]. The virus's ability to establish lifelong latency and undergo periodic reactivation presents a formidable public health challenge.
Current antiviral therapies, including acyclovir (ACV), penciclovir, valacyclovir, famciclovir, foscarnet, and cidofovir, primarily function by inhibiting viral DNA synthesis via targeting the viral DNA polymerase [15]. However, these treatments do not completely eradicate the infection but help reduce the severity and frequency of its reactivation episodes. The emergence of drug-resistant HSV-1 strains, particularly in immunocompromised patients, coupled with concerns of drug efficacy, highlight the dire need for new antiviral agents with novel mechanisms of action [18].
The traditional drug discovery process is lengthy, costly, and characterized by a high attrition rate. In recent years, Computer-Aided Drug Design (CADD) has emerged as a powerful approach to expedite this process. CADD utilizes computational methods to predict the interactions between drug candidates and their biological targets, streamlining the identification of potential therapeutic compounds [19]. Molecular docking, a pivotal component of CADD, plays a crucial role in predicting the most favorable binding orientations, conformations, and affinities of small molecules to specific target proteins. The stability of these protein-ligand complexes can be further evaluated through Molecular Dynamics (MD) simulations, offering insights into the dynamic behavior of the complexes under physiological conditions.
In addition to molecular docking, computational ADME (Absorption, Distribution, Metabolism, and Excretion) studies are indispensable in drug discovery and development. These studies can be integrated with other computational techniques like QSAR (Quantitative Structure-Activity Relationship) and pharmacophore modeling, providing a comprehensive understanding of a drug candidate’s efficacy and safety profile. The integration of molecular docking and ADME calculations offers a holistic view of a drug candidate’s potential, providing insights into both its inhibition efficacy and safety profile.
Despite the availability of several open-source GUI-based programs for molecular docking [20], most require the use of multiple tools during the pre- or post-processing stages, necessitating advanced computational expertise. This complexity can be a significant barrier for researchers without extensive computational backgrounds. To address these challenges, we have developed HerpDock, a windows-based software tailored for HSV-1 research. HerpDock simplifies the process of molecular docking and high-throughput virtual screening of HSV-1 viral proteins and host proteins involved in HSV-1 infection. With preprocessed target proteins, users can perform docking and basic ADME studies with just a few clicks by providing ligand files or PubChem IDs and selecting the target protein via a user-friendly dropdown menu. By integrating these capabilities, HerpDock aims to accelerate the discovery of novel antiviral compounds and the development of effective therapies for HSV-1 infections.
2. Materials and methods
2.1. HerpDock pipeline
HerpDock is a GUI-based pipeline written using Python that automates the entire process of molecular docking, virtual screening and ADME studies with just a few clicks. HerpDock can be run as a standalone desktop application on Microsoft Windows 10 or higher and requires Open Babel [21] to be preinstalled on the system. HerpDock already includes structures of many HSV-1 proteins and other host proteins which play a significant role in herpes infection. These proteins have been optimized beforehand using Schrödinger Protein Preparation Wizard [22] This wizard checks each protein for inconsistencies and broken chains. It repairs them, adds polar hydrogens, and finally minimizes the protein using OPLS forcefield [23], [24]. Search space configuration (grid) parameters have also been generated for these proteins centered on their active site (coordinates provided in Table 1). These proteins can readily be used for molecular docking studies later.
Table 1.
Host and Viral Proteins along with their Active Sites Used in HerpDock.".
| Positive Control | |||||
|---|---|---|---|---|---|
| Protein | Reference for HSV−1 | Reference for Docking | PDB ID | Active Site | Sorafenib |
| STAT3 | [32] | [39] | 1BG1 | Arg609, Arg595, Lys591, Arg609, Ser611, Glu612, Ser613, Gln633, Ile634, Gln635, Ser636, Val637, Gln638, Met586, Gly587, Phe588, Ilu589, and Ser590 | Rapamycin |
| mTOR(rapamycin binding site) | [33] | [40] | 4DRI | PHE2108,TYR2105,PHE77,LYS88,TYR113,VAL86,SER118,ASP68,PHE2039,PHE130 | OGT 2115 |
| Heperanase | [34] | [34] | 5E9C | Thr97, Asn224, His296, Gln383, Arg93 and Lys232 | Stevioside |
| AKT | [35] | [41] | 3QKM | HIS134,LYS276,GLU278 | Penciclovir |
| HSV−1 thymidine kinase | [36] | [36] | 1KI3 | Arg176, Tyr172, Gln125, His58 and Glu83 | Acyclovir |
| HSV−1 DNA polymerase | [37] | [37] | 2GV9 | ASP717,PHE718,TYR722,PRO723,TYR818,VAL817 | Not Available |
| gD (HVEM binding site) | [38] | [42] | 1JMA | Ala7, Ser8, Leu9, Lys10, Met11, Ala12, Asp13, Pro14, Asn15, Val24, Leu25, Asp26, Gln27, Leu28, Thr29, Asp30, Pro31, Pro32 | Not Available |
| Nectin−1 (gD binding site) | [38] | [42] | 3U82 | Ser59, Lys61, Thr63, Gln64, Thr66, Gln68, Lys75, Gln76, Asn77, Ile80, Tyr81, Asn82, Met85, Gly86, Val87, Ser88, Leu90, Glu125, Ala127, Thr128, Phe129, Pro130, Thr131, Gly132, and Asn133 | Not Available |
| gD (Nectin−1 binding site) | [38] | [42] | 3U82 | Pro23, Leu25, Gln27, Arg36, Val37, Tyr38, His39, Gln132, Val214, Asp215, Ser216, Ile217, Gly218, Met219, Leu220, Pro221, Arg222, Phe223, Thr230, Val231, Tyr234 | Not Available |
| HVEM (gD binding site) | [38] | [42] | 1JMA | Pro17, Lys18, Cys19, Ser20, Pro21, Gly22, Tyr23, Arg24, Val25, Lys26, Gly30, Glu31, Leu32 Thr33, Gly34, Thr35, Val36, Cys37, Glu38, Pro39, Ser74, Arg75, and Thr76 | Not Available |
HerpDock incorporates several other programs such as Open Babel, AutoDock Vina, Rasmol, and ChEMBL. Open Babel is used for the conversion of different molecular file formats and minimizing ligand files. The latest version of AutoDock Vina (version:1.2.5) [25], [26] is used for molecular docking. Rasmol 2.7.2.1.1 [27] is used for visualization of complexes. ChEMBL [28] is used for obtaining ADME properties for the ligand molecules.
2.2. Cells and Viruses
HCE cells (RCB1834 HCE-T) were obtained from K. Hayashi (National Eye Institute, Bethesda, MD) and were cultured in minimum essential medium (MEM) (Life Technologies, Carlsbad, CA) with 10 % fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO) and 1 % penicillin-streptomycin (P/S) (Life Technologies, Carlsbad, CA). African green monkey kidney (Vero) cells were provided by P. G. Spear (Northwestern University). Vero cells were passaged in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with 10 % FBS and P/S.
HSV-1 (KOS-WT), GFP–HSV-1 (K26-GFP), and β-galactosidase–expressing HSV-1 (gL86), used in this study were provided by P. Spear’s laboratory at Northwestern University. Virus stocks were propagated and tittered on Vero cells and stored at −80 °C.
2.3. Plaque assay
HCE cells infected with HSV-1 (KOS-WT) were collected in Corning™ CellStripper Dissociation Reagent and subsequently resuspended in 1 ml of Opti-MEM (Life Technologies). The samples underwent sonication for 10 s at 70 % amplitude using a probe sonication system, with the probe surface cleaned with 70 % ethanol between each sample. The resulting cell lysates and supernatant were serially diluted in Opti-MEM and then added to Vero cell plates at 90–100 % confluence. After a 2-hour incubation at 37 °C with the diluted virus, the media was replaced with 0.05 % methylcellulose in DMEM. The plates were incubated for 72 h at 37 °C. Following incubation, 500 μl of 100 % methanol was added to each well for 1 h to fix the cells. The methanol was then replaced with crystal violet to visualize the plaques.
2.4. Viral entry assay
Monolayer HCE cells were cultured in a 12-well plate. The cells were pretreated with Rilapladib at different concentrations (12.5 µM,3.1 µM, and 0.31 µM) for 24 h. After 24 h Rilapladib pre-treatment, cells were incubated with 10 MOI of HSV-1 (KOS-WT) virus for 30 min at 4 °C, followed by a 30-min incubation in 37 °C with 5 % CO2. Non-internalized virions were removed by vigorous washing with Phosphate-buffered saline (PBS) multiple times. The cells were then collected Corning™ CellStripper Dissociation Reagent and processed for western blot assay.
2.5. Entry assay imaging
HCE cells were plated in imaging dishes. The cells were pretreated with Rilapladib at 12.5 µM concentration for 24 h. The next day, cells were incubated with 10 MOI of HSV-1 K26-GFP (Green Fluorescent Protein) virus for 30 min at 4 °C, followed by a 30-min incubation in 37 °C with 5 % CO2. Non-internalized virions were removed by vigorous washing with PBS.
The cells were fixed in 4 % paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA), permeabilized with 0.01 % Triton-X (Thermo Fisher Scientific), and stained with rhodamine-phalloidin (Invitrogen, Thermo Fisher Scientific). We used 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies) as a counterstain. The images were captured under 63x objectives using an observer microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) with a spinning disk (CSU-X1; Yokogawa, Tokyo, Japan). We used digital imaging software (Axiovision; Carl Zeiss Microscopy GmbH) to quantify fluorescence intensity.
2.6. Drugs
All the drugs used in this study were procured from MedChem express. The drugs were diluted to stock concentrations (1000-fold higher than working concentration) then aliquoted into 50 μl vials and stored at −80 °C until use.
2.7. Cell viability assay
Cell viability assay using various concentrations of Rilapladib and Bizelesin was performed on HCE cells plated at a density of 1 × 104 per well in 96-well plates overnight. Concentrations starting at 50 µM and 1 µM for Rilapladib and Bizelesin, respectively, were twofold serially diluted in whole media and added to cell monolayers for a period of 24 h. At the end of incubation, 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT; BioVision, USA) at a concentration of 0.5 mg/ml in whole media was added to cells and incubated for a period of 3 h to allow crystal formation. Acidified isopropanol (1 % glacial acetic acid v/v) was added to cells to dissolve the formazan crystals. Dissolved violet crystals were transferred to new 96-well plates and analyzed through a microplate reader for absorption at 562 nm.
2.8. Viral β-galactosidase entry assay
β-Galactosidase–expressing viruses HSV-1 gL86 at MOI 10 was used to study viral entry into cells. HCEs were plated at a density of 1 × 104 in 96-well plates overnight before use. HSV-1 strain gL86 was mixed with multiple concentrations of Rilapladib and Bizelesin in MEM media and overlaid on HCEs to infect the cell monolayers for 6 h, after which the cells were washed with PBS twice and 100 μl of soluble substrate o-nitrophenyl-β-d-galactopyranoside (ONPG; 3 mg/ml; Thermo Fisher Scientific) was added to the cells along with 0.5 % NP-40 (USB Corporation, Cleveland, OH, USA) in PBS. Enzymatic activity was measured via a microplate reader at 405 nm.
2.9. In-vitro antiviral-GFP screening
HCE cells were plated at a density of 1 × 104 per well in 96-well plates overnight. We have performed two kinds of screening studies. In the pre-treatment study, HCE cells were pre-treated with Rilapladib and Bizelesin at different concentrations starting from 25 µM and 1 µM, respectively, for 24 h followed by infection with HSV1 K26-GFP at 0.1 MOI for 24 h. After 24 h post-infection (HPI), the GFP intensity was read using microplate reader at 479 nm and 520 nm. In the post-treatment study, HCE cells were first infected with HSV1 K26-GFP at 0.1 MOI for 2 h followed by treatment with Rilapladib and Bizelesin at different concentrations starting from 25 µM and 1 µM, respectively, for 22 h. After 24 HPI, the GFP intensity was read using microplate reader at 479 nm and 520 nm.
3. Results
3.1. Application overview
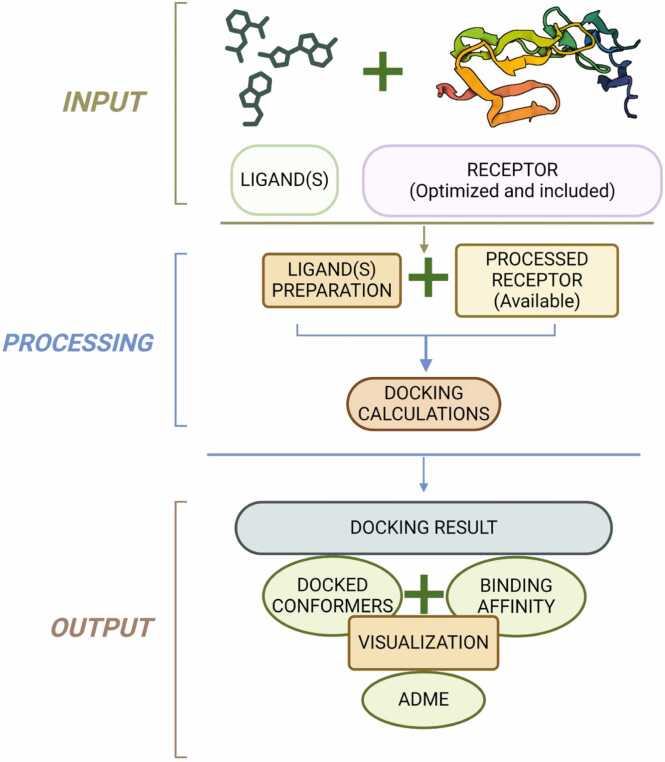
The workflow of HerpDock is summarized in Fig. 1 and can be broadly divided into three major steps:
-
1.
Ligand selection and target protein preparation: Users begin by providing the ligands and selecting the appropriate target protein. The target proteins included with HerpDock have been pre-optimized using Schrödinger’s Protein Preparation Wizard, ensuring they are ready for docking.
-
2.
Ligand optimization and molecular docking: In the next step, the provided ligands are optimized to their global minimum energy conformer and converted to pdbqt format using Open Babel. Molecular docking is then performed between the selected target protein and each ligand, one at a time, using AutoDock Vina.
-
3.
Output processing: Finally, the output files for each ligand are split into conformers using “vina_split”, a tool included with the AutoDock Vina distribution. Protein-ligand complexes are visualized using Rasmol. Additionally, ChEMBL is used for retrieval of ADME properties for small molecules.
Fig. 1.
Workflow of the HerpDock program that is divided into three major steps: (i) input of receptor and ligand(s) (ii) docking simulations of the receptor–ligand system using AutoDock Vina and (iii) output containing docking and ADME results.
3.2. Stepwise application workflow and usage
Using HerpDock is straightforward and requires only the preinstallation of Open Babel. Users can install Open Babel by navigating to the "Help" tab in the HerpDock application and selecting "OpenBabel," which directs them to the OpenBabel GitHub page for downloading and installation. Below are the step-by-step instructions for using HerpDock, illustrated with an example:
-
1.
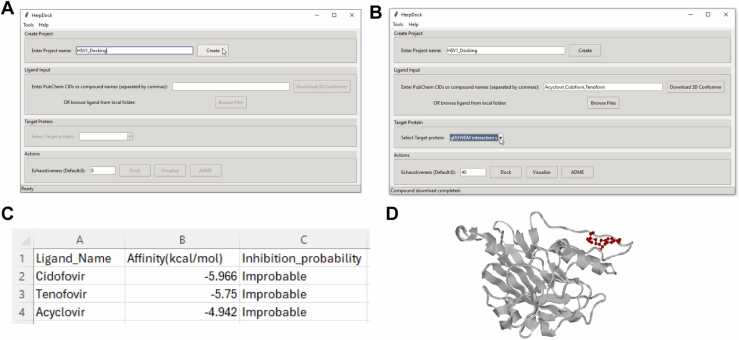
Project creation: The user starts by entering a project name and clicking the "Create" button. This action generates a project folder in the same directory as the HerpDock application. For instance, in the example shown in Fig. 2(A), the project was named “HSV-1 gD docking HVEM site”. Pressing the "Create" button created a corresponding folder.
-
2.
Ligand selection and target protein choice: After creating the project folder, the user can enter PubChem CIDs or compound names and press the "Download" button, which creates a "ligands" folder where all downloaded files are stored. Alternatively, users can select ligand files from their local system by clicking the "Browse" button, which also copies the selected files into the "ligands" folder. After selecting the ligands, the user must choose the target protein from the dropdown menu. In the example shown in Fig. 2(B), the names of three nucleotide analogs—cidofovir, tenofovir, and acyclovir—were entered, separated by commas. Pressing the "Download" button stored the files in the "ligands" folder, and “gD (HVEM interaction site)” was selected as the target protein.
-
3.
Docking and result analysis: When the "Dock" button is clicked, all ligands in the "ligands" folder are first optimized, converted to pdbqt format, and moved to a new "ligands_pdbqt" folder within the project directory. The molecular docking process between the target protein and ligands is then carried out using AutoDock Vina. Once docking is complete, the results are saved in the "Results" folder within the project directory, including a "Docking_results.csv" file and protein-ligand complexes stored in the "Ligand_Complex_PDB" folder. After docking, the "Visualize" and “ADME” buttons become accessible, allowing users to visualize the protein-ligand complexes and perform ADME studies on top-ranking ligands. ADME results are also stored in the "Results" folder. HerpDock performs ADME calculations using the ChEMBL only for ligands that are downloaded from Pubchem. In the example docking results shown in Fig. 2(C), the protein-ligand complex (gD-cidofovir) obtained after docking was visualized, Fig. 2(D).
Fig. 2.
Different steps to operate HerpDock: (A) Create a project (B) Input the name of compounds to be tested followed by clicking on “Download 3D Conformer” and selecting the target protein to inhibit (C) Click "Dock" to obtain the docking result/analysis file and (D) Protein(Grey) –Ligand(Red) complex (gD-cidofovir) file will also be generated.
3.3. In-silico screening utilizing herpdock gives rilapladib and bizelesin as candidates to block HSV-1 viral entry
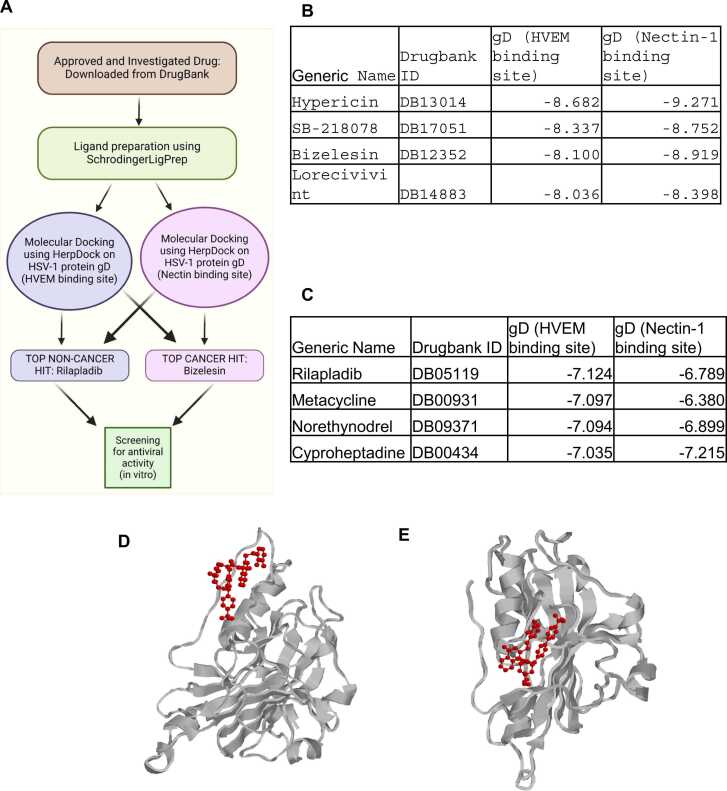
The interaction between glycoprotein D (gD) and its two primary cellular receptors, nectin-1 and HVEM (herpesvirus entry mediator), is characterized by non-reciprocal competitive binding. When gD binds to HVEM, it adopts a hairpin structure that occludes the binding site for nectin-1 [29]. Conversely, binding to nectin-1 blocks the HVEM binding sites on gD. Given the distinct amino acid residues involved in the binding of each receptor, we conducted targeted molecular docking on each site independently. Initial docking was performed against the HVEM binding site of gD, followed by a refined docking of compounds against the nectin-1 binding site. A schematic diagram illustrating the virtual screening process, and outcomes is provided in Fig. 3(A).
Fig. 3.
This case study uses HerpDock to reveal which drugs would inhibit gD interaction with HVEM or Nectin1: (A) Flow chart giving an overview of the case study (B) Top 4 cancer drugs that can inhibit gD interaction with HVEM and Nectin-1 (C) Top 4 non-cancer drugs that can inhibit gD interaction with HVEM and Nectin-1 (D) Visualization of top hit Rilapladib’s interaction with gD at it’s HVEM binding site (E) Visualization of top hit Rilapladib’s interaction with gD at it’s Nectin1 binding site.
HerpDock, utilizing AutoDock Vina, was employed to estimate the theoretical affinity between the HVEM binding site of gD and a library of over 5000 compounds. The docking scores are based on the change in Gibbs free energy between bound and unbound states. More negative scores reflect higher putative affinity. To assess inhibitory cross-reactivity with the nectin-1 binding site of gD, we performed additional docking analyses using HerpDock on compounds that showed a binding score of −7 or lower for the HVEM binding site.
Our screenings revealed that many of the top hits were compounds currently used in cancer treatment Fig. 3(B). For subsequent in vitro antiviral testing, we selected Rilapladib, the top non-cancer drug Fig. 3(C), to avoid the challenges associated with repurposing cancer drugs for viral infections. Cancer drugs are typically optimized for specific targets related to cancer and are not ideal for treating viral infections like herpes due to differences in disease mechanism, potential for severe side effects, lack of efficacy, and ethical concerns. However, despite these considerations, we included Bizelesin—an anticancer drug and a top hit for both HVEM and nectin-1 binding sites of gD—in further in-vitro antiviral testing due to its dual-site targeting potential.
3.4. Investigating the cell viability and antiviral activity of Rilapladib and Bizelesin By In-Vitro screening
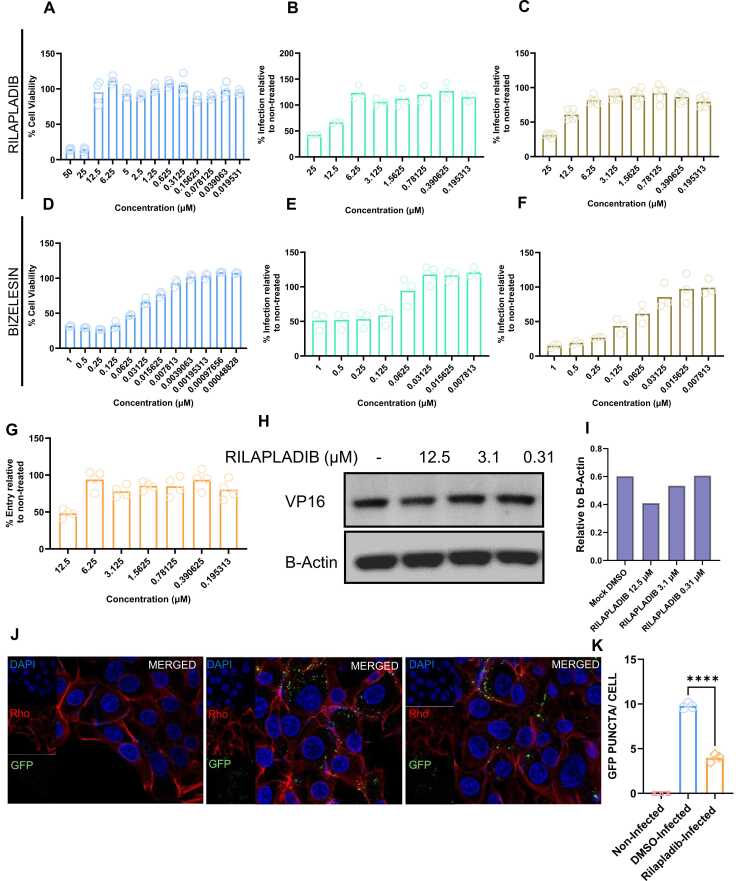
The in-vitro cytotoxicity of Rilapladib and Bizelesin was evaluated on HCE cells to identify non-toxic concentrations suitable for the subsequent evaluation of in-vitro antiviral activity. An MTT (3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide) assay was performed for different concentrations of Rilapladib Fig. 4(A) and Bizelesin Fig. 4(D). Our results showed more than 90 % cell viability at concentrations of 12.5 µM or lower for Rilapladib and 0.007813 µM or lower for Bizelesin. Based on the cell viability data, we tested the antiviral activity of Rilapladib (starting concentration of 25 µM then decreasing by half incrementally) and Bizelesin (starting concentration of 1 µM then decreasing by half incrementally) using a microplate reader in the GFP range.
Fig. 4.
Antiviral screening assay for top hits procured from HerpDock:. (A) MTT assay of the drug Rilapladib performed on HCE cells (B) Fluorescence intensity was measured using a plate reader where cells were pre-treated for 24 h with Rilapladib followed by 24 h infection (C) Fluorescence intensity was measured using a plate reader where cells were infected, and drug was administered 2 hpi and cells were collected 24 hpi (D) MTT assay of the drug Bizelesin performed on HCE cells (E) Fluorescence intensity was measured using a plate reader where cells were pre-treated for 24 h with Bizelesin followed by 24 h infection (F) Fluorescence intensity was measured using a plate reader where cells were infected, and drug was administered 2 hpi and cells were collected 24 hpi (G) Beta galactosidase assay of the drug Rilapladip performed on HCE cells that were pre-incubated with the drug for 24 h (H) Western blot assay showing viral entry at different concentrations of Rilapladib performed on pre-treated HCE cells (I) Densitmetric quantification of western blot shown in image H (J) Immunofluroscence images showing viral entry of Rilapladib performed on pre-treated HCE cells compared to DMSO treated (control) (K) Quantification of the images shown in figure J.
We conducted two types of in vitro screenings. In the pre-treatment studies, the HCE cells were first treated with drugs for 24 h then infected with HSV-1. Rilapladib exhibited antiviral activity at concentrations of 25 µM and 12.5 µM (Fig. 4(B)). Bizelesin showed antiviral activity at a concentration of 0.0625 µM or higher (Fig. 4(E)). From these pre-treatment studies and MTT assays, we concluded that 12.5 µM is the optimal concentration of Rilapladib for further studies, as it shows antiviral activity with low toxicity. For Bizelesin, there was no optimal concentration since no antiviral activity was observed at the non-toxic concentration of 0.007813 µM or below.
In the post-treatment screening, where HCE cells were first infected with HSV-1 then treated with the drugs at 2HPI, Rilapladib again showed antiviral activity at concentrations of 25 µM and 12.5 µM (Fig. 4(C)), similar to the pre-treatment screenings. Bizelesin exhibited antiviral activity at concentrations of 0.03125 µM and above, which is again higher than the non-toxic concentration (Fig. 4(F)). Based on the post-treatment antiviral and MTT assay data, we reached the same conclusion as in the pre-treatment assay: 12.5 µM is the optimal concentration for Rilapladib, whereas Bizelesin does not show antiviral activity at non-toxic concentrations (0.007813 µM or lower). Therefore, Rilapladib at 12.5 µM is a promising candidate for future studies, while Bizelesin was not pursued further due to its toxicity and lack of antiviral efficacy at safe concentrations.
3.5. Assessing the potential of Rilapladib in inhibiting the HSV1 viral entry
Based on the results of the pre-treatment screening studies, we hypothesized that Rilapladib interferes with viral entry. To test this hypothesis, we conducted several experiments. In the first experiment, HCE cells were pre-treated with different concentrations of Rilapladib starting at 12.5 µM and incrementally decreasing by half, followed by the addition of a β-galactosidase-producing reporter virus, gL86, for 6 h. The results showed that Rilapladib pre-treatment at 12.5 µM reduced HSV-1 entry by approximately 50 % (Fig. 4(G)).
In a different experiment, cells were pre-treated with Rilapladib at 12.5 µM, 3.1 µM, 0.31 µM, or mock-treated with DMSO for 24 h, then infected with HSV-1 (KOS-WT) at an MOI of 10 at 4 °C for 30 min. Following a 30-minute incubation period at 37 °C, which allows virion particles to enter the cells, samples were then vigorously washed with PBS to remove residual virions that had attached to, but not entered, the cell. The cells were then immunoblotted for HSV-1 VP16 protein to determine the extent of viral entry (Fig. 4(H)). The densitometric quantification of Fig. 4(H) is shown as (Fig. 4(I)). We observed that viral entry was significantly lower in cells treated with 12.5 µM Rilapladib as compared to DMSO treated cells.
In yet another experiment, cells were pre-treated with Rilapladib at 12.5 µM or mock-treated with DMSO for 24 h, then infected with HSV-1 K26-GFP at an MOI of 10 at 4 °C for 30 min. Following a 30-minute incubation period at 37 °C, the cells were then fixed and stained with Rhodamine-phalloidin and DAPI after vigorous washing with PBS, instead of processed for immunoblotting. The samples were then imaged with a confocal microscope at the same magnification (63x) and exposure (Fig. 4(J)). The number of GFP puncta, denoting tagged tegument HSV-1 protein, was observed to be significantly lower in cells treated with 12.5 µM Rilapladib compared to mock DMSO-treated cells. The quantification of GFP puncta is shown in (Fig. 4(K)).
3.6. Time of addition assay for Rilapladib indicates the restriction of HSV-1 infection
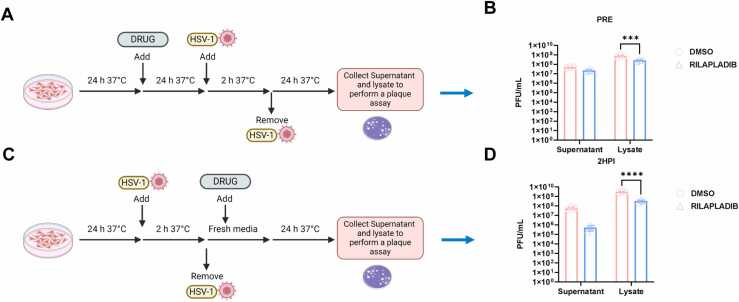
To determine whether prophylactic treatment with Rilapladib can restrict HSV-1 replication inside HCE cells, we treated cells with 12.5 μM Rilapladib and incubated them for 24 h. After washing the cells multiple times with PBS to remove any traces of the drug, cells were infected with HSV-1 KOS-WT at 0.1 MOI. No additional Rilapladib was added after infection (Fig. 5(A)). Subsequent plaque assay data (Fig. 5(B)) revealed a lower viral count in the Rilapladib-treated cells compared to DMSO-treated controls, which may be attributed to reduced viral entry as earlier observed.
Fig. 5.
Time of addition study for Rilapladib: (A) Schematic diagram of the plaque assay performed by pre-treating the HCE cells with the drug for 24 h followed by HSV-1 infection for 24 h (B) Results shown for the pre-treatment of the drug as compared to DMSO treated (control) (C) Schematic diagram of the plaque assay performed for post-entry antiviral effect of the drug where the drug was added 2-hour post-infection and Plaque results are shown in (D).
Next, to assess the effect of Rilapladib post-viral entry, we infected HCE cells at 0.1 MOI and allowed the infection to proceed for 2 h to ensure viral entry. After 2 HPI, we added 12.5 µM Rilapladib for an additional 22 h to observe its effect on viral replication (Fig. 5(C)). Plaque assay results (Fig. 5(D)) showed a lower viral count in Rilapladib-treated cells compared to DMSO-treated cells. These pre- and post-treatment studies indicate that Rilapladib effectively hinders both viral entry and replication.
4. Discussion
In this study, we developed a Python-based software integrating AutoDock Vina for molecular docking and high-throughput virtual screening, specifically targeting HSV-1 viral proteins and host proteins involved in HSV-1 infection. The primary goal of the software is to streamline the process of molecular docking and ADME (Absorption, Distribution, Metabolism, and Excretion) calculation, enabling researchers to perform these tasks with minimal effort. By integrating AutoDock Vina for docking and ChEMBL for ADME properties, our software provides a comprehensive tool for antiviral drug discovery, focusing on HSV-1 infection.
This software was applied in a case study involving more than 5000 ligands, which were screened against the HSV-1 glycoprotein D (gD) using the built-in dropdown menu to select the target protein. Glycoprotein D is a critical component in the viral entry process, mediating the interaction between the virus and its cellular receptors, nectin-1 and HVEM. The ability of gD to bind to these receptors is essential for HSV-1 to enter host cells and initiate infection.
Our screening identified Rilapladib as the top hit among non-cancer-related compounds. Rilapladib is known as an inhibitor of lipoprotein-associated phospholipase A2 (Lp-PLA2) [30], but its potential antiviral properties have not been previously explored. Subsequent experimental validation through viral entry assays and plaque assays demonstrates that Rilapladib significantly reduced HSV-1 infection when administered either as pre-treatment or post-infection.
The inhibition of gD binding to nectin-1 and HVEM by Rilapladib is particularly significant. These interactions are pivotal for the initiation of HSV1 infection, as gD binding to these receptors triggers the fusion of the viral envelope with the host cell membrane, allowing the viral genome to enter the host cell [31]. By blocking these interactions, Rilapladib effectively hinders the initial step of HSV-1 infection, thereby reducing viral entry and subsequent replication.
Furthermore, our therapeutic studies revealed an intriguing finding: even when Rilapladib was administered 2 h post viral entry, there was still a significant decrease in viral count. This suggests that Rilapladib might also interfere with viral replication through a mechanism independent of its ability to block gD receptor binding, indicating a potential dual mode of action that warrants further investigation.
Despite having many strengths HerpDock has few weaknesses also. HerpDock is currently only available for windows. Small molecules without 3D structures in PubChem cannot be processed directly for molecular docking in HerpDock. Users must manually download these molecules, convert their 2D structures into 3D using external software, and then utilize the 'Browse' function in HerpDock for docking. HerpDock’s ADME functionality relies on ChEMBL, which limits it to providing basic ADME properties for small molecules.
5. Conclusion
This study demonstrates the effectiveness of a novel Python-based software (HerpDock) that integrates molecular docking and ADME for the discovery of antiviral agents targeting HSV-1. By leveraging AutoDock Vina for high-throughput virtual screening and ChEMBL for ADME analysis, our software provides a streamlined and accessible tool for researchers, facilitating the identification of promising drug candidates with minimal effort. In our case study, Rilapladib emerged as a top candidate from a library of over 5000 ligands screened against HSV-1 glycoprotein D (gD). Experimental validation confirmed that Rilapladib significantly reduces HSV-1 infection by inhibiting gD's critical interactions with its cellular receptors, nectin-1 and HVEM, which are essential for viral entry into host cells. Notably, our findings also suggest that Rilapladib may possess additional antiviral properties beyond blocking viral entry, as evidenced by its efficacy when administered after viral entry had already occurred. This dual mode of action highlights Rilapladib's potential as a versatile therapeutic agent against HSV-1. Overall, our software not only identified a novel antiviral candidate but also demonstrated its potential to accelerate and optimize the drug discovery process by incorporating key computational tools into a unified platform. Further research will be focused on elucidating the mechanisms underlying Rilapladib's antiviral effects and assessing its efficacy in-vivo, paving the way for potential clinical applications in the treatment of HSV-1 infections.
Authors statement
As the author of the manuscript titled “HerpDock: A GUI-Based Gateway to HSV-1 Molecular Docking Insights,” I confirm that this work reflects our original research conducted on HerpDock, an open-source Graphical User Interface (GUI) program designed for molecular docking, high-throughput virtual screening, and absorption, distribution, metabolism, and elimination (ADME) studies. We believe that HerpDock stands out as a unique, user-friendly pipeline that extends beyond the capabilities of existing docking programs based on AutoDock Vina. It requires only the ligand structure from the user, as it already includes optimized structures of numerous HSV-1 viral proteins and relevant host proteins. With just a few clicks, HerpDock performs comprehensive docking studies, result analysis, and visualization, making it particularly valuable for Herpes researchers without a bioinformatics background. HerpDock is freely available at https://github.com/Sudhk24/Herpdock. We have provided a video tutorial to use the software on the same link and the authors are thankful to Theresa P. Sengpraseuth for the voiceover. We have adhered to all ethical guidelines in conducting our research, and we declare that there are no conflicts of interest. This manuscript has not been published elsewhere and is not under consideration by any other journal. We appreciate the opportunity to submit our work to CSBJ and look forward to the feedback from the reviewers. Thank you for considering our manuscript.
Institutional review board statement
Not Applicable.
Informed consent statement
Not Applicable.
Funding
This work was supported by NIH RO1 grants EY029426, EY024710 and R24 EY033598 (to D.S.).
CRediT authorship contribution statement
Deepak Shukla: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Sudhanshu Kumar Singh: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Divya Kapoor: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation.
Declaration of Competing Interest
The authors declare no conflict of interest.
Data Availability
Data available on request due to restrictions.
References
- 1.Scott E., Burkhart C. Herpes simplex virus: an immunopathological review. JAAD Rev. 2024 [Google Scholar]
- 2.Gilbert C., Bestman-Smith J., Boivin G. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist Updat. 2002;5(2):88–114. doi: 10.1016/s1368-7646(02)00021-3. PMID: 12135584. [DOI] [PubMed] [Google Scholar]
- 3.Karasneh G.A., Kapoor D., Bellamkonda N., Patil C.D., Shukla D. Protease, growth factor, and heparanase-mediated syndecan-1 shedding leads to enhanced HSV-1 egress. Viruses. 2021;13(9):1748. doi: 10.3390/v13091748. PMID: 34578329; PMCID: PMC8473078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P., Kapoor D., Shukla D. Role of heparanase and syndecan-1 in HSV-1 release from infected cells. Viruses. 2022;14(10):2156. doi: 10.3390/v14102156. PMID: 36298711; PMCID: PMC9612286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canova P.N., Charron A.J., Leib D.A. Models of herpes simplex virus latency. Viruses. 2024;16(5):747. doi: 10.3390/v16050747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patil C.D., Suryawanshi R., Ames J., Koganti R., Agelidis A., Kapoor D., et al. Intrinsic antiviral activity of optineurin prevents hyperproliferation of a primary herpes simplex virus type 2 infection. J Immunol. 2022;208(1):63–73. doi: 10.4049/jimmunol.2100472. Epub 2021 Dec 8. PMID: 34880107; PMCID: PMC9015683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapoor D., Shukla D. Neutrophil extracellular traps and their possible implications in ocular herpes infection. Pathogens. 2023;12(2):209. doi: 10.3390/pathogens12020209. PMID: 36839481; PMCID: PMC9958879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley H., Markowitz L.E., Gibson T., McQuillan G.M. Seroprevalence of herpes simplex virus types 1 and 2—United States, 1999–2010. J Infect Dis. 2014;209(3):325–333. doi: 10.1093/infdis/jit458. [DOI] [PubMed] [Google Scholar]
- 9.Patil C.D., Suryawanshi R.K., Kapoor D., Shukla D. Postinfection metabolic reprogramming of the murine trigeminal ganglion limits herpes simplex virus-1 replication. mBio. 2022;13(5) doi: 10.1128/mbio.02194-22. Epub 2022 Aug 31. PMID: 36043789; PMCID: PMC9600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadavalli T., Sharma P., Wu D., Kapoor D., Shukla D. CREB3 plays an important role in HPSE-facilitated HSV-1 release in human corneal epithelial cells. Viruses. 2022;14(6):1171. doi: 10.3390/v14061171. PMID: 35746643; PMCID: PMC9227461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S., Zhou M., Ji S., Ma D., Zhu S. Recent advances in the study of alphaherpesvirus latency and reactivation: novel guidance for the design of herpesvirus live vector vaccines. Pathogens. 2024;13(9):779. doi: 10.3390/pathogens13090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadavalli T., Patil C., Sharma P., Volety I., Borase H., Kapoor D., et al. Unique attributes of guinea pigs as new models to study ocular herpes pathophysiology and recurrence. Invest Ophthalmol Vis Sci. 2023;64(14):41. doi: 10.1167/iovs.64.14.41. PMID: 38015175; PMCID: PMC10691389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagan S., Khapuinamai A., Kapoor D., Sharma P., Yadavalli T., Joseph J., et al. Exploring heparanase levels in tears: insights from herpes simplex virus-1 keratitis patients and animal studies. Invest Ophthalmol Vis Sci. 2024;65(3):7. doi: 10.1167/iovs.65.3.7. PMID: 38466284; PMCID: PMC10929742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil C.D., Borase H., Gagan S., Sharma P., Kapoor D., Yadavalli T., et al. Rapid NETosis is an effector mechanism to combat ocular herpes infection. Invest Ophthalmol Vis Sci. 2024;65(6):36. doi: 10.1167/iovs.65.6.36. PMID: 38916883; PMCID: PMC11210628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor D., Sharma P., Shukla D. Emerging drugs for the treatment of herpetic keratitis. Expert Opin Emerg Drugs. 2024;29(2):113–126. doi: 10.1080/14728214.2024.2339899. [DOI] [PubMed] [Google Scholar]
- 16.Piperi E., Papadopoulou E., Georgaki M., Dovrat S., Bar Illan M., Nikitakis N.G., et al. Management of oral herpes simplex virus infections: The problem of resistance. A narrative review. Oral Dis. 2024;30(3):877–894. doi: 10.1111/odi.14635. [DOI] [PubMed] [Google Scholar]
- 17.Iqbal A., Suryawanshi R., Yadavalli T., Volety I., Shukla D. BX795 demonstrates potent antiviral benefits against herpes simplex Virus-1 infection of human cell lines. Antivir Res. 2020;180 doi: 10.1016/j.antiviral.2020.104814. Epub 2020 May 5. Erratum in: Antiviral Res. 2022 Apr;200:105293. doi: 10.1016/j.antiviral.2022.105293. PMID: 32380150; PMCID: PMC7387215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y.C., Feng H., Lin Y.C., Guo X.R. New strategies against drug resistance to herpes simplex virus. Int J Oral Sci. 2016;8(1):1–6. doi: 10.1038/ijos.2016.3. PMID: 27025259; PMCID: PMC4822185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baig M.H., Ahmad K., Rabbani G., Danishuddin M., Choi I. Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr Neuropharmacol. 2018;16(6):740–748. doi: 10.2174/1570159X15666171016163510. PMID: 29046156; PMCID: PMC6080097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sousa S.F., Ribeiro A.J., Coimbra J.T., Neves R.P., Martins S.A., Moorthy N.S., et al. Protein-ligand docking in the new millennium--a retrospective of 10 years in the field. Curr Med Chem. 2013;20(18):2296–2314. doi: 10.2174/0929867311320180002. PMID: 23531220. [DOI] [PubMed] [Google Scholar]
- 21.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J Chemin- 2011;3:33. doi: 10.1186/1758-2946-3-33. PMID: 21982300; PMCID: PMC3198950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. Epub 2013 Apr 12. PMID: 23579614. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- 24.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 25.Eberhardt J., Santos-Martins D., Tillack A.F., Forli S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61(8):3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert J.Bernstein. (2009). RasMol. Retrieved from http://www.rasmol.org/.
- 28.Gaulton A., Bellis L.J., Bento A.P., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazear E., Whitbeck J.C., Zuo Y., Carfí A., Cohen G.H., Eisenberg R.J., et al. Induction of conformational changes at the N-terminus of herpes simplex virus glycoprotein D upon binding to HVEM and nectin-1. Virology. 2014;448:185–195. doi: 10.1016/j.virol.2013.10.019. Epub 2013 Oct 29. PMID: 24314649; PMCID: PMC3979344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilensky R., Shi Y., Mohler E., et al. Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nat Med. 2008;14:1059–1066. doi: 10.1038/nm.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly S.A., Jackson J.O., Jardetzky T.S., Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol. 2011;9(5):369–381. doi: 10.1038/nrmicro2548. Epub 2011 Apr 11. PMID: 21478902; PMCID: PMC3242325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okemoto K., Wagner B., Meisen H., Haseley A., Kaur B., Chiocca E.A. STAT3 activation promotes oncolytic HSV1 replication in glioma cells. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071932. PMID: 23936533; PMCID: PMC3732216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabiner B.C., Nardi V., Birsoy K., Possemato R., Shen K., Sinha S., et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4(5):554–563. doi: 10.1158/2159-8290.CD-13-0929. Epub 2014 Mar 14. PMID: 24631838; PMCID: PMC4012430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chopra P., Yadavalli T., Palmieri F., Jongkees S.A.K., Unione L., Shukla D., et al. Synthetic heparanase inhibitors can prevent herpes simplex viral spread. Angew Chem Int Ed Engl. 2023;62(41) doi: 10.1002/anie.202309838. Epub 2023 Sep 6. PMID: 37555536. [DOI] [PubMed] [Google Scholar]
- 35.Liu X., Cohen J.I. The role of PI3K/Akt in human herpesvirus infection: from the bench to the bedside. Virology. 2015;479-480:568–577. doi: 10.1016/j.virol.2015.02.040. Epub 2015 Mar 20. PMID: 25798530; PMCID: PMC4424147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A., De S., Moharana A.K., et al. Inhibition of herpes simplex virus-1 infection by MBZM-N-IBT: in silico and in vitro studies. Virol J. 2021;18:103. doi: 10.1186/s12985-021-01581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., Knafels J.D., Chang J.S., Waszak G.A., Baldwin E.T., Deibel M.R., et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem. 2006;281(26):18193–18200. doi: 10.1074/jbc.M602414200. Epub 2006 Apr 24. PMID: 16638752. [DOI] [PubMed] [Google Scholar]
- 38.Madavaraju K., Koganti R., Volety I., Yadavalli T., Shukla D. Herpes simplex virus cell entry mechanisms: an update. Front Cell Infect Microbiol. 2021;10 doi: 10.3389/fcimb.2020.617578. PMID: 33537244; PMCID: PMC7848091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rafiq H., Hu J., Hakami M.A., Hazazi A., Alamri M.A., Alkhatabi H.A., et al. Identification of novel STAT3 inhibitors for liver fibrosis, using pharmacophore-based virtual screening, molecular docking, and biomolecular dynamics simulations. Sci Rep. 2023;13(1) doi: 10.1038/s41598-023-46193-x. PMID: 37978263; PMCID: PMC10656421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Torres V., Losada-Echeberría M., Herranz-López M., Barrajón-Catalán E., Galiano V., Micol V., et al. New mammalian target of rapamycin (mTOR) modulators derived from natural product databases and marine extracts by using molecular docking techniques. Mar Drugs. 2018;16(10):385. doi: 10.3390/md16100385. PMID: 30326670; PMCID: PMC6213183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijayalakshmi Deenadayalan A., Janaki S., Jayaraman C.S. S. Molecular docking analysis of stevioside with Akt and PPARγ. Bioinformation. 2021;17(1):283–288. doi: 10.6026/97320630017283. PMID: 34393447; PMCID: PMC8340692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J., Power H., Miranda-Saksena M., et al. Identifying HSV-1 inhibitors from natural compounds via virtual screening targeting surface glycoprotein D. Pharmaceuticals. 2022;15(3):361. doi: 10.3390/ph15030361. Published 2022 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request due to restrictions.