Summary
There is an intimate relationship between pain hypersensitivity and attention deficit hyperactivity disorder (ADHD); however, the underlying mechanisms are still elusive. Individuals carrying the mutation in CRY1 (c. 1657 + 3A > C), which leads to deletion of exon 11 expression in the CRY1 protein (CRY1Δ11), exhibit ADHD symptoms. Here, we demonstrate that the responses to thermal and mechanical stimuli were amplified in the Cry1Δ11 mice. RNA-sequencing analysis identified protein kinase A (PKA) signaling as being overactive in the spinal cords of Cry1Δ11 mice. The neuronal excitability was significantly enhanced in the spinal cords of Cry1Δ11 mice as determined by in vitro electrophysiology. The PKA inhibitor H89 normalized hyperalgesia in Cry1Δ11 mice, underscoring the causative effect of overactive PKA signaling. Our results thus point to the PKA signaling pathway as the underlying mechanism and a potential therapeutic target for pain hypersensitivity in a validated ADHD mouse model.
Subject areas: Molecular biology, Neuroscience, Behavioral neuroscience, Molecular neuroscience, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Cry1Δ11 mutation disrupted the circadian rhythm of pain sensation in mice
-
•
Cry1Δ11 mice showed hypersensitivity to noxious heat, cold stimuli, and mechanical stimuli
-
•
Overactive cAMP-PKA pathway in the spinal cord underlies the hyperalgesia in Cry1Δ11 mice
Molecular biology; Neuroscience; Behavioral neuroscience; Molecular neuroscience; Omics; Transcriptomics
Introduction
Attention-deficit and hyperactivity disorder (ADHD) is a highly heritable neuropsychiatric disorder characterized by symptoms of inattention and/or hyperactivity/impulsivity.1,2 It is one of the most common psychiatric disorders in children, with an estimated prevalence of 5.3% worldwide. The ADHD symptoms persist into adulthood in up to two-thirds of patients.1,2 ADHD patients are likely to exhibit psychiatric comorbidity and generally have poor academic, occupational, and social functioning.3
Interestingly, ADHD is commonly associated with pain hypersensitivity.4,5,6,7 ADHD adults tend to perceive pain more intensely or are more likely to develop a pain disorder.7,8 Additionally, ADHD patients are more prone to use pain medication.9 Conversely, chronic pain can lead to cognitive impairments and exacerbate ADHD symptoms in both humans,10,11 and animal models.12 Nevertheless, the mechanisms underlying the relationship between pain hypersensitivity and ADHD are not yet fully understood.
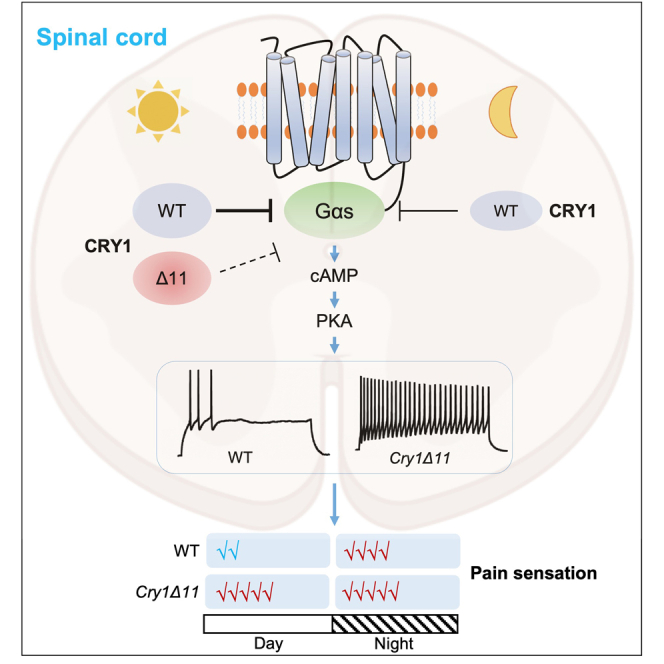
A recent study links a mutation in the core circadian clock gene CRY1 (c. 1657 + 3A > C) to ADHD.13 This specific mutation leads to the loss of exon 11 expression in the CRY1 protein (CRY1Δ11), resulting in the absence of 24 amino acids in the CRY1 protein’s C-terminal region.13 We generated Cry1Δ11 (c. 1717 + 3A > C) mice using CRISPR-Cas9 technology and observed that these mice exhibited ADHD-like symptoms, including hyperactivity, impulsivity, and learning and memory deficits. Our findings also highlight hyperactive dopamine D1 (DRD1) signaling as the mechanism behind the ADHD symptoms induced by the CRY1Δ11 mutation. We found that the wild type (WT) CRY1 protein interacts with the Gαs protein and inhibits DRD1 signaling. Yet, the CRY1Δ11 protein lacks the ability to interact with Gαs protein, resulting in hyperactive DRD1 signaling.2 However, the impact of the Cry1Δ11 mutation on pain sensation has remained unexplored until now.
In this study, we show that Cry1Δ11 mice exhibit hyperalgesia behaviors and demonstrate that the hyperactive cAMP-dependent protein kinase A (PKA) signaling pathway may be the underlying pathological mechanism and therapeutic target for pain induced by the Cry1Δ11 mutation.
Results
Cry1Δ11 mutation disrupted the circadian rhythm of pain sensation in mice
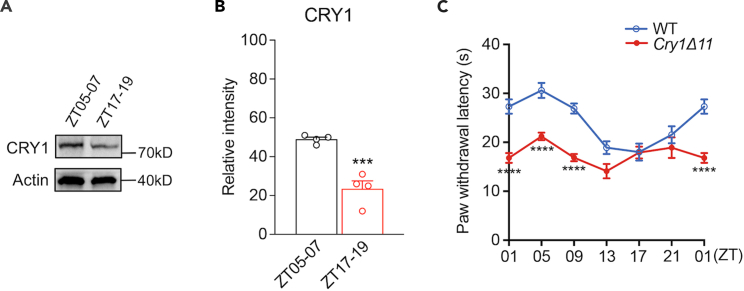
CRY1 is a key component of the circadian oscillator and shows circadian expression in multiple tissues.14 We first analyzed Cry1 protein levels in the spinal cord of WT mice taken at zeitgeber time (ZT) 05–07 and ZT17-19, respectively (ZT is measured in hours after the light has been turned on in a 12 h light:12 h dark cycle). As shown in Figures 1A and 1B, the Cry1 protein level at ZT05-07 was significantly higher than that at ZT17-19.
Figure 1.
The circadian rhythm of pain sensation
(A) Representative western blot for CRY1 protein in the spinal cords of WT mice taken at ZT05-07 and ZT 17–19, respectively.
(B) Quantification of western blot shown in (A). n = 4 mice/time point. ∗∗∗p < 0.001, unpaired t-test.
(C) The paw withdrawal latency of mice placed on a plate of 50oC at different circadian times. Data are presented as mean ± SEM (n = 5 mice/group/time point). ∗∗∗∗p < 0.0001; two-way ANOVA with post hoc Bonferroni analysis.
Then we assessed the response of WT and Cry1Δ11 mice to heat stimuli (50°C) at six different time points (ZT01, 05, 09, 13, 17, and 21). As shown in Figures 1C and S1, the pain sensitivity in WT mice exhibited a circadian rhythm; i.e., the shortest withdrawal latency was seen at ZT13-17, whereas the longest withdrawal latency was seen at ZT01-05 (F[5, 53] = 11.91, p < 0.0001, One-way analysis of variance [ANOVA]). Although Cry1Δ11 mice still showed time-dependent variation in the pain sensation (F[5, 54] = 3.220, p = 0.0129, One-way ANOVA); however, this circadian rhythm was significantly attenuated (F[1, 125] = 85.27, p < 0.0001, two-way ANOVA). The withdrawal latency was significantly reduced in Cry1Δ11 mice as compared with WT mice during ZT01-09, coincident with the higher expression of Cry1 protein during this period in the spinal cord.
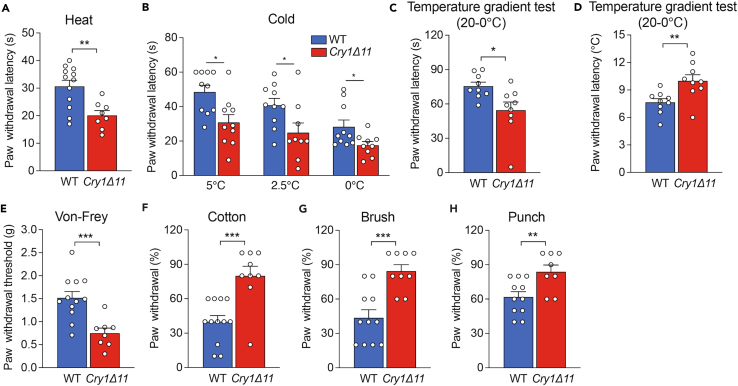
Hypersensitivity of Cry1Δ11 mice to noxious heat and cold stimuli
Next, we systematically measured the responses of WT and Cry1Δ11 mice to thermal and mechanical stimuli at ZT01-05. Consistent with the data shown in Figure 1C, Cry1Δ11 mice showed a reduced latency to withdraw on the hot plate (50°C) as compared to their littermate controls (Figure 2A). We also evaluated the response of animals to three different low temperatures, using a cold plate set at 5°C, 2.5°C, or 0°C. As shown in Figure 2B, Cry1Δ11 mice showed reduced withdrawal latency compared to littermate controls at all three of these temperatures.
Figure 2.
Hypersensitivity of Cry1Δ11 mice to thermal and mechanical stimuli
(A) The paw withdrawal latency for WT and Cry1Δ11 mice on a heat plate set at 50oC.
(B) The paw withdrawal latency for WT and Cry1Δ11 mice on cold plates set at 0, 2.5, and 5oC.
(C and D) The paw withdrawal latency (C) and temperature (D) for WT and Cry1Δ11 mice in the temperature gradient test.
(E) Paw withdrawal threshold for WT and Cry1Δ11 mice in a von Frey test.
(F and H) Paw withdrawal percentages for WT and Cry1Δ11 mice in the cotton swab test (F), brush test (G), and punch test (H). Data are presented as means ± SEM (n = 8–12/group), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t-test.
We also performed a temperature gradient test, in which the plate surface temperature was gradually dropped from 20°C to 0°C. As shown in Figures 2C and 2D, Cry1Δ11 mice showed reduced withdrawal latency than that of littermate controls, and Cry1Δ11 mice withdrew their paws at a higher temperature as compared with WT mice. Taken together, Cry1Δ11 mice were hypersensitive to both noxious heat and cold stimuli.
Hypersensitivity of Cry1Δ11 mice to mechanical stimuli
We also examined pain-related reactivity to mechanical stimuli by using a noxious/innocuous approach. In the von Frey filament assessment, as shown in Figure 2E, Cry1Δ11 mice showed a significantly reduced withdrawal threshold compared to their littermate controls. Furthermore, Cry1Δ11 mice showed higher withdrawal percentages than WT controls in the cotton, brush, and punch tests (Figures 2F–2H). These findings demonstrate that Cry1Δ11 mice were hypersensitive to mechanical stimuli.
Hyperactive cAMP-PKA signaling in the spinal cord of Cry1Δ11 mice
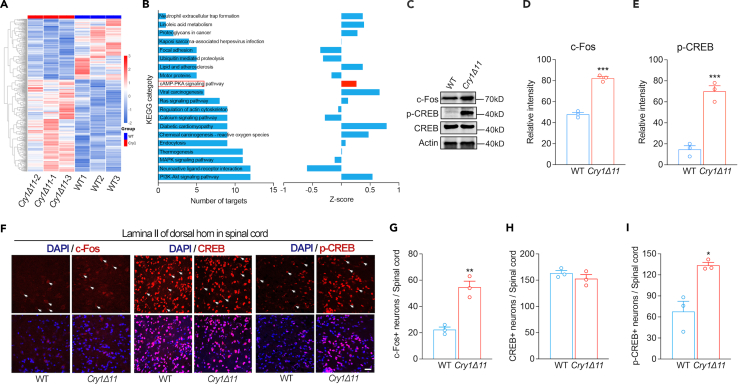
To assess the molecular basis underlying hyperalgesia in Cry1Δ11 mice, we used RNA sequencing to compare the gene expression profile of Cry1Δ11 mice with the profile of WT controls in the 3L–6L lumbar region of the spinal cord taken at ZT01-05. As a result, we identified 549 upregulated and 366 downregulated genes in the spinal cord of Cry1Δ11 mice (Figure 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed enrichment of differentially expressed genes in several pathways (Figure 3B). Among these, the cAMP-PKA signaling pathway is particularly interesting, as it is involved in nociception and is regulated by CRY1.2,15,16 Several genes in the cAMP-PKA signaling pathway, such as Ppplr16b, Ppplr2, Grin2d, and Atp10a were significantly upregulated in the Cry1Δ11 mice. We then measured the levels of phosphorylated cAMP response element-binding protein (p-CREB) and c-Fos to verify that the cAMP-PKA signaling pathway indeed had been altered (Figure 3C). As shown in Figures 3D and 3E, p-CREB and c-Fos were significantly upregulated in spinal cords taken from Cry1Δ11 mice compared with those from WT littermate controls. Immunofluorescence analysis confirmed a significant increase in p-CREB- and c-Fos-positive neurons in spinal cords from Cry1Δ11 mice (Figures 3F–3I). Although p-CREB was identified in both VGLUT1-positive excitatory neurons and VGAT-positive inhibitory neurons, significant increases of p-CREB were only observed in the excitatory neurons (Figures S2A and S2B). Our findings unveiled an overactive cAMP-PKA signaling pathway in the spinal cord of Cry1Δ11 mice.
Figure 3.
Hyperactive cAMP-PKA signaling pathway in the spinal cord of Cry1Δ11 mice
(A) Heatmap of gene expression in the spinal cord of WT and Cry1Δ11 mice as assayed by RNA-seq.
(B) KEGG pathway analysis of differentially expressed genes. Left, number of targets; right, Z score analysis. The cAMP-PKA signaling pathways are indicated by a red boxes.
(C) Representative western blots of p-CREB and c-Fos in the spinal cords of WT and Cry1Δ11 mice.
(D and E) Quantification of western blots of p-CREB and c-Fos in the spinal cords of WT and Cry1Δ11 mice.
(F) Representative immunofluorescence images of p-CREB, CREB, and c-Fos-positive neurons in the lamina II of the dorsal horn in the spinal cords of WT and Cry1Δ11 mice. Scale bar: 10 μm.
(G–I) Representative positive neurons are indicated with arrows. Quantification of p-CREB, CREB, and c-Fos-positive neurons in the spinal cords of WT and Cry1Δ11 mice. Data are presented as means ± SEM. n = 3 mice/genotype; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t-test.
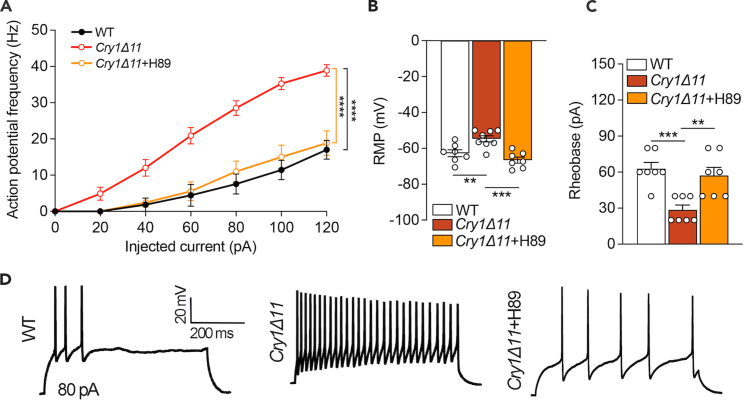
Hyperexcitability of the spinal cord neurons from Cry1Δ11 mice
Next, we performed whole-cell patch clamp recordings in lamina II neurons in the spinal cord. As shown in Figures 4A–4D, neurons from Cry1Δ11 mice exhibited a significantly higher action potential (AP) frequency and depolarized resting membrane potential (RMP) than those from WT controls. Further analysis in spinal cord neurons of Cry1Δ11 mice also revealed a decrease of rheobase, indicating hyperexcitability of these neurons. This hyperexcitability is likely the result of an overactive cAMP-PKA pathway, as excitability was normalized by the PKA inhibitor H89, which was perfused at a concentration of 10μM during the electrophysiological recording (Figures 4A–4D).
Figure 4.
Hyperexcitability in neurons from the spinal cords of Cry1Δ11 mice
(A) Frequency of action potentials of spinal cord neurons induced by 120-pA current steps. ∗∗∗∗p < 0.001, two-way ANOVA with post hoc Bonferroni analysis.
(B) Resting membrane potential of spinal cord neurons. ∗∗p < 0.01. ∗∗∗p < 0.001, one-way with post hoc Bonferroni analysis.
(C) The rheobase of spinal cord neurons was significantly lower in Cry1Δ11 mice than in WT mice. Data are presented as means ± SEM. n = 3 mice/genotype; ∗∗p < 0.01. ∗∗∗p < 0.001, one-way ANOVA with post hoc Bonferroni analysis.
(D) Representative action potentials of spinal cord neurons induced by 80-pA current steps.
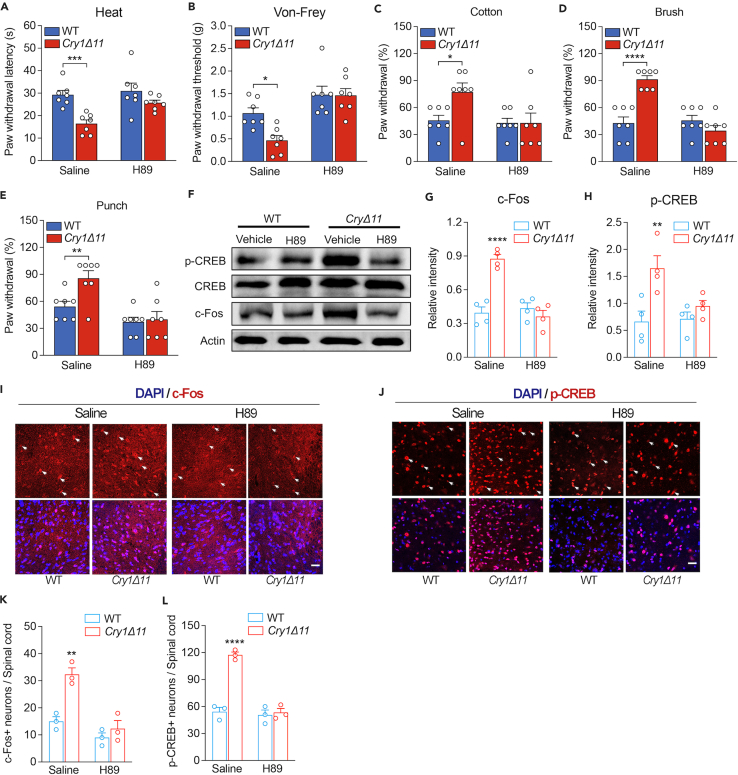
PKA inhibitor H89 rescued the hyperalgesia in Cry1Δ11 mice
To investigate whether hyperactive PKA signaling is responsible for the hyperalgesia in Cry1Δ11 mice, we intrathecally injected mice with the PKA inhibitor H89 (15μg/Kg) and measured their behavioral response 1 h later. All experiments were carried out during ZT01-05 when hyperalgesia behaviors were consistently observed in Cry1Δ11 mice. Consistent with the data presented in Figure 2, the responses to heat and mechanical stimuli were significantly higher in Cry1Δ11 mice compared with their littermate WT controls when treated with saline; however, there was no genotypic difference in the responses to these stimuli after H89 treatment (Figures 5A–5E), suggesting a causative effect of overactive PKA signaling in the hyperalgesia induced by Cry1Δ11 mutation. Nevertheless, intrathecal injection of H89 did not influence the ADHD-related behaviors in Cry1Δ11 mice, including hyperactivity (Figure S4A), reduced anxiety-like behaviors (Figure S4B), reduced depressive-like behavior (Figure S4C), and impaired learning and memory (Figure S4D).
Figure 5.
Inhibition of PKA signaling rescued the hyperalgesia behaviors in Cry1Δ11 mice
Saline or PKA inhibitor H89 (15μg/Kg) was injected intrathecally during ZT01-05, and behaviors were measured at 60 min after injection.
(A–E) H89 normalized the hyperactivity to heat (A), von Frey (B), cotton (C), brush (D), and punch (E) stimuli. Data are presented as means ± SEM. n = 7 mice/genotype; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, two-way ANOVA followed by Bonferroni analysis. (F–L) H89 normalized the over-activated cAMP-PKA pathway.
(F) Representative Western blots of p-CREB and c-Fos in the spinal cord.
(G and H) Quantification of western blots of c-Fos (G) and p-CREB (H) in the spinal cord.
(I and J) Representative immunofluorescence images of c-Fos (I) and p-CREB in the lamina II of the dorsal horn in the spinal cords. Representative positive neurons are indicated with arrows. Scale bar: 10 μm.
(K and L) Quantification of c-Fos-positive (K) and p-CREB-positive (L) neurons in the spinal cord. Data are presented as means ± SEM. n = 3 mice/genotype; ∗p < 0.05, ∗∗p < 0.01, two-way ANOVA followed by Bonferroni analysis.
We also measured the levels of p-CREB and c-Fos to verify that the cAMP-PKA signaling pathway had been normalized by H89. As shown in Figures 5F–5H, both p-CREB and c-Fos expressions were significantly downregulated in Cry1Δ11 mice after treatment with H89 to levels comparable to their littermate controls.
Similarly, immunofluorescence analysis revealed a significant reduction of p-CREB and c-Fos positive neurons in the spinal cord of Cry1Δ11 mice treated with H89 (Figures 5I–5L). On the other hand, unphosphorylated CREB expression remained unaltered in saline and H89 treatments for both Cry1Δ11 mice and WT littermates as indicated in Figures S3A and S3B. These results suggest that PKA inhibitors may have potential as a therapeutic approach for hyperalgesia induced by Cry1Δ11 mutations.
Alteration of protein phosphorylation in the spinal cord of Cry1Δ11 mice
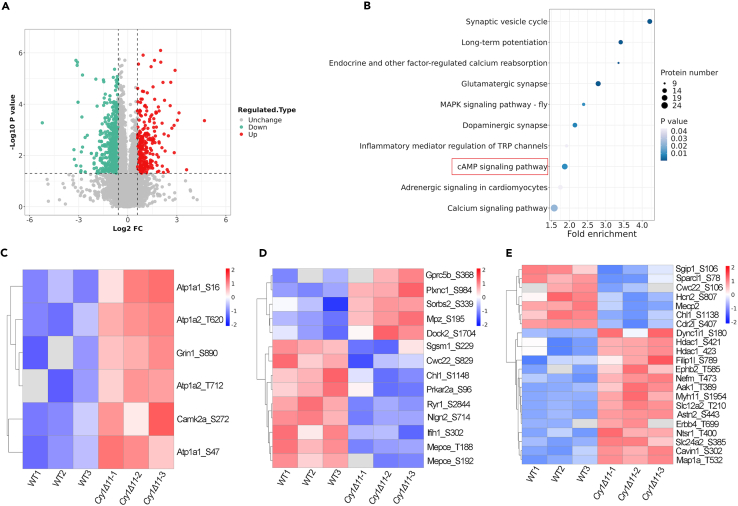
Since protein phosphorylation plays an important role in pain sensation, we performed phospho-proteomics on the spinal cords of WT and Cry1Δ11 mice. 228 protein phosphorylation sites on 80 proteins were upregulated, whereas 576 protein phosphorylation sites on 119 proteins were downregulated in the Cry1Δ11 mice (Figure 6A). KEGG analysis revealed enrichment of several pain-related pathways, including glutamatergic synapse, regulation of TRP channels, calcium signaling pathway, Mitogen-activated protein kinase (MAPK) signaling pathway, and cAMP signaling pathway (Figure 6B).
Figure 6.
Phospho-proteomics analysis on the spinal cords from WT and Cry1Δ11 mice
(A) Volcano plot of the differentially regulated phosphorylation sites.
(B) The enriched pathways as analyzed by KEGG.
(C) Heatmap for the upregulated phosphorylation sites in the cAMP-PKA pathway.
(D) Heatmap for the putative PKA phosphorylation sites involved in pain sensation.
(E) Heatmap for the non-PKA phosphorylation sites involved in pain sensation. Upregulation is indicated in red, and downregulation is indicated in blue.
Confirming that the cAMP-PKA pathway is overactivated in Cry1Δ11 mice, a heatmap revealed upregulated phosphorylation of several proteins in the cAMP-PKA pathway, including Atp1a1, Grin1, Atp1a2, and Camk2a (Figure 6C). In addition, we also plotted a heatmap for altered phosphorylation in proteins involved in pain sensation. Some of these proteins have altered phosphorylation on the consensus PKA recognition sequence (Arg/Lys-X-Ser/Thr, wherein Ser/Thr is the phosphorylation site) (Figure 6D; Tables S1 and S2); however, many altered protein phosphorylation sites occurred outside of this sequence (Figure 6E). These results suggest that the overactivated PKA pathway may elicit hyperalgesia by directly or indirectly altering the phosphorylation of multiple pain-related protein pathways.
Discussion
In this study, we demonstrated that Cry1Δ11 mice showed hyperalgesia behaviors. We further identified that hyperactive cAMP-PKA signaling may be the underlying pathological mechanism for the hypersensitivity to pain. Importantly, the PKA inhibitor H89 rescued hyperalgesia behaviors in Cry1Δ11 mice, supporting PKA pathway inhibition as a potential therapeutic target for pain induced by the Cry1Δ11 mutation.
Circadian rhythms, which are biological processes with a roughly 24-h cycle, are observed at both the molecular and behavioral levels.17 These rhythms are regulated by a cell-autonomous mechanism known as the circadian clock.18 The core of this clock is a transcription-translation feedback loop. In mammals, the proteins CLOCK and BMAL1 form a heterodimer that activates the transcription of genes PERIOD1-3 (PER1-3) and CRYPTOCHROME1-2 (CRY1-2) by binding to the E-box element in their promoters.18,19 PER and CRY proteins then dimerize and inhibit the CLOCK/BMAL1 activity, thereby suppressing their transcription.20 Additionally, REV-ERBs and retinoic acid receptor-related orphan receptor (ROR) proteins contribute to an extra regulatory loop by negatively and positively regulating BMAL1 transcription, respectively.20
Emerging evidence suggests that the circadian clock also influences the processing of nociceptive information.21 Clinical observations reveal that several pain conditions, such as toothaches, follow a circadian pattern, often intensifying in the early morning and disrupting nocturnal sleep.21 Similarly, pain intensity in mice displays a circadian rhythm. Research findings, including those from this study, show that mice demonstrate lower pain sensitivity during the early light hours and increased sensitivity during the early dark hours.22
Notably, our study revealed a negative correlation between the expression of the Cry1 protein and pain sensitivity in mice. Mice exhibited heightened pain sensitivity when Cry1 expression was low and increased resilience to pain when Cry1 levels were high. This suggests that Cry1 acts as a suppressor of pain sensation and is involved in the circadian regulation of this sensation. Supporting this, Cry1Δ11 mice displayed hyperalgesia during ZT01-09, but no significant genotypic difference was noted at ZT13-21 when the Cry1 protein level was relatively lower in the spinal cord.
Activation of the cAMP pathway is a significant cellular process following the binding of ligands to G-protein coupled receptors (GPCRs). The pathway starts with the synthesis of cAMP from ATP by adenylate cyclase enzymes. Once synthesized, cAMP activates PKA, leading to the phosphorylation of various proteins, including neurotransmitter receptors and transcription factors.23 The cAMP-PKA pathway may mediate hyperalgesia induced by GPCR ligands like adenosine, prostaglandin E2, and serotonin. Conversely, inhibiting the cAMP-PKA pathway can reduce hyperalgesia in various pain models, including inflammatory, non-inflammatory, and neuropathic pain.23
In this study, we demonstrated an overactive cAMP-PKA signaling pathway in the spinal cords of Cry1Δ11 mice through transcriptomic analysis. Importantly, the rescuing effect of the PKA inhibitor H89 strongly suggests that the overactive cAMP-PKA pathway plays a critical role in the hyperalgesia observed in Cry1Δ11 mice. Zhang et al. have shown that the CRY1 protein interacts with the Gαs protein and inhibits the accumulation of cAMP in response to GPCR activation.16 However, as demonstrated in our recent study,2 the CRY1Δ11 mutation disrupts the interaction between CRY1 and Gαs proteins, which may contribute to the elevated cAMP-PKA signaling observed in Cry1Δ11 mice. One of the notable effects of the cAMP-PKA pathway is on the cAMP-responsive element binding protein (CREB).24 Gene transcription initiated by phosphorylation of CREB can elevate cellular proteins mediating pain. On the other hand, the phosphorylation of receptor subunits by PKA may also mediate pain-related responses. For instance, PKA phosphorylates the NR1 subunit of the glutamate receptor at Ser-897 and the GluR1 subunit of the AMPA receptor at Ser-845, promoting receptor trafficking and potentiating ion-channel function.25,26,27 In this study, the Cry1Δ11 mutation altered phosphorylation in a panel of proteins; many of these are involved in pain sensation. Specifically, we found increased phosphorylation at S890 in NR1 (Grin1), which may contribute, at least partially, to the neuronal hyperexcitability and hyperalgesia in Cry1Δ11 mice.
Cry1Δ11 mutation also leads to increased phosphorylation on ATP1A1 and ATP1A2, two subunits of Na+/K+-ATPase. ATP1A1 has been reported in association with headaches and paresthesias,28 whereas ATP1A2 has been linked to familial hemiplegic migraine type 2 (FHM2), a condition characterized by intense migraines and, in certain instances, pain syndromes.29 The genetic alterations often result in a reduction in the efficiency of ion pumps. Specifically, mutations in ATP1A2 can hinder the removal of glutamate and potassium in the spaces between nerve cells, making the brain more susceptible to conditions like migraines and potentially pain.29 CaMKIIα is another protein with increased phosphorylation in Cry1Δ11 mice. Studies have demonstrated that stimulation of CaMKIIα in nociceptive neurons can affect their responsiveness and the magnitude of pain signals they convey.30 In the spinal cord, CaMKIIα promotes the transfer of pain signals from peripheral nerves to the brain. Further, nerve injury activates CaMKIIα, which leads to the excitability of neurons and heightened pain induction.31,32 It will be intriguing to investigate the increased phosphorylation of these proteins in the hyperalgesia induced by Cry1Δ11 mutation.
Limitations of the study
In this study, we demonstrated that overactive PKA signaling underlies the hyperalgesia in Cry1Δ11 mice by using pharmacological rescue with a PKA antagonist H89. It has been well documented that the cAMP-PKA pathway may contribute to sensory sensitization at peripheral, spinal, and cortical levels.33,34,35,36,37 We speculate that the rescuing effect of H89 may be a combinatory effect of more than one level as Cry1 is ubiquitously expressed in these systems. However, we only measured the PKA signaling pathway in the spinal cord in this study; it will be intriguing to dissect the contribution of PKA signaling at different levels to the hyperalgesia in Cry1Δ11 mice in the future.
The other limitation of this study is that we only used male mice, as the pain sensitivity of female mice may be confounded by the estrous cycles.38,39 In the future, we will pursue a detailed comparison of the pain sensitivity of WT and Cry1Δ11 mice at different estrous cycles.
Resource availability
Lead contact
Requests for resources and reagents should be directed to the lead contact Jia-Da Li (lijiada@sklmg.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
-
•
Data: RNA-seq data have been deposited at NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report the original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We would like to thank Edwards Sharman for language editing. We also thank the members of the Li Lab for fruitful discussions. The project is aided financially by the following funding agencies: National Natural Science Foundation of China of grants 31972913 (J.D.L.), 32371218 (J.D.L.), 81770780 (J.D.L.), and 8271250 (M.D.); Key Research and Development Programs from Hunan Province of grant 2021DK2001 (J.D.L.); Guangdong Key Project in “Development of New Tools for Diagnosis and Treatment of Autism” of grant 2018B030335001 (J.D.L.); of the Postgraduate Scientific Research Innovation Project of Hunan Province of grant CX20220318 (D.F.L.); Hunan Provincial Natural Science Foundation of China, of grant 2023JJ0082 (M.D.) and Independent Exploration and Innovation Project for Postgraduate of Central South University of grant 2022ZZTS0868 (M.D.).
Author contributions
J.D.L., M.D., D.F.L., and D.O.K. conceptualized the main research idea. J.D.L., M.D., D.O.K., D.F.L., and Y.Y. designed the methodology and conducted the investigation. J.D.L., M.D., D.F.L., D.O.K., and Y.Y. completed formal raw data analyses and curated the raw data. P.Y.G. offered a supplementary experiment. J.D.L. and M.D. provided insight and reagents. All authors analyzed the processed data. J.D.L. and D.O.K. wrote the original draft which was reviewed and edited by all authors. J.D.L. and M.D. supervised the project.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Cry1 antibody | Abcam | Cat#ab104736; RRID: AB_10710308 |
| Anti-c-Fos antibody | Abcam | Cat#ab209794; RRID: AB_2905616 |
| Anti-CREB antibody | Cell Signaling Technology | Cat#9197; RRID:AB_331277 |
| Anti-p-CREB antibody | Cell Signaling Technology | Cat#9198; RRID:AB_659957 |
| Anti-β-actin antibody | Abcam | Cat#ab8226; RRID: AB_306371 |
| Anti-VGLUT1 antibody | Abcam | Cat#ab227805; RRID: AB_2868428 |
| Anti-VGAT antibody | Abcam | Cat#ab308062; RRID: N/A |
| Chemicals, peptides, and recombinant proteins | ||
| H89 | Selleck | Cat#S1582 |
| Critical commercial assays | ||
| RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific | Cat#K1622 |
| SYBR™ Green PCR Master Mix | Thermo Scientific | Cat#4309155 |
| Trichrome Stain Kit (Connective Tissue Stain) | Abcam | Cat#ab150686 |
| Deposited data | ||
| RNA sequencing data | This paper | NCBI: PRJNA1136699 |
| Original western blot images | This paper | Mendeley Data: https://doi.org/10.17632/c3h9gpxnj9.1 |
| Experimental models: Organisms/strains | ||
| Cry1Δ11 mouse | Liu et al.37 | N/A |
| Software and algorithms | ||
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| MultiClamp 700B amplifier | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/multiclamp-patch-clamp#ad-700b |
| Digidata 1550B with pCLAMP 10.6 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/digidata-digitizers |
| Hisat2 (v2.0.5) | Johns Hopkins University | https://daehwankimlab.github.io/hisat2/ |
| featureCounts (v1.5.0-p3) | Subread | http://subread.sourceforge.net |
| Spectronaut (v17.0) | Biognosys | https://www.biognosys.com/products/software/spectronaut/ |
Experimental model and study participant details
Cry1Δ11 mice were generated using CRISPR/Cas9 technology as described.2 Mice of the same sex were group-housed. They were kept under a temperature of 20°C ± 2°C, 50-60% relative humidity, and 12h light: 12h dark cycle. They were also provided with free access to food and water. This study was approved by the IACUC of the Central South University of China (Study approval number: DWSY-2021-0507).
Method details
Behavioral tests
Male Cry1Δ11 mice and their wild-type littermates, at the age of 8-16 weeks, were used to carry out the following behavioral tests.
Heat test
Each mouse was individually placed on a plate heated to a temperature of 50°C and keenly observed until it started licking its hind paw or withdrawing the paw from the hot plate. The mouse was then removed from the plate, and the time taken until withdrawal was recorded in seconds.
Cold test
This test was conducted at three temperatures: 5°C, 2.5°C and 0°C. Each mouse was individually placed on the plate and keenly observed until it started licking its hind paw or withdrawing the paw from the cold plate. The mouse was then removed, and the time taken until withdrawal was recorded in seconds.
Temperature gradient test
This test was conducted by progressively lowering the temperature of a plate from 20°C to 0°C. The temperature and time-point at which the mouse first responded, as in the heat/cold test, were recorded.
Von frey tests
The mouse was placed in a Perspex box on top of a wire mesh and allowed to acclimate for 30 minutes. A von Frey filament with a particular force (0.16, 0.4, 0.6, 1.0, 1.4, or 2.0 g) was applied perpendicularly to the plantar surface of a hind paw. Upon applying the filament, the plantar surface was pressed until the filament was bent into an S shape for 3-5 seconds, and the behavioral response of the mouse was noted. A positive response is recorded if the animal exhibits any withdrawal behaviors either during application of the stimulus or immediately after the filament is removed. If there was no response, the next filament with a higher force was applied; if there was a response, the next lower-force filament was applied, with an interval of 1 minute between tests for each mouse. This continued until six readings were obtained, and the sequence of outcomes (no response or response) was used to calculate the 50% withdrawal threshold using the formula described by Deuis et al.,.40
Cotton swab assay
This test was conducted by gently stroking a cotton swab on the plantar surface of a hind paw. The swab was shaped as a Q-tip by pulling manually to puff it out. Mice were placed in the von Frey chamber to acclimate for 1 hour. The cotton swab was then gently stroked on the plantar surface of a hind paw. A paw withdrawal in response to a stroke was scored as positive. The stroking was repeated five times with 1-min intervals, and the percentage withdrawal for each mouse was calculated out of five trials.
Brush swabs
Mice were acclimated in a von Frey chamber for 1 hour. A 5/0 brush was gently stroked on the lateral side of a hind paw from heel to toe. A paw withdrawal in response to a stroke was scored as positive. The stroking was repeated five times with 1-min intervals, and the percentage withdrawal for each mouse was calculated out of five trials.
Punch
Mice were acclimated in a von Frey chamber for 1 hour. A 27 gauge needle was gently applied to the glabrous skin of a hind paw, taking care not to pierce the skin. A paw withdrawal in response to a punch was scored as positive. The punch was repeated five times with 1-min intervals, and the percentage withdrawal for each mouse was calculated out of five trials.
Open field test (OFT)
Each animal was placed in the center of the arena (72 × 72 × 36 cm) and the movement track was video recorded for 10 minutes. The total distance travelled (locomotion activity) and the duration at the central zone (36 × 36 cm, anxiety-like behavior) were calculated.
Tail suspension test (TST)
TST was used to assess depressive-like behavior. Animals were individually suspended by the tail in a white box (40 × 20 × 8 cm) for 6 minutes. The time spent immobile during the last 4 minutes was quantified. Immobility was defined as the absence of volitional body or limb movement.
Y-maze
The Y-maze apparatus consisted of three opaque arms (30 × 5 × 15 cm) that radiate from the center in a Y shape. The behavioral test was initiated by placing the mouse in the center of the Y, allowing free access to any of the three arms. The movement of the mouse was tracked by a video camera for 10 minutes. An arm entry was counted when all four paws of the mouse entered the arm; and an “alternation” was defined as a set of choices of consecutive arms without a repeated entry.
Western blot
Tissue samples were obtained from the dorsal horn region of the spinal cord and lysed in 2× SDS lysis buffer (2% SDS, 63 mM Tris-HCl, and 10% glycerol). Protein concentration was measured using the BCA assay method. Samples were added with 5×loading buffer (7.5% wt/vol SDS, 30% vol/vol glycerol, 200 mM DTT, 50 mM Tris, bromophenol blue, pH 6.8) and incubated at 65°C for 10 min. Proteins in lysates were then separated using SDS-PAGE, transferred to a PVDF membrane, and underwent immunoblotting with the corresponding antibodies overnight at 4°C. Membranes were then washed and incubated with HRP-conjugated secondary antibodies. The visualizing of proteins was done using the Pierce ECL Western Blotting Substrate kit (Thermo Fisher Scientific, 32106). Quantification of band intensities was performed by ImageJ (NIH).
Immunofluorescent staining
Mice were perfused intracardially with pre-chilled phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. The spinal cord was removed, postfixed, and cryoprotected in 30% sucrose overnight. The 30% sucrose was replaced by 40% sucrose solution for extended dehydration for another 48 hours. Finally, the spinal cord was transferred into pre-cooled isopentane and stored at -80°C. The spinal was removed from OCT and placed in the cryostat for 10 minutes for cryosectioning. Sections of 30 μm thickness were made from the lamina II of the dorsal horn in the spinal cord in the lumbar region, washed three times with PBS, and then treated with PBST (PBS plus 4% Triton) for 45 min before being incubated with primary antibodies to c-Fos (diluted 1:400; Abcam, ab209794), or p-CREB (diluted 1:400; CST, 9198), and/or VGLUT1 (diluted 1:400; Abcam, ab227805) and VGAT (diluted 1:400; Abcam, ab308062) overnight. The slices were then incubated in the dark with Alexa Fluor-conjugated secondary antibodies. Nuclei were stained with DAPI. Slices were imaged using a confocal microscope (TCS SP5; Leica).
Drug administration
H89 (2 mg/mL), dissolved in saline and 10% DMSO, was injected intrathecally at an hour prior to assessing behaviours and dissection of tissue, as described by Zhou et al., and Hosseini-Zare et al..41,42 A 15 μl microsyringe was inserted into the subarachnoid space through the L4 and L5 intervertebral space and the reagent was delivered to the cerebrospinal fluid. A typical appendix reflex (sudden lateral movement of the tail) confirmed that the syringe was successfully inserted.
Electrophysiological tests
Electrophysiological experiments were performed in spinal cord slices containing lamina II from 6- to 8-week-old male mice. The mice were deeply anesthetized by sodium pentobarbital (50 mg/kg, i.p.). Following decapitation, the spinal cord removed and was rapidly immersed in an ice-cold sucrose-based artificial cerebrospinal fluid (ACSF) containing 2.5 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 26 mM NaHCO3, 1.25 mM NaH2PO4·H2O, 10 mM dextrose, 213 mM sucrose, and pH 7.2-7.3. Spinal cords were sliced to 300-350 μm in this ice-cold ACSF. After slicing, they were immediately transferred to an incubation chamber filled with normal ACSF (126 mM NaCl, 2.5 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 26 mM NaHCO3, 1.25 mM NaH2PO4, 25 mM dextrose, pH 7.2-7.3) and maintained first at 35.5°C for 45-60 minutes and then at room temperature before use. Slices were perfused with carbogenated normal ACSF at 34°C-35°C, at a rate of 2 mL/min. For whole-cell recording, patch pipettes with an impedance of 4-7 MΩ were filled with normal internal solution containing 140 mM potassium gluconate, 3 mM KCl, 2 mM MgCl2, 0.2 mM EGTA, 10 mM HEPES, and 2 mM Na2ATP (285-295 mOsm, pH 7.2-7.25). Neurons within lamina II were visually identified under a microscope equipped with IR-DIC optics (BX-51WI, Olympus). Recordings with a series of resistance of less than 20 MΩ were included for data analysis. Current clamp recordings were achieved using a MultiClamp 700B amplifier (Molecular Devices). Digidata 1550B with pCLAMP 10.6 was used for signal acquisition. Signals were filtered at 10 kHz (voltage clamp) or 2.2 kHz (current clamp) and sampled at 50 kHz. Positive (20 pA/step, 500 ms in duration) current pulses were given to examine electrophysiological properties such as the F-I curve and input resistance. The cell membrane potential without any current injection was defined as resting membrane potential (RMP). The liquid junction potential (∼15 mV) was not corrected in the data analysis process. A successful action potential (AP) was defined when the amplitude surpassed 30 mV. The rheobase was defined as the minimum current pulse size when AP was initiated.
RNA-sequencing
Spinal cords from male mice at the age of 6 weeks were lysed with TRIzol reagent (Thermo Fisher Scientific, 15596026) according to the manufacturer’s instructions. Total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies). mRNA was purified from total RNA by using poly-T oligonucleotide-attached magnetic beads. After PCR amplification, the PCR product was purified by AMPure XP beads to obtain the library. The library was initially quantified by a Qubit 2.0 fluorometer and then sequenced with an Illumina NovaSeq 6000. Image data measured by the high-throughput sequencer were converted into sequence data (reads) by CASAVA base recognition. The reference genome index was built using Hisat2 (v2.0.5), and featureCounts (v1.5.0-p3) was used to count the read numbers mapped to each gene. Differential expression analysis was performed using the DESeq2 R package (1.20.0). The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the FDR. Padj of 0.05 or less and log 2 (fold change) of 1 or greater were set as the thresholds for significantly differential expression. We used the cluster Profiler R package (v3.8.1) to test the statistical enrichment of differentially expressed genes in KEGG pathways. Reactome pathways with corrected P-values less than 0.05 were considered significantly enriched by differentially expressed genes. All original RNA-Seq data have been deposited in the NCBI [PRJNA1136699].
Protein phosphoproteomics analysis
Spinal cords from male mice at the age of 6 weeks were grounded into cell powder in liquid nitrogen, and then four volumes of lysis buffer (8 M urea, 1% protease inhibitor cocktail) were added. After sonication for three minutes using a high-intensity ultrasonic processor, the lysates were centrifuged at 12000 g at 4°C for 10 min to remove the remaining debris. Finally, the supernatant was collected and the protein concentration was determined with a BCA kit according to the manufacturer’s instructions. After digestion with trypsin overnight, the samples were reduced with 5 mM dithiothreitol and alkylated with 11 mM iodoacetamide at room temperature in darkness. The resulting peptides were desalted by a Strata X SPE column, and the peptide mixture was then incubated in IMAC microspheres. The elution buffer containing NH4OH was used to elute phosphopeptides, which were then collected and lyophilized for LC-MS/MS analysis. The full MS scan was set to 100-1700 (MS/MS scan range) and 22 PASEF (MS/MS mode), and the isolation window was set to 20 m/z. In building the Spectral Library, the DDA data were processed using Spectronaut (v .17.0) software coupled with the Pulsar search engine. Tandem mass spectra were searched against Mus_musculus_10090_SP_20230103.fasta (17132 entries) concatenated with the reverse decoy database. The number of max missing cleavages was set to 2. Phosphorylation (S/T) were specified as variable modifications. The false discovery rate (FDR) of protein, peptide, and PSM was adjusted to < 1%. The corresponding spectral library was imported into Spectronaut (v. 17.0) software to predict the retention time by nonlinear correction and searched against DIA data.
Quantification and statistical analysis
Statistical analysis was done using GraphPad Prism version 9.0 software. Two-tailed Student’s t-tests were used for statistical comparison between 2 groups and two-way analysis of variance (ANOVAs) or multiple-comparison tests were utilized with Bonferroni’s post hoc test when conditions were 3 or more. A P value of less than 0.05 (P < 0.05) was considered statistically significant.
Published: October 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111110.
Contributor Information
Dengfeng Liu, Email: dengfeng_l@163.com.
Meichun Deng, Email: dengmch@csu.edu.cn.
Jia-Da Li, Email: lijiada@sklmg.edu.cn.
Supplemental information
References
- 1.Faraone S.V. The pharmacology of amphetamine and methylphenidate: Relevance to the neurobiology of attention-deficit/hyperactivity disorder and other psychiatric comorbidities. Neurosci. Biobehav. Rev. 2018;87:255–270. doi: 10.1016/j.neubiorev.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu D., Xie Z., Gu P., Li X., Zhang Y., Wang X., Chen Z., Deng S., Shu Y., Li J.D. Cry1Delta11 mutation induces ADHD-like symptoms through hyperactive dopamine D1 receptor signaling. JCI Insight. 2023;8 doi: 10.1172/jci.insight.170434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posner J., Polanczyk G.V., Sonuga-Barke E. Attention-deficit hyperactivity disorder. Lancet. 2020;395:450–462. doi: 10.1016/S0140-6736(19)33004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erskine H.E., Ferrari A.J., Nelson P., Polanczyk G.V., Flaxman A.D., Vos T., Whiteford H.A., Scott J.G. Epidemiological modelling of attention-deficit/hyperactivity disorder and conduct disorder for the Global Burden of Disease Study 2010. JCPP (J. Child Psychol. Psychiatry) 2013;54:1263–1274. doi: 10.1111/jcpp.12144. [DOI] [PubMed] [Google Scholar]
- 5.Polanczyk G.V., Willcutt E.G., Salum G.A., Kieling C., Rohde L.A. ADHD prevalence estimates across three decades: an updated systematic review and meta-regression analysis. Int. J. Epidemiol. 2014;43:434–442. doi: 10.1093/ije/dyt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stickley A., Koyanagi A., Takahashi H., Kamio Y. ADHD symptoms and pain among adults in England. Psychiatr. Res. 2016;246:326–331. doi: 10.1016/j.psychres.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Bouchatta O., Aby F., Sifeddine W., Bouali-Benazzouz R., Brochoire L., Manouze H., Fossat P., Ba-M'hamed S., Bennis M., Landry M. Pain hypersensitivity in a pharmacological mouse model of Attention-Deficit/Hyperactivity Disorder (ADHD) Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2114094119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerekes N., Sanchéz-Pérez A.M., Landry M. Neuroinflammation as a possible link between attention-deficit/hyperactivity disorder (ADHD) and pain. Med. Hypotheses. 2021;157 doi: 10.1016/j.mehy.2021.110717. [DOI] [PubMed] [Google Scholar]
- 9.Vingilis E., Erickson P.G., Toplak M.E., Kolla N.J., Mann R.E., Seeley J., vanderMaas M., Daigle D.S. Attention Deficit Hyperactivity Disorder Symptoms, Comorbidities, Substance Use, and Social Outcomes among Men and Women in a Canadian Sample. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/982072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veldhuijzen D.S., Kenemans J.L., de Bruin C.M., Olivier B., Volkerts E.R. Pain and attention: attentional disruption or distraction? J. Pain. 2006;7:11–20. doi: 10.1016/j.jpain.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Moore D.J., Meints S.M., Lazaridou A., Johnson D., Franceschelli O., Cornelius M., Schreiber K., Edwards R.R. The Effect of Induced and Chronic Pain on Attention. J. Pain. 2019;20:1353–1361. doi: 10.1016/j.jpain.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Higgins G.A., Silenieks L.B., Van Niekerk A., Desnoyer J., Patrick A., Lau W., Thevarkunnel S. Enduring attentional deficits in rats treated with a peripheral nerve injury. Behav. Brain Res. 2015;286:347–355. doi: 10.1016/j.bbr.2015.02.050. [DOI] [PubMed] [Google Scholar]
- 13.Patke A., Murphy P.J., Onat O.E., Krieger A.C., Özçelik T., Campbell S.S., Young M.W. Mutation of the Human Circadian Clock Gene CRY1 in Familial Delayed Sleep Phase Disorder. Cell. 2017;169:203–215.e13. doi: 10.1016/j.cell.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura R., Yoshimi K., Sawai Y., Yasumune N., Kajihara K., Maejima T., Koide T., Node K., Akashi M. The role of cell-autonomous circadian oscillation of Cry transcription in circadian rhythm generation. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110703. [DOI] [PubMed] [Google Scholar]
- 15.Narasimamurthy R., Hatori M., Nayak S.K., Liu F., Panda S., Verma I.M. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. USA. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang E.E., Liu Y., Dentin R., Pongsawakul P.Y., Liu A.C., Hirota T., Nusinow D.A., Sun X., Landais S., Kodama Y., et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 18.Hirano A., Fu Y.H., Ptáček L.J. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016;23:1053–1060. doi: 10.1038/nsmb.3326. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang Y., Li Z., Xiong S., Sun C., Li B., Wu S.A., Lyu J., Shi X., Yang L., Chen Y., et al. Circadian clocks are modulated by compartmentalized oscillating translation. Cell. 2023;186:3245–3260.e23. doi: 10.1016/j.cell.2023.05.045. [DOI] [PubMed] [Google Scholar]
- 20.Rijo-Ferreira F., Takahashi J.S. Genomics of circadian rhythms in health and disease. Genome Med. 2019;11:82. doi: 10.1186/s13073-019-0704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu Y., He H., Liu Q., Jia S., Fan W., Huang F. The Circadian Clocks, Oscillations of Pain-Related Mediators, and Pain. Cell. Mol. Neurobiol. 2023;43:511–523. doi: 10.1007/s10571-022-01205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palada V., Gilron I., Canlon B., Svensson C.I., Kalso E. The circadian clock at the intercept of sleep and pain. Pain. 2020;161:894–900. doi: 10.1097/j.pain.0000000000001786. [DOI] [PubMed] [Google Scholar]
- 23.Skyba D.A., Radhakrishnan R., Bement M.K.H., Sluka K.A. The cAMP pathway and pain: Potential targets for drug development. Drug. Discov. Today Dis. Model. 2004;1:115–119. doi: 10.1016/j.ddmod.2004.07.003. [DOI] [Google Scholar]
- 24.Hoeger-Bement M.K., Sluka K.A. Phosphorylation of CREB and mechanical hyperalgesia is reversed by blockade of the cAMP pathway in a time-dependent manner after repeated intramuscular acid injections. J. Neurosci. 2003;23:5437–5445. doi: 10.1523/JNEUROSCI.23-13-05437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song I., Huganir R.L. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- 26.Ji R.R., Kohno T., Moore K.A., Woolf C.J. Central sensitization and LTP: do pain and memory share similar mechanisms? Trends Neurosci. 2003;26:696–705. doi: 10.1016/j.tins.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Otaño I., Ehlers M.D. Learning from NMDA receptor trafficking: clues to the development and maturation of glutamatergic synapses. Neurosignals. 2004;13:175–189. doi: 10.1159/000077524. [DOI] [PubMed] [Google Scholar]
- 28.Kutlar A., Wells L.G., Xu H., Bowman L., Bora P., Chand A.R., Clair B., Meiler S.E., Wang X., Natrajan K., et al. Genetic Association Studies of Pain Phenotypes in Adult SCD Patients. Blood. 2014;124:2708. doi: 10.1182/blood.V124.21.2708.2708. [DOI] [Google Scholar]
- 29.Li Y., Tang W., Kang L., Kong S., Dong Z., Zhao D., Liu R., Yu S. Functional correlation of ATP1A2 mutations with phenotypic spectrum: from pure hemiplegic migraine to its variant forms. J. Headache Pain. 2021;22:92. doi: 10.1186/s10194-021-01309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Zhang Y., Ma N., Zhao W., Ren X., Sun Y., Zang W., Cao J. SIRT1 mediates the excitability of spinal CaMKIIα-positive neurons and participates in neuropathic pain by controlling Nav1.3. CNS Neurosci. Ther. 2024;30 doi: 10.1111/cns.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo S.-H., Lin J.-P., Huang L.-E., Yang Y., Chen C.-Q., Li N.-N., Su M.-Y., Zhao X., Zhu S.-M., Yao Y.-X. Silencing of spinal Trpv1 attenuates neuropathic pain in rats by inhibiting CAMKII expression and ERK2 phosphorylation. Sci. Rep. 2019;9:2769. doi: 10.1038/s41598-019-39184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y.B., Jia G.L., Wang J.W., Ye X.Y., Lu J.H., Chen J.L., Zhang M.B., Xie C.S., Shen Y.J., Tao Y.X., et al. Activation of CaMKII and GluR1 by the PSD-95-GluN2B Coupling-Dependent Phosphorylation of GluN2B in the Spinal Cord in a Rat Model of Type-2 Diabetic Neuropathic Pain. J. Neuropathol. Exp. Neurol. 2020;79:800–808. doi: 10.1093/jnen/nlaa035. [DOI] [PubMed] [Google Scholar]
- 33.Chen W., McRoberts J.A., Ennes H.S., Marvizon J.C. cAMP signaling through protein kinase A and Epac2 induces substance P release in the rat spinal cord. Neuropharmacology. 2021;189 doi: 10.1016/j.neuropharm.2021.108533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao W.L., Qin Q.R., Li Q., Hao J.W., Wei S., Li X.M., Liu T.T., Qiu C.Y., Hu W.P. Group II metabotropic glutamate receptor activation suppresses ATP currents in rat dorsal root ganglion neurons. Neuropharmacology. 2023;227 doi: 10.1016/j.neuropharm.2023.109443. [DOI] [PubMed] [Google Scholar]
- 35.Kohno T., Wang H., Amaya F., Brenner G.J., Cheng J.-K., Ji R.-R., Woolf C.J. Bradykinin Enhances AMPA and NMDA Receptor Activity in Spinal Cord Dorsal Horn Neurons by Activating Multiple Kinases to Produce Pain Hypersensitivity. J. Neurosci. 2008;28:4533–4540. doi: 10.1523/JNEUROSCI.5349-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miletic G., Pankratz M.T., Miletic V. Increases in the phosphorylation of cyclic AMP response element binding protein (CREB) and decreases in the content of calcineurin accompany thermal hyperalgesia following chronic constriction injury in rats. Pain. 2002;99:493–500. doi: 10.1016/S0304-3959(02)00242-7. [DOI] [PubMed] [Google Scholar]
- 37.Cao H., Gao Y.J., Ren W.H., Li T.T., Duan K.Z., Cui Y.H., Cao X.H., Zhao Z.Q., Ji R.R., Zhang Y.Q. Activation of extracellular signal-regulated kinase in the anterior cingulate cortex contributes to the induction and expression of affective pain. J. Neurosci. 2009;29:3307–3321. doi: 10.1523/JNEUROSCI.4300-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocks D., Cham H., Kundakovic M. Why the estrous cycle matters for neuroscience. Biol. Sex Differ. 2022;13:62. doi: 10.1186/s13293-022-00466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman J.J., LeResche L. Does experimental pain response vary across the menstrual cycle? A methodological review. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R245–R256. doi: 10.1152/ajpregu.00920. [DOI] [PubMed] [Google Scholar]
- 40.Deuis J.R., Dvorakova L.S., Vetter I. Methods Used to Evaluate Pain Behaviors in Rodents. Front. Mol. Neurosci. 2017;10:284. doi: 10.3389/fnmol.2017.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X.L., Wang Y., Zhang C.J., Yu L.N., Cao J.L., Yan M. PKA is required for the modulation of spinal nociceptive information related to ephrinB-EphB signaling in mice. Neuroscience. 2015;284:546–554. doi: 10.1016/j.neuroscience.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 42.Hosseini-Zare M.S., Salehi F., Seyedi S.Y., Azami K., Ghadiri T., Mobasseri M., Gholizadeh S., Beyer C., Sharifzadeh M. Effects of pentoxifylline and H-89 on epileptogenic activity of bucladesine in pentylenetetrazol-treated mice. Eur. J. Pharmacol. 2011;670:464–470. doi: 10.1016/j.ejphar.2011.09.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: RNA-seq data have been deposited at NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report the original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.