Abstract
Taf14 is a subunit of multiple fundamental complexes implicated in transcriptional regulation and DNA damage repair in yeast cells. Here, we investigate the association of Taf14 with the consensus sequence present in other subunits of these complexes and describe the mechanistic features that affect this association. We demonstrate that the precise molecular mechanisms and biological outcomes underlying the Taf14 interactions depend on the accessibility of binding interfaces, the ability to recognize other ligands, and a degree of sensitivity to temperature and chemical and osmotic stresses. Our findings aid in a better understanding of how the distribution of Taf14 among the complexes is mediated.
Keywords: Taf14, ET domain, YEATS, Tfg1, Ino80, Snf5, Taf2, Sth1, Mechanism
1. Introduction
Transcription initiation factor subunit 14 (Taf14) is a multifunctional yeast protein identified in a diverse set of chromatin associating complexes implicated in transcriptional regulation, DNA damage repair, chromosome maintenance, and cell cycle progression (Fig. 1a). Taf14 is an integral component of the general transcription factor (TF) complex TFIID, which binds to gene promoters and plays a critical role in the assembly of the preinitiation complex (PIC), and the TFIIF complex, which recruits RNA polymerase II (RNA Pol II) to PIC [1–5]. Taf14 is also present in the histone acetyltransferase complex NuA3 that acetylates lysine 14 of histone H3 and facilitates transcription elongation [6], and in the ATP-dependent chromatin-remodeling complexes INO80, SWI/SNF and RSC [3,7,8]. In addition, Taf14 is functionally linked to the transcript elongation factor TFIIS, which promotes elongation by RNA Pol II [9], and to the mediator complex [10]. Taf14 is required for the mitotic cell cycle progression, and Taf14 depleted cells are characterized by cytoskeletal and morphological defects, diminished transcription in vivo, and are thermosensitive [2,11].
Fig. 1.

EBM binding site of Taf14ET. (a) Taf14 is a component of a diverse set of yeast complexes. (b) Domain architecture of full length Taf14 (Taf14FL). (c) Structural overlay of Taf14ET (grey) in complex with Sth1EBM (green) (PDB 6LQZ) and Taf14ET (wheat) in complex with Taf2EBM (yellow) (PDB 7UHE). (d) Alignment of the EBM (ET domain binding motif) sequences from Ino80, Taf2, Tfg1, Snf5 and Sth1 proteins. In the consensus sequence, h is a hydrophobic residue, + is a positively charged residue, and x is any residue. Conserved hydrophobic and lysine residues are orange and blue, respectively. (e) Superimposed 1H,15N HSQC spectra of Taf14ET recorded in the presence of increasing amounts of Tfg1EBM, Ino80EBM and Snf5EBM. The spectra are colour coded according to the protein:peptide molar ratio. (f) Structural overlay of Sth1EBM (green) from the Taf14ET-Sth1EBM complex, Taf2EBM (dark yellow) from the Taf14ET-Taf2EBM complex, and Snf5EBM (salmon) from Snf5FL generated by AlphaFold. Taf14ET (wheat) only from the Taf14ET-Taf2EBM complex is shown as a ribbon for clarity. Residues of EBM peptides are shown as sticks and labeled. (g) Superimposed 1H,15N HSQC spectra of Taf14ET recorded in the presence of increasing amounts of the indicated mutants of Tfg1EBM. The spectra are colour coded according to the protein:peptide molar ratio.
Taf14 contains two folded domains, the YEATS (Yaf9, ENL, AF9, Taf14, Sas5) domain and the ET (extra-terminal) domain, coupled via a short linker (Fig. 1b). The YEATS domain of Taf14 is a well-established reader of acylated lysine 9 of histone H3 (H3K9acyl), and this function is essential in transcriptional regulation and DNA-damage response [12–15]. The unstructured linker is implicated in DNA binding, and this activity depends on dynamic equilibrium between autoinhibited and open states of Taf14 [16]. The ET domain has been shown to recognize the ‘hxh + h’ motif [where h is a hydrophobic residue, + is a positively charged residue, and x is any residue] and named EBM (ET binding motif). EBM is present in co-factors, such as Sth1, a subunit of the RSC complex, and Taf2, a subunit of TFIID [16,17]. Interestingly, the ET domain of Taf14 undergoes liquid-liquid phase separation (LLPS) with the cofactor binding stimulating LLPS [17]. The LLPS capability was proposed to concentrate the Taf14-containing complexes at specific chromatin loci and drive efficient gene expression [17]. Because Taf14 is present in multiple and distinctly different yeast complexes, its precise biological role within each complex remains difficult to define.
In this study, we use structural, biochemical and mutagenesis tools to identify mechanistic features that affect the Taf14 engagement with co-factors from different complexes. Our genetic analysis using mutated co-factors provides an approach to disrupt the Taf14 association with individual co-factors and therefore dissect the role of Taf14 independently in each complex.
2. Results and discussion
2.1. EBMs occupy a conserved binding site of Taf14ET
Structural analysis of the Taf14 ET domain (Taf14ET) in complex with Sth1 peptide or Taf2 peptide shows that the peptides are bound in a cleft formed by α1 helix and β-sheet of the protein [16,17] (Fig. 1c). Both Sth1 and Taf2 contain the hxh + h sequence, or EBM, which is also present in Tfg1 (Tfg1EBM), a catalytic subunit of TFIIF, in Ino80 (Ino80EBM), a catalytic subunit of the INO80 complex, and in Snf5 (Snf5EBM), a catalytic subunit of the SWI/SNF complex (Fig. 1d). We used NMR spectroscopy to assess whether the EBM recognition mode is conserved for other ligands of Taf14ET. We generated uniformly 15N-labeled Taf14ET and recorded its 1H,15N heteronuclear single quantum coherence (HSQC) spectra as unlabeled Tfg1EBM (aa 614–625), Ino80EBM (aa 368–379) and Snf5EBM (aa 782–793) peptides were gradually added to the protein samples (Fig. 1e). Large chemical shift perturbations (CSPs) induced by either Tfg1EBM, Ino80EBM or Snf5EBM in Taf14ET indicated formation of the complexes (Fig. 1e). A number of amide resonances of Taf14ET that were observed in the apo-state disappeared upon addition of the peptides, and another set of crosspeaks corresponding to the EBM-bound state of Taf14ET appeared. The slow and slow-to-intermediate exchange regime on the NMR timescale suggested tight binding, and an overall similar pattern of CSPs induced by each peptide, as well as Taf2EBM [16], indicated that these ligands are bound in the same binding site of Taf14ET.
2.2. Contribution of hydrophobic and ionic contacts
The structure of the Taf14ET-Taf2EBM complex reveals that Taf2EBM forms the third anti-parallel β-strand, pairing with the second β-strand of Taf14ET, and that the complex is stabilized through extensive intermolecular contacts [16] (Fig. 1f). These include the backbone-backbone hydrogen bonds, hydrophobic contacts involving the side chains of V1397 and I1399 of Taf2EBM that are buried in the hydrophobic cleft of Taf14ET, as well as electrostatic interactions involving R1400 and K1398 of Taf2EBM. AlphaFold derived models of full length Tfg1, Ino80 and Snf5 show that Tfg1EBM, Ino80EBM and Snf5EBM adopt an extended conformation even in the apo-states of these proteins (Suppl. Fig. 1). Notably, Snf5EBM and Tfg1EBM superimpose with Taf2EBM or Sth1EBM bound to Taf14ET with an rmsd (root-mean-square deviation) of 0.9 Å and 2.8 Å, respectively (Fig. 1f). We also found that hydrophobic contacts are required for binding of these ligands, as a single point mutation of either I618 or L620 in Tfg1EBM to an aspartate abolished the inter-action with Taf14ET (Fig. 1g).
To delineate the contribution of hydrophobic and ionic contacts, we tested formation of the Taf14ET-Sth1EBM complex in variable salt concentrations using pull-down assays. GST-Taf14ET and Sumo-Sth1EBM were incubated with glutathione Sepharose beads at 4 °C or room temperature (RT) and Sumo-Sth1EBM bound Taf14ET was detected by electrophoresis (Fig. 2a, b). As shown in Fig. 2a, an increase in concentration of NaCl from 300 mM to 800 mM resulted in a reduced binding of Sth1EBM, indicating that electrostatic interactions are essential for the formation of the tight complex which can be regulated by osmotic pressure (Fig. 2c). Still, the binding was not completely abolished even at a salt concentration of 1 M, indicating that hydrophobic contacts are the driving force of this interaction (Fig. 2b).
Fig. 2.
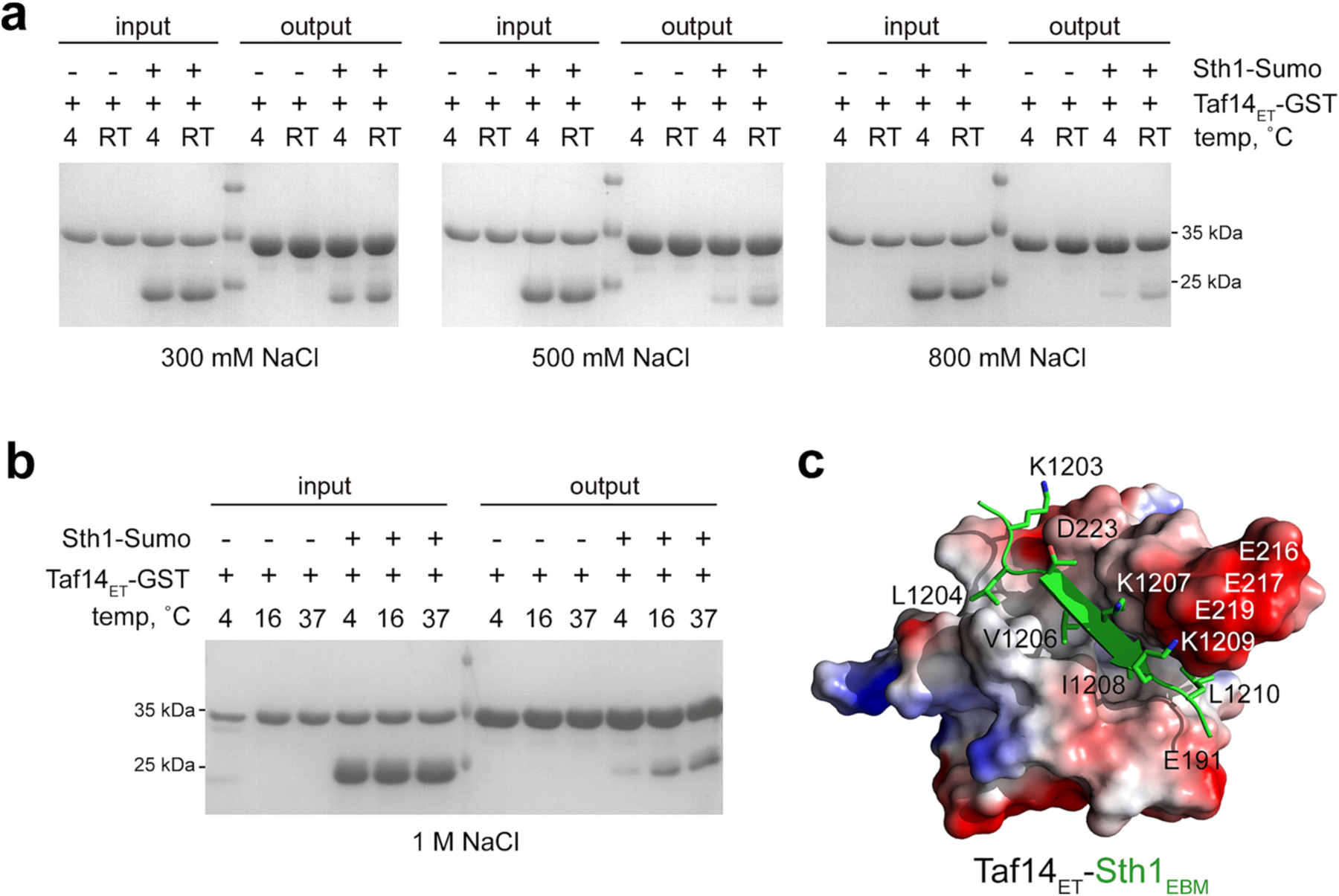
Effects of hydrophobic and ionic contacts. (a, b) SDS-PAGE gels showing binding of SUMO-Sth1EBM to GST-Taf14ET on glutathione Sepharose 4B beads at indicated temperature and NaCl concentrations. (c) The structural model of the Taf14ET-Sth1EBM complex. Taf14ET is shown as a surface model colored according to its electrostatic potential (positive potential is blue and negative potential is red). Residues of the Sth1EBM peptide are shown as green sticks.
Interestingly, we found that in contrast to the binding of Ino80EBM or Snf5EBM (data not shown), the interaction between Taf14ET and Sth1EBM is temperature sensitive. GST-Taf14ET pulled down more Sth1EBM with the increase of the reaction temperature (Fig. 2a, b). Together, these data suggest that the degree of temperature and osmotic adaptation could mediate the ability of Taf14 to form complexes to ensure the optimal growth and proliferation of the mesophilic yeast.
2.3. Diverse mechanisms of the Taf14 complex formation
We have previously shown that both domains of Taf14, Taf14ET and Taf14YEATS, are capable of binding to Taf2EBM and proposed a model in which Taf2EBM is sandwiched between Taf14ET and Taf14YEATS [16]. Upon Taf2EBM binding, a contiguous twisted β-sheet encompassing both domains of Taf14 could be created to release the linker region of Taf14 for binding to DNA (Fig. 3a, b). As shown in Fig. 3c, full length Taf14FL maintained the ability to bind EBM peptides in NMR titration experiments. Much like Taf2EBM, Ino80EBM, Snf5EBM, and Tfg1EBM induced CSPs in 15N-labeled Taf14FL (Fig. 3c). However, CSPs in 1H,15N HSQC spectra of the isolated 15N-labeled Taf14YEATS were caused only by Taf2EBM (Fig. 3d). The inability of Ino80EBM, Snf5EBM, and Tfg1EBM to induce significant resonance changes indicated that in contrast to Taf2EBM, these EMBs do not interact with Taf14YEATS and pointed to the unique binding mechanism that differentiates the Taf14-Taf2 complex formation.
Fig. 3.

Unique mechanisms of the Taf14 complex formation. (a) Schematic of Taf14FL in complex with H3K9acyl and Taf2EBM. (b) Overlay of the crystal structures of the Taf14YEATS:H3K9cr complex (light green:grey), Taf14ET:Taf2EBM complex (wheat:yellow), and the cryo-EM structure of Taf14FL in the RNA Pol II TFIIF complex (5SVA, not shown) [4]. (c, d) Superimposed 1H,15N HSQC spectra of Taf14FL (c) or Taf14YEATS (d) recorded in the presence of increasing amounts of the indicated EBMs. Taf2EBM data are from [16]. The spectra are colour coded according to the protein:peptide molar ratio.
The dual domain engagement with Taf2EBM in Taf14FL led to a ~ two-fold tighter binding compared to the interaction of an isolated Taf14ET with Taf2EBM [16] (Fig. 4a). However, the full-length Taf14 protein has been shown to exist in a dynamic equilibrium between the autoinhibited state, in which Taf14ET associates with the linker and therefore is not easily accessible, and the open state, in which Taf14ET is more freely available (Fig. 4b). To test whether the Taf14FL autoinhibition affects interactions with Ino80EBM, Snf5EBM, Sth1EBM and Tfg1EBM, we measured binding affinities by microscale thermophoresis (MST) (Fig. 4a, c and Suppl. Fig. 2). The measurements showed that compared to the isolated Taf14ET [17], Taf14FL associated five-fold weaker with Ino80EBM and seven-fold weaker with Snf5EBM, but its binding to Sth1EBM and Tfg1EBM was unaffected. Much like the isolated domain, the full length protein had highest binding affinity toward Sth1EBM and Ino80EBM (dissociation constants (Kds) of 0.07 μM and 1.2 μM, respectively) (Fig. 4a).
Fig. 4.

Full length Taf14FL binds EBM. (a) Binding affinities to indicated EBMs measured by MST. Kd values represent average of three independent measurements ± SEM (two for Snf5). Taf2EBM data are from [16]. (b) A model of the autoinhibited apo-state of Taf14FL. (c) MST binding curve for the interaction of Taf14FL with Tfg1EBM. The Kd value represents average of three independent measurements ± SEM, and error bars represent SEM for each point. n = 3 (d) AlphaFold generated models of the structures of the EBM-containing proteins in apo-states. The EBM sequence is red.
Another aspect of the Taf14-EBM complex formation that needs to be taken into account is the accessibility of EBM sequences in full length proteins which can fine tune the assembly of the complexes. Even though Taf2EBM is located in the C-terminal tail of the protein, the AlphaFold model of Taf2FL suggests that this region is involved in intramolecular contacts that may impede binding to Taf14 (Fig. 4d). Similarly, it is unclear how accessible Ino80EBM, Snf5EBM, Sth1EBM and Tfg1EBM are in the context of full length Ino80, Snf5, Sth1 and Tfg1 proteins (Fig. 4d) and whether this accessibility is regulated by interactions with other ligands and/or conformational changes. Clearly, the binding capability of Taf14 have to be re-examined in the context of full length EBM-containing proteins.
2.4. EBM recognition is linked to distinct biological outcomes
To explore the ways to independently characterize the functional significance of EBM recognition by Taf14, we investigated the phenotypes of yeast strains in which binding of Snf5 and separately binding of Sth1 to Taf14 was disrupted. We generated yeast strains harboring L1204A/V1206A/I1208A/L1210A mutant of Sth1 (sth1M4) and L785A/L787A/I789A/L791A mutant of Snf5 (snf5M4) that are impaired in binding to Taf14ET [17]. The plasmids carrying SNF5 or snf5M4 were transformed into a snf5Δ strain. As sth1 is an essential gene, we transformed the diploid strain BY4743 with plasmids carrying either wt STH1 or sth1M4 and isolated spore clones carrying STH1 or sth1M4 plasmids. The sth1M4 strain with the Taf14-binding deficient mutant of Sth1 displayed heat-sensitivity and failed to grow at 34 °C or 37 °C (Fig. 5a). In contrast, the snf5M4 strain with the Taf14-binding deficient mutant of Snf5 grew faster than wt at all temperatures tested (Fig. 5b). Collectively, these results, together with the findings that the deletion of Taf2EBM leads to a mild slow growth phenotype while preventing the stable association of Taf14 with TFIID [18], pointed to distinct roles of the EBM recognition which can be examined through impairing EBMs. Our data, for example, suggested that, unlike binding of Sth1, binding of Snf5 to Taf14 might be important for repressive action of the SWI/SNF complex. However, we cannot exclude the possibility that once the SWI/SNF complex is depleted of Taf14 due to defective Snf5EBM, more Taf14 is available to other complexes, and such a redistribution of Taf14 could also perturb functions of these complexes.
Fig. 5.

Genetic analyses of the Taf14-Sth1 and Taf14-Snf5 interactions. (a) Spotting assays with sth1Δ strains transformed with plasmids expressing full-length wild-type Sth1 or M4 mutant (L120A/V1206A/I1208A/L1210A) at different temperatures. (b) Spotting assays with snf5Δ strain transformed with plasmids expressing full-length wild-type Snf5 or M4 mutant (L785A/L787A/I789A/L791A) at different temperatures. (c) Spotting assays with sth1Δ strains transformed with plasmids expressing full-length wild-type Sth1 or M4 mutant on plates containing 20 mM hydroxyurea (HU), 0.015 % methyl methane sulfonate (MMS), and 10 mg/ml phleomycin (Phle) at 30 °C. (d) Spotting assays with snf5Δ strains transformed with plasmids expressing full-length wild-type Snf5 or M4 mutant on plates containing 20 mM hydroxyurea (HU), 0.015 % methyl methane sulfonate (MMS), and 10 mg/ml phleomycin (Phle) at 30 °C.
Comparison of the phenotypes of wt STH1 and sth1M4 yeast strains and wt SNF5 and snf5M4 yeast strains in response to DNA-damage agents, including methyl methanesulfonate (MMS), phleomycin (Phle), and hydroxyurea (HU) further confirmed the effectiveness of this approach in studying biological effects associated with the recognition of diverse EBM ligands. The sth1M4 yeast strain showed slight growth defects compared to the wt STH1 yeast strain (Fig. 5c), but the snf5M4 yeast strain grew much better than wt SNF5 strain in the presence of these drugs (Fig. 5d).
2.5. Concluding remarks
The findings that Taf14ET associates with several essential co-factors through recognizing their EBM sequences raises the question as to what are the mechanisms that control the distribution of Taf14 among the co-factor-containing complexes in yeast cells. In this study, we show that while isolated EBM peptides tested are readily recognized by Taf14ET, the molecular mechanisms and biological outcomes of the full length Taf14 interactions differ and depend on a variety of factors, including the ability of EBMs to alleviate the Taf14 autoinhibitory state and interact with other ligands. In addition, the Taf14-containing complexes can be characterized by a different degree sensitivity to temperature, nutrient, chemical and osmotic stresses and likely depend on the accessibility of the EBM sequences in the context of full length ligand proteins. The latter could have a profound impact on Taf14 incorporation into the complexes, necessitating thorough investigation of the interactions between full length binding partners.
We further demonstrate the feasibility of characterization of the role of Taf14 in specific complexes through a design of EBM-impaired co-factors. This approach expands our previous work showing that the disruption of Taf14-EBM interaction within the RSC complex results in yeast cells incapable of growing at high temperature and alters the transcription of genes involved in heat-response and catabolic metabolism [17]. Although some Taf14-mediated interactions have been characterized at the atomic resolution level [12,14–17], to better understand the differences in binding co-factors, structures of other complexes need to be elucidated. Additionally, cryo-EM studies could provide a comprehensive view of the entire complexes and may reveal additional contacts of Taf14 with other subunits to fine tune the complex assemblies and functions.
3. Methods
3.1. Protein purification
The S. cerevisiae GST- and His-tagged Taf14FL (aa 1–244), Taf14YEATS (aa 1–137), and Taf14ET (aa 168–243 and aa 162–243 K163N/R164Q/K166N) were expressed in Escherichia coli Rosetta-2 (DE3) pLysS cells grown in either Terrific Broth, Luria Broth or minimal media supplemented with 15NH4Cl (Sigma). Protein production was induced with 0.3 mM IPTG for 18 h at 16 °C. Bacteria were harvested by centrifugation and lysed by sonication. GST-fusion proteins were purified on glutathione agarose 4B beads (Thermo Fisher Sci). The GST-tag was cleaved with either PreScission or tobacco etch virus (TEV) protease. His-fusion proteins were purified on a Ni-NTA resin (ThermoFisher) and eluted with a gradient of imidazole. The His-tag was cleaved with TEV protease and removed by dialysis. Unlabeled proteins were further purified by column chromatography and concentrated in Millipore concentrators (Millipore). For pull-down assays, Sth1EBM-containing fragment (aa 1183–1240) was expressed as a 6xHis-Sumo-tagged protein and purified as above.
3.2. NMR experiments
Nuclear magnetic resonance (NMR) experiments were performed at 298 K on Varian INOVA 900 MHz and 600 MHz spectrometers equipped with cryogenic probes. The NMR samples contained 0.1–0.3 mM uniformly 15N-labeled Taf14FL, Taf14YEATS, or Taf14ET in 50 mM Tris-HCl pH 6.6–7.5, 150 mM NaCl, 0–5 mM DTT or 0.2 mM TCEP or in PBS pH 6.6–7.5, 0–5 mM DTT and 8–10 % D2O. Binding was characterized by monitoring chemical shift changes in 1H,15N HSQC spectra of the proteins induced by the addition of Tfg1EBM (aa 614–625), Ino80EBM (aa 368–379), Snf5EBM (aa 782–793) and Taf2EBM (aa 1392–1402) peptides (synthesized by Synpeptide). NMR data were processed and analyzed as described [19].
3.3. GST pull-down assays
For pull-down assays, 50 μg of GST-tagged Taf14ET and 50 μg Sumo-Sth1EBM were mixed with 10 μL of glutathione Sepharose 4B beads in 100 μL of binding buffer (25 mM Tris-HCl, pH 8.0, 300 mM/ 500 mM/ 800 mM /1 M NaCl, and 2 mM DTT). 10 μL reaction mixtures were taken out as input controls. After incubation at the indicated temperature for 3 h, the beads were washed four times with 500 μL of the binding buffer. The bound samples were eluted with 25 μL elution buffer (15 mM reduced glutathione, 25 mM Tris-HCl, pH 8.0, 300 mM/500 mM/800 mM/1 M NaCl, and 2 mM DTT). The input and eluted samples were analyzed by SDS-PAGE.
3.4. MST
Microscale thermophoresis (MST) experiments were performed using a Monolith NT.115 instrument (NanoTemper). All experiments were performed using SEC purified His-Taf14FL protein in a buffer containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0–0.2 mM TCEP. His-Taf14FL was labeled using a His-Tag Labeling Kit RED-tris-NTA (2nd Generation, Nanotemper), and concentration of each labeled protein was kept at 10 nM. Dissociation constants were determined using a direct binding assay in which increasing amounts of unlabeled binding partner (Tfg1EBM, Snf5EBM, Sth1EBM or Ino80EBM peptides) were added stepwise. The measurements were performed at 40 % LED and medium MST power with 3 s steady state, up to 20 s laser on time and 1 s off time. The Kd values were calculated using MO Affinity Analysis software (NanoTemper) (n = 3; 2 for Snf5). Plots were generated in GraphPad PRISM.
3.5. Stress resistance assay and growth assay
Yeast strains were cultured in YC medium at 30 °C and suspended in sterile water at a final concentration of OD600 ~ 0.3. Five-fold serial dilutions were plated onto YC medium containing the different concentrations of HU, MMS, and Phle. Plates were incubated at different temperatures for 2–3 days and photographed. Each assay was repeated three times, and all showed consistent results, therefore one representative result is shown.
Supplementary Material
Acknowledgements
We thank R. Singh for help with experiments. This work was supported in part by grants from the NIH: GM125195 and GM135671 to T. G.K., the Shanghai Pilot Program for Basic Research from Chinese Academy of Science (JCYJ-SHFY-2022-008) to Y.C., and the Shanghai Sailing Program (22YF1453800) and China Postdoctoral Science Foundation (2022M710145) to D.W.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbagrm.2023.194961.
CRediT authorship contribution statement
M.C.N., D.W. and B.J.K. performed experiments and together with Y. C. and T.G.K. analyzed the data. T.G.K. wrote the manuscript with input from all authors.
Declaration of competing interest
The authors declare the following financial interests/personal reationships which may be considered as potential competing interests: Tatiana G Kutateladze reports financial support was provided by National Institutes of Health. Tatiana G Kutateladze reports a relationship with NIH that includes: funding grants.
Data availability
Data will be made available on request.
References
- [1].Poon D, Bai Y, Campbell AM, Bjorklund S, Kim YJ, Zhou S, Kornberg RD, Weil PA, Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A 92 (18) (1995) 8224–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Henry NL, Campbell AM, Feaver WJ, Poon D, Weil PA, Kornberg RD, TFIIF-TAF-RNA polymerase II connection, Genes Dev 8 (23) (1994) 2868–2878. [DOI] [PubMed] [Google Scholar]
- [3].Kabani M, Michot K, Boschiero C, Werner M, Anc1 interacts with the catalytic subunits of the general transcription factors TFIID and TFIIF, the chromatin remodeling complexes RSC and INO80, and the histone acetyltransferase complex NuA3, Biochem. Biophys. Res. Commun 332 (2) (2005) 398–403. [DOI] [PubMed] [Google Scholar]
- [4].Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD, Structure of a complete mediator-RNA polymerase II pre-initiation complex, Cell 166 (6) (2016), 1411–1422 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kolesnikova O, Ben-Shem A, Luo J, Ranish J, Schultz P, Papai G, Molecular structure of promoter-bound yeast TFIID, Nat. Commun 9 (1) (2018) 4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].John S, Howe L, Tafrov ST, Grant PA, Sternglanz R, Workman JL, The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF (II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex, Genes Dev 14 (10) (2000) 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- [7].Shen X, Preparation and analysis of the INO80 complex, Methods Enzymol 377 (2004) 401–412. [DOI] [PubMed] [Google Scholar]
- [8].Cairns BR, Henry NL, Kornberg RD, TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9, Mol. Cell. Biol 16 (7) (1996) 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fish RN, Ammerman ML, Davie JK, Lu BF, Pham C, Howe L, Ponticelli AS, Kane CM, Genetic interactions between TFIIF and TFIIS, Genetics 173 (4) (2006) 1871–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD, A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II, Cell 77 (4) (1994) 599–608. [DOI] [PubMed] [Google Scholar]
- [11].Dahan O, Kupiec M, The Saccharomyces cerevisiae gene CDC40/PRP17 controls cell cycle progression through splicing of the ANC1 gene, Nucleic Acids Res 32 (8) (2004) 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shanle EK, Andrews FH, Meriesh H, McDaniel SL, Dronamraju R, DiFiore JV, Jha D, Wozniak GG, Bridgers JB, Kerschner JL, Krajewski K, Martin GM, Morrison AJ, Kutateladze TG, Strahl BD, Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response, Genes Dev 29 (17) (2015) 1795–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Andrews FH, Shanle EK, Strahl BD, Kutateladze TG, The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation, Transcription 7 (1) (2016) 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi XB, Strahl BD, Kutateladze TG, The Taf14 YEATS domain is a reader of histone crotonylation, Nat. Chem. Biol 12 (6) (2016) 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Klein BJ, Ahmad S, Vann KR, Andrews FH, Mayo ZA, Bourriquen G, Bridgers JB, Zhang J, Strahl BD, Cote J, Kutateladze TG, Yaf9 subunit of the NuA4 and SWR1 complexes targets histone H3K27ac through its YEATS domain, Nucleic Acids Res 46 (1) (2018) 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Klein BJ, Feigerle JT, Zhang J, Ebmeier CC, Fan L, Singh RK, Wang WW, Schmitt LR, Lee T, Hansen KC, Liu WR, Wang YX, Strahl BD, Anthony Weil P, Kutateladze TG, Taf2 mediates DNA binding of Taf14, Nat. Commun 13 (1) (2022) 3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen G, Wang D, Wu B, Yan F, Xue H, Wang Q, Quan S, Chen Y, Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation, Nat. Commun 11 (1) (2020) 4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feigerle JT, Weil PA, The C terminus of the RNA polymerase II transcription factor IID (TFIID) subunit Taf2 mediates stable association of subunit Taf14 into the yeast TFIID complex, J. Biol. Chem 291 (43) (2016) 22721–22740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gatchalian J, Futterer A, Rothbart SB, Tong Q, Rincon-Arano H, Sanchez de Diego A, Groudine M, Strahl BD, Martinez AC, van Wely KH, Kutateladze TG, Dido3 PHD modulates cell differentiation and division, Cell Rep 4 (1) (2013) 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


