ABSTRACT
Genomic surveillance detected clonal Escherichia coli sequence type-361 isolates carrying blaNDM-5, blaKPC-3, blaCTX-M-15, and rmtB1 from a patient in Ukraine and four wounded foreign soldiers evacuated to Germany. Isolates were non-susceptible to carbapenems, aminoglycosides, and cefiderocol and aztreonam/avibactam due to a PBP3 YRIN insertion and the blaCMY-145 AmpC β-lactamase. Coordinated surveillance efforts across civilian, military, and veteran healthcare systems are essential to prevent further spread as international volunteers return home after medical evacuation from Ukraine.
KEYWORDS: Ukraine, antibiotic resistance, carbapenem resistance, genomic surveillance
INTRODUCTION
Carbapenem-resistant Enterobacterales (CRE) are considered as critical priority pathogens by the WHO (1). CREs mainly result from the production of carbapenem-hydrolyzing enzymes such as serine carbapenemases (e.g., KPCs and OXA-48) or metallo-β-lactamases (MBLs) (2). MBLs are of particular concern as most clinically available β-lactams/β-lactamase inhibitor (BL/BLI) combinations, the main treatment for CREs, are not active (3). For these strains, polymyxins are one of the last treatment options available, along with the new siderophore cephalosporin antibiotic, cefiderocol or aztreonam/avibactam, a BL/BLI combination that shows excellent activity against MBL producers (4).
Extensively drug-resistant (XDR) Escherichia coli sequence type (ST)-361 carrying the blaNDM-5 MBL has recently been reported in multiple European countries (5–7), including in isolates resistant to cefiderocol (6). Here, between May 2023 and January 2024, five E. coli ST-361 co-carrying blaNDM-5 and blaKPC-3 were cultured from a war wound of a hospitalized patient in Ukraine and peri-rectal swabs of four international soldiers wounded in Ukraine and evacuated to a US military treatment facility (MTF) in Germany.
To compare, 23 historical E. coli ST-361 isolates were identified from the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN) isolate collection. These were cultured from 23 patients between 2010 and 2022 being treated in MTFs in the continental US (n = 12), Germany (n = 8), or Afghanistan (n = 3) and had no known association with Ukraine (Table S1). Whole genome sequencing (Illumina) was performed on all isolates, and the five isolates linked to Ukraine (Fig. 1) were also sequenced by long-read sequencing (Oxford Nanopore Technologies), as previously described (8). Finally, to correlate genotype and phenotype, antibiotic susceptibility testing was performed using a customized Sensititre panel (Thermo Scientific) and broth microdilution for aztreonam/avibactam and cefiderocol, with breakpoints interpreted using 2023 CLSI guidelines where available. For aztreonam/avibactam, a tentative breakpoint of S ≤ 8 µg/mL was applied based on the recent study by Sader and colleagues (9).
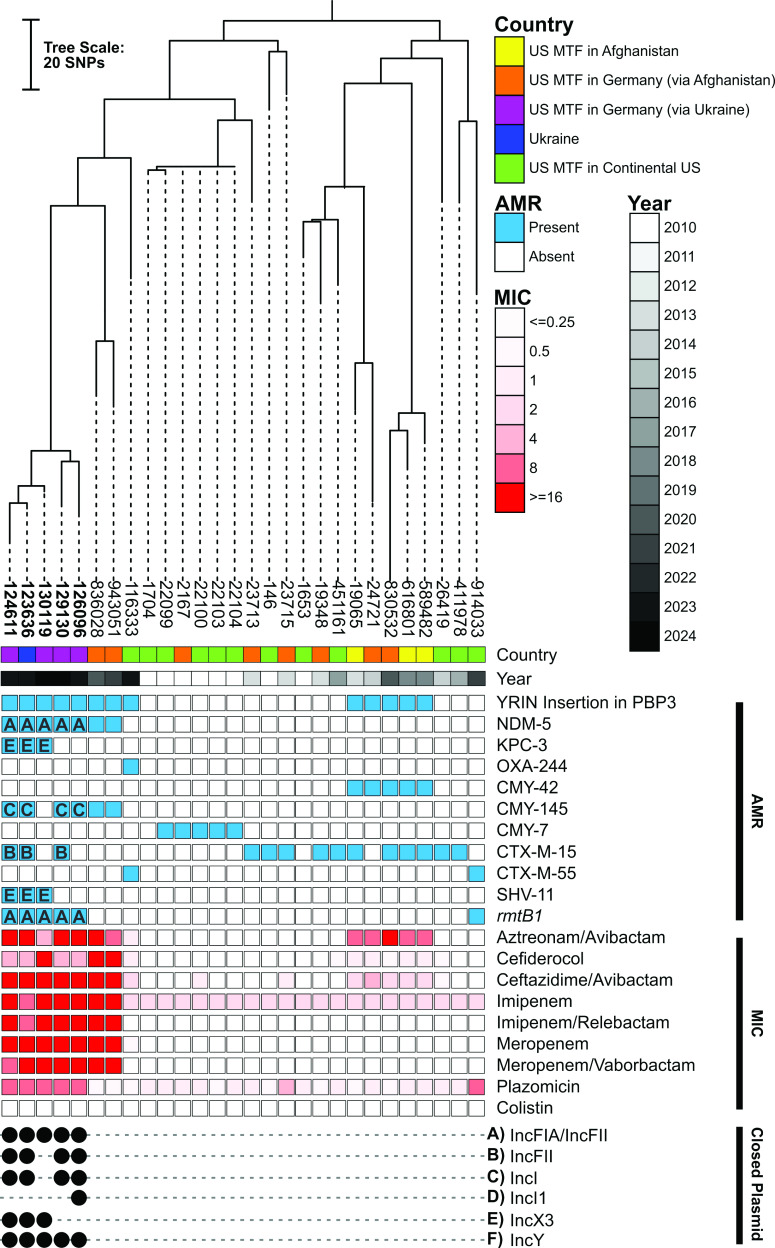
Fig 1.
A core genome phylogenetic tree of ST-361 E. coli (n = 28) from the US Military Health System and Ukraine. Country of origin, year of collection, and presence (closed square) or absence (open square) of selected antimicrobial resistance genes and minimum inhibitory concentrations of selected antibiotics. Replicon typing (A–F) of closed plasmids is indicated by the closed black circle. For closed sequences (label in bold font), the genetic location of antimicrobial resistance genes is indicated by the letter of the replicon type identified in the plasmid. The midpoint was used as a root for the phylogenetic tree.
Phylogenomic analysis of all 28 ST-361 isolates revealed that none of the 18 isolates collected pre-2020 carried an acquired carbapenemase compared to 8 out of 10 collected after 2020 (Fig. 1; Table S1). Despite being monophyletic and genetically related (single nucleotide polymorphism [SNP] counts ranging from 11 to 183), the eight carbapenemase producers did not harbor the same carbapenemase genes. A single isolate from the US in 2023 carried blaOXA-244 (OXA-48-like) chromosomally located within a truncated Tn51098, similar to previous observations (10). The remaining seven isolates carried blaNDM-5. These included five clonal isolates (11–57 SNPs), from the patients associated with Ukraine in this study, for which long-read sequencing was performed (Fig. 1; Table S1). Analysis of the complete genomes revealed these five isolates harbored an IncFIA/IncFII-type plasmid co-carrying the 16S rRNA methyltransferase gene rmtB1 and blaNDM-5 (Fig. S1A). The latter was found on a 10.8-kb IS26-formed composite transposon, which also carried a typical class 1 integron containing resistance genes sul1, qacE, aadA5, and dfrA17, as previously described (Fig. S1B) (11). Interestingly, isolate 130119 uniquely carried four identical copies of the transposable element carrying blaNDM-5. Besides this exception, the IncFIA/IncFII plasmids were virtually identical (>99.9% nucleotide identity over the full length) to each other and to p1606a first detected from an ST-361 isolate in Switzerland in 2020 (5). Finally, three of the five isolates also carried a virtually identical IncX3 plasmid co-harboring blaKPC-3 and blaSHV-11 (Fig. S1C), similar to previously described p1606b (5).
Phenotypically, all blaNDM-5 carrying isolates were non-susceptible to imipenem (≥8 µg/mL), meropenem (≥16 µg/mL), imipenem/relebactam (≥8 µg/mL), meropenem/vaborbactam (≥8 µg/mL), and ceftazidime/avibactam (≥16 µg/mL) (Fig. 1; Table S1). These also had high minimum inhibitory concentrations (MICs) against aztreonam/avibactam (from 4 to ≥16 µg/mL) and cefiderocol (from 4 to 16 µg/mL) (Fig. 1; Table S1). Similar to findings from Simner and colleagues (12), the increased copy number of blaNDM likely explained the increased cefiderocol MIC in isolate 130119 (16 µg/mL) compared to the four clonal isolates (4 µg/mL) with a single copy of the gene. Further, six isolates without blaNDM-5 also had increased MICs to cefiderocol, albeit more moderate (0.5 to 1 µg/mL). Increased cefiderocol MICs correlated exactly (13 of 13 isolates) with the presence of a four-amino acid insertion (YRIN at position 333) within PBP3, a feature previously linked to cefiderocol resistance (6). Interestingly, insertions in PBP3 also cause a reduced affinity for aztreonam resulting in a moderate to high (in association with an acquired AmpC β-lactamases) MIC increase for aztreonam/avibactam in E. coli (13). Here, 13 isolates had increased MIC for aztreonam/avibactam: 2 had a moderate increase (1–4 µg/mL) and only carried the PBP3 YRIN insertion and 11 had a high increase (8–16 µg/mL) and additionally carried an acquired blaCMY-42 or blaCMY-145. The latter included four of the five isolates linked to Ukraine, which carried blaCMY-145 on an IncI plasmid (Fig. 1).
In conclusion, upon admission to a US MTF in Germany, four international soldiers wounded in Ukraine were colonized with a dual-carbapenemase-producing E. coli with reduced cefiderocol susceptibility. The same strain was also cultured in Ukraine from a wounded Ukrainian soldier. This follows the recent detection of six distinct XDR strains/species from a single patient at the same German-based US MTF (14), which began receiving wounded foreign and US volunteers from Ukraine in June 2022. The potential spread of XDR E. coli ST-361, carrying multiple carbapenemases and mutations conferring resistance to last-line treatment options, is concerning. Joint surveillance efforts in civilian, active military, and veteran healthcare systems are necessary to prevent further spread as these international soldiers return home.
ACKNOWLEDGMENTS
The authors are thankful to all the staff of the MRSN. The manuscript has been reviewed by the Walter Reed Army Institute of Research, and there is no objection to its presentation. The views expressed herein are those of the author(s) and do not necessarily reflect the official policy or position of the Defense Health Agency, the Department of Defense, or any agencies under the US Government.
This study was funded by the Defense Health Program (DHP) Operation & Maintenance (O&M).
M.J.M., T.L.L., P.T.M., and F.L. designed the research; M.J.M., T.L.L., A.C.O., and Y.I.K. performed the research. M.J.M., T.L.L., J.W.B., P.T.M., and F.L. analyzed the research; H.D.D., Va.K., Vi.K., I.K., Y.I.K., B.J.P., J.M.K., C.P.A., J.R.S., J.S.H., P.T.M., and J.W.B. contributed clinical metadata and isolates. M.J.M., P.T.M., and F.L. wrote the paper with input from all authors.
Contributor Information
Patrick T. McGann, Email: Patrick.t.mcgann4.civ@health.mil.
Francois Lebreton, Email: francois.lebreton.ctr@health.mil.
Pranita D. Tamma, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
ETHICS APPROVAL
The isolates and clinical information were collected as part of the public health surveillance activities of the MRSN, as determined by the WRAIR branch director and Human Subjects Protection Branch (HSPB), which granted ethical approval. Informed patient consent was waived as samples were taken under a hospital surveillance framework for routine sampling. The research conformed to the principles of the Helsinki Declaration.
DATA AVAILABILITY
Genomes described herein have been deposited at GenBank under BioProject PRJNA1116075.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.01090-24.
Comparisons of plasmids.
List of isolates and metadata.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. World Health Organization . 2024. WHO bacterial priority pathogens list, 2024: bacterial pathogens of public health importance, to guide research, development, and strategies to prevent and control antimicrobial resistance. World Health Organization. Available from: https://iris.who.int/handle/10665/376776 [Google Scholar]
- 2. Iovleva A, Doi Y. 2017. Carbapenem-resistant Enterobacteriaceae. Clin Lab Med 37:303–315. doi: 10.1016/j.cll.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tompkins K, van Duin D. 2021. Treatment for carbapenem-resistant Enterobacterales infections: recent advances and future directions. Eur J Clin Microbiol Infect Dis 40:2053–2068. doi: 10.1007/s10096-021-04296-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackow NA, van Duin D. 2024. Reviewing novel treatment options for carbapenem-resistant Enterobacterales. Expert Rev Anti Infect Ther 22:71–85. doi: 10.1080/14787210.2024.2303028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sadek M, Saad AM, Nordmann P, Poirel L. 2022. Genomic characterization of an extensively drug-resistant extra-intestinal pathogenic (ExPEC) Escherichia coli clinical isolate co-producing two carbapenemases and a 16S rRNA methylase. Antibiotics (Basel) 11:1479. doi: 10.3390/antibiotics11111479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jousset AB, Bouabdallah L, Birer A, Rosinski-Chupin I, Mariet J-F, Oueslati S, Emeraud C, Girlich D, Glaser P, Naas T, Bonnin RA, Dortet L. 2023. Population analysis of Escherichia coli sequence type 361 and reduced cefiderocol susceptibility, France. Emerg Infect Dis 29:1877–1881. doi: 10.3201/eid2909.230390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Linkevicius M, Bonnin RA, Alm E, Svartström O, Apfalter P, Hartl R, Hasman H, Roer L, Räisänen K, Dortet L, et al. 2023. Rapid cross-border emergence of NDM-5-producing Escherichia coli in the European Union/European economic area, 2012 to June 2022. Euro Surveill 28:2300209. doi: 10.2807/1560-7917.ES.2023.28.19.2300209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russo TA, Alvarado CL, Davies CJ, Drayer ZJ, Carlino-MacDonald U, Hutson A, Luo TL, Martin MJ, Corey BW, Moser KA, Rasheed JK, Halpin AL, McGann PT, Lebreton F. 2024. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. MBio 15:e0286723. doi: 10.1128/mbio.02867-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sader HS, Carvalhaes CG, Arends SJR, Castanheira M, Mendes RE. 2021. Aztreonam/avibactam activity against clinical isolates of Enterobacterales collected in Europe, Asia and Latin America in 2019. J Antimicrob Chemother 76:659–666. doi: 10.1093/jac/dkaa504 [DOI] [PubMed] [Google Scholar]
- 10. Emeraud C, Girlich D, Bonnin RA, Jousset AB, Naas T, Dortet L. 2021. Emergence and polyclonal dissemination of OXA-244-producing Escherichia coli, France. Emerg Infect Dis 27:1206–1210. doi: 10.3201/eid2704.204459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. 2019. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simner PJ, Mostafa HH, Bergman Y, Ante M, Tekle T, Adebayo A, Beisken S, Dzintars K, Tamma PD. 2022. Progressive development of cefiderocol resistance in Escherichia coli during therapy is associated with an increase in blaNDM-5 copy number and gene expression. Clin Infect Dis 75:47–54. doi: 10.1093/cid/ciab888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Livermore DM, Mushtaq S, Vickers A, Woodford N. 2023. Activity of aztreonam/avibactam against metallo-β-lactamase-producing Enterobacterales from the UK: impact of penicillin-binding protein-3 inserts and CMY-42 β-lactamase in Escherichia coli. Int J Antimicrob Agents 61:106776. doi: 10.1016/j.ijantimicag.2023.106776 [DOI] [PubMed] [Google Scholar]
- 14. Mc Gann PT, Lebreton F, Jones BT, Dao HD, Martin MJ, Nelson MJ, Luo T, Wyatt AC, Smedberg JR, Kettlewell JM, Cohee BM, Hawley-Molloy JS, Bennett JW. 2023. Six extensively drug-resistant bacteria in an injured soldier, Ukraine. Emerg Infect Dis 29:1692–1695. doi: 10.3201/eid2908.230567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparisons of plasmids.
List of isolates and metadata.
Data Availability Statement
Genomes described herein have been deposited at GenBank under BioProject PRJNA1116075.